Abstract
Free full text

Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Abstract
An epidemic of an avian-origin H7N9 influenza virus has recently emerged in China, infecting 134 patients of which 45 have died. This is the first time that an influenza virus harboring an N9 serotype neuraminidase (NA) has been known to infect humans. H7N9 viruses are divergent and at least two distinct NAs and hemagglutinins (HAs) have been found, respectively, from clinical isolates. The prototypes of these viruses are A/Anhui/1/2013 and A/Shanghai/1/2013. NAs from these two viruses are distinct as the A/Shanghai/1/2013 NA has an R294K substitution that can confer NA inhibitor oseltamivir resistance. Oseltamivir is by far the most commonly used anti-influenza drug due to its potency and high bioavailability. In this study, we show that an R294K substitution results in multidrug resistance with extreme oseltamivir resistance (over 100 000-fold) using protein- and virus-based assays. To determine the molecular basis for the inhibitor resistance, we solved high-resolution crystal structures of NAs from A/Anhui/1/2013 N9 (R294-containing) and A/Shanghai/1/2013 N9 (K294-containing). R294K substitution results in an unfavorable E276 conformation for oseltamivir binding, and consequently loss of inhibitor carboxylate interactions, which compromises the binding of all classical NA ligands/inhibitors. Moreover, we found that R294K substitution results in reduced NA catalytic efficiency along with lower viral fitness. This helps to explain why K294 has predominantly been found in clinical cases of H7N9 infection under the selective pressure of oseltamivir treatment and not in the dominant human-infecting viruses. This implies that oseltamivir can still be efficiently used in the treatment of H7N9 infections.
Introduction
Influenza A virus contains two major surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). HA initiates viral fusion by binding to sialic acid-containing receptors on the host cell and NA facilitates viral release by hydrolyzing the ketosidic bond of sialic acid. The balance of HA and NA activities is thought to be critical for influenza virus infectivity and transmissibility1. As an enzyme presented on the surface of the virus, NA is an ideal target for the design of competitive inhibitors2. Accordingly, there are currently four anti-NA inhibitors, oseltamivir, zanamivir, laninamivir and peramivir, which are approved for use in various countries2,3,4. Except NA-like molecule N10 from bat-origin influenza-like virus and influenza B NA, the nine remaining NA serotypes are classified into two groups according to their primary sequences5. N1, N4, N5 and N8 belong to group 1, while N2, N3, N6, N7 and N9 belong to group 26. N1 and N2 are the two most common serotypes found in epidemic and pandemic influenza viruses. In contrast, N9 has never before been observed in a human case of influenza virus infection.
Our previous studies reported that the NA gene of the novel H7N9 virus belongs to the Eurasian lineage7. Amino acid sequence alignment of A/Anhui/1/2013 N9 and A/Shanghai/1/2013 N9 with the previously characterized A/Tern/Australia/G70C/1975(H11N9) N9 illustrates that H7N9 N9 is missing five amino acids in the stem region (Figure 1). This deletion has previously been found to be important for virus adaptation from migratory to terrestrial birds8. Moreover, there are 23 amino acid polymorphisms between the A/Anhui/1/2013 N9 and A/Tern/Australia/G70C/1975 N9 head regions. However, none of these variant amino acids are located in or nearby the N9 enzymatic active site.

Sequence alignment of the neuraminidase N9. Amino acid sequence alignment of N9 from A/Tern/Australia/G70C/1975, A/Shanghai/1/2013 and A/Anhui/1/2013. Residues highlighted in red are completely conserved, and residues in blue boxes are highly conserved (> 80%). The green box indicates the missing five amino acids in the stem region of H7N9 N9. Residue 294 is highlighted with a blue asterisk.
There are only two residues that are different between A/Shanghai/1/2013 N9 and A/Anhui/1/2013 N9 (Figure 1). One polymorphism (residue 40) is located in the stem region and the other (residue 294 (equivalent to residue 292 in N2)) is one of three highly-conserved arginine residues that interact with the sialic acid carboxylate and are critical for substrate binding and catalysis9,10. Specifically, A/Anhui/1/2013 N9 contains R294 while A/Shanghai/1/2013 N9 has K2949. The R294K substitution has later been reported in 3 other H7N9 patients that were treated with oseltamivir9,11. Therefore, we presumed that A/Shanghai/1/2013 N9 must have arisen from oseltamivir treatment although the full clinical data are not available.
Previous studies have found that N2 and in vitro generated avian H11N9 NA carrying the R294(292)K substitution are resistant to multiple NA inhibitors12,13,14,15. However, naturally-occurring N9 carrying the R294K substitution has never been found in nature before. Furthermore, viruses carrying R294K exhibit compromised growth and fitness in vitro and were found to revert back to wild-type NA (R294) following multiple cycles of replication16. Moreover, these mutant viruses have been found to be 2-3 logs less infectious than wild-type virus and significantly less transmissible in a ferret model system17, suggesting that this substitution impairs NA activity and limits transmission ability.
The novel H7N9 virus has caused ongoing highly pathogenic influenza outbreaks throughout China. However, the structural and functional features of this naturally-occurring H7N9 NA remain unknown. Here, we report the crystal structure and functional characterization of two distinct human-derived influenza virus N9 proteins (from R294-containing A/Anhui/1/2013 virus and K294-containing A/Shanghai/1/2013 virus, respectively). Our findings demonstrate that the H7N9 R294K substitution not only confers multidrug resistance, but also decreases NA activity and impairs virus replication. This explains why H7N9 carrying R294K (A/Shanghai/1/2013 virus as its prototype) failed to be the dominant virus in this outbreak.
Results
The R294K substitution impairs H7N9 virus fitness and N9 enzymatic activity
To investigate the effect of the R294K substitution on H7N9 virus replication, we used reverse genetics to generate various viruses containing NA and HA from either A/Anhui/1/2013 or A/Shanghai/1/2013. We first generated wild-type A/Anhui/1/2013 H7N9 (hereafter referred to as “Anhui virus”), A/Anhui/1/2013 H7N9 with A/Shanghai/1/2013 HA (hereafter referred to as “Anhui-SH HA virus”), A/Anhui/1/2013 H7N9 with A/Shanghai/1/2013 NA (hereafter referred to as “Anhui-SH NA virus”), and A/Anhui/1/2013 H7N9 with A/Shanghai/1/2013 HA and NA (hereafter referred to as “Shanghai virus”). The TCID50 values (Figure 2) for the Anhui virus and Anhui-SH NA virus were the highest and lowest, respectively. The TCID50 values for the Anhui-SH HA virus and Shanghai virus were intermediate, with the former higher than the later (except 48 h after infection) (Figure 2A). These growth curves clearly indicate that K294-containing N9 impairs the replication ability of H7N9 virus. Moreover, to exclude any influence by internal genes, we also tested TCID50 values for H9N2 virus with A/Anhui/1/2013 HA and NA, and H9N2 virus with A/Shanghai/1/2013 HA and NA. Similar result was observed (Figure 2B), further confirming that K294-containing N9 impairs virus fitness.
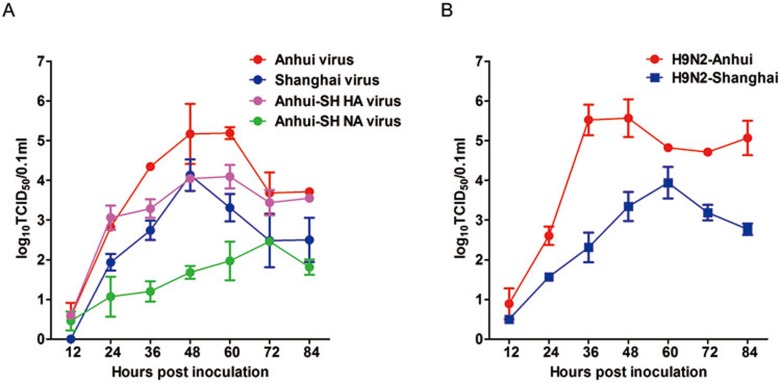
TCID50 growth curves for viruses containing Anhui or Shanghai HA and NA. (A) TCID50 growth curves of wild-type A/Anhui/1/2013 virus (Anhui virus, red), A/Anhui/1/2013 containing Shanghai HA (Anhui-SH HA virus, pink), A/Anhui/1/2013 containing Shanghai NA (Anhui-SH NA virus, green), and A/Anhui/1/2013 containing Shanghai HA and NA (Shanghai virus, blue). (B) TCID50 growth curves of H9N2 containing Anhui HA and NA (H9N2-Anhui, red) and H9N2 containing Shanghai HA and NA (H9N2-Shanghai, blue).
To provide more insights into the mechanism of how K294-containing N9 decreases viral fitness, we determined the relative activity and Michaelis-Menten (Km) constants of soluble recombinant N9 proteins and viruses generated by reverse genetics. In both systems, A/Shanghai/2013 N9 (K294-containing) shows lower activity and much higher Km values relative to A/Anhui/1/2013 N9 (R294-containing) (Table 1 and Figure 3). The significant reduction in NA catalytic efficiency is most likely the major factor underlying the impaired viral fitness of A/Shanghai/1/2013.
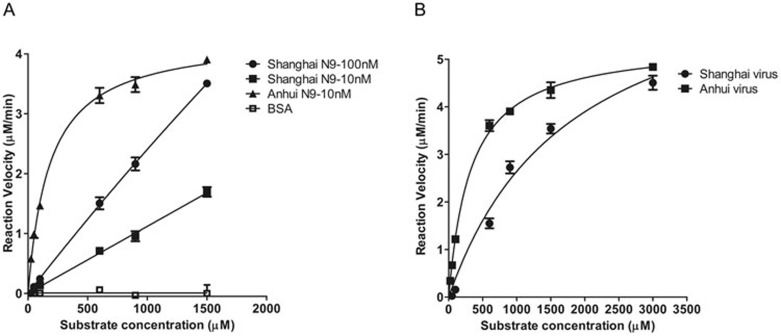
Kinetic analysis of H7N9 NA activity. (A) Comparison of the reaction velocity of Shanghai N9 and Anhui N9 is shown based on substrate conversion. 10 nM Anhui N9 (triangle), 10 nM Shanghai N9 (black square) and 100 nM Shanghai N9 (black dot) show distinct enzymatic activities. BSA (open square) was used as a negative control. (B) Comparison of the reaction velocity of viruses generated by reverse genetics. Anhui H7N9 activity is shown as black squares, and Shanghai H7N9 activity is shown as black dots. Mean values were determined from at least three duplicates and are presented with SDs indicated by error bars.
Table 1
| | | IC50 (nM) | |||
|---|---|---|---|---|---|
| Inhibitor | Km (μM) | Oseltamivir Carboxylate | Zanamivir | Peramivir | Laninamivir |
| Shanghai virus | 1,799 | 149,467 (112,750-198,142) | 58.5 (31.4-109) | 165 (108-254) | 53.4 (36.4-78.2) |
| Anhui virus | 325.7 | 1.13 (0.91-1.40) | 1.27 (0.91-1.77) | 0.40 (0.29-0.55) | 2.04 (1.19-3.50) |
| Shanghai N9 | 16,018 | 214,770 (126,699-364,063) | 75.7 (57.4-99.7) | 184.7 (129-264) | 89.6 (71.1-113) |
| Anhui N9 | 181.9 | 0.79 (0.51-1.23) | 0.41 (0.10-1.72) | 0.40 (0.25-0.67) | 3.24 (2.48-4.23) |
The R294K substitution in N9 from epidemic H7N9 confers multidrug resistance
The sensitivity of N9 to four classical NA inhibitors was determined using both soluble recombinant N9 and H7N9 viruses. Remarkably, Shanghai virus (K294-containing) displayed extreme resistance to oseltamivir carboxylate (271 860-fold). In the case of zanamivir, laninamivir and peramivir, the R294K mutation conferred less resistance relative to oseltamivir carboxylate (Table 1). For purified Shanghai N9, the IC50 values for zanamivir, laninamivir and peramivir were 75.7 nM, 89.6 nM and 184.7 nM, respectively. More potent inhibition of Anhui N9 was apparent with the IC50 values of 0.41 nM, 3.24 nM and 0.40 nM for zanamivir, laninamivir and peramivir, respectively. The inhibition results of the H7N9 virus-based assay agreed well with those of the recombinant protein-based assay. The Shanghai virus was 132 271 times more resistant to oseltamivir carboxylate relative to the Anhui virus. The IC50 values for inhibition of Shanghai virus by zanamivir, laninamivir and peramivir were 58.5 nM, 53.4 nM and 165 nM, respectively. In contrast, the corresponding IC50 values for the Anhui virus were 1.27 nM, 2.04 nM and 0.40 nM, respectively.
Structural basis of multidrug resistance in the N9 of human-infecting H7N9
To gain insight into the structural basis of the N9-R294K drug-resistant substitution, high-resolution crystal structures of Anhui N9 and Shanghai N9 were solved. N9 crystals were also soaked with classical NA inhibitors to solve the N9-inhibitor complex structures. In the uncomplexed Shanghai N9 and Anhui N9 structures, all active site residues adopt the same typical conformations except E276 and K/R294 (Figure 4). E276 is a flexible residue that adopts two distinct conformations in known NA structures, one oriented towards the center of the NA active site and the other oriented toward R224. In Anhui N9, E276 forms a salt bridge with R224, the conformation favorable for the binding of the oseltamivir carboxylate hydrophobic pentyloxy group. In contrast, Shanghai N9 contains a K294, which forms a salt bridge (2.95 Å in the uncomplexed Shanghai N9 structure, 3.12 Å in the Shanghai N9-oseltamivir carboxylate complex) with E276, forcing E276 to adopt a conformation that interferes with the binding of the oseltamivir pentyloxy group.
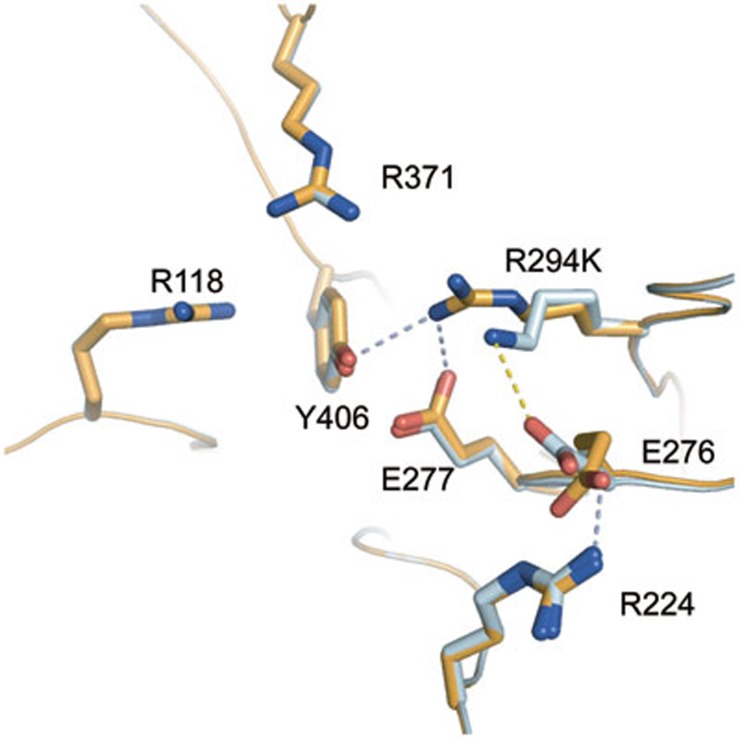
Comparison of Anhui N9 and Shanghai N9 uncomplexed active sites. In Anhui N9 (bright orange), R294 is able to hydrogen-bond with the key catalytic residues, Y406 and E277. However, R294 is out of range to interact with E276, which is oriented toward R224, a conformation favorable for oseltamivir carboxylate binding. E276 forms a salt bridge with R224. In Shanghai N9 (palecyan), K294 is out of range to form any typical hydrogen bonds with Y406 or E277. Instead, K294 is able to form a salt bridge with E276, which is oriented toward K294, a conformation unfavorable for oseltamivir carboxylate binding.
In the Shanghai and Anhui N9-oseltamivir carboxylate complex structures, E276 adopts the same conformations as in the uncomplexed structures (Figure 5A). Therefore, Anhui N9 is able to accommodate oseltamivir carboxylate well with a hydrophobic pocket formed by the favorable conformation of E276. However, in Shanghai N9, E276 is oriented toward the oseltamivir carboxylate hydrophobic pentyloxy group. This unfavorable interaction pushes the oseltamivir carboxylate pentyloxy group away from the active site (by 2.98 Å).
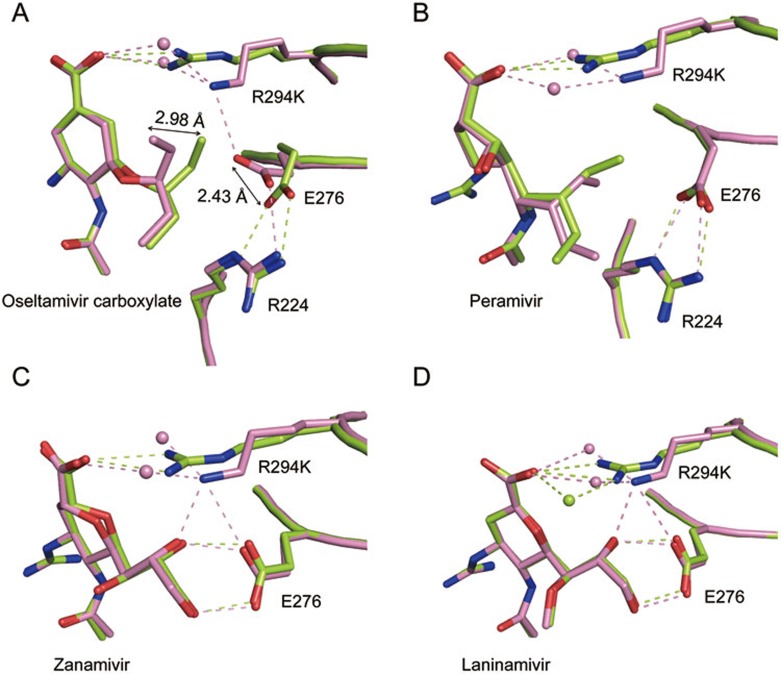
Structural analysis of inhibitor binding to Anhui N9 and Shanghai N9. (A) Comparison of the oseltamivir binding modes in Shanghai N9 (pink) and Anhui N9 (limon). Differences in the Anhui N9- and Shanghai N9-oseltamivir carboxylate complex structures are indicated by arrows. (B) Comparison of the peramivir binding modes in Shanghai N9 (pink) and Anhui N9 (limon). (C) Comparison of the zanamivir binding modes in Shanghai N9 (pink) and Anhui N9 (limon). (D) Comparison of the laninamivir binding modes in Shanghai N9 (pink) and Anhui N9 (limon). Inhibitors are shown using stick representation. Residues are shown using cartoon and stick representation. Water molecules are represented as spheres.
Interestingly, despite containing a pentyloxy group like oseltamivir, in the Shanghai N9-peramivir complex structure, E276 is still able to adopt the optimal conformation where it forms a salt bridge with R224 (Figure 5B). This is in agreement with our inhibition data showing that peramivir resistance is not as severe as oseltamivir resistance. We speculate that this is due to a smaller cyclopentane ring that affords more distance between the peramivir pentyloxy group and E276.
In addition, Shanghai N9 K294 is approximately 1.5 Å further from the inhibitor carboxylate, further contributing to a weaker binding mode. The smaller lysine residue in position 294 also results in the addition of two water molecules between K294 and the inhibitor carboxylate (Figure 5). This loss of a direct inhibitor carboxylate-K294 interaction is also observed in the Shanghai N9 complex structures with zanamivir, laninamivir and peramivir, explaining why Shanghai N9 also displays mild resistance to these inhibitors. Furthermore, the tri-arginyl-carboxylate interaction is thought to be a major factor for distorting the sialic acid pyranose ring from a chair to a boat conformation. Current data indicate that influenza NA first binds α2,3- or α2,6-linked glycoconjugates in a 2C5 chair conformation; the substrate then undergoes a conformation change to form a boat conformation and Tyr406 attacks the anomeric center, leading to glycosidic bond cleavage18. Therefore, R294 is likely a key residue in the NA catalytic mechanism and it is not surprising that N9 from A/Shanghai/1/2013 H7N9, which cannot form a K294-carboxylate interaction, has much lower NA activity compared to A/Anhui/1/2013.
Discussion
Except N7, crystal structures for all typical influenza A NA serotypes are currently available in the Protein Data Bank (www.pdb.org). Some interesting structural differences between each serotype have been revealed through structural analysis. For example, group 1 influenza A NAs (N1, N4, N5 and N8) have been found to contain an additional cavity (150-cavity) adjacent to their active site, whereas all group 2 NAs with known structures (N2, N3, N6 and N9) lack this cavity6. Yet, although the N1s from H5N1 and 1918 H1N1 contain a 150-cavity, the N1 from the 2009 pandemic H1N1 does not19. This illustrates that there may be significant structural differences even within each NA serotype and highlights the importance of the structural analysis of novel NA proteins like N9 from H7N9.
Influenza NA inhibitor development is regarded as a classical example of structure-based drug design and the continued structural analysis of influenza NAs has contributed greatly to the design and evaluation of effective anti-influenza agents. As vaccines are still under development for the H7N9 virus, NA inhibitors are very important options for the treatment of H7N9 patients. In fact, oseltamivir treatment within 48 h of the disease onset has been recommended by the Chinese Center for Disease Control and Prevention. This practice has saved many lives. Therefore, the structural analysis of H7N9 NA is crucial for directing drug development and clinical administration of NA inhibitors for H7N9 patients.
It is clear that N9 proteins from early clinical isolates are divergent and an oseltamivir-resistant K294-containing N9 was identified from the first few isolates. N9 from A/Shanghai/1/2013 is the prototype in this regard although the history of inhibitor use for this patient is not clear. It is reported that in some clinical cases of H7N9 infection treated with oseltamivir, viruses carrying an R294K multidrug-resistant substitution in their N9 proteins have been isolated11. R294 is one of the three key conserved active site arginines that surround the carboxylate group of sialic acid. This tri-arginyl cluster is thought to be a major factor for distorting the sialic acid pyranose ring from a chair to a boat conformation, a critical step for influenza NA to catalyze the hydrolytic cleavage of terminal sialic acid from α2,3- or α2,6-linked glycoconjugates10,18. Therefore, it is not surprising that N9 from A/Shanghai/1/2013 H7N9 has much lower NA activity and that R294K has never been found before in a wild-type N9 variant, though laboratory artificial drug-present passages yielded an R294K N920.
The knowledge that the Shanghai H7N9 viruses may contain an R294K substitution in their NA protein is critical for directing future drug administration in H7N9 patients. In these cases, oseltamivir should be used with caution, as it is > 100 000 times less effective against the R294K-containing H7N9. This is at a level much higher than even the prevalent H274Y oseltamivir-resistant substitution found in N1 viruses, which results in up to 1 200-fold resistance21. Of the 4 reported patients with the R294K substitution, 2 have died and 1 has remained hospitalized9,11. All of these patients were only reported to be treated with oseltamivir and not with any of the other available classical NA inhibitors. Therefore, the drug-resistant mutations should be monitored vigilantly in H7N9 viruses in order to help administer the most effective drugs available and help to prevent unnecessary loss of human lives. Our results of low replication rate of the viruses with K294-containing N9 demonstrate that these viruses are less fit and cannot survive long, therefore oseltamivir is still a good inhibitor for the treatment of H7N9 patients.
Materials and Methods
Generation and growth of recombinant viruses
Recombinant viruses were generated by reverse genetics using previously described methods22,23. For the Anhui virus, all eight segments stemmed from the isolated virus. For the Shanghai virus, HA and NA segments were from A/Shanghai/1/2013 virus, while the other internal gene segments were the same as the Anhui virus. For the H9N2 systems, A/Chicken/Shandong/lx1023/2007 (H9N2) internal genes were used as the backbone24, while HA and NA segments were from A/Anhui/1/2013 virus (H9N2-Anhui) or from A/Shanghai/1/2013 virus (H9N2-Shanghai). Replication curves were generated by inoculating MDCK cells at a multiplicity of infection (MOI) of 0.001 50% tissue culture infective doses (TCID50) per cell. Supernatants were collected at 12, 24, 36, 48, 72 and 84 h post infection and the virus titers were determined by the Reed-Muench method25.
Neuraminidase production and crystallization experiments
Purified N9 was prepared in an insect/baculovirus expression system as previously reported4,19,26,27. Crystallization conditions were screened using sitting drop vapor diffusion technique, with 1 μL of 8 mg/ml N9 protein mixed with 1 μL of reservoir solution. Anhui N9 crystals were obtained in the condition of 0.1 M MES monohydrate pH 6.0 and 14% w/v Polyethylene glycol 4000. Shanghai N9 crystals were obtained in the condition of 5% v/v(±)-2-Methyl-2,4-pentanediol, 0.1 M HEPES pH 7.5 and 10% Polyethylene glycol 10000. NA crystals were incubated in the mother liquor containing 20 mM inhibitor (oseltamivir carboxylate, zanamivir or laninamivir) or 10 mM peramivir, and then flash-cooled at 100 K.
N9 crystals were cryoprotected and then flash-cooled at 100 K. Diffraction data were collected at Shanghai Synchrotron Radiation Facility beamline BL17U. Diffraction data were processed and scaled using HKL200028. The structure of Anhui N9 was solved by molecular replacement using the A/tern/Australia/G70C/1975 H11N9 N9 molecule (PDB 7NN9) as the search model with Phaser29 in the CCP4 program suite30. The structure of Shanghai N9 was determined by molecular replacement using the solved Anhui N9 structure. Extensive model building was performed manually with COOT31, and restrained refinement was performed using REFMAC532. Further rounds of refinement were performed using the phenix.refine program implemented in the PHENIX package with energy minimization, isotropic ADP refinement and bulk solvent modeling33. The stereochemical quality of the final model was assessed with PROCHECK34. Crystallographic data and refinement statistics are shown in Table 2.
Table 2
| Parameter | Shanghai N9 | Anhui N9 | Shanghai N9-Oseltamivir carboxylate | Shanghai N9-Zanamivir | Shanghai N9-Laninamivir | Shanghai N9-Peramivir | Anhui N9-Oseltamivir carboxylate | Anhui N9-Zanamivir | Anhui N9-Laninamivir | Anhui N9-Peramivir |
|---|---|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||||
| Space group | I432 | I432 | I432 | I432 | I432 | I432 | I432 | I432 | I432 | I432 |
| Unit cell | 181.0, | 180.8, | 180.9, | 181.5, | 181.5, | 180.8, | 181.9, | 181.5, | 180.9, | 180.5, |
| dimensions | 181.0, | 180.8, | 180.9, | 181.5, | 181.5, | 180.8, | 181.9, | 181.5, | 180.9, | 180.5, |
| (a, b, c) | 181.0 | 180.8 | 180.9 | 181.5 | 181.5 | 180.8 | 181.9 | 181.5 | 180.9 | 180.5 |
| Unit cell | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, |
| dimensions | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, | 90.00, |
| (α, β, γ) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| Resolution | 50.00-1.80 | 50.00-1.80 | 50.00-1.90 | 50.00-1.80 | 50.00-1.80 | 50.00-2.10 | 50.00-2.00 | 50.00-1.80 | 50.00-1.80 | 50.00-2.00 |
| range (Å) | (1.86-1.80) | (1.86-1.80) | (1.97-1.90) | (1.86-1.80) | (1.86-1.80) | (2.18-2.10) | (2.07-2.00) | (1.86-1.80) | (1.86-1.80) | (2.07-2.00) |
| Rmerge (%) | 14.1 (47.1) | 11.8 (36.5) | 16.5 (55.9) | 11.7 (37.5) | 13.8 (41.7) | 14.0 (59.5) | 16.2 (51.8) | 13.1 (60.7) | 11.8 (32.5) | 12.3 (55.3) |
| I/σ | 20.1 (6.2) | 23.4 (8.1) | 17.3 (4.9) | 25.1 (8.0) | 21.4 (7.5) | 18.6 (5.0) | 16.3 (5.1) | 24.0 (4.8) | 24.6 (9.4) | 26.0 (6.3) |
| Redundancy | 13.9 (13.9) | 13.7 (13.6) | 11.5 (11.5) | 13.4 (13.5) | 13.6 (13.3) | 12.3 (12.4) | 10.7 (10.6) | 14.1 (14.2) | 13.4 (12.9) | 16.4 (16.6) |
| Completeness % | 100 (100) | 100 (99.9) | 100 (100) | 100 (100) | 100 (100) | 96.9 (99.9) | 99.9 (100) | 100 (100) | 100 (100) | 100 (100) |
| Refinement | ||||||||||
| Resolution (Å) | 42.7-1.80 | 48.3-1.80 | 35.5-1.90 | 42.7-1.80 | 42.8-1.80 | 42.8-2.10 | 35.7-2.00 | 42.8-1.80 | 42.6-1.80 | 48.2-2.00 |
| Rwork/Rfree | 13.7/15.6 | 14.3/16.4 | 14.4/16.6 | 13.7/15.7 | 13.7/15.4 | 16.7/20.6 | 15.0/17.1 | 14.7/17.3 | 14.2/16.7 | 14.8/17.7 |
| RMSDs | ||||||||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.005 | 0.006 | 0.006 | 0.006 | 0.005 | 0.005 | 0.006 | 0.005 |
| Bond angles (°) | 1.086 | 1.134 | 1.083 | 1.126 | 1.141 | 1.092 | 1.072 | 1.066 | 1.106 | 1.064 |
Enzymatic activity and inhibition assays
N9 activity was determined using 4-MUNANA (4-methylumbelliferyl-N-acetylneuraminic acid) as a fluorescent NA substrate according to previously reported methods4,18,35. All virus assays were performed under BSL-3 laboratory conditions with 105.75 (EID50) units of virus for each condition. The fluorescence intensity of the released product (Ex. 355 nM, Em. 460 nM) was measured on a microplate reader (SpectrMax M5, Molecular Devices). All assays were done in triplicates and the Km and IC50 values were calculated using GraphPad Prism.
PDB accession codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (www.pdb.org) with the following PDB codes: Anhui N9, 4MWJ; Shanghai N9, 4MWL; Anhui N9-oseltamivir carboxylate, 4MWQ; Anhui N9-zanamivir, 4MWR; Anhui N9-laninamivir, 4MWU; Anhui N9-peramivir, 4MWV; Shanghai N9-oseltamivir carboxylate, 4MWW; Shanghai N9-zanamivir, 4MWX; Shanghai N9-laninamivir, 4MWY; Shanghai N9-peramivir, 4MX0.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC; 81301465 and 81341002) and the intramural special grant for influenza virus research from the Chinese Academy of Sciences (KSZD-EW-Z-002). GFG is a leading principal investigator of the NSFC Innovative Research Group (81021003). CJ Vavricka is supported by the NSFC Research Fund for Young International Scientists (31150110147). We acknowledge the assistance of the staff at Shanghai Synchrotron Radiation Facility. We are also grateful to the Genewiz Corporation for the rapid synthesis of NA genes.
References
- Xu R, Zhu X, McBride R, et al. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol. 2012;86:9221–9232. [Europe PMC free article] [Abstract] [Google Scholar]
- Moscona A. Medical management of influenza infection. Annu Rev Med. 2008;59:397–413. [Abstract] [Google Scholar]
- Ison MG. Clinical use of approved influenza antivirals: therapy and prophylaxis. Influenza Other Respi Viruses. 2012;7:7–13. [Europe PMC free article] [Abstract] [Google Scholar]
- Vavricka CJ, Li Q, Wu Y, et al. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog. 2011;7:e 1002249. [Europe PMC free article] [Abstract] [Google Scholar]
- Li Q, Sun X, Li Z, et al. Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc Natl Acad Sci USA. 2012;109:18897–18902. [Europe PMC free article] [Abstract] [Google Scholar]
- Russell RJ, Haire LF, Stevens DJ, et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–49. [Abstract] [Google Scholar]
- Liu D, Shi W, Shi Y, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. [Abstract] [Google Scholar]
- Sorrell EM, Song H, Pena L, Perez DR. A 27-amino-acid deletion in the neuraminidase stalk supports replication of an avian H2N2 influenza A virus in the respiratory tract of chickens. J Virol. 2010;84:11831–11840. [Europe PMC free article] [Abstract] [Google Scholar]
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. [Abstract] [Google Scholar]
- Varghese JN, McKimm-Breschkin JL, Caldwell JB, Kortt AA, Colman PM. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins. 1992;14:327–332. [Abstract] [Google Scholar]
- Hu Y, Lu S, Song Z, et al. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet. 2013;381:2273–2279. [Abstract] [Google Scholar]
- Varghese JN, Smith PW, Sollis SL, et al. Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure. 1998;6:735–746. [Abstract] [Google Scholar]
- Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759–765. [Abstract] [Google Scholar]
- Tai CY, Escarpe PA, Sidwell RW, et al. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS 4071. Antimicrob Agents Chemother. 1998;42:3234–3241. [Europe PMC free article] [Abstract] [Google Scholar]
- Gubareva LV, Robinson MJ, Bethell RC, Webster RG. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol. 1997;71:3385–3390. [Europe PMC free article] [Abstract] [Google Scholar]
- Yen HL, Herlocher LM, Hoffmann E, et al. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob Agents Chemother. 2005;49:4075–4084. [Europe PMC free article] [Abstract] [Google Scholar]
- Carr J, Ives J, Kelly L, et al. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res. 2002;54:79–88. [Abstract] [Google Scholar]
- Vavricka CJ, Liu Y, Kiyota H, et al. Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors. Nat Commun. 2013;4:1491. [Abstract] [Google Scholar]
- Li Q, Qi J, Zhang W, et al. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat Struct Mol Biol. 2010;17:1266–1268. [Abstract] [Google Scholar]
- McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72:2456–2462. [Europe PMC free article] [Abstract] [Google Scholar]
- Hurt AC, Chotpitayasunondh T, Cox NJ, et al. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis. 2012;12:240–248. [Abstract] [Google Scholar]
- de Wit E, Spronken MI, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 2004;103:155–161. [Abstract] [Google Scholar]
- Sun Y, Qin K, Wang J, et al. High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc Natl Acad Sci USA. 2011;108:4164–4169. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang Y, Yin Y, Bi Y, et al. Molecular and antigenic characterization of H9N2 avian influenza virus isolates from chicken flocks between 1998 and 2007 in China. Vet Microbiol. 2012;156:285–293. [Abstract] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hygiene. 1938;27:493–497. [Google Scholar]
- Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol. 2008;82:10493–10501. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu Y, Qin G, Gao F, et al. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep. 2013;3:1551. [Europe PMC free article] [Abstract] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. [Abstract] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. [Abstract] [Google Scholar]
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. [Abstract] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. [Abstract] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. [Abstract] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. [Abstract] [Google Scholar]
- Laskowski R MM, Moss D, Thornton J. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal Biochem. 1979;94:287–296. [Abstract] [Google Scholar]
Articles from Cell Research are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/cr.2013.144
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3847574?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/cr.2013.144
Article citations
MedChemExpress compounds prevent neuraminidase N1 via physics- and knowledge-based methods.
RSC Adv, 14(27):18950-18956, 12 Jun 2024
Cited by: 0 articles | PMID: 38873542
Targeting the Human Influenza a Virus: The Methods, Limitations, and Pitfalls of Virtual Screening for Drug-like Candidates Including Scaffold Hopping and Compound Profiling.
Viruses, 15(5):1056, 26 Apr 2023
Cited by: 1 article | PMID: 37243142 | PMCID: PMC10221779
Detecting the Neuraminidase R294K Mutation in Avian Influenza A (H7N9) Virus Using Reverse Transcription Droplet Digital PCR Method.
Viruses, 15(4):983, 17 Apr 2023
Cited by: 1 article | PMID: 37112963 | PMCID: PMC10146270
A systemic review on medicinal plants and their bioactive constituents against avian influenza and further confirmation through in-silico analysis.
Heliyon, 9(3):e14386, 10 Mar 2023
Cited by: 1 article | PMID: 36925514 | PMCID: PMC10011005
Review Free full text in Europe PMC
In Silico Studies Reveal Peramivir and Zanamivir as an Optimal Drug Treatment Even If H7N9 Avian Type Influenza Virus Acquires Further Resistance.
Molecules, 27(18):5920, 12 Sep 2022
Cited by: 1 article | PMID: 36144655 | PMCID: PMC9503969
Go to all (65) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (Showing 11 of 11)
-
(1 citation)
PDBe - 4MWVView structure
-
(1 citation)
PDBe - 4MWWView structure
-
(1 citation)
PDBe - 4MWXView structure
-
(1 citation)
PDBe - 4MWYView structure
-
(1 citation)
PDBe - 4MWJView structure
-
(1 citation)
PDBe - 4MWLView structure
-
(1 citation)
PDBe - 7NN9View structure
-
(1 citation)
PDBe - 4MX0View structure
-
(1 citation)
PDBe - 4MWQView structure
-
(1 citation)
PDBe - 4MWRView structure
-
(1 citation)
PDBe - 4MWUView structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A novel pyrosequencing assay for the detection of neuraminidase inhibitor resistance-conferring mutations among clinical isolates of avian H7N9 influenza virus.
Virus Res, 179:119-124, 05 Nov 2013
Cited by: 9 articles | PMID: 24211668 | PMCID: PMC7126809
Resistance to neuraminidase inhibitors conferred by an R292K mutation in a human influenza virus H7N9 isolate can be masked by a mixed R/K viral population.
mBio, 4(4):e00396-13, 16 Jul 2013
Cited by: 50 articles | PMID: 23860768 | PMCID: PMC3735122
Characterization of substitutions in the neuraminidase of A(H7N9) influenza viruses selected following serial passage in the presence of different neuraminidase inhibitors.
Antiviral Res, 168:68-75, 25 May 2019
Cited by: 2 articles | PMID: 31132385
Discovery of potential drugs for human-infecting H7N9 virus containing R294K mutation.
Drug Des Devel Ther, 8:2377-2390, 01 Dec 2014
Cited by: 10 articles | PMID: 25489236 | PMCID: PMC4257025




