Abstract
Free full text

Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode
Significance
Carbohydrates (glycans) are complex cell surface molecules that control multiple aspects of cell biology, including cell–cell communication, cancer metastasis, and inflammation. Glycan biosynthesis requires the coordination of many enzymes, but how this is regulated is not well understood. Herein we show that microRNA (miRNA), small noncoding RNA, are a major regulator of cell surface glycosylation. We map miRNA expression onto carbohydrate signatures obtained by using lectin microarrays, a glycan analysis method. We identify and validate several miRNA–glycan networks, including a major decision point in N-linked glycan biosynthesis. Overall, our work provides insights into the “black box” of carbohydrate control mechanisms.
Abstract
Cell surface glycans form a critical interface with the biological milieu, informing diverse processes from the inflammatory cascade to cellular migration. Assembly of discrete carbohydrate structures requires the coordinated activity of a repertoire of proteins, including glycosyltransferases and glycosidases. Little is known about the regulatory networks controlling this complex biosynthetic process. Recent work points to a role for microRNA (miRNA) in the regulation of specific glycan biosynthetic enzymes. Herein we take a unique systems-based approach to identify connections between miRNA and the glycome. By using our glycomic analysis platform, lectin microarrays, we identify glycosylation signatures in the NCI-60 cell panel that point to the glycome as a direct output of genomic information flow. Integrating our glycomic dataset with miRNA data, we map miRNA regulators onto genes in glycan biosynthetic pathways (glycogenes) that generate the observed glycan structures. We validate three of these predicted miRNA/glycogene regulatory networks: high mannose, fucose, and terminal β-GalNAc, identifying miRNA regulation that would not have been observed by traditional bioinformatic methods. Overall, our work reveals critical nodes in the global glycosylation network accessible to miRNA regulation, providing a bridge between miRNA-mediated control of cell phenotype and the glycome.
Cell surface glycans form a critical interface with the biological milieu, informing diverse processes from cellular migration to pathogen–cell interactions. Assembly of discrete carbohydrate structures requires coordination of a complex repertoire of proteins, including glycosyltransferases, glycosidases, and sugar nucleotide transporters, acting in tandem (1, 2). Little is known about the regulation of the complex biosynthetic networks required for glycosylation. MicroRNAs (miRNAs) are small noncoding RNAs that can bind the 3′-UTR of mRNA and inhibit mRNA stability or translation (3). Recent work points to a role for miRNA in modulating the levels of glycan biosynthetic enzymes (glycogenes) (4–11) with profound biological consequences, including promotion of tumor metastasis (5) and regulation of neuronal migration (9). Enrichment of glycogene mRNA in miRNA/mRNA/RNA-induced silencing complex-complexes during Caenorhabditis elegans development hints that miRNA may be a major regulator of glycosylation (12). Studying the relationship between the glycome and miRNA is complicated by the low abundance of glycogene transcripts, resulting in inaccuracies in microarray-based expression analysis, the basis for most miRNA target prediction algorithms (13, 14). These algorithms predict hundreds to thousands of gene targets with a precision between ~30% and 50% (15, 16). Target prioritization is often based on mRNA conservation across species (15), which presents a second issue for glycogenes as glycosylation is a rapidly evolving system (17). We reasoned that a systems-based approach integrating glycosylation patterns, the functional outcome of mRNA regulation, with miRNA expression would allow us to map miRNA onto glycan biosynthetic pathways. Harnessing the power of lectin microarrays, our glycomic platform, we demonstrate that miRNAs are critical modulators of the human glycome and identify miRNA regulation of glycogenes elusive to current prediction algorithms.
Results
Glycomic Analysis of the NCI-60 Reveals Tissue Type-Specific Glycan Signatures.
Lectin microarrays, in which carbohydrate-binding proteins are probes for glycan structure, provide a systems-level view of the glycome (Fig. 1A) (18–20). These microarrays give specific information on the repertoire of glycans present, e.g., high mannose epitopes, branching patterns, and terminal α-2,3- or α-2,6-sialic acids, in a high-throughput format. Unlike other glycomic methods, both the N- and O-linked glycome are observed (19), and the data are in the same format as mRNA and miRNA datasets. We analyzed the NCI-60, a cancer cell line panel containing 59 cell lines from nine tissue types, using our lectin microarray technology. This cell panel is a model for integrated analysis across multiple data types because of the availability of chemosensitivity profiles for these cells (21–23). The miRNA and mRNA profiles of the NCI-60 show the most coherent signatures for four tissue types: colon, leukemia, melanoma, and renal (22, 23). We first profiled the glycome of cells from these four tissue types for integration with miRNA datasets and observed clear segregation by tissue of origin (SI Appendix, Fig. S1). An expanded analysis of the cell panel displayed similar results and is shown in Fig. 1. Glycosylation patterns were confirmed by fluorescence microscopy (SI Appendix, Fig. S2). For both datasets, normalized data were subjected to hierarchical clustering by using the Pearson correlation coefficient (PearCC) as the distance metric and average linkage analysis. Melanoma and renal cell lines showed enrichment in high mannose [ASA, AMA, UDA, scytovirin (SVN), Calsepa, HHL; see SI Appendix, Tables S1 and S2 for full lectin names] but were distinguished by differing levels of complex multiantennary N-glycans (PHA-L, PHA-E; renal, high; melanoma, low). In contrast, leukemia and colon had lower levels of high mannose and divergent levels of complex multiantennary glycans (colon, high; leukemia, low) and β-GalNAc epitopes (BPA, SBA; colon, high; leukemia, low). In the larger NCI-60 dataset, no discrete glycan signatures were observed for the remaining cell types. Our data are consistent with previous mRNA- and miRNA-based profiles (22, 24). Overall, our data suggest that the glycome of a cell is a direct representation of the complex flow of genetic information that encodes cell type.
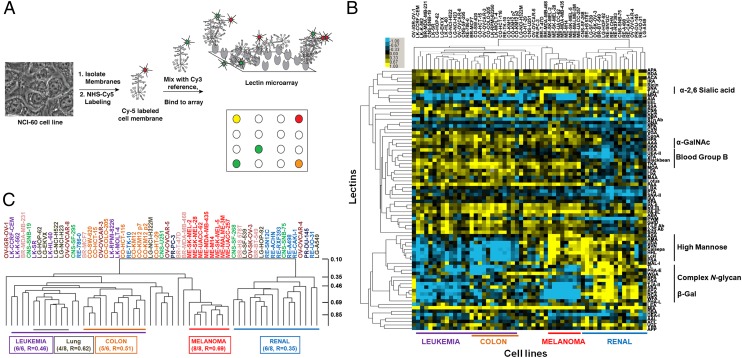
Ratiometric comparison of NCI-60 cell lines. (A) Experimental scheme. Equal amounts of Cy5-sample (S) and Cy3-reference (R) were analyzed on the lectin microarray. (B) Median-normalized log2 ratios (S/R) for 55 cell lines of the NCI-60 were hierarchically clustered by using centered PearCC as the distance metric and average linkage analysis (n = 76 lectins). Heat map is shown. Yellow, log2(S/R) > log2(Smedian/Rmedian); blue, log2(Smedian/Rmedian) > log2(S/R). (C) Dendrogram from B. As expected, biological replicates for KM12 clustered tightly. PearCC scale is shown at right.
Multidimensional Singular Value Decomposition Analysis Identifies Regulatory miRNA/Glycosylation Networks.
To examine the relationship between miRNAs and the observed glycan signatures, we integrated data from our original glycomic analysis (SI Appendix, Fig. S1) with the miRNA expression data from Liu et al. (22) by using singular value decomposition (SVD), an unsupervised matrix method (25). SVD decomposes datasets into unique “eigenarrays” and “eigengenes,” related by a significance matrix, which can map onto biological phenotype. We observed five eigengenes (E2–E6; Fig. 2A), reflecting cell type-dependent patterns, which, together with the first eigengene (E1), account for 94% of the variation in the data. Projection of the lectins and miRNA onto individual eigengenes resulted in sets of lectins with mixed specificities, confounding our mapping of miRNA/glycan associations. We reasoned that miRNA that tightly associate with a particular glycan structure via regulation of underlying glycogenes should covary with lectin binders in their contributions to the eigengenes. Therefore, we performed a multidimensional analysis of miRNA and lectin contributions to the eigengenes by hierarchically clustering projection scores for the first six eigengenes by using PearCC as the distance metric and average linkage analysis (Fig. 2B). This improved segregation of lectins and identified multiple miRNA/lectin clusters that map onto biosynthetic pathways. We identified regulatory networks for high mannose (Fig. 2 C and D and SI Appendix, Fig. S3A), fucose (Fig. 2E and SI Appendix, Fig. S3B), terminal β-GalNAc (Fig. 2F and SI Appendix, Fig. S3C), Tn and T-antigens (SI Appendix, Figs. S3D and S4A), hybrid-N-glycans (SI Appendix, Figs. S3E and S4B), and blood group B (SI Appendix, Figs. S3F and S4C), suggesting widespread control of glycan biosynthetic pathways by miRNA. We validated three of these networks—high mannose, fucose, and terminal β-GalNAc—as subsequently discussed in detail, identifying seven previously unknown miRNA/glycogene interactions and providing strong evidence that combining glycomic outcomes with miRNA expression levels is a powerful systems-level approach for finding glycan regulatory relationships.
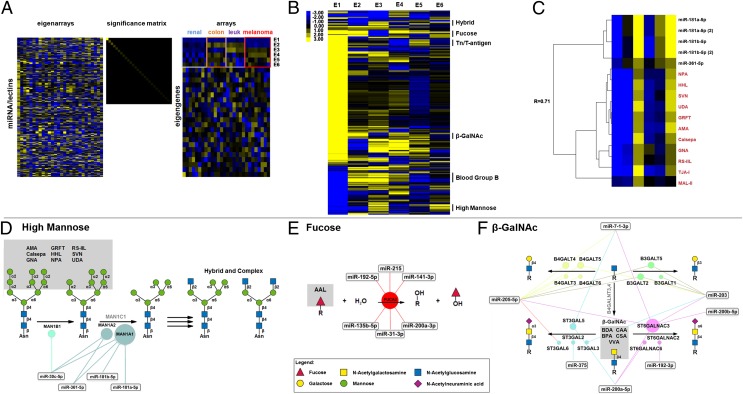
Mapping of miRNA/glycosylation networks. (A) SVD of lectin and miRNA data for renal, colon, leukemia (leuk), and melanoma cell lines. The first six eigengenes (E1–E6) are indicated. (B) Hierarchical clustering of the projection correlation values for E1–E6 using uncentered PearCC as the distance metric and average linkage analysis. Select clusters are annotated by glycan specificity. (C) A detailed representation of the high mannose cluster (R = 0.71; one-tailed P = 0.06; n = 6). Lectins are shown in red. (D–F) Networks derived from lectin/miRNA clusters: (D) high mannose network, (E) fucose network, (F) β-GalNAc network. Gray boxes indicate epitopes recognized by lectins. Bubbles indicate predicted miRNA targets, with bubble size reflecting number of miRNA targeting gene. Lines connect miRNA with targets; genes in silver are genes in the pathway that are not targeted.
High Mannose Network.
More than half of all proteins are predicted to contain N-linked glycosylation, which governs functions as diverse as protein folding, trafficking, and activity (26). We observed an association between lectins that recognize high mannose (AMA, Calsepa, GNA, griffithsin, HHL, NPA, RS-IIL, SVN, and UDA) and a network of miRNAs (miR-30c, -181a-5p, -181b-5p, -361–5p) predicted to target the α-mannosidase I (MAN1) family of enzymes (Fig. 2 C and D and SI Appendix, Fig. S3A). These glycosidases are involved in trimming of Man9GlcNAc2 to Man5GlcNAc2, a critical prerequisite for maturation of complex N-linked carbohydrates (1). Down-regulation of MAN1 results in increased high mannose. We transfected HT-29, a colon cell line with intermediate high mannose levels, with miRNA mimics and visualized glycans by lectin staining (HHL, PSA, GNA, NPA, with LcH, a core fucose lectin, as a control) and fluorescence microscopy. The miRNA from the high-mannose cluster increased binding of high-mannose lectins by twofold, suggesting a direct effect of the miRNA on MAN1 family enzymes (Fig. 3 A and B and SI Appendix, Fig. S5).

Validation of the high mannose network. (A and B) Representative fluorescence images of HT-29 cells stained with high mannose lectins (A) HHL or (B) PSA following 96 h treatment with miR-30c, -181b-5p, -361–5p, or scramble mimics. Monosaccharide control for staining is shown. Data are representative of three biological replicates. Statistical analysis of the staining and additional lectin data for GNA, NPA, and the LcH control is shown in SI Appendix, Fig. S5. (C–F) Real-time qPCR analysis of indicated glycogenes in HT-29 cells treated with indicated miRNA mimics (50 nM; C and E) or corresponding inhibitors (anti-, 25 nM; D and F) for 72 h. Scrambled sequences are used as a control (scramble). Graphs show average relative expression normalized to GAPDH of three biological replicates. (C and D) MAN1A1. (E and F) MAN1A2. (G and H) Western blot analysis of MAN1A2 samples treated as described with (G) miRNA mimics or (H) inhibitors. Graphs are of average signal normalized to GAPDH for three biological replicates. Representative images corresponding to the graphs are shown. (I) Graphical representation of luciferase activity from MAN1A2 constructs cotransfected with miR-30c, -181b-5p, -361–5p, or scramble mimics (60 nM) in HEK-293T/17 cells. Mut, miR-30 mutant or miR-361–5p mutant construct as indicated (SI Appendix, Fig. S8A and Table S3). Luciferase data were normalized to scramble control. Error bars denote SD (*P < 0.05, Student t test).
All miRNAs in the high-mannose network were predicted by the MIRANDA algorithm (microRNA.org) to target MAN1A1 (14, 27). We treated HT-29 with mimics and inhibitors of miRNA in the cluster and examined MAN1A1 expression levels by real-time quantitative PCR (qPCR) and Western blot analysis (Fig. 3 C and D and SI Appendix, Fig. S6 A and B). No change was observed. Analysis of MAN1 transcript levels in four NCI-60 cell lines (Sk-Mel-5, SN12C, HT-29, HCT-116) identified MAN1A2 as the predominant mRNA (SI Appendix, Fig. S7). We observed strong down-regulation at the transcript and protein levels for MAN1A2 in response to miR-30c, -181b-5p, and -361–5p mimics (Fig. 3 E and G), but no response was seen for miR-181a-5p (Fig. 3E and SI Appendix, Fig. S6C). The reciprocal effect was observed for miRNA inhibitors (Fig. 3 F and H and SI Appendix, Fig. S6C). Similar results were obtained in the renal cell line SN12C (SI Appendix, Fig. S6 D and E).
To determine whether miR-181b-5p, -361–5p, and -30c target MAN1A2 expression through direct binding to the 3′-UTR, we used a luciferase-MAN1A2-3′-UTR reporter assay (Fig. 3I and SI Appendix, Fig. S8A). Mimics of miR-30c and -361–5p inhibited luciferase expression (Fig. 3I). In contrast, miR-181b-5p, which is not predicted to target MAN1A2, did not affect luciferase levels. This miRNA was identified as regulating high mannose through our glycomic integration analysis and may be affecting MAN1A2 expression levels through an indirect mechanism or by targeting a region other than the 3′-UTR of the mRNA (3, 15). Mutation of the predicted binding sites of miR-30c or -361–5p abrogated the effect of these miRNA on MAN1A2-3′-UTR reporter, confirming their sites of action (Fig. 3I). Overall, our network identified predicted and unpredicted miRNA regulators of a key node in the N-linked pathway responsible for the balance of high-mannose and complex glycans.
Fucose Network.
Fucosylation is a key terminal modification involved in processes from the inflammatory cascade to microbial adhesion (28). We observed all five members of the miR-200 family [miR-200f: miR-141, -200a-3p (200a), -200b-3p (200b), -200c-3p (200c), -429] in a cluster with the fucose lectin AAL (SI Appendix, Fig. S3B) (29). The 200 family of miRNA is strongly associated with an epithelial phenotype and is down-regulated in epithelial to mesenchymal transition, a process in which epithelial cells transform into migratory mesenchymal cells (30, 31). This change in cell state is a critical process normally occurring in development and wound healing, but dysregulated in cancer metastasis and fibrosis (32). Previous analysis of miRNA in the NCI-60 by Park et al. identified a clear correlation between miR-200f levels and epithelial phenotype in this cell set (30). We examined the binding of AAL and UEA-I, another well-known fucose lectin, as a function of epithelial/mesenchymal status in the expanded NCI-60 dataset (Fig. 4A). Overall, fucosylation was enriched in the epithelial cell type, correlating with known miR-200f levels (30).
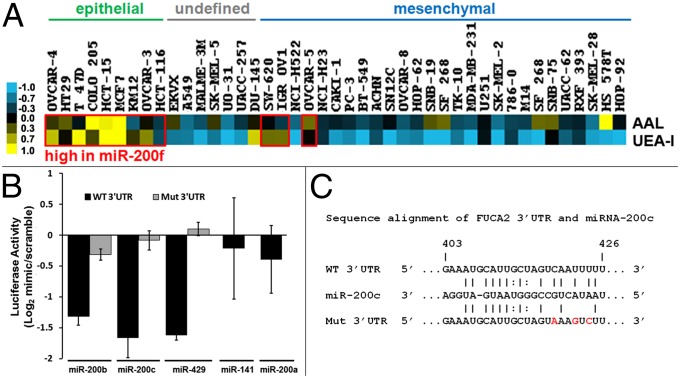
Validation of the fucose network. (A) Fucose lectin data from Fig. 1B (AAL, UEA-I) arranged to reflect phenotype as previously described (30). Cell lines with high expression of miR-200f are boxed in red (30). (B) Graphical representation of luciferase activity from FUCA2 constructs cotransfected with miR-200b, -200c, -429, or scramble mimics. Mut, FUCA2 mutant as indicated in C. Data were generated as in Fig. 3I. (C) Sequence alignment of miR-200c and FUCA2-3′UTR; mutated bases are shown in red.
We identified α-l-fucosidase-2 (FUCA2), a secreted enzyme that removes fucose, altering cell surface fucosylation levels (33), as a common predicted target for 6 of the 13 miRNA in the cluster, including miR-200f members miR-141 and -200a (Fig. 2E and SI Appendix, Fig. S3B). MiR-141/200a and miR-200b/200c/429 form two distinct groups of miR-200f and have overlapping and discrete targets (31). We examined the effects of miR-200f on FUCA2 expression by using a luciferase-FUCA2-3′-UTR reporter assay (Fig. 4B and SI Appendix, Fig. S8B). Of the five family members tested, only miR-200b, -200c, and -429 mimics showed inhibition of the luciferase signals. Mimics of miR-141 and -200a, which are predicted by microRNA.org to target FUCA2, showed no effect. miRNA target prediction algorithms allow for only a single mismatch between the seed region of the miRNA and the target sequence (14, 16). These requirements may be too stringent, precluding important sites. A scan of eight prediction programs using miRWalk revealed no binding sites for miR-200b, -200c, or -429 (34), but manual alignment of the miRNAs with the 3′-UTR of FUCA2 revealed a potential binding site with a 7-bp seed region and two mismatches flanked by multiple additional matched base pairs (Fig. 4C). Mutation of this site abrogated the effect of miRNA mimics on luciferase expression, validating it as the binding site (Fig. 4 B and C). We confirmed the differential effects of miR-200f members on FUCA2 in the HT-29 cell line, observing down-regulation of mRNA levels by real-time qPCR for miR-200b, -200c, and -429, but not for miR-141 and -200a, in line with our luciferase assays (Fig. 5A and SI Appendix, Fig. S9A). Mimics of miR-200b, -200c, and -429 also increased binding of AAL to HT-29, confirming their ability to modulate fucosylation in line with the NCI-60 analysis (Fig. 5B and SI Appendix, Fig. S9B). Taken together, these data validate our fucose network, demonstrating that fucosylation is controlled by the same miRNA switch responsible for epithelial cell status and may be a marker for this cell subtype.

MiR-200f members down-regulate FUCA2 expression and increase fucosylation in HT-29. (A) Real-time qPCR analysis of FUCA2 mRNA expression in cells treated with miR-200b-3p (200b), -200c-3p (200c), -429, or scramble mimic. Data were generated as in Fig. 3 C and E. (B) Representative fluorescence images of cells treated as in Fig. 3 A and B with indicated miR-200f mimics or scramble and stained with AAL. Monosaccharide inhibition control is shown (Ctrl). Data are representative of three biological replicates; SI Appendix, Fig. S9B provides statistical quantification of staining.
β-GalNAc Network.
Terminal GalNAc-β1,4-GlcNAc epitope (β-GalNAc) is found on a select subset of glycoproteins and glycolipids and correlates with neuroblastoma malignancy in humans (35). We observed a strong association between terminal β-GalNAc binding lectins (BDA, BPA, CAA, CSA, VVA) and miRNA predicted to target glycosyltransferases that modulate terminal β-GalNAc levels (R = 0.92, P = 0.005; Fig. 2F and SI Appendix, Fig. S3C). ST6GALNAC3, an α-2,6-sialyltransferase that catalyzes addition of sialic acid to β-GalNAc, is a predicted target of five of the seven miRNAs in this complex network. By using an ST6GALNAC3-3′-UTR luciferase reporter assay, we examined the ability of three miRNAs from the cluster [miR-200a-5p (200a*), 200b-5p (200b*), and miR-205–5p] to regulate enzyme expression (Fig. 6 and SI Appendix, Fig. S8C). Of the three mimics tested, only miR-200b* inhibited luciferase expression. No effect was observed for miR-200a* or miR-205–5p. miR-200a* and -200b* have identical seed regions but differ in their flanking bases, arguing that these are important in determining target specificity. Mutation of the miR-200b* binding region abrogated the effect of mimic on luciferase expression, validating the predicted binding site (Fig. 6). The miR-200b* binding site in ST6GALNAC3 is not conserved across species and would not be prioritized by prediction algorithms. By integrating miRNA and glycomic data, our analysis prompted us to validate this glycogene as a target of miR-200b*.

Validation of β-GalNAc network. Graphical representation of luciferase activity from ST6GALNAC3 constructs cotransfected with miR-200a-5p, -200b-5p, -205, or scramble mimics. Mut, ST6GALNAC3 mutant (SI Appendix, Table S3). Data were generated as in Fig. 3I.
Conclusions
The sheer complexity of the glycome, with its multiple potentially redundant enzymes, low glycogene expression levels, rapid evolution, and dendritic structures, makes identification of critical control mechanisms difficult. Herein we harness the power of our lectin microarray platform to demonstrate that the glycome communicates the genomic information flow governing tissue type and cell state at the surface of the cell. miRNAs are master regulators of this information flow and control the glycan biosynthetic network through direct interaction with glycogene transcripts, as we show in this work. miRNAs are known to target multiple genes concurrently, opening up the possibility that the glycome may be modulated along several pathways (e.g., N-linked, O-linked, glycolipid) simultaneously by the same miRNA. Although cancer cells were used in this work, miRNAs regulate normal biological processes ranging from T-cell development to embryogenesis, opening a window into glycogene regulation and involvement in these systems. Overall, our work begins to map glycans onto critical regulatory networks controlling cell phenotype, providing a means to deconstruct the glycocode.
Materials and Methods
Cell Lines.
The NCI-60 cell set was from the Division of Cancer Treatment and Diagnosis Tumor Repository (National Cancer Institute, Frederick, MD). Cell lines were grown in RPMI-1640 (Lonza) supplemented with 10% (vol/vol) FBS (Atlanta Biologics) and 2 mM l-glutamine (Lonza) at 37 °C in 5% CO2.
Lectin Microarray.
Sample preparation.
Cell samples were prepared and Cy5-labeled as previously described (36). Reference was prepared by mixing equal amounts (by protein) of membrane samples from HS-578T, PC-3, OVCAR-3, OVCAR-4, LOX IMVI, and SNB-19 before labeling with Cy3.
Printing, hybridization, and analysis.
Lectins were purchased from E. Y. Laboratories or Vector Laboratories with the following exceptions: recombinant cyanovirin, SVN and griffithsin were gifts from B. O’Keefe (Frederick National Laboratory for Cancer Research, Frederick, MD); recombinant Gaf-D, PA-IL, PA-IIL, and RS-IIL were made as previously described (37); TJA-I and TJA-II were from NorthStar Bioproducts. All antibodies were purchased from Abcam. SI Appendix, Tables S1 and S2, summarize the print lists and buffers. Printing, hybridization and data analysis were performed as previously described (20, 36). Our data were normally distributed as determined by the Lilliefors test in MATLAB. Lectins were excluded from analysis if they did not meet our minimal threshold for activity (20).
SVD and Multidimensional Analysis.
The normalized miRNA dataset from Liu et al. (22) was downloaded from CellMiner [RNA; Agilent Human microRNA (V2); http://discover.nci.nih.gov/cellminer/]. miRNA and lectin microarray data (SI Appendix, Fig. S1) were combined and ordered by tissue of origin for the four tissue types. SVD and the projection correlation using the dot product were performed by using the built-in “SingularValueDecomposition” function in Mathematica 8 on the collated dataset. For multidimensional analysis, projection correlation values for each lectin or miRNA vector were calculated by taking the dot product of the expression values with each of the first six eigengenes, which account for 94% of data variance. These correlation values were hierarchically clustered by using the PearCC (uncentered) as the distance metric and average linkage analysis in Cluster 3.0 and visualized in Java TreeView. Clusters were chosen for annotation if they met the following criteria: (i) >50% of lectins in the cluster had overlapping specificity toward a defined epitope, (ii) PearCC > 0.51 (P ≤ 0.15, single-tailed t test), and (iii) miRNAs in the cluster were annotated in microRNA.org and mapped onto a clear biosynthetic pathway. The centered PearCC as the distance metric gave different results, generally segregating lectins from miRNA; however, the high mannose cluster was observed and included miR-30c, which we validated (SI Appendix, Fig. S3A).
Mapping miRNA Onto Glycosylation Pathways.
Biosynthetic pathways for relevant glycan epitopes were mapped by using the Kyoto Encyclopedia of Genes and Genomes (38) and the work of Nairn et al. (2). The pathway data were then compared with the complete set of all predicted glycogene targets from the downloaded microRNA.org database (14) for miRNA within the corresponding cluster. miRNA glycogene targets were overlaid onto the appropriate pathways.
Transfection of miRNA Mimics and Inhibitors.
Cells were seeded in 12-well or 35-mm glass-bottom dish (1 × 105 cells), cultured 24 h, and transfected with miRNA mimics (50 nM) or inhibitors (25 nM, miRIDIAN; Dharmacon; SI Appendix, Table S3) with Lipofectamine 2000 (Invitrogen). Samples were analyzed 72 h (real-time qPCR and Western) or 96 h (microscopy) posttransfection.
Fluorescence Microscopy.
Fluorescence microscopy was used to confirm lectin microarray and miRNA data as it causes minimal perturbation to adherent cells. Cells were cultured in glass-bottom dishes, fixed in 4% (wt/vol) paraformaldehyde in HBSS (5 min, room temperature) and incubated with biotinylated lectins (10 µg/mL in PBS solution, 45 min, 37 °C; Vector Laboratories), followed by streptavidin-Cy5 staining (1:100 vol/vol in PBS solution, 30 min, 37 °C; Invitrogen) in the dark. NCI-60 validation samples were stained with DAPI (600 nM) to test for permeabilization, which was not observed under these fixation conditions. miRNA mimic-treated samples were not stained with DAPI. Samples were imaged by fluorescence microscopy (60× PlanFluor objective, NA 0.3, Eclipse TE 2000-U; Nikon) and a minimum of 10 fluorescence (excitation/emission, 625–650 nm/670 nm) and bright-field (phase contrast) image pairs were obtained. All samples stained with the same lectin were imaged under identical conditions. Lectins were preincubated with monosaccharide inhibitors (30 min, 200 mM mannose for HHL, PSL, GNA, NPA, LcH; 200 mM fucose for AAL) as a staining control. Images were analyzed in MetaMorph (Molecular Devices). For statistical analysis, images were background-subtracted and six random areas per image were selected in bright field. Regions with aggregated lectin or internal staining (as observed by bright fluorescent aggregates and/or DAPI staining where applicable) were excluded from the statistical analysis and alternatives regions selected. For NCI-60 validation studies, the average fluorescence of six areas was normalized to cell count (>29 cells per image, bright field) for four random images and averaged to generate graphs. For miRNA validation, data from 10 images per replicate (three biological replicates, 30 images total, 180 random areas) were averaged to generate graphs.
RNA Extraction and Real-Time qPCR.
Total RNA was extracted from samples (miRNeasy Mini Kit; Qiagen), quantified by using a NanoDrop ND-1000 device, and reverse-transcribed (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems). Transcripts were quantified by real-time qPCR using Power SYBR Green PCR Master Mix (Applied Biosystems) in a LightCycler 480 (Roche). Primers were designed by using PrimerSelect (SI Appendix, Table S3). Cycle threshold values were normalized to the housekeeping gene GAPDH. The average for three biological replicates was plotted as relative transcript abundance.
Western Blotting.
Cells were lysed in cold RIPA buffer supplemented with protease inhibitors. Equal amounts of protein were resolved by 10% SDS/PAGE, transferred onto nitrocellulose membranes, and blocked in block buffer [5% (wt/vol) BSA, PBST (PBS, pH 7.4, 0.05% Tween-20), 1 h, room temperature]. Antibodies were diluted in block buffer; primary antibodies were as follows: α-MAN1A2 (1:1,000; Novagen), α-MAN1A1 (1:1,000; Abcam), α-GAPDH (1:5,000; Abcam); secondary antibodies corresponding to the primary antibodies were as follows: α-mouse or α-rabbit-HRP (1:5,000; Bio-Rad). Blots were developed by using SuperSignal West Pico (Thermo Scientific).
Luciferase Reporter Assay.
MAN1A2-3′UTR and ST6GALNAC3-3′UTR reporter plasmids were obtained from SwitchGear Genomics. FUCA2-3′UTR was cloned from HT-29 cDNA and inserted into pLightSwitch-MT vector (SwitchGear Genomics). Mutagenesis of seed regions was performed with Phusion Hot Start Flex (New England Biolabs) and 5′-phosphorylated primers (SI Appendix, Table S3, shows primers and constructs). Plasmids were purified using EndoFree Plasmid Maxi Kit (Qiagen) for transfection. Each plasmid (250 ng DNA) was cotransfected with 60 nM miRNA or scramble mimic in HEK 293T/17 cells in a 96-well plate by using Lipofectamine 2000 (Life Technologies). After 24 h, luminescence was developed by using LightSwitch Assay Reagent (SwitchGear Genomics) and read on a SynergyHT microplate reader. All luciferase data were normalized to scramble control.
Acknowledgments
We thank Drs. E. Hernando-Monge and J. Ribeiro for critical reading of the manuscript and Boval Biosolutions for lyophilized protease- and IgG-free bovine serum albumin (no. LY-0081). This work was supported by National Institutes of Health Grant 7 DP2 OD004711-02.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1321524111/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1321524111
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/111/11/4338.full.pdf
Citations & impact
Impact metrics
Article citations
ST3GAL1 Promotes Malignant Phenotypes in Intrahepatic Cholangiocarcinoma.
Mol Cell Proteomics, 23(9):100821, 26 Jul 2024
Cited by: 0 articles | PMID: 39069074 | PMCID: PMC11385758
The modulation of the hexosamine biosynthetic pathway impacts the localization of CD36 in macrophages.
Acta Biochim Pol, 71:13004, 08 Jul 2024
Cited by: 0 articles | PMID: 39041003 | PMCID: PMC11261345
Protein glycosylation in cardiovascular health and disease.
Nat Rev Cardiol, 21(8):525-544, 18 Mar 2024
Cited by: 1 article | PMID: 38499867
Review
The forecasting power of the mucin-microbiome interplay in livestock respiratory diseases.
Vet Q, 44(1):1-18, 12 Apr 2024
Cited by: 0 articles | PMID: 38606662 | PMCID: PMC11018052
Deep learning explains the biology of branched glycans from single-cell sequencing data.
iScience, 25(10):105163, 19 Sep 2022
Cited by: 4 articles | PMID: 36217547 | PMCID: PMC9547197
Go to all (45) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Insights into miRNA regulation of the human glycome.
Biochem Biophys Res Commun, 445(4):774-779, 23 Jan 2014
Cited by: 20 articles | PMID: 24463102 | PMCID: PMC4015186
Reading and Writing the Human Glycocode.
Annu Rev Biochem, 93(1):529-564, 02 Jul 2024
Cited by: 0 articles | PMID: 38669516
Review
A computational framework for the automated construction of glycosylation reaction networks.
PLoS One, 9(6):e100939, 30 Jun 2014
Cited by: 25 articles | PMID: 24978019 | PMCID: PMC4076241
miRNA proxy approach reveals hidden functions of glycosylation.
Proc Natl Acad Sci U S A, 112(23):7327-7332, 26 May 2015
Cited by: 25 articles | PMID: 26015571 | PMCID: PMC4466752
Funding
Funders who supported this work.
NIH HHS (2)
Grant ID: DP2 OD004711
Grant ID: 7 DP2 OD004711-02





