Abstract
Purpose of review
To provide an update on neutralizing antibody targets in the context of the recent HIV-1 envelope trimer structure, describe new antibody isolation technologies, and discuss the implications of these data for HIV-1 prevention and therapy.Recent findings
Recent advances in B-cell technologies have dramatically expanded the number of antibodies isolated from HIV-infected donors with broadly neutralizing plasma activity. These, together with the first high-resolution crystal and cryo-electron microscopy (cryo-EM) structures of a cleaved, prefusion HIV-1 trimer, have defined new regions susceptible to neutralization. This year, three epitopes in the gp120-gp41 interface were structurally characterized, highlighting the importance of prefusion gp41 as a target. Similar to many other broadly neutralizing antibody epitopes, these new antibodies define a target that is also highly glycan dependent. Collectively, the epitopes for broadly neutralizing antibodies now reveal a continuum of vulnerability spanning the length of the HIV-1 envelope trimer.Summary
Progress in the last year has provided support for the use of rationally stabilized whole HIV-1 trimers as immunogens for eliciting antibodies to multiple epitopes. Furthermore, the increasing number of broad and potent antibodies with the potential for synergistic/complementary combinations opens up new avenues for preventing and treating HIV-1 infection.Free full text

HIV broadly neutralizing antibody targets
Abstract
Purpose of review
To provide an update on neutralizing antibody targets in the context of the recent HIV-1 envelope trimer structure, describe new antibody isolation technologies, and discuss the implications of these data for HIV-1 prevention and therapy.
Recent findings
Recent advances in B-cell technologies have dramatically expanded the number of antibodies isolated from HIV-infected donors with broadly neutralizing plasma activity. These, together with the first high-resolution crystal and cryo-electron microscopy (cryo-EM) structures of a cleaved, prefusion HIV-1 trimer, have defined new regions susceptible to neutralization. This year, three epitopes in the gp120–gp41 interface were structurally characterized, highlighting the importance of prefusion gp41 as a target. Similar to many other broadly neutralizing antibody epitopes, these new antibodies define a target that is also highly glycan dependent. Collectively, the epitopes for broadly neutralizing antibodies now reveal a continuum of vulnerability spanning the length of the HIV-1 envelope trimer.
Summary
Progress in the last year has provided support for the use of rationally stabilized whole HIV-1 trimers as immunogens for eliciting antibodies to multiple epitopes. Furthermore, the increasing number of broad and potent antibodies with the potential for synergistic/complementary combinations opens up new avenues for preventing and treating HIV-1 infection.
INTRODUCTION
The HIV-1 envelope (Env) glycoprotein spike mediates viral entry, and is the sole target of neutralizing antibodies. The entry-mediating form exists as a trimer composed of three host receptor binding gp120 molecules, noncovalently linked to three gp41 transmembrane fusion proteins. gp120 is heavily glycosylated and shielded by the hypervariable regions (loops V1–V5, the α2 helix, and β14 sheet), whereas gp41 is more conserved, less solvent exposed, and less glycosylated. As a result of host immune pressures Env is the most diverse of all HIV proteins with up to 30% variation between different genetic subtypes. Amino acid substitution, insertions/deletions, and glycan shifting occur predominantly in the variable regions that are most easily accessed by neutralizing antibodies. The dominant neutralizing antibody response is therefore strain specific, however over the course of HIV-1 infection most individuals develop antibodies with some level of cross-reactivity [1]. Those with the greatest breadth have been the source of new broadly neutralizing antibodies (bNAbs) [2]. Characterizing the epitopes of these bNAbs has led to high resolution structures of the HIV-1 Env trimer [3,4,5■■], allowing us to more clearly define sites of vulnerability that might be exploited for HIV-1 vaccine design and antibody mediated therapy.
TECHNOLOGIES FOR THE ISOLATION OF NEW BROADLY NEUTRALIZING ANTIBODIES
The first bNAbs to HIV-1 (b12, 2G12, 2F5, and 4E10) were isolated by phage display or B-cell immortalization, selected for binding to Env peptides or monomeric proteins, and generally limited in breadth and/or potency. The ability to culture memory B cells, together with high-throughput neutralization assays that allowed for direct functional screening, led to the isolation of several new antibodies targeting novel quaternary structure specific epitopes, as well as more potent antibodies to previously identified sites [6-8,9■■,10■■]. New bNAbs to previously known targets (but possessing greater breadth and potency) have also been identified using structure-guided methods to design sorting antigens for labelling B cells by flow cytometry [11,12]. Unlike B-cell culture this technique does not rely on potent neutralization to identify bNAbs, but it is limited by the specific mode of recognition. More recently, quaternary structure specific bNAbs have been isolated using native, cleaved, prefusion trimers as sorting antigens, which appear to preferentially bind neutralizing antibodies [13■■].
The successful isolation of bNAbs has been aided by first mapping the neutralization specificities in donor plasma, to tailor an appropriate selection technique [14,15]. In addition bioinformatics approaches have been used to predict specificities and design targeted approaches for the isolation of bNAbs [16-18]. Once a B-cell lineage has been identified the use of next-generation sequencing to mine the repertoire allows for literally hundreds of related variants to be identified [19-22]. A major obstacle of next-generation sequencing however is the inability to identify naturally occurring heavy-chain and light-chain antibody pairs. This was overcome when Georgiou et al. devised a method of pairing heavy-chain and light-chain PCR products prior to sequencing [23]. Information on the targets for bNAbs, as well as neutralization, sequence, and structural data on the monoclonal antibodies (mAbs) that have been isolated is being extensively catalogued into two new publically available databases: CATNAP on the LANL website (http://www.hiv.lanl.gov/components/sequence/HIV/neutralization/main.comp) and bNAber [24], providing useful resources for the field.
BROADLY NEUTRALIZING ANTIBODY TARGETS
The isolation of exceptionally broad and potent bNAbs has enabled the identification of five roughly defined targets on the HIV-1 Env, such as the V2 site, the N332 supersite, the CD4 binding site (CD4bs), the gp120–gp41 interface, and the membrane proximal external region (MPER). Identifying multiple bNAbs with similar epitopes has pinpointed minimal sites of vulnerability, whose recognition confers the greatest neutralization breadth. However as discussed below, new bNAbs with novel epitopes have revised our understanding of how these distinct sites partially merge in the context of the trimer.
THE V2 SITE
The V2 site at the trimer apex is formed from the converging, sequence conserved regions of the V1V2 domain and the V3 loop [3,4,25]. It is protected by densely packed glycans (particularly those at positions N156 and N160) and the hypervariable loops V1 and V2 [26]. Access to the underlying peptide epitope is only possible by antibodies with unusually long (between 26 and 39 amino acids), anionic heavy chain complementarity determining region loop three (CDR-H3) [7,14,26]. Anti-V2 bNAbs generally bind poorly to monomeric gp120 or scaffolded V1V2s [7,14]. In the case of the prototypical V2 antibody PG9, this quaternary specificity was partially explained by the fact that the antibody binds to N160 glycans from two separate protomers [27,28]. However, for some relatives of the CAP256-VRC26 lineage which targets a similar epitope, broad neutralization was not dependent on the N160 glycan [14]. Despite these differences, the actual peptide epitope determined by mutagenesis is minimal for both the PG9 and CAP256-VRC26 antibodies, made up of a short mostly cationic stretch of seven amino acids (position 165–171). For PG9 the underlying peptide comprises less than 25% of the epitope, with the rest of the epitope predominantly formed by the glycans at N156 and N160 [7,14,26,29]. The conserved nature of these glycans, and the small peptide footprint, likely contributes to the breadth of this class of antibodies.
THE N332 SUPERSITE
The N332 supersite is composed of a number of overlapping glycan-dependent epitopes [30]. V3 epitopes lie structurally proximal to the V2 site [4], and are the most well described within the N332 supersite. Antibodies targeting V3 show a similar mechanism to V2 site recognition, in that they access a minimal eight residue peptide epitope between positions 323 and 330 via long (20–26 amino acids) CDR-H3s [31]. Two such antibodies, PGT121 and PGT128, are highly dependent on the glycans at positions N301 and N332 [6], but somatic variants of PGT121 also depend on glycans in V1 (N137) and V2 (N156) [4]. In this way PGT121-like antibodies can recognize a different side of the N156 glycan that is critical to most anti-V2 bNAbs (Fig. 1). PGT130 was isolated from the same donor as PGT128, but represents an alternate branch of the B-cell lineage that preferentially recognizes a glycan at N334 [6,32]. The N334 and N332 glycans are mutually exclusive, and thus in both donors somatic variants have evolved to recognize different immunotypes of the V3 site, indicative of the role for viral diversification in driving bNAb maturation, that ultimately accounts for total plasma neutralization breadth [32,33].
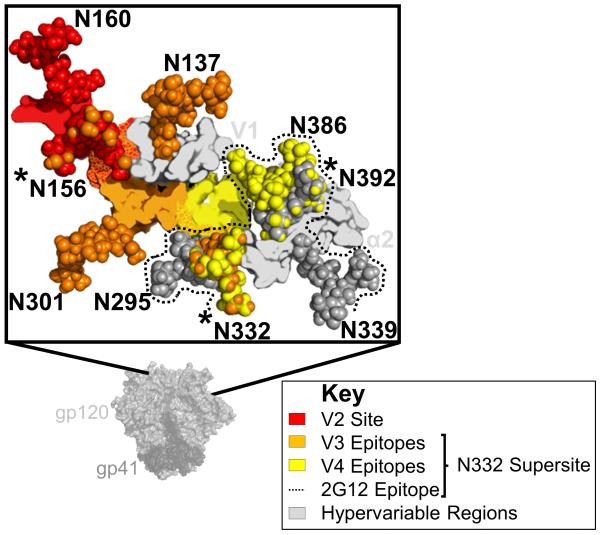
The V2 site overlaps with the N332 supersite. The HIV-1 envelope (Env) trimer is shown in light (gp120) and dark (gp41) grey surface view. An expanded graphic of the V2 and N332 sites is shown in the zoomed out box. Epitopes in V2 (red), V3 (orange), or V4 (yellow) are defined as residues within 5 Å of broadly neutralizing antibodies (bNAbs) PG9, PGT122/PGT128, or PGT135, respectively. Regions of overlap between epitopes are shown with mottled colours red/orange for V2-V3, and orange/yellow for V3–V4. Key glycans are shown as spheres, labelled, and coloured according to their epitopes. Glycans predicted to be a part of the 2G12 epitope are coloured dark grey, and bordered with a dotted line. Glycans N156, N332, and N392 that are bound by bNAbs targeting different epitopes are asterisked. Proximal hypervariable regions V1 and α2 are shown in light grey. Figure based on protein databank accession code 4TVP.
bNAb PGT135 defines a second epitope in the N332 supersite that does not involve V3, but rather contacts amino acids at the base of the V4 loop [30]. This mAb binds predominantly to the apolar face of the N332 glycan (unlike PGT128), and recognizes additional glycans at positions N295, N386, and N392 [30]. A third epitope, defined by the bNAb 2G12 overlaps the PGT135 epitope but unusually does not involve any peptide contact (Fig. 1). Rather, 2G12 uses a rare variable heavy (VH) chain domain swap to create a large paratope capable of binding to the terminal sugars of four glycans on the silent face of gp120 (N295, N332, N339, and N392) [34]. Thus, while 2G12 does not actually penetrate the glycan shield, it does share the recognition of key glycans with PGT135. The isolation of additional bNAbs with specificities similar to PGT135 and 2G12 will help to better define the vulnerabilities within this supersite.
THE CD4 BINDING SITE
In contrast to bNAbs targeting V2 or N332, antibodies targeting the CD4bs generally make minimal glycan contacts. Only one mode of recognition at the CD4bs has been extensively described, that of VH1-2 or VH1-46 derived bNAbs, typified by VRC01 and 8ANC131. The germline precursors of these bNAbs have specific genetic signatures that mimic CD4 binding, such as R71 in the heavy chain that, like R59 of CD4, interacts with D368 in the highly conserved CD4 binding loop of gp120 [12,35]. VH1-2 bNAbs also have unusually short CDR-L3 loops to avoid clashes with the glycan at position N276 in the D loop of gp120 [36-38]. Conversely the bNAb HJ16 derived from the VH3-30 gene does not interact with D368, and is entirely dependent on the glycan at position N276 for neutralization, defining a second subsite within the CD4bs [39,40]. This glycan is sometimes bound by VRC01-like antibodies, but is not critical to their neutralization [41]. HJ16 and other non-VH1-2/1-46 derived CD4bs bNAbs have long CDR-H3 loops important for their binding, but are still limited in their angle of approach to gp120 by extensive glycosylation and the quaternary nature of the trimer [15,42]. Cryo-electron microscopy (cryo-EM) structures have shown that some CD4bs bNAbs such as PGV04 may actually interact with the glycans that closely border the CD4bs (N276, N363, and N386) as well as the N301 glycan from the V3 loop of an adjacent protomer [3]. These data also suggest contact between the positively charged amino acids in V3, and an anionic insertion in the PGV04 heavy-chain framework region 3 (FWR3). The contribution of these additional contacts to neutralization is unclear considering many CD4bs bNAbs induce a conformation of gp120 that would rearrange the V1V2 and V3 loops. Nonetheless the complexity of the CD4bs has previously been underestimated.
THE gp120–gp41 INTERFACE
The N276 glycan described above is also the target for bNAb 8ANC195 (Fig. 2), recently shown to bind an epitope in the gp120–gp41 interface [43■■]. This mAb is dependent on glycans at positions N234 and N276, and uses a four amino acid insertion to thrust the Fab heavy-chain FWR3 between these two glycans, contacting R456 at the distal most tip [43■■]. The 8ANC195 CDR-H3 is 22 amino acids long, but is not used to penetrate the glycan shield. Rather the CDR-H3 folds back towards the light chain forming part of a wider paratope that makes significant contact with the 7-stranded β-sandwich (the gp41 interacting region) of gp120 [43■■]. The affinity for gp120 is sufficient that 8ANC195 was isolated using a stabilized gp120 core, but docking of the Fab–gp120 complex into the prefusion trimer revealed potential interactions between the 8ANC195 light chain and gp41 near the N637 glycan. Deep sequencing in the 8ANC195 donor showed that the stabilized core specifically selected for a single branch of the bNAb lineage. More genetically diverse antibodies, created by pairing related heavy-chain and light-chain variants, exhibited increased potencies that were attributed almost entirely to light chain–gp41 interactions [43■■]. Thus, 8ANC195 was the first of several recently identified bNAbs targeting epitopes in the gp120–gp41 interface [9■■,10■■,43■■,44]
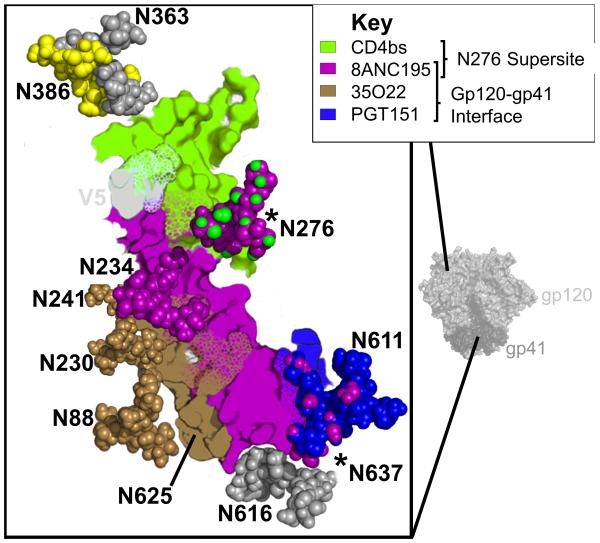
The CD4bs adjoins the gp120–gp41 interface. The HIV-1 envelope trimer is shown in light (gp120) and dark (gp41) grey surface view. An expanded graphic of the CD4bs and gp120–gp41 interface sites are shown in the zoomed out box. Epitopes for VRC01 (green), 8ANC195 (purple), 35O22 (brown), or PGT151 (blue) are defined as residues within 5 Å of each respective broadly neutralizing antibody (bNAb). Key glycans are shown as spheres, labelled, and coloured according to the epitopes they comprise. Regions that make up more than one epitope are shown with mottled colours, and glycans that are bound by bNAbs targeting different epitopes are asterisked. The hypervariable region V5 is shown in light grey.
Two new antibodies published in 2014, PGT151 and 35O22, were also shown to target the gp120–gp41 interface [5■■,9■■,10■■,45■■]. Unlike 8ANC195, these bNAbs do not bind monomeric gp120. bNAb 35O22 (isolated from the same donor as MPER bNAb 10E8) binds immediately adjacent to 8ANC195 near N234 (Fig. 2), close enough to contact the seven-stranded β-sandwich via an eight amino acid FWR3 insertion [5■■]. This bNAb epitope is remarkably low on the trimer, seemingly incompatible with the presence of the viral membrane, and it is speculated that certain rearrangements induced by CD4 are required for its binding [9■■]. Neutralization by 35O22 was incompatible with glycosylation of the N625 sequon, but still critically dependent on the N625 residue [5■■]. bNAb PGT151 binds between the 8ANC195 and 35O22 epitopes, over a cavity between gp160 protomers [45■■]. Unlike 35O22, it has a specific requirement for cleaved trimers, and a maximum observed stoichiometry of two antigen-binding fragments (Fabs) per trimer, suggesting an allosteric alteration of the third binding site after the first two Fabs have bound [45■■]. This binding mechanism also allows PGT151 to stabilize native, cleaved Env trimers from cell membranes. Both 35O22 and PGT151 have highly glycan-dependent epitopes, with 35O22 neutralization dependent on the glycans at N88, N230, and N241 in gp120, and neutralization of PGT151 requiring gp41 glycans N611 and N637 [9■■,10■■]. Based on proximity, PGT151 was also predicted to interact with glycans in gp120 (N262, N276, N230, N234, N241, and N448) though these interactions are not critical for neutralization [45■■]. This glycan dependence results in varied neutralization plateaus for both bNAbs. For 35O22, neutralization plateaus were improved (higher levels of maximum inhibition indicating enhanced potency) when pseudoviruses were grown in kifunensine, indicating a role for high mannose glycans in its epitope [9■■]. In contrast, PGT151 could not neutralize kifunensine or 293S-grown pseudoviruses (glycosylated only with mannose rich glycans), and bound directly to triantennary and tetraantennary complex glycans [10■■,45■■]. As virus grown in peripheral blood mononuclear cells appears to have greater proportions of complex glycan, PGT151-like antibodies may exhibit better potencies in vivo [10■■].
THE MEMBRANE PROXIMAL EXTERNAL REGION
Epitopes in the MPER of gp41 are almost exclusively contained within a linear, α-helical stretch of amino acids that links the transmembrane domain to the ectodomain of gp41. The MPER can be divided into an N-terminal and C-terminal helix around a kink at position 674, used to define various overlapping epitopes. bNAb 2F5 binds to the N-terminal helix, Z13e1 binds to the elbow between helices, while 4E10 and 10E8 bind to an epitope in the C-terminal helix. The hydrophobic C-terminus of the MPER is partially buried in the viral membrane and is thought to play a critical role in fusion with the host membrane. It is therefore highly conserved, and antibodies 4E10 and 10E8 are some of the broadest yet identified [8,46]. bNAbs targeting the MPER often interact with the viral membrane via long CDR-H3 loops, which may promote higher than normal levels of autoreactivity for these types of antibodies [47]. The precise location and conformation of the MPER in the prefusion HIV-1 trimer is not yet known, and further characterization will be necessary to determine the proximity of, and relationship between, epitopes in this site and those in the gp120–gp41 interface.
A CONTINUUM OF VULNERABILITY
The increased availability of bNAbs targeting novel sites, or variants of previously described sites, has blurred the definition of distinct bNAb epitopes. For instance CD4bs antibodies, once almost exclusively classified by their sensitivity to mutations at D368, now include a new group typified by HJ16 that are largely glycan dependent and insensitive to mutations at D368 [39,40]. Similarly V2 antibodies, previously defined as being N160 glycan dependent, now also include antibodies such as CAP256-VRC26 that target the same protein epitope but are not necessarily sensitive to glycan deletion [14]. These antibodies might be described as targeting subsites within the greater CD4bs or V2 site of vulnerability. This paradigm currently does not apply to the N332-dependent antibodies PGT128, PGT135, and 2G12, whose epitopes have mostly nonoverlapping peptide components (Figs. 1 and and3).3). One way to characterize these larger epitope clusters has been to define glycan supersites [30]. Based on these criteria, the glycan at position N276, which is the target for CD4bs antibodies (such as HJ16) and certain gp120–gp41 interface antibodies (e.g. 8ANC195) might now also be classified as a glycan supersite (Figs. 2 and and3).3). Other glycans, such as those at positions N156, N386, and in the newly described gp120–gp41 epitope cluster, are bound by antibodies targeting two distinct epitopes, but only form a critical component for one (Figs. 1 and and2).2). These glycans do not yet meet the requirements of a supersite (though isolation of additional mAbs may change this), but they may contribute towards the affinity maturation of bNAbs at both sites. Thus, it seems that bNAb epitopes form a continuum of vulnerability that includes conserved residues stretching from the trimer apex to the MPER (Fig. 3). The glycans which protect these regions, divide up the continuum into sites of vulnerability, but also because of their conservation become targets for neutralization themselves.

A continuum of broadly neutralizing antibody (bNAb) targets. A graphic of the HIV-1 envelope (Env) trimer is shown and surface coloured for broadly defined epitopes in V2 (red), V3 (orange), V4 (yellow), the CD4bs (green), and the gp120–gp41 interface (brown, purple, and blue for 8ANC195, 35O22, and PGT151, respectively). Overlap between these epitopes through recognition of common glycans or amino acids is indicated with mottling of the relevant colours. The highly accessible positions of hypervariable regions V1, V2, α2, V4, β 14, and V5 (coloured in cyan) are indicated with arrows. The first residue of the MPER (truncated in the HIV-1 trimer structure 4TVP) is indicated with an asterisk.
IMPROVED NEUTRALIZATION COVERAGE
Vaccine-induced bNAbs ideally need to protect against infection from all viral subtypes, and therefore those that target highly conserved epitopes are likely to be most effective. First-generation bNAbs, such as b12, 2F5, and 2G12, had fairly restricted breadth, and were often more effective against subtype-matched heterologous viruses [46]. More recently isolated bNAbs show much greater breadth [2], although even among the broadest bNAbs coverage (and/or potency) varies between subtypes [7,9■■,14]. Subtype preference was also seen using large panels of chronic sera, which showed increased potency of plasmas against subtype-matched viruses [48]. Notably, however, this effect was reduced in older and more diversified subtypes, suggesting that the significance of this observation for vaccine design will decrease overtime [48]. Differential coverage may occur for other reasons – bNAbs to the N332-supersite are less effective against viruses with the glycan at 334 despite some promiscuity in recognition [49], a finding that may be important as the 332 glycan is less common in subtype C founder viruses [50]. However, as a single antibody lineage could evolve to recognize both N332 and N334 variants (such as in the case of PGT128 and PGT130), inclusion of multiple immunotypes into vaccine design strategies may enhance coverage.
The availability of a large number of new more potent and cross-reactive mAbs has also enhanced prospects for passive immunotherapy. Early studies demonstrated that bNAbs could protect against infection in nonhuman primates and humanized mice [51-56]. However, these first-generation bNAbs were only able to transiently reduce viral loads in infected subjects [57-59]. The latest mAbs (mostly in combinations) resulted in more durable control of viremia while the mAbs were present, and in some cases thereafter [60-62]. Passively infused bNAbs will probably be used in combination. Indeed, mAbs targeting four different sites in double, triple, and quadruple combinations showed 98–100% coverage [63]. In addition to coverage, potency is likely to be important, with bNAbs engineered for greater in-vitro potency associated with protection at lower plasma levels [64]. Lack of potency maybe a particular concern for bNAbs with relatively low neutralization plateaus (incomplete neutralization even at high concentrations) such as 35O22, PGT151, and many V2-dependent antibodies. Ongoing human clinical trials will allow assessment of the dose of bNAb that is required (for protection and immunotherapy) and whether systemic infusion protects mucosal portals of infection.
CONCLUSION
The last year has brought significant advances in our understanding of the HIV-1 Env structure and its neutralization targets. New isolation technologies have allowed for less biased identification of bNAbs targeting increasingly complex epitopes. These new antibodies have provided important insights, culminating in the near complete structural delineation of the HIV-1 Env spike. However, in many instances only a single mAb targeting a given site is available (e.g. PGT135, 2G12, 8ANC195, PGT151, and 35O22), and these often demonstrate some level of clade preference. Many more antibodies will need to be identified to properly characterize each of these new HIV-1 vulnerabilities. Additional bNAb–trimer complex structures will be necessary to define ‘hotspots’ or supersites for neutralization. Using the structural data from multiple bNAb–antigen complexes for a single target site will facilitate rational approaches to display specific sites of vulnerability, as is currently being done for the CD4bs and MPER epitope scaffolds. However, the extensive overlap between bNAb targets suggests that immunogen design may benefit from efforts to include whole, stabilized, cleaved HIV-1 trimers – perhaps rationally designed to expose the entire continuum of bNAb vulnerabilities while minimizing antigenicity to the hypervariable structures. The number of new bNAb targets, and our understanding of their structure have provided novel opportunities for preventing and treating HIV-1.
Acknowledgements
The authors thank Jinal Bhiman and Jay Gorman for helpful discussions.
Financial support and sponsorship
The authors receive research funding from the Centre for the AIDS Programme of Research (CAPRISA), the South African Medical Research Council (MRC) through the Flagship and SHIP programs, the National Institutes of Health (NIH) through an R01 Grant (R01 AI104387-01A1) and a U01 grant (AI116086-01), the Center for HIV/AIDS Vaccine Immunology (CHAVI grant AI067854), the Center for AIDS Vaccine Discovery (CAVD) of the Bill and Melinda Gates Foundation, the South African HIV/AIDS Research and Innovation Platform of the South African Department of Science and Technology and by a HIVRAD NIH grant (AI088610). CAPRISA was supported by NIAID, NIH, US Department of Health and Human Services (Grant U19 AI51794). P.L.M. is a Wellcome Trust Intermediate Fellow in Public Health and Tropical Medicine (Grant 089933/Z/09/Z). C.K.W. is supported by the Poliomyelitis Research Foundation (PRF) of South Africa and was the recipient of a Fogarty AITRP Fellowship.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
Full text links
Read article at publisher's site: https://doi.org/10.1097/coh.0000000000000153
Read article for free, from open access legal sources, via Unpaywall:
https://journals.lww.com/co-hivandaids/Fulltext/2015/05000/HIV_broadly_neutralizing_antibody_targets.3.aspx
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1097/coh.0000000000000153
Article citations
Editorial: Cryogenic electron microscopy of infectious diseases.
Front Mol Biosci, 11:1506197, 16 Oct 2024
Cited by: 0 articles | PMID: 39479500 | PMCID: PMC11521902
Membrane HIV-1 envelope glycoproteins stabilized more strongly in a pretriggered conformation than natural virus Envs.
iScience, 27(7):110141, 28 May 2024
Cited by: 2 articles | PMID: 38979012 | PMCID: PMC11228805
Contemporary HIV-1 consensus Env with AI-assisted redesigned hypervariable loops promote antibody binding.
Nat Commun, 15(1):3924, 09 May 2024
Cited by: 0 articles | PMID: 38724518 | PMCID: PMC11082178
Phase 1 trial evaluating safety and pharmacokinetics of HIV-1 broadly neutralizing mAbs 10E8VLS and VRC07-523LS.
JCI Insight, 9(7):e175375, 08 Apr 2024
Cited by: 1 article | PMID: 38587079 | PMCID: PMC11128198
Antiviral Protein-Protein Interaction Inhibitors.
J Med Chem, 67(5):3205-3231, 23 Feb 2024
Cited by: 2 articles | PMID: 38394369 | PMCID: PMC10945500
Review Free full text in Europe PMC
Go to all (97) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(2 citations)
PDBe - 4TVPView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.
PLoS Pathog, 13(1):e1006074, 11 Jan 2017
Cited by: 26 articles | PMID: 28076415 | PMCID: PMC5226681
Potent Induction of Envelope-Specific Antibody Responses by Virus-Like Particle Immunogens Based on HIV-1 Envelopes from Patients with Early Broadly Neutralizing Responses.
J Virol, 96(1):e0134321, 20 Oct 2021
Cited by: 9 articles | PMID: 34668778 | PMCID: PMC8754226
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.
J Virol, 90(7):3446-3457, 13 Jan 2016
Cited by: 23 articles | PMID: 26763999 | PMCID: PMC4794693
Challenges for structure-based HIV vaccine design.
Curr Opin HIV AIDS, 4(5):431-440, 01 Sep 2009
Cited by: 76 articles | PMID: 20048708
Review
Funding
Funders who supported this work.
NIAID NIH HHS (9)
Grant ID: P01 AI088610
Grant ID: R01 AI104387
Grant ID: U01 AI116086
Grant ID: U19 AI067854
Grant ID: U01 AI116086-01
Grant ID: R01 AI104387-01A1
Grant ID: U01 AI067854
Grant ID: U19 AI051794
Grant ID: U19 AI51794
Wellcome Trust (1)
Grant ID: 089933/Z/09/Z





