Abstract
Free full text

Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation
Associated Data
Abstract
The transcription of genomic information in eukaryotes is regulated in large part by chromatin. How a diverse array of chromatin regulator (CR) proteins with different functions and genomic localization patterns coordinates chromatin activity to control transcription remains unclear. Here we take a synthetic biology approach to decipher the complexity of chromatin regulation by studying emergent transcriptional behaviors from engineered combinatorial, spatial, and temporal patterns of individual CRs. We fuse 223 yeast CRs to programmable zinc finger proteins. Site-specific and combinatorial recruitment of CRs to distinct intra-locus locations reveals a range of transcriptional logic and behaviors, including synergistic activation, long-range and spatial regulation, and gene expression memory. Comparing these transcriptional behaviors with annotated CR complex and function terms provides design principles for the engineering of transcriptional regulation. This work presents a bottom-up approach to investigating chromatin-mediated transcriptional regulation, and introduces new, chromatin-based components and systems for synthetic biology and cellular engineering.
Introduction
Eukaryotic genomes are packaged into chromatin, a higher-order structure of DNA, histones, and associated proteins. A diverse array of chromatin regulators (CRs) form complexes that act on and modify chromatin in unique combinatorial, spatial, and temporal patterns, thereby regulating how the underlying genomic information is transcribed and vastly extending the information potential of the genome (Figure 1) (Li et al., 2007; Narlikar et al., 2002; Ram et al., 2011). Yet, despite being the subject of extensive studies, the relationships between CRs and gene regulation remain unclear.
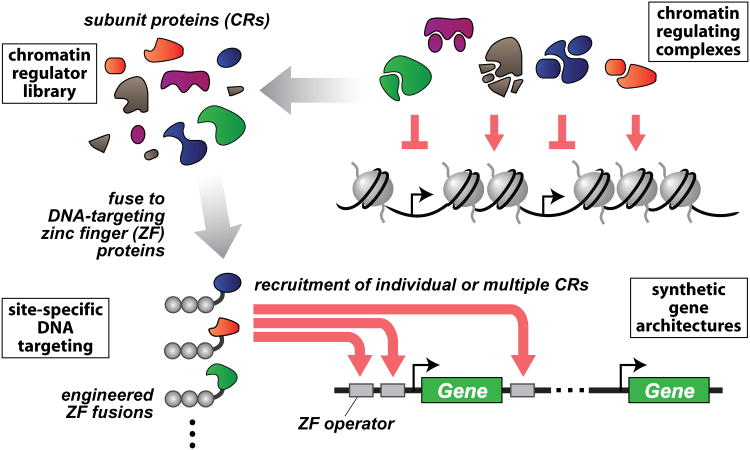
Eukaryotic gene transcription is regulated by diverse chromatin-regulating complexes and networks (top right). The complexes were decomposed into a library of subunit chromatin regulator (CR) proteins (top left). These subunits were fused to engineered zinc finger (ZF) proteins to enable site-specific spatial and combinatorial targeting to designed gene loci (bottom). This modular framework allows the direct functional characterization of individual CRs as transcriptional regulators, and for designing locus architectures that recruit different combinations of CRs to explore and engineer complex spatial and combinatorial transcriptional regulation.
There are a number of hypothesized mechanisms by which CRs modulate and control gene transcription. First, at each gene, chromatin can be combinatorially regulated by numerous CR proteins with different functions (Li et al., 2007; Ram et al., 2011; Venters et al., 2011). Thus, processes ranging from forming and recruiting preinitiation complexes, remodeling and assembling nucleosomes, increasing chromatin accessibility through histone modifications, and promoting transcriptional elongation may act in concert to generate a wide range of transcriptional outputs and logic (Lam et al., 2008; Mirny, 2010; Narlikar et al., 2002).
Relatedly, histone tails have numerous residues that can be decorated by a wide assortment of biochemical modifications. Genome-wide and gene expression profiling studies have correlated specific combinations of modifications (Liu et al., 2005; Zhou et al., 2011) and associated CRs (Ram et al., 2011; Venters et al., 2011) with chromatin structure and gene expression state. These findings have lent support to the “histone code” hypothesis, which posits that specific combinations of histone tail modifications serve to recruit proteins that establish or alter transcriptional activity (Strahl and Allis, 2000). Uncovering the distinction between the simple presence of and the causal transcriptional function of chromatin marks (and CRs) remains an active area of investigation (Henikoff and Shilatifard, 2011).
In addition to the combinatorial patterning of chromatin modifications, histones (Zhou et al., 2011) and CRs (Ram et al., 2011; Venters et al., 2011) are found in distinct spatial patterns around and throughout genes, raising the compelling possibility that spatial organization underlies transcriptional control (Li et al., 2007; Pokholok et al., 2005; Weinberger et al., 2012). Methods for directly linking transcriptional function with the localization of CRs within and around genes are needed to establish these principles.
Finally, spatial changes in chromatin modifications, such as spreading of DNA methylation and histone hypoacetylation marks, are believed to give rise to stable epigenetic states (Dodd et al., 2007; Hathaway et al., 2012). Identifying specific CRs and conditions that drive these epigenetic changes is critical for understanding how gene expression memory is established and how genes and loci are stably activated or repressed during developmental or disease processes.
Understanding these regulatory principles requires systematic approaches for investigating CR function, for example, to determine: (i) which CRs (or classes of CRs) can activate or repress transcription; (ii) what forms of transcriptional logic are obtained from combinatorial regulation by multiple CRs at a single gene; (iii) how transcriptional regulatory information is encoded in the spatial organization of CRs and genes; and (iv) what potential epigenetic properties are associated with CRs.
Current approaches to study chromatin function are largely based on pharmacological and genetic perturbations combined with genome-wide measurements of gene expression and chromatin state. These approaches have yielded fundamental insights (Lenstra et al., 2011; Ram et al., 2011), but are limited in their ability to directly test CR function because of global and pleiotropic effects and context-dependent recruitment of CRs to different genomic loci. Furthermore, correlative measurements of chromatin structure and function make it difficult to distinguish downstream from causative perturbations (Henikoff and Shilatifard, 2011; Ptashne, 2013).
To address these limitations, synthetic biology approaches may provide unique and complementary advantages, such as the ability to decompose these complex systems into well-understood components and to directly test CR function through site-specific perturbations. Moreover, with the recent advent of programmable DNA-targeting platforms, CRs can be site-specifically recruited to defined genomic sequences, a feature that has been exploited to develop “epigenome editing” tools for altering DNA methylation states and histone modifications (Hathaway et al., 2012; Konermann et al., 2013; Maeder et al., 2013a; Mendenhall et al., 2013).
Here, we take a synthetic biology approach to study and classify transcriptional behaviors emerging from engineered combinatorial, spatial, and temporal patterns of targeted CRs. Specifically, programmable zinc fingers (ZFs) are fused to a library of 223 yeast CR proteins encompassing 45 known chromatin complexes (Figure 1). First, this library is site-specifically targeted to a minimal gene locus to identify factors that activate or repress transcription. CRs are clustered by gene ontology annotations in order to classify chromatin complexes and protein functions that causatively regulate transcription. We then recruit CRs in combination with the VP16 transactivator to reveal different forms of transcriptional logic. Spatially recruiting CRs in distinct patterns and locations within single- and multi-gene loci identifies classes of engineered CRs capable of regulation from (non-promoter) downstream positions, long-range transcriptional regulation, and gene expression memory. Taken together, our work motivates bottom-up experimental approaches for assigning CR function and uncovering rules governing chromatin-based gene regulation. This work also presents a new class of regulatory components, locus architectures, and design principles for synthetic biology applications (Fischbach et al., 2013; Khalil and Collins, 2010; Purnick and Weiss, 2009; Weber and Fussenegger, 2010; Ye et al., 2013).
Results
Targeted transcriptional regulation at a synthetic reporter locus
We introduced a synthetic transcriptional reporter into the Saccharomyces cerevisiae genome, in which expression of yEGFP is controlled by a minimal CYC1 promoter harboring upstream, tandem operator sites recognized by an engineered ZF protein (43-8, GAGTGAGGA) (Figure 2A, top) (Khalil et al., 2012). pCYC1 (-183 TSS +66) was chosen for its intermediate basal level of expression (Blount et al., 2012; Gari et al., 1997; Khalil et al., 2012). The core CYC1 promoter has also been used to identify both transcriptional repressors and activators (Martens et al., 2001). Furthermore, the depletion of histone H4 has been shown to activate the core promoter 94-fold, indicating the importance of basal chromatin in its regulation and thus its potential utility in this study (Han and Grunstein, 1988). Finally, this minimal promoter lacks endogenous upstream regulatory sequences, including both the heme-responsive activating sequence and the glucose-mediated repression site (Guarente et al., 1984; Olesen et al., 1987), thus reducing the effects of signaling crosstalk, non-coding RNAs, and endogenous recruitment of synthetic CRs. This reporter construct and others described below were genomically integrated into the URA3 locus. We also integrated copies of the reporter into the HIS3 and LEU2 loci to confirm that our results were similar in different genomic loci (see Figures S1, S3, S5E-F, S7A, S7D and Tables S1 and S4). In order to test the ability of this reporter to recruit regulators and report on transcriptional activity, we fused canonical transcriptional activating (VP16) and repressing (Mig1, aa481-503) domains to the targeting ZF protein (43-8) as well as to a non-specific ZF (42-10, GACGCTGCT) (Khalil et al., 2012). Expression of these fusion proteins was driven by a small-molecule inducible version of the GAL1 promoter. Upon expression, only the targeted factors activated or repressed the locus (Figure 2A, bottom).
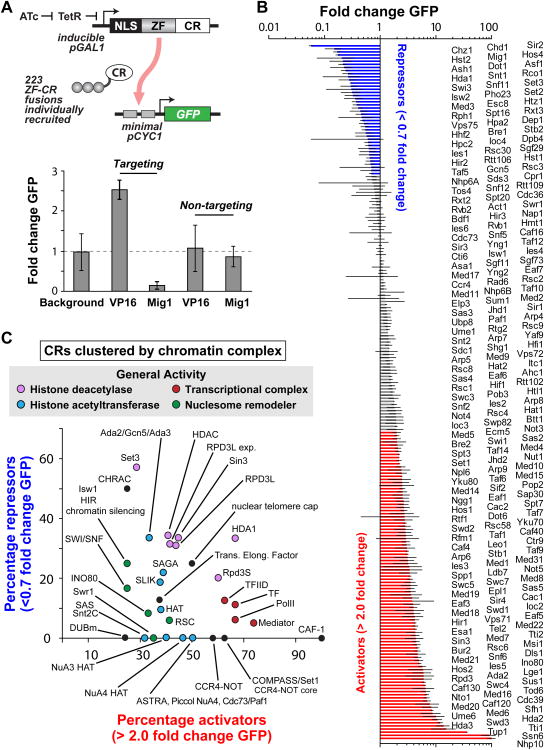
(A, top) 223 CR proteins were fused to an engineered ZF protein (or non-targeting ZF) and placed under the control of an inducible GAL1 promoter. Each fusion protein was individually recruited to operators placed upstream of a minimal CYC1 promoter driving the expression of GFP. NLS, nuclear localization signal. (A, bottom) Fold change in GFP expression induced by VP16 activation and Mig1 repression domains fused to targeting or non-targeting ZF proteins. (B) Fold change in GFP expression for the library of 223 ZF-CR fusions (normalized to uninduced levels). Repressors (blue bars) were classified as having < 0.7 fold change, while activators (red bars) have > 2 fold change. (C) CRs grouped by complex and plotted according to the percentage of activators and repressors in each complex. Dot colors correspond to the general activities of each complex. Error bars are standard deviations of three isogenic strains. See also Figures S1 and S2.
Identifying functional transcriptional regulators from a library of targeted CRs
A large body of work has identified correlations between the expression of specific CRs and global transcriptional activity. However, it is often unclear which CRs are causative of or merely associated with changes in transcriptional activity at specific loci. Therefore, we fused a library of 223 full-length putative CRs, comprising 45 chromatin-regulating complexes (Lenstra et al., 2011), to the targeting ZF and individually tested each protein's ability to activate or repress transcription from the pCYC1 reporter (Figure 2A, top). As shown in Figure 2B, numerous repressors and activators were identified from the library, spanning approximately 20-fold changes in repression and activation (Figures 2B, S1A, S1D and Table S1). We also observed expected changes in histone modifications at the reporter locus, measured by chromatin immunoprecipitation-quantitative PCR, upon expression of 17 CRs chosen for their predicted histone modifying catalytic domains/activities (Figure S2B). To confirm the changes in reporter expression were not simply a product of CR overexpression, we fused the 27 strongest repressors, 48 strongest activators, and all CRs with histone-modifying catalytic domains to a truncated, non-binding ZF protein. The vast majority of these fusions generated negligible changes in yEGFP expression (Figure S6B). Rsc3, Ldb7, Sum1 and Tod6 were exceptions, exhibiting changes in yEGFP levels regardless of targeting, suggesting either ZF-independent recruitment to the locus or global transcriptional regulation. We observed no correlation between transcriptional activation and the level of CR expression as measured by western blot (Figure S2C).
Clustering CRs by chromatin complex and function
We next asked if the targeted library could identify relationships between transcription and CR protein functions or complexes. Using gene ontology annotations, we first clustered all CRs by macromolecular complex (Table S2). Individual CRs were then conservatively classified as activators (Figure 2B, red bars, > 2 fold change yEGFP) and repressors (blue bars, < 0.7 fold change yEGFP). This classification excluded all non-targeted CRs, aside from the exceptions noted above (Figure S6B). When the percentage of activators in each chromatin complex was plotted against the percentage of repressors (Figure 2C, Table S2, Figure S1B), we discovered a number of clear patterns. Histone acetyltransferase (blue dots), H3K4 methyltransferase (“COMPASS/Set1”), and RNA PolII transcription-related complexes (red dots) were mostly composed of activating CRs. Histone deacetylase complexes were primarily composed of repressive CRs (pink dots). Nucleosome remodeling complexes trended weakly towards having more activators than repressors (green dots). When clustered by protein function terms (Figure S2A, Table S2, Figure S1C), groups associated with the transcriptional complex (red dots), histone acetyltransferase (blue dot), and histone methyltransferase (brown dots) terms contained primarily activators, while groups associated with chromatin binding (orange dots) and histone deacetylase (pink dots) terms contained primarily repressors. These results largely agree with the regulatory roles assigned to various complexes and protein functions through previous genome-wide and knockout/mutant strain studies (Lenstra et al., 2011; Ram et al., 2011; Venters et al., 2011).
Engineering combinatorial transcriptional logic
Native genes are simultaneously regulated by multiple proteins with different functions and activities, often giving rise to combinatorial transcriptional logic. Therefore, we next explored how co-recruitment of factors affects transcription. In particular, we were interested in how different CRs modulate the activity of a co-recruited VP16 domain. We fused VP16 to a second, orthogonal ZF protein (97-4, TTATGGGAG) (Khalil et al., 2012), which could be independently recruited to an operator placed directly downstream of the ZF-CR operator (Figure 3A). As expected, upon co-recruitment of VP16 with the ZF-CR library, we found that transcriptional outputs generally increased as compared to recruitment of CRs alone (Table S3). The CRs divided into six distinct classes of combinatorial regulators based on transcriptional logic: CRs capable of (1) dominant (Sir2 and Mig1) and (2) partial (Ash1 and Dot1) inhibition of VP16-mediated activation; CRs with no regulatory roles on their own and either (3) no (Eaf7 and Rvb2) or (4) enhanced (Ies6 and Cdc73) effect on VP16-mediated activation; finally, CRs that act (5) additively (Taf14 and Med4) or (6) synergistically (Cac2 and Set1) with VP16 to increase yEGFP expression (Figures 3B, S3A, and S3B). Synergy is the “cooperation” of factors to produce a total output, and here it was defined as the fraction of total output in excess of summing the outputs from the individual components (Figures 3C, 3D, S3C, and S3D).
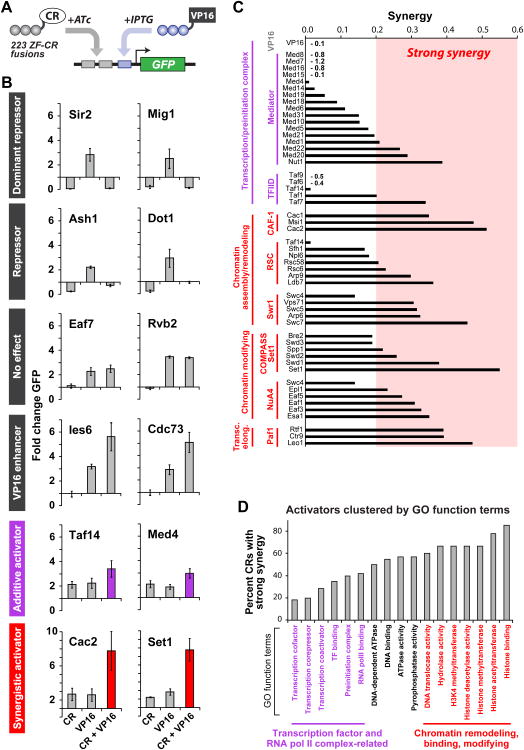
(A) An engineered two-input system enabling the co-recruitment of CRs and VP16 transactivating domain (ZF 43-8, grey; ZF 97-4, blue) (Khalil et al., 2012). (B) Representative transcriptional logic outputs of the two-input system divide CRs into six distinct classes (top to bottom): VP16-independent dominant repressors, repressors, CRs with no effect, VP16 enhancers, additive activators (purple), and synergistic activators (red). (C) Activating CRs clustered by complex and plotted by level of transcriptional synergy. Transcription/preinitiation complex regulators generated weak synergy, while chromatin assembly/remodeling, chromatin-modifying, and transcription-elongating regulators generated strong synergy. Synergy is the “cooperation” of factors to produce a total output, and here is defined as the fraction of total output not accounted for by summing the outputs from the individual components. Synergy = [(A – 1) – (B – 1) – (C – 1)]/(A – 1) where A = CR and VP16, B = CR only, and C = VP16 only. (D) Activators clustered by gene ontology function terms and plotted as percentage of CRs in each term group with “strong synergy” (greater than the average synergy of 0.2). Error bars are standard deviations of three isogenic strains. See also S3.
To develop insight into CR functions that may underlie these different, combinatorial logic behaviors, we clustered CRs by complex (Figures 3C and S3C) and function (Figures 3D and S3D) and calculated the percentage of CRs in each cluster with strong synergy. When clustered by complex, we found that the majority of Mediator and TFIID subunits exhibited weak synergy with VP16 (Figures 3C and S3C, purple bars). In contrast, complexes that remodel and assemble chromatin (Swr1, RSC, CAF-1), promote transcriptional elongation (Paf1), or modify histones to open chromatin structure (NuA4, Set1) were comprised primarily of CRs that synergistically enhanced activation (Figures 3C and S3C, red bars).
We observed the same general trend when we clustered activating CRs by function, as opposed to complex (Figures 3D and S3D, Table S4); that is, CRs related to transcription factor and RNA PolII terms exhibited weak synergy with VP16 (purple), while those associated with chromatin remodeling, modifying, and binding exhibited strong synergy (red). VP16 is believed to activate transcription by recruiting preinitiation and transcription complex factors along with the Mediator complex (Milbradt et al., 2011). Thus, additive activation might occur through a co-recruited CR that functions similarly to VP16 or is part of either the transcription complex or Mediator. Importantly, we observed a simple additive relationship when we co-recruited two identical VP16 domains (Figure 3C, “VP16”). In contrast, other functions such as remodeling nucleosomes, modifying histones to alter chromatin accessibility, and promoting transcriptional elongation may synergistically amplify the output by increasing access of transcriptional machinery to DNA (Lam et al., 2008; Mirny, 2010).
Revealing spatially-encoded regulatory modes
While transcriptional regulation is canonically focused at promoter regions, there is also considerable evidence for chromatin-mediated regulation at other locations relative to ORFs: (1) nucleosomes are arrayed over entire genes with distinct positioning at promoter and terminator regions (Lam et al., 2008); (2) native CRs and transcription factors are often localized to spatially-specific, non-promoter regions to regulate genes (Groner et al., 2010); and (3) histone mark gradients have been observed over genes (Li et al., 2007; Pokholok et al., 2005). These observations suggest that CRs may asymmetrically and differentially regulate genes depending on their relative location to an ORF.
We sought to explore spatially-dependent regulatory behaviors using site-specific CR recruitment. We moved the ZF operators in our reporter locus from upstream of the coding sequence to downstream of the terminator (Figures 4 and S4). Our library of ZF-CRs was then inducibly recruited to the downstream element (Figure S4, blue and gold bars). No CRs were able to activate transcription from the downstream position, suggesting the importance of preinitiation/transcription complex assembly at promoters for activation. However, many CRs were able to repress transcription from the downstream position. Interestingly, several of these CRs exhibited “asymmetric” regulatory modes; in other words, they had opposite regulatory functions when targeted upstream versus downstream (Figure S4, grey bars). To develop insight into CR functions that may underlie these spatially-encoded behaviors, we grouped CRs by their spatial regulatory profile (i.e., upstream-activating or -repressive and downstream-activating or -repressive) and obtained associated gene ontology function terms for each group (Figure 4, Table S5). Subsets of these terms were unique to each grouping (Figure 4). Interestingly, while many upstream-activating/downstream-neutral CRs were associated with regulation of the transcriptional complex, factors that were upstream-activating/downstream-repressive appeared enriched in ATPase remodeling and DNA translocase activity. This suggests that remodeling activities can influence transcription from both ends of a gene, potentially by increasing RNA PolII accessibility at upstream regions while disrupting transcriptional elongation at downstream regions.
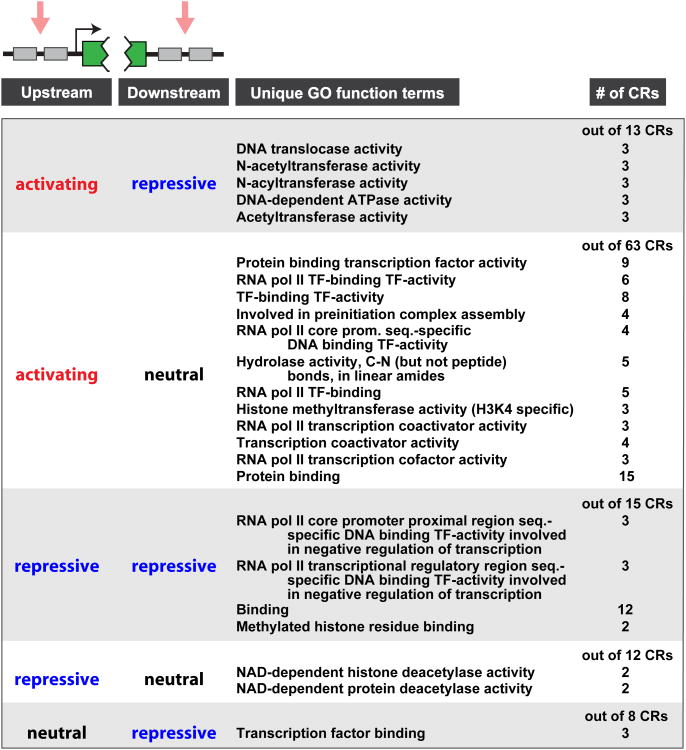
A gene locus was engineered to recruit 223 CR fusions to operators either upstream or downstream (downstream of a CYC1 terminator) of a reporter gene. CRs were grouped according to their upstream- and downstream-targeted regulatory profiles. Gene ontology function terms unique to each group are listed along with the number of CRs in the group associated with each term. See also Figure S4.
Simultaneous and differential regulation of multiple genes
The spatial qualities of chromatin-based regulation could be exploited to engineer the simultaneous regulation of multiple genes. For example, based upon our comparison of upstream versus downstream targeting of the CR library (Figure 4), a single CR might simultaneously (and differentially) regulate two genes if recruited upstream of one ORF and downstream of another. To identify CRs capable of such simultaneous regulation, we constructed a dual-gene reporter system (Figure 5A) and recruited a small subset of our ZF-CRs to it. The CRs exhibited a variety of dual-gene regulation profiles (Figures 5A, S5A, and S5E). Notably, these included factors that could activate expression of one reporter gene while repressing the other. To confirm that the local contexts of the genes were not responsible for the observed behaviors, the ZF operators were swapped between upstream and downstream positions at both genes (Figures 5B, S5B, and S5F). As expected, the regulatory profiles were correspondingly inverted. Moreover, fusions of CRs to non-binding ZFs did not appreciably modulate transcription, strongly suggesting that these engineered regulatory modes are the result of site-specific targeting (Figures S5A, S5B, S5E, and S5F, left).
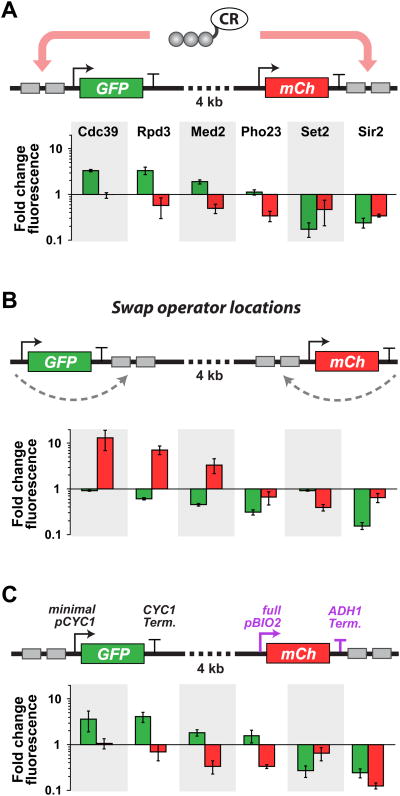
(A) Schematic of the engineered, dual-gene reporter locus (CYC1 promoters and terminators used throughout) (top). Fold change in GFP (green bars) and mCherry (red bars) expression for six targeted CR fusions (bottom). (B) Swapping operator locations results in inversion of transcriptional outputs. (C) Schematic of the same locus architecture as in (A) but containing two different promoters and terminators (BIO2 promoter and ADH1 terminator in purple). Error bars are standard deviations of three isogenic strains. See also Figure S5.
To test if we could shift the dynamic ranges of both reporter genes while qualitatively maintaining the same dual-gene regulatory profiles, we co-recruited VP16 to upstream positions in the dual-gene architecture (Figure S5, right columns). We were able to engineer increased dynamic ranges of both reporter genes while maintaining similar regulatory trends of the CRs. Since future applications may require the simultaneous regulation of two distinct promoters, we next added a second, different promoter to the reporter construct. Specifically, we replaced the downstream CYC1 promoter that drives the expression of mCherry with the full-length BIO2 promoter, which has a similar intermediate basal level of expression as the CYC1 promoter (Figures 5C, S5C, and S5D) (Blount et al., 2012). Overall, we found that the regulatory output profiles were consistent with those from the reporter harboring two repeated CYC1 promoters, suggesting conservation in these forms of regulation.
Long-range and multi-gene regulation
Our results support the notion that spatial location and patterning of CRs influences regulatory function. To further explore spatial effects, we next asked if CRs could regulate genes from longer distances. Heterochromatic structures are known to spread over large regions of the genome through hypothesized self-reinforcing mechanisms (Bi et al., 2004; Dodd et al., 2007; Hathaway et al., 2012). We sought to harness this potential by constructing a three-color reporter system that could be used to identify factors capable of long-range transcription control (Figure 6A). We recruited a set of the strongest repressors and activators upstream of the first gene (Figures 6, S6C, and S7A). Most CRs modulated expression of only the proximal gene (yEGFP) without affecting downstream genes (Figure 6B), while non-targeting controls did not affect expression of any of the reporter genes (Figures 6B, S6B, and S7A). However, two CRs (Sir2 and Rph1) were able to robustly repress all three genes in the cluster. Intriguingly, Sum1 also showed evidence for multi-gene regulation but through a distinctive spatial pattern, in which repression was strongest for the most distal gene and weakest for the proximal gene. Yet, it should be noted that Sum1 was also unique in that it showed some (weak) repressive abilities in an inverted spatial pattern (strongest repression for the proximal gene and weakest for the distal gene) when fused to a non-targeting ZF (Figures 6B and S7A).

(A) Schematic of the engineered, multi-gene reporter locus. The 27 strongest repressors and 48 strongest activators identified from the full ZF-CR library as well as all CRs with histone-modifying catalytic domains were targeted upstream of the first gene. (B) Heat map of the fold change in fluorescence for GFP, mCherry, and BFP, revealing classes of CRs that regulate only the proximal gene (left and middle) or that repress all three genes in the locus (right). See also Figure S6.
Long-range regulation and epigenetic memory are both hypothesized to rely on self-reinforcing mechanisms that enable spreading of chromatin modifications from nucleosome to nucleosome (Dodd et al., 2007). To explore the engineering of memory via our targeted CRs, we chose three representative regulators (Med16, Isw2, Sir2) and tested their ability to sustain gene expression changes. We performed induction/wash-out experiments for these CRs and measured reporter output over time. While outputs for the activator Med16 and repressor Isw2 returned to basal levels post-washout, Sir2 was able to stably repress the proximal gene (yEGFP) for 24 hr post-washout (Figure 7A). Interestingly, the reactivation rate of the downstream genes appeared to correlate with distance from the position of CR binding. To test for the possibility that ZF-Sir2 was long-lived and still present post-washout, we measured ZF-Sir2 occupancy at its operator, H4K16 acetylation levels at the yEGFP promoter, and yEGFP expression at several time points (Figure S7B and S7C). At 12 hours post-washout, we observed ZF-Sir2 occupancy had returned to pre-induction levels while yEGFP expression and H4K16 acetylation remained repressed for several cell divisions (between 24-30 hrs), suggesting heritable reporter repression and histone modification.
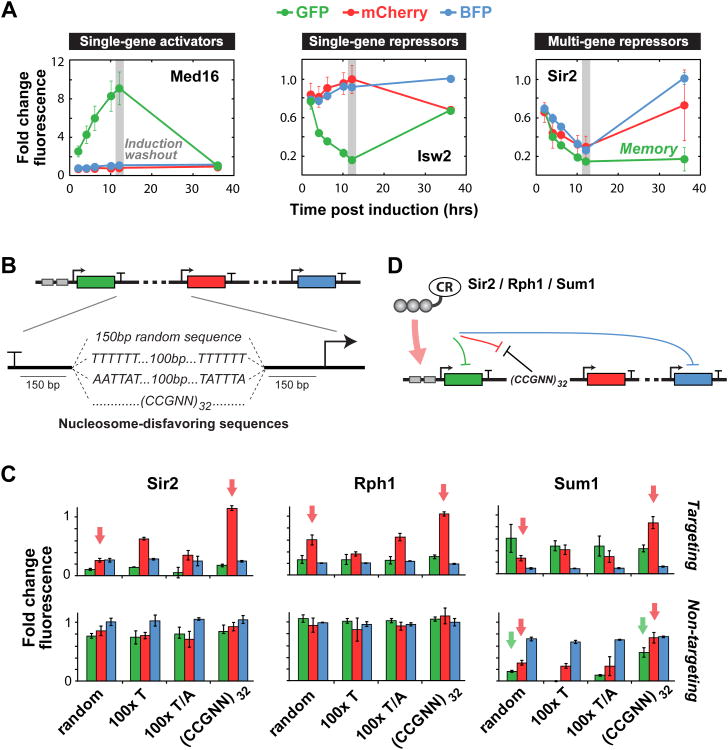
(A) Time courses of induction/wash-out experiments for three CRs. CR fusions were expressed at t = 0 hr by the addition of the small molecule ATc, which was subsequently washed out at t = 12 hr (grey bars). Med16 and Isw2 show reversible activation and repression of GFP, respectively. Sir2 maintains full repressive memory of the proximal gene and partial repressive memory of the downstream gene. (B) Nucleosome-disfavoring sequences inserted between the GFP and mCherry genes as putative barrier or insulator elements. (C) Fold change in fluorescence for GFP, mCherry, and BFP induced by targeting (top) or non-targeting (bottom) multi-gene repressors (Sir2, Rph1, Sum1 fusions). The (CCGNN)32 sequence robustly insulates only the middle gene (mCherry) from repression by the CRs. (D) Schematic of the multi-gene regulatory circuit. Error bars are standard deviations of three isogenic strains. See also Figure S7.
DNA sequences have been identified that block heterochromatin spreading by disfavoring nucleosome-binding through DNA conformation preferences and binding thermodynamics (Bi et al., 2004; Raveh-Sadka et al., 2012). These include a stretch of 100 deoxythymidines, a mix of 100 deoxythymidines and deoxyadenines, and 32 repeats of CCGNN (where N is any deoxynucleotide). We asked if these sequence elements could be inserted into our triple reporter locus to insulate specific genes from long-range repression by Sir2, Rph1, and Sum1 (Figure 7B). We found that only CCGNN repeats were able to fully block repression (Figures 7C and S7D). Moreover, they could relieve repression of the proximal downstream gene (mCherry), but not the distal downstream gene (BFP). Similar effects were observed with Rph1 and Sum1. Thus (CCGNN)32 could be used to insulate genes, even those in the middle of an expression cassette, from the effects of long-range repressors (Figure 7D).
Discussion
Hundreds of CR proteins act on chromatin in complex and combinatorial ways to regulate gene transcription. Here we took a synthetic biology approach to study and classify transcriptional behaviors emerging from engineered combinatorial, spatial, and temporal patterns of CRs. Our results provide us with new components that can be used in synthetic biology and chromatin biology: (1) functional activators and repressors; (2) six classes of combinatorial regulators for programming multi-input logic: dominant repressors, repressors, neutral factors, VP16 enhancers, additive activators, and synergistic activators; (3) distinct classes of spatially-encoded regulators (e.g., “asymmetric” regulators), including CRs that can repress transcription from a downstream position; (4) CRs capable of regulating only proximal genes, as well as CRs capable of regulating all genes simultaneously (long-range regulators), which in one case also produced robust gene expression memory.
New parts for synthetic biology and cellular engineering
Synthetic biology offers a bottom-up approach for exploring the design and function of biological systems, and for engineering cells and organisms to address a range of biomedical and industrial applications (Fischbach et al., 2013; Khalil and Collins, 2010; Purnick and Weiss, 2009; Weber and Fussenegger, 2010; Ye et al., 2013). Here we decomposed chromatin-based transcriptional regulation into minimal components – minimal promoters and individually-targeted CRs – to provide a useful framework of parts and behaviors for broad applications in synthetic biology and cellular engineering.
Targeted CRs could be used as synthetic transcriptional activators and repressors in eukaryotic organisms. Many CRs matched or exceeded the activation or repression levels achieved by commonly used regulatory domains, such as VP16 and Mig1 (Figure 2A and 2B). While the behavior of any individual CR may vary for different genomic contexts, the general regulatory properties revealed by this library-based approach will streamline selection and testing of relevant CRs. Moreover, we observed strong correlation between the relative activities of CRs in alternative loci (Figures S1, S3, S5E-F, S7A, S7D and Tables S1 and S4), suggesting some conservation or robustness in the function of these factors across different genomic contexts.
This work also has interesting implications for the design of new synthetic gene circuits. First, our work demonstrates that chromatin-based components can vastly extend the regulatory potential of an individual genetic locus, thus expanding the regulatory possibilities of circuit nodes. A diverse range of transcriptional logic can be programmed by designing a genetic locus to recruit different combinations of CRs. As a result, circuits composed of CRs may represent a more efficient solution to information processing than those composed of canonical transcriptional components, like bacterial transcription factors. In other words, a minimal number of CRs targeted to a single locus may perform similar logical or computational tasks as a network composed of many interacting transcription factors. This feature could be useful for biotechnology applications by helping to reduce the size of gene expression cassettes to be delivered into a cellular host. Yields from bioprocesses could also be increased by the reduction in metabolic load on production organisms. Second, quantitative control of transcriptional outputs, including the ability to program synergistic activation, could be useful in controlling the expression levels of enzymes in engineered metabolic pathways and of regulatory proteins in synthetic circuits. Chromatin-based control schemes could be used to tune the sensitivity of cellular sensors to multiple environmental factors, or to tune the expression range of signaling factors such as chimeric antigen receptors in T-cell adoptive immunotherapy. The properties of synthetic circuits such as induction threshold, cycle period, and entrainment strength are known to be sensitive to expression levels (Atkinson et al., 2003), which could in principle be tuned through co-recruitment of synergistic CRs. Finally, epigenetic regulation of specific sets of genomic loci fundamentally underlies the transition between distinct cellular states, including in response to stress (Crews et al., 2012) or differentiation into cells of distinct tissue types (Meissner, 2010). The ability to establish epigenetic states at defined loci may enable construction of simplified synthetic systems to study the regulatory principles governing these processes.
Chromatin-based systems also enable multi-gene regulation, providing interesting new strategies for precisely addressing individual genes within a locus. For example, an asymmetric spatial regulator could be used to simultaneously repress one gene while activating another, a property that could serve as the foundation for new bistable genetic switches. Furthermore, some of the components presented here (CRs, nucleosome-disfavoring sequences, etc.) may be used to mitigate undesired context effects of placing genes and regulatory elements in proximity to one another. Finally, long-range CR repressors could be deployed to stably silence entire genomic regions, for example, to inactivate a synthetic circuit or to regulate an entire secondary metabolite production cassette. Quantitative measurement of properties, such as the kinetics of activation or repression, distance-dependence of spatial regulators, and spreading kinetics of long-range regulators, would greatly enhance the utility of CRs for these purposes.
Finally, recent advances in programmable DNA targeting technologies are providing new opportunities for inducing epigenomic alterations at any desired locus, for example, to correct disease-associated epigenomic changes. ZFs, transcription activator-like effector (TALE) repeat domains, and the recently described CRISPR/Cas system (Cong et al., 2013; Jinek et al., 2012) each provide unique benefits, and all are compatible with the approach outlined here. For example, ZFs are highly-specific, small, and efficient for gene/DNA delivery applications (Urnov et al., 2010). TALE proteins are easier to engineer, have a larger targeting range, and have been shown to enable the targeting of CR domains (Konermann et al., 2013; Mendenhall et al., 2013). Lastly, the CRISPR/Cas system can be used to promote multiplex recruitment of effectors to numerous loci simultaneously (Cong et al., 2013; Maeder et al., 2013b; Perez-Pinera et al., 2013).
Bottom-up approaches for chromatin biology
In addition to applications in cellular engineering, the bottom-up approach presented here may complement current methods in chromatin biology, by providing new tools and approaches to directly test the functional role of chromatin states in gene expression. Current methods for testing causality employ perturbations that globally affect activities of CRs (knock-down, overexpression, and chemical inhibition) with potential pleiotropic effects. Thus, these methods do not directly assess causal functional roles for CRs at specific loci. By targeting CRs to specific gene loci, we provide functional evidence supporting the causative roles of certain chromatin complexes in regulating transcription, including activation by H3K4 methyltransferases and histone acetyltransferases, and repression by histone deacetylases. Our approach could, in principle, be used to study the effects of DNA and histone modifications at specific endogenous loci, and could be applied to the study of chromatin regulation in mammalian cells (Konermann et al., 2013; Maeder et al., 2013a; Mendenhall et al., 2013).
An interesting result that emerged from our CR library screens is that transcription can be repressed but not activated from downstream of a gene. Furthermore, activators do not appear to display the long-range properties that some repressors do, at least not within the spatial contexts studied (CRs targeted to enhancers may exhibit different properties (Mendenhall et al., 2013)). This highlights an interesting “asymmetric” property of transcriptional regulation by some CRs. Activation is generally controlled at specific locations (e.g., promoters and enhancers), while repression can be controlled throughout a gene presumably though spreading mechanisms or disrupting the synthesis of full-length transcripts. Site-specific targeting of CRs could also be a useful tool in elucidating the mechanisms underlying long-range repression and spreading of chromatin modifications (Hathaway et al., 2012; Moazed, 2011). In conjunction with chromatin modification mapping and protein domain knockouts, site-specific targeting of CRs could provide additional insight into the domains and protein activities required for heterochromatic spreading.
Targeting specific domains that comprise CRs may also be useful in understanding the importance of protein-protein interactions and protein complex recruitment in chromatin-based regulation. In addition, use of minimal chromatin-modifying catalytic domains could provide supporting evidence of a histone code. In this study, we focused on targeting full-length proteins because it enabled the use of gene ontology annotations to garner insights into chromatin-based transcriptional regulation, such as the classification of distinct sets of combinatorial regulators in programming transcriptional logic. We found that CRs with distinct regulatory mechanisms from VP16 were able to generate synergy. This suggests a general design principle in which protein complexes with distinct functions may interact to produce emergent properties, and may be combined to execute myriad regulatory decisions. Future work may reveal many novel behaviors arising from the large interaction space between two or more chromatin complexes with distinct mechanisms of action.
The complexity of chromatin arises from the large number of regulating complexes and their combinatorial and spatial modes of action. We show here that decomposing chromatin regulation into modular elements benefits our understanding of the function of individual components and complexes. Furthermore, diverse combinatorial and spatiotemporal regulatory modes can be encoded within synthetic gene architectures and executed by the site-specific recruitment of engineered chromatin regulators. This bottom-up approach may be a useful platform for both untangling and harnessing the complexities of chromatin control over cellular behaviors.
Experimental Procedures
Strains and Media
The host strain used for all experiments in this study was S. cerevisiae YPH500 (α, ura3-52, lys2-801, ade2-101, trp1Δ63, his3Δ200, leu2Δ1) (Stratagene). Culturing and genetic transformation were done as previously described (Khalil et al., 2012) using either the URA3, HIS3, or LEU2 genes as selectable markers.
Plasmid Construction
Reporter plasmids were constructed from integrative plasmid pRS406 (Stratagene) by cloning ZF (43-8 and/or 97-4) binding sequences at various locations within a previously described reporter construct (Khalil et al., 2012). ZF-CR and VP16 fusion proteins were expressed from previously described TetR- or LacI-regulated GAL1 promoters (Khalil et al., 2012). The ZF-CR expression constructs were cloned into single-integrating plasmid pNH603 (HIS3), and the VP16 fusion expression constructs into single-integrating plasmid pNH605 (LEU2).
Our host strain was generated by cloning an expression cassette that constitutively expresses TetR, LacI, and GEV into single-integrating plasmid pNH607 (HO). Constitutive expression of the repressors in glucose-containing media ensures low basal levels of expression of ZF-CRs from the engineered GAL1 promoters, which can be relieved by the respective addition of the chemical inputs, ATc and IPTG, along with beta-estradiol to the medium. The negative control, truncated (non-binding) ZF amino acid sequence is PRHLKTHLR. pNH603, pNH605, pNH607, and BFP were kind gifts from the Lim Lab (Zalatan et al., 2012).
Library Construction
Primer sequences were obtained from the Saccharomyces Genome Database (Cherry et al., 2012) (Table S6), synthesized (Integrated DNA Technologies), and used to amplify full length CR ORFs from wild-type yeast (BY4742). SbfI and NotI flanking restriction sites were used to ligate PCR products C-terminal to (3×FLAG) – (nuclear localization sequence) – (zinc finger array) – (17 amino acid glycine-serine linker).
Induction Experiments
Three single yeast colonies for each strain were picked after genomic integration and used to inoculate 500 μl of SD- media (synthetic drop-out media containing 2% glucose with defined amino acid mixtures) in Costar 96-well assay blocks (V-bottom; 2 ml max volume; Fisher Scientific). The cultures were grown at 30°C with 900 rpm shaking for 24-48 hr. Cultures, with and without inducers, were inoculated in SD-complete media to an OD600 of 0.05-0.1 and grown at 30°C with 900 rpm shaking for 12 hr. Cells were treated with 10 μg/mL cycloheximide to inhibit protein synthesis, and then assayed for yEGFP, mCherry, and BFP expression by flow cytometry.
Flow Cytometry and Data Analysis
For all experiments, 5,000–10,000 events were acquired using a BD LSRFortessa equipped with a High Throughput Sampler (BD Biosciences). Events were gated by forward and side scatter, and geometric means of the fluorescence distributions were calculated in FlowJo. The autofluorescence value of S. cerevisiae YPH500 cells harboring no genomic integrations was subtracted from these values. “Fold activation” values were calculated as the ratio of fluorescence values from induced cells to those from uninduced cells. All values obtained were the means of three isogenic strains. BFP and mCherry expression, driven by the CHO1 and BIO2 promoters, respectively, remained largely invariant between induced and uninduced cultures (Figure S6C); thus GFP values are not expected to vary significantly with any growth rate differences in strains.
Gene ontology queries were submitted to the SGD database between July 10, 2013 and August 17, 2013 (Cherry et al., 2012). Cluster and background frequencies are in the Supplementary Tables.
Acknowledgments
We thank members of the Collins and Khalil labs for helpful discussions. Emily Egan and the Moazed Lab provided valuable technical advice on ChIP experiments. We thank Sylvain Meylan and Kwan T. Chow for engaging discussions and manuscript feedback. This work was supported by a NIH-NIGMS Ruth L. Kirschstein Postdoctoral Fellowship (1F32GM105272-01A1) (A.J.K.), a Boehringer Ingelheim Fonds PhD Fellowship (S.K.), a Defense Advanced Research Projects Agency grant (DARPA-BAA-11-23) (A.S.K. and J.J.C.), startup funds from the Department of Biomedical Engineering at Boston University (A.S.K.), a National Science Foundation CAREER Award (MCB-1350949) (A.S.K.), the Wyss Institute for Biologically Inspired Engineering (PRJ-0329) (J.J.C.), and the Howard Hughes Medical Institute (J.J.C.).
Footnotes
Extended Experimental Procedures are available online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. [Abstract] [Google Scholar]
- Bi X, Yu Q, Sandmeier JJ, Zou Y. Formation of Boundaries of Transcriptionally Silent Chromatin by Nucleosome-Excluding Structures. Molecular and Cellular Biology. 2004;24:2118–2131. [Europe PMC free article] [Abstract] [Google Scholar]
- Blount BA, Weenink T, Vasylechko S, Ellis T. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS One. 2012;7:e33279. [Europe PMC free article] [Abstract] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Research. 2012;40:D700–705. [Europe PMC free article] [Abstract] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. [Europe PMC free article] [Abstract] [Google Scholar]
- Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9143–9148. [Europe PMC free article] [Abstract] [Google Scholar]
- Dodd IB, Micheelsen Ma, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. [Abstract] [Google Scholar]
- Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Science Translational Medicine. 2013;5:179ps177. [Europe PMC free article] [Abstract] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. [Abstract] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genetics. 2010;6:e1000869. [Europe PMC free article] [Abstract] [Google Scholar]
- Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. [Abstract] [Google Scholar]
- Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. [Abstract] [Google Scholar]
- Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and Memory of Heterochromatin in Living Cells. Cell. 2012;149:1447–1460. [Europe PMC free article] [Abstract] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends in Genetics. 2011;27:389–396. [Abstract] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. [Abstract] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature Reviews Genetics. 2010;11:367–379. [Europe PMC free article] [Abstract] [Google Scholar]
- Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. [Europe PMC free article] [Abstract] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. [Europe PMC free article] [Abstract] [Google Scholar]
- Lam FH, Steger DJ, O'Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. [Europe PMC free article] [Abstract] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NACH, Margaritis T, van de Pasch LAL, van Heesch SAAC, Brok MO, Groot Koerkamp MJA, et al. The specificity and topology of chromatin interaction pathways in yeast. Molecular Cell. 2011;42:536–549. [Europe PMC free article] [Abstract] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. [Abstract] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biology. 2005;3:e328. [Abstract] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nature Biotechnology 2013a [Europe PMC free article] [Abstract] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nature Methods. 2013b;10:977–979. [Europe PMC free article] [Abstract] [Google Scholar]
- Martens C, Krett B, Laybourn PJ. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Molecular Microbiology. 2001;40:1009–1019. [Abstract] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nature Biotechnology. 2010;28:1079–1088. [Abstract] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nature Biotechnology 2013 [Europe PMC free article] [Abstract] [Google Scholar]
- Milbradt AG, Kulkarni M, Yi T, Takeuchi K, Sun ZY, Luna RE, Selenko P, Naar AM, Wagner G. Structure of the VP16 transactivator target in the Mediator. Nature Structural & Molecular Biology. 2011;18:410–415. [Europe PMC free article] [Abstract] [Google Scholar]
- Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22534–22539. [Europe PMC free article] [Abstract] [Google Scholar]
- Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. [Europe PMC free article] [Abstract] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. [Abstract] [Google Scholar]
- Olesen J, Hahn S, Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987;51:953–961. [Abstract] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nature Methods. 2013;10:239–242. [Europe PMC free article] [Abstract] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. [Abstract] [Google Scholar]
- Ptashne M. Epigenetics: core misconcept. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7101–7103. [Europe PMC free article] [Abstract] [Google Scholar]
- Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nature Reviews Molecular and Cell Biology. 2009;10:410–422. [Abstract] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. [Europe PMC free article] [Abstract] [Google Scholar]
- Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nature Genetics. 2012;44:743–750. [Abstract] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. [Abstract] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics. 2010;11:636–646. [Abstract] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang CZ, Hemeryck-Walsh C, et al. A Comprehensive Genomic Binding Map of Gene and Chromatin Regulatory Proteins in Saccharomyces. Molecular Cell. 2011;41:480–492. [Europe PMC free article] [Abstract] [Google Scholar]
- Weber W, Fussenegger M. Synthetic gene networks in mammalian cells. Current Opinion in Biotechnology. 2010;21:690–696. [Abstract] [Google Scholar]
- Weinberger L, Voichek Y, Tirosh I, Hornung G, Amit I, Barkai N. Expression noise and acetylation profiles distinguish HDAC functions. Molecular Cell. 2012;47:193–202. [Europe PMC free article] [Abstract] [Google Scholar]
- Ye H, Aubel D, Fussenegger M. Synthetic mammalian gene circuits for biomedical applications. Current Opinion in Chemical Biology. 2013;17:910–917. [Abstract] [Google Scholar]
- Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science. 2012;337:1218–1222. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Reviews Genetics. 2011;12:7–18. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2014.04.047
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867414006758/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cell.2014.04.047
Article citations
Utilization of CRISPR-Cas genome editing technology in filamentous fungi: function and advancement potentiality.
Front Microbiol, 15:1375120, 28 Mar 2024
Cited by: 0 articles | PMID: 38605715 | PMCID: PMC11007153
Review Free full text in Europe PMC
AI in cellular engineering and reprogramming.
Biophys J, 123(17):2658-2670, 04 Apr 2024
Cited by: 1 article | PMID: 38576162
Review
Using High-Throughput Measurements to Identify Principles of Transcriptional and Epigenetic Regulators.
Methods Mol Biol, 2842:79-101, 01 Jan 2024
Cited by: 0 articles | PMID: 39012591
A modular dCas9-based recruitment platform for combinatorial epigenome editing.
Nucleic Acids Res, 52(1):474-491, 01 Jan 2024
Cited by: 7 articles | PMID: 38000387 | PMCID: PMC10783489
Fluorescent Reporter Systems to Investigate Chromatin Effector Proteins in Living Cells.
Methods Mol Biol, 2842:225-252, 01 Jan 2024
Cited by: 0 articles | PMID: 39012599
Go to all (81) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A high-throughput synthetic biology approach for studying combinatorial chromatin-based transcriptional regulation.
Mol Cell, 84(12):2382-2396.e9, 01 Jun 2024
Cited by: 0 articles | PMID: 38906116
Transcriptional networks: reverse-engineering gene regulation on a global scale.
Curr Opin Microbiol, 7(6):638-646, 01 Dec 2004
Cited by: 39 articles | PMID: 15556037
Review
Transcriptional regulatory network shapes the genome structure of Saccharomyces cerevisiae.
Nucleus, 4(3):216-228, 01 May 2013
Cited by: 8 articles | PMID: 23674068 | PMCID: PMC3720752
Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast.
J Mol Biol, 360(1):213-227, 03 May 2006
Cited by: 130 articles | PMID: 16762362
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIGMS NIH HHS (2)
Grant ID: F32 GM105272
Grant ID: 1F32GM105272-01A1





