Abstract
Free full text

Oncogenic Mutant p53 Gain of Function Nourishes the Vicious Cycle of Tumor Development and Cancer Stem-Cell Formation
Abstract
More than half of human tumors harbor an inactivated p53 tumor-suppressor gene. It is well accepted that mutant p53 shows an oncogenic gain-of-function (GOF) activity that facilitates the transformed phenotype of cancer cells. In addition, a growing body of evidence supports the notion that cancer stem cells comprise a seminal constituent in the initiation and progression of cancer development. Here, we elaborate on the mutant p53 oncogenic GOF leading toward the acquisition of a transformed phenotype, as well as placing mutant p53 as a major component in the establishment of cancer stem cell entity. Therefore, therapy targeted toward cancer stem cells harboring mutant p53 is expected to pave the way to eradicate tumor growth and recurrence.
During the past three and a half decades of p53 research, ample data have accumulated pertaining to the role of p53 in the regulation of various cellular processes and in preventing cancer development (Levine and Oren 2009; Bieging et al. 2014). Hence, the wild-type p53 (WT-p53) protein is considered the guardian of the genome (Lane 1992) and a key regulator of homeostasis (Vousden and Lane 2007) that exerts its activities both at the cell-autonomous and -nonautonomous levels (Lujambio et al. 2013). In fact, mutations in the p53 (mut-p53) gene are found in more than half of human cancer cases (Sigal and Rotter 2000; Brosh and Rotter 2009). It was widely shown that mutations in p53 result in loss of its tumor-suppressive function. Importantly, not only does mut-p53 interfere with the remaining WT-p53 protein via dominant negative mechanism, but also mutations empower p53 with oncogenic gain-of-function (GOF) effects that endow cells with tumorigenic potential (Brosh and Rotter 2009; Muller and Vousden 2014). This unique feature of mut-p53 in facilitation of malignant transformation indicates that p53 may be regarded as a proto-oncogene tumor suppressor.
The mutant p53 GOF characteristics and mechanisms of action have been broadly described (Brosh and Rotter 2009; Muller and Vousden 2014). Here, we will provide an update on the latest developments in this field and address the growing understanding of the complexity and heterogeneity of tumors, zooming in on the subpopulation of cancer stem cells (CSCs) within the tumors. Finally, we will discuss how mut-p53’s unique features set it as a prime candidate for therapeutic targeting and how this relates to the latest developments in mut-p53 targeted therapy.
THE EVOLUTION OF THE mut-p53 GOF CONCEPT
Remarkably, one of the earliest observations in the field was that mut-p53 promotes cellular malignant transformation (Eliyahu et al. 1984; Jenkins et al. 1984; Parada et al. 1984; Wolf et al. 1984). This initial experimental evidence was obtained by using p53 cDNAs that originated from transformed cells that incorporated a mutant form of p53 (Wolf et al. 1984; Halevy et al. 1990; Shaulsky et al. 1991). Yet, these first reports did not consider the fact that cDNA may represent WT-p53 or mut-p53 sequences. The notion that a missense mutation in p53 confers oncogenic GOF that enhances tumorigenic potential was formally introduced several years later by Dittmer and colleagues: Overexpressed mut-p53 in cells lacking endogenous p53 expression resulted in malignant transformation of these cells both in vitro and in vivo (Dittmer et al. 1993). This was further confirmed by analysis of several experimental cellular models using either knockdown or overexpression of mut-p53. However, the final clincher in the establishment of the mut-p53 GOF concept was obtained through studies of transgenic mice expressing endogenous mut-p53. Although both mut-p53 and null-p53 mice showed facilitated tumor development, mice harboring mut-p53 displayed significantly higher incidence of metastasis (Lang et al. 2004; Olive et al. 2004). Further analysis of the aforementioned p53 models uncovered another important feature of the GOF mechanism of action—the requirement of mut-p53 to accumulate to exert its oncogenic function. Apparently, in normal mouse tissue, under physiological conditions, mut-p53, similarly to WT-p53, is maintained at low levels. However, on cellular insults, such as oncogene activation, DNA damage, or high reactive oxygen species (ROS) levels, mut-p53 undergoes constitutive stabilization and accumulates in the cell. In contrast to the WT-p53 protein, once mut-p53 is stabilized, it does not undergo degradation, mainly because of the inability of MDM2, a negative regulator of WT-p53, to polyubiquitinate mut-p53 (Frum and Grossman 2014). Indeed, augmented levels of p53 protein, manifested by mut-p53 accumulation in tumors and tumor lines, were suggested to serve as a clinical marker >30 years ago (Rotter 1983; Soussi and Beroud 2001).
Many tumors have shown an apparent dependency on specific gene expression to retain the malignant phenotype (Weinstein and Joe 2008). This phenomenon is referred as the Weinstein hypothesis of “oncogene addiction.” Recently, by establishment of an elegant, novel system of mice harboring mut-p53, Alexandrova et al. (2015), showed that ablation of mut-p53 substantially diminished tumor burden, suggesting that mut-p53 stabilization confers tumor cells with an oncogenic addiction. This finding suggests that it might be sufficient to ablate the mutant p53 protein to obtain the required therapeutic outcome.
DIFFERENT p53 MUTATIONS SHOW VARIATIONS IN THEIR GOF
Approximately 2000 different mutations in the p53 gene were detected in sporadic tumors (Petitjean et al. 2007). Yet, only a small subset of missense mutations in the DNA-binding domain of p53 is modified frequently. These mutations, which lead to the formation of full-length protein, are referred as “hot-spot” mutations. They can be categorized into two groups: the conformational mutations such as R175H, G245S, R249S, and R282W and the DNA-contact mutations represented by R248Q and R273H (Fig. 1) (Petitjean et al. 2007). In addition to the occurrence of p53 mutations in somatic cells, germline mutations are associated with the rare familial cancer predisposition termed Li–Fraumeni syndrome (LFS). The LFS patients develop early onset of a wide spectrum of tumors (Malkin et al. 1990). These patients are heterozygous for mut-p53 (WT/mut-p53) in every cell of their body. Initial analysis of tumors derived from LFS patients showed that ~60% of tumors lost the remaining WT-p53 allele in a process termed loss of heterozygosity (LOH) (Varley et al. 1997). Interestingly, a recent study that performed p53-based genomic and transcriptomic meta-analyses using data from the Cancer Genome Atlas estimated that >93% of sporadic tumors with mut-p53 undergo p53 LOH (Parikh et al. 2014). These findings support the concept that p53 is a recessive tumor suppressor and loss of the remaining WT allele is required for tumor development.
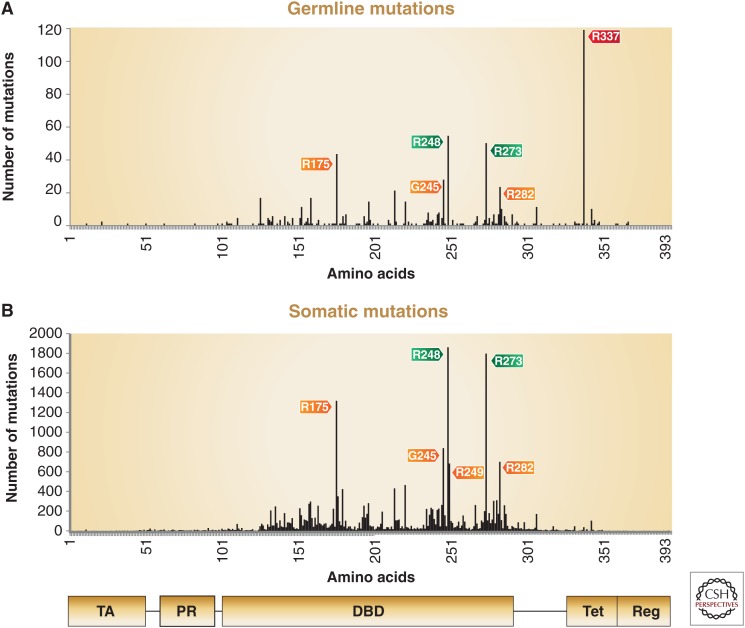
Distribution of somatic and germline p53 mutations. The distribution of reported missense mutations across 393 amino acids of the p53 protein. (A) The six most frequent “hot-spot” mutations detected in tumors obtained from Li–Fraumeni syndrome (LFS) patients (N = 636). (B) The six most frequent hot-spot mutations occurring in sporadic tumors (N = 24,210). The domain architecture of p53 is aligned below. Hot-spot mutations are highlighted in orange for conformational mutations, in green for DNA-contact mutations, and in red for tetramization mutation. Notably, five of the six hot-spot mutations are shared between somatic and germline mutations, indicating that they confer properties that contribute to their selection. R249 is unique to somatic mutations because of the mutagenic effect of aflatoxin B1 that is associated with food contamination. In contrast, R337 is unique to germline mutations because of a founder effect of a single source to its progeny. TA, Transactivation domain; PR, proline-rich domain; DBD, DNA-contact domain; Tet, tetramization domain; Reg, carboxy-terminal regulatory domain. (Data derived from the IARC TP53 mutation database, version R17, November 2013.)
Notably, five of the six hot-spot mutations, R175H, G245S, R248Q, R273H, and R282W, are shared between sporadic tumors and tumors obtained from LFS patients. This observation may suggest that the GOF effect of these p53 mutants predominantly contributes to their selection during tumorigenesis. In contrast, the conformational hot-spot p53 mutation R249S that is often induced by aflatoxin B1 was preferentially found in somatic liver tumors (Aguilar et al. 1993). As this mutation is induced by food contamination, an environmental factor, it is rarely observed in germline-derived tumors. On the other hand, the R337H-specific germline mutation associated with multiple cancers of the LFS spectrum in the population of Southern Brazil is a result of a founder effect (Fig. 1) (see Garritano et al. 2010; Achatz and Zambetti 2016).
The effect and the magnitude of oncogenic GOF acquired by the different p53 mutants is a function of the mutation site, the type of nucleotide substitution, and the specific cell type, thus making the relationship between genotype and phenotype extremely complex (Freed-Pastor and Prives 2012; Bisio et al. 2014; Xu et al. 2014). Accordingly, we were able to show that, in human fibroblasts, various hot-spot p53 mutants cooperate in different ways with constitutively active H-RAS to promote cellular transformation. More specifically, the conformational mutants (R175H and H179R) induced a unique pattern of a cancer-related gene signature by elevating H-RAS activity through perturbation of BTG2, whereas DNA-contact mutants (R248Q and R273H) prompted cancer-related gene expression by cooperating with NF-κB. Notably, the L3 loop region conformational mutant G245S did not show an oncogenic GOF effect in this system (Solomon et al. 2012). In contrast to the latter, a recent study showed an oncogenic GOF effect of the G245S p53 mutation in osteosarcomas that were developed following reprogramming of fibroblasts obtained from LFS patients into induced pluripotent stem cells (iPSCs) that were further differentiated into osteoblasts. The mechanism of this GOF was manifested by suppressing the expression of the imprinted gene H19 during osteogenesis (Lee et al. 2015). Another interesting example of the variation in the p53 GOF effect resulting from different substitutions of a single amino acid at the same location of the p53 gene is represented in humanized p53 knockin (HUPKI) mouse models. Mice carrying the R248W mutation showed GOF in a broader spectrum of tumor types as well as more metastasis compared with p53-null mice, with no difference in their life span (Song et al. 2007), whereas mice harboring the R248Q mutation showed earlier tumor onset and shorter survival compared with their p53-null counterparts, thus showing significantly stronger GOF (Hanel et al. 2013).
MOLECULAR MECHANISMS UNDERLYING mut-p53 GOF
WT-p53 functions as a transcription factor (TF) that exerts the transactivation of its target genes via direct binding to its specific responsive elements entailed within the target gene loci (Raycroft et al. 1990; el-Deiry et al. 1993). In contrast, the majority of mut-p53 proteins lose the WT-p53 transactivation capacity because of alterations in their DNA-binding domain or conformation state that prevents their binding to the canonical responsive element of WT-p53. Therefore, mut-p53 function is often conveyed through protein–protein interactions (Brosh and Rotter 2009; Oren and Rotter 2010). One of the well-studied mechanisms underlying mut-p53 GOF effects can be attributed to mut-p53-dependent inactivation of its family members’ p63 and p73 (Irwin 2004; Lunghi et al. 2009). In addition, it has been shown that the GOF effects of mut-p53 may be mediated by its interaction with numerous TFs, including SP1, NF-Y, VDR, SREBP1, Twist1, E2F family, and the ETS family, which serve also as WT-p53-interacting partners (Fig. 2) (Brosh and Rotter 2009; Menendez et al. 2009). Importantly, however, their cooperation with either WT- or mut-p53 usually leads to an opposite cell-fate outcome. For example, Di Agostino and colleagues have shown that mut-p53 interacts with the TF NF-Y to recruit histone acetyltransferase (p300) instead of the histone deacetylases (HDACs) commonly used by WT-p53 (Di Agostino et al. 2006; Oren and Rotter 2010). The mut-p53/NF-Y protein complex provokes aberrant transactivation of NF-Y target genes that eventually induces facilitated proliferation (Di Agostino et al. 2006). This is a perfect example emphasizing a mechanism frequently underlying the mut-p53 GOF effect, in which the WT-p53-interacting partner is “hijacked” by mut-p53 and used to support transformation by altering the epigenetic response.
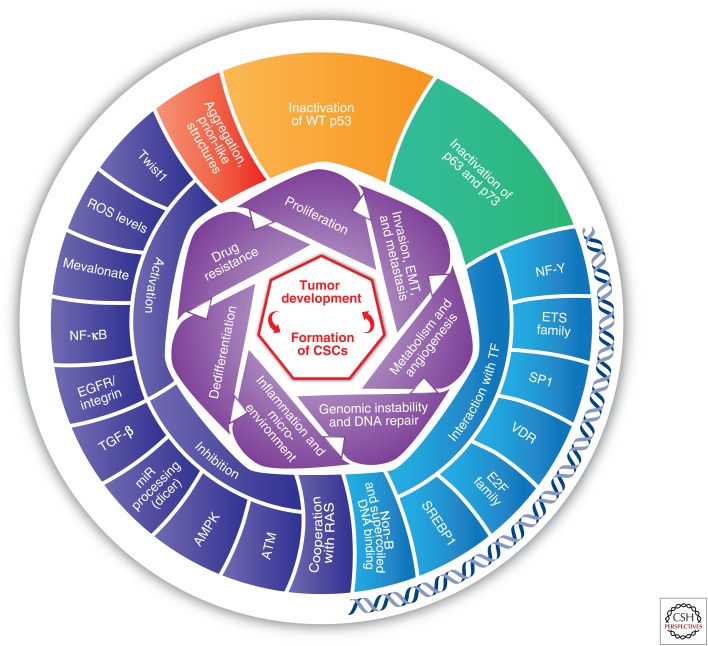
Selected phenotypes and mechanisms underlying oncogenic mut-p53 GOF (gain of function). The inner level (purple) represents the oncogenic GOF phenotypes associated with mut-p53. The outer level illustrates the pivotal mechanisms of mut-p53 GOF. Importantly, each phenotype can be associated with several underlying mechanisms. Mechanisms of mut-p53 interactions with DNA are depicted near a double-strand DNA. Because cancer stem cells (CSCs) are suggested to be the cell of origin of many human cancers, their formation constitutes the cornerstone of mut-p53 GOF in tumor initiation and progression capabilities. AMPK, 5′ AMP-activated protein kinase; ATM, ataxia telangiectasia mutated; EGFR, epidermal growth factor receptor; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; ROS, reactive oxygen species; SREBP-1, sterol regulatory element-binding protein; TGF-β, transforming growth factor β; VDR, vitamin D receptor.
Another facet associated with the understanding of mut-p53 GOF is related to microRNAs (miRs) (Donzelli et al. 2014), in which mut-p53 was found to affect Dicer, a pivotal regulator and processer of miRs (Muller et al. 2014) and noncoding RNAs, such as H19 (Lee et al. 2015). Furthermore, mut-p53 can bind to non-B DNA structure and supercoiled DNA with high affinity (Gohler et al. 2005; Brazdova et al. 2013) and thus might affect transcription by binding to DNA motifs. Finally, mut-p53 was shown to aggregate and to form prion-like structures (Ano Bom et al. 2012; Rangel et al. 2014). The importance of this phenomenon requires further investigation to determine how and whether it confers cancer-related properties. Yet, it opens new horizons in the continuously evolving p53 field.
mut-p53 DEREGULATES CELL CYCLE AND ENHANCES PROLIFERATION
WT-p53 is a pivotal cell-cycle regulator. Indeed, it was shown that WT-p53 regulates the G1/S checkpoint, by activating specific target genes, mainly CDKN1A (p21Waf1) and GADD45α (Kastan et al. 1992; el-Deiry et al. 1993). Accordingly, loss of WT-p53 activity abrogates the normal cell-cycle control that may lead to facilitated proliferation, a known cancer hallmark (Hanahan and Weinberg 2011). In an effort to unravel the basis for the mut-p53 oncogenic GOF, it became evident that mut-p53 disrupts the normal cell-cycle pattern. This appears to be the most prominent feature of mut-p53 GOF that constitutes a common denominator across the mutation spectrum (Brosh and Rotter 2010). Therefore, it is not surprising that mutations in p53 coincide with the Ki-67 proliferation marker (Olivier et al. 2005). Yet, although Ki-67 is only a single marker, it was shown that many of mut-p53 isoforms affect a cluster of cell-cycle-associated genes that has been termed “core proliferation signature.” This set of genes is involved in DNA replication, spindle assembly and checkpoint, chromosome segregation, and mitotic processes (Whitfield et al. 2006). Several mechanisms underlying the molecular basis of mut-p53 GOF regarding enhanced proliferation were suggested. The transforming growth factor β (TGF-β), a key regulator of proliferation control, was proposed as an important candidate contributing to mut-p53 oncogenic GOF. In fact, TGF-β has a dual effect on cancer progression. While in early stages of cancer, TGF-β serves as an antiproliferation barrier in epithelial cells through interaction with the Smad pathway; at advanced stages, it promotes invasion and metastasis by enhancing proteolytic activity and the expression of cell-adhesion molecules (Blobe et al. 2000). In our previous studies, we found that mut-p53 reduces the expression of TGF-β receptor type II, thereby hampering the TGF-β/Smad pathway (Kalo et al. 2007). In contrast, Adorno and colleagues showed that mut-p53 increased TGF-β-mediated invasiveness and metastasis ability, by counteracting p63/Smad complex assembly (Adorno et al. 2009). This apparent discrepancy in the effects of mut-p53 on TGF-β action can be explained by the timing of p53 mutation appearance during the course of tumor development. In the event p53 mutation occurs early in tumorigenesis, when TGF-β serves as antiproliferation barrier, mut-p53 provides cells with clonal advantage via perturbing TGF-β function. However, if p53 mutation takes place in later stages of tumor progression, when TGF-β does not attenuate the cell cycle, cooperation between mut-p53 and TGF-β will induce invasiveness and metastasis. Recently, another molecular mechanism pertaining mut-p53 GOF was uncovered. Regγ is a proteasome 20S inducer that upon activation enhances proliferation via degradation of cell-cycle inhibitors such as p21Waf1, p16, and WT-p53. Regγ itself is a target of WT-p53 and TGF-β signaling. mut-p53 was shown to prevent TGF-β-mediated response by inhibiting Smad3 recruitment to the Regγ promoter in a GOF manner permitting the interaction with p300 to induce proliferation and drug resistance (Ali et al. 2013; Wang et al. 2015). Similarly, mut-p53 was shown to induce histone acetylation on the Axl promoter, a tyrosine kinase receptor that is involved in the stimulation of cell proliferation. Thus, these signals lead to a higher proliferation rate and increased motility (Vaughan et al. 2012b). In all, mut-p53 not only loses its normal role as cell-cycle regulator but also gains proliferative activities that facilitate tumorigenesis.
mut-p53 FACILITATES INVASION, EPITHELIAL–MESENCHYMAL TRANSITION, AND METASTASIS
The ability of the cancer cells to invade and metastasize is one of the typical hallmarks of malignant transformation. The plasticity of cancer cells permits them to undergo epithelial–mesenchymal transition (EMT) and thereby gain mesenchymal properties required to invade the surrounding tissue and metastasize. A set of TFs, including Snail, Slug, Twist1, and Zeb1/2, orchestrates the EMT and related migratory processes during embryogenesis and tumorigenesis (Singh and Settleman 2010). Several reports indicated that mut-p53 shows an oncogenic GOF by promoting the EMT process in various cancer cells by enhancing the expression of EMT inducers. In agreement with this notion, we have shown that, in prostate cells, mut-p53 up-regulates a key activator of EMT, Twist1, via reduction of BMI-1-mediated methylation of the Twist1 promoter. This resulted in higher expression of mesenchymal markers, lower expression of epithelial markers, and enhanced invasive properties in vitro (Kogan-Sakin et al. 2011). Additionally, it was shown that mut-p53 induces EMT through miR-130b, a negative regulator of Zeb1 (Dong et al. 2013). An alternative mechanism used by mut-p53 GOF to promote EMT is attenuation of EMT suppressors. Indeed, Ali et al. (2013) showed that, in metastatic breast cancer cells, mut-p53 suppresses the expression of Klf17, a negative regulator of metastasis and EMT, leading to EMT-associated gene transcription and enhanced cancer progression.
Furthermore, mut-p53 enhances invasiveness and metastasis by exerting an oncogenic GOF involving the constitutive activation of EGFR/integrin signaling by inhibition of p63 (Adorno et al. 2009; Muller et al. 2009), as well as by up-regulation of Pla2g16 (Xiong et al. 2014). Additional mechanisms suggested for this phenomenon are mut-p53-mediated down-regulation of Dicer through p63-dependent and -independent means (Muller et al. 2014), inhibition of PDGFRβ by blocking the p73/NF-Y complex (Weissmueller et al. 2014), and interactions with NRG1 and Pin1 (Girardini et al. 2011; Coffill et al. 2012).
mut-p53 AFFECTS TUMOR STROMA AND PROMOTES CHRONIC INFLAMMATION
A tumor is a complex tissue composed of proliferating cancerous cells that reside in a rich microenvironment provided by resident fibroblasts and additional various nonmalignant cell types. This tumor microenvironment was shown to contribute substantially to the malignant process (Hanahan and Weinberg 2011). We have shown an interesting cross talk between the tumor and its microenvironment with respect to mut-p53. Lung carcinoma cells affect their cocultured cancer-associated fibroblasts to secrete IFN-β, which normally prevents cancer cell migration. Notably, mut-p53-expressing cancer cells attenuate this response via SOCS1-mediated inhibition of STAT1 phosphorylation in a negative feedback loop. Remarkably, IFN-β reduces mut-p53 RNA levels by restricting WIG1, suggesting that patients with mut-p53 might benefit from IFN-β therapy (Madar et al. 2013). Mutations in p53 were also reported in tumor stroma of breast cancer patients (Patocs et al. 2007). Albeit there is some controversy as to their prevalence (Campbell et al. 2008; Roukos 2008; Zander and Soussi 2008), it was shown that mut-p53-expressing stromal cells promote tumorigenesis in prostate cancer cells better than their p53-null counterparts (Addadi et al. 2010).
Chronic inflammation has a role in tumorigenesis by creating a vicious cycle between the tumor and its microenvironment. Indeed, inflammation was recognized as one of the hallmarks of cancer (Hanahan and Weinberg 2011). One of the canonical pathways of inflammatory response is the tumor necrosis factor (TNF)-α/NF-κB pathway (Lawrence 2009). The role of NF-κB in cancer is complex; on the one hand, in some models NF-κB activation blocks tumor development, whereas in others it inhibits apoptosis and favors cell proliferation (Pikarsky and Ben-Neriah 2006). Activation of NF-κB by TNF-α can drive cancer progression in the context of chronic inflammation (Pikarsky et al. 2004). Several studies linked mut-p53 and chronic inflammation (Cooks et al. 2014). We have reported that, in cancer cells, mut-p53 induces NF-κB in response to TNF-α treatment. Accordingly, down-regulation of mut-p53 sensitized cancer cells to the apoptotic effects of TNF-α (Weisz et al. 2007). Recently, it was shown that mut-p53-mediated NF-κB activation by TNF-α is based on mut-p53-dependent inhibition of the tumor suppressor DAB2IP in the cytoplasm (Di Minin et al. 2014). Another study, aiming to uncover the role of mut-p53 in inflammation-associated tumors, showed that mice heterozygous for mut-p53 (WT/mut-p53R172H), but not hemizygous mice (WT/null-p53), are prone to develop invasive carcinoma upon induction of stress in their colons. This was a result of chronic inflammation and augmented NF-κB activation promoted by mut-p53 (Cooks et al. 2013). This correlates with the finding that in colitis-associated colorectal cancer, mutations in p53 are an early event (Cooks et al. 2013). Furthermore, we have shown that in non-small-cell lung carcinoma cells, mut-p53 cooperates with constitutively active H-RAS to up-regulate a proinflammatory gene signature, which leads to aggressive transformed phenotypes in vivo (Buganim et al. 2010; Solomon et al. 2012). Additional inflammatory mechanisms that mut-p53 isoforms use to promote carcinogenesis are suppression of the secreted IL1 receptor antagonist by binding to its promoter with the corepressor MAFF (Ubertini et al. 2015), induction of NF-κB2 activation via recruitment of CBP and STAT2 to acetylate NF-κB2 promoter (Vaughan et al. 2012a), and up-regulation of CXC chemokines (Yan and Chen 2009; Yeudall et al. 2012).
In conclusion, mut-p53 does not only facilitate invasion and metastasis in a cell-autonomous fashion but also affects the tumor microenvironment and contributes to the vicious cycle of chronic inflammation and tumorigenesis.
mut-p53 INDUCES CANCER-PROMOTING METABOLIC SHIFT AND ANGIOGENESIS
The initial observation that the majority of cancer cells display alterations in glucose processing was made in the beginning of the previous century. The term “Warburg effect” describes the phenomenon that cancer cells predominantly use glycolysis for energy production from glucose, instead of the oxidative phosphorylation used by normal cells. Despite being considerably less efficient, glycolysis enables the tumor cells to gain valuable building blocks to sustain their rapid proliferation rate (Vander Heiden et al. 2009). Accumulating data suggest that, in addition to alterations in glucose processing, tumor cells modify other metabolic pathways to maximize the malignant potential (Hsu and Sabatini 2008). As a part of its tumor-suppressive activities, WT-p53 was shown to directly influence various metabolic pathways, enabling cells to respond to metabolic stress (Vousden and Ryan 2009; Maddocks and Vousden 2011; Goldstein and Rotter 2012; Berkers et al. 2013; Maddocks et al. 2013). In contrast, mut-p53 facilitates cancer-promoting metabolic shift. For example, in head and neck, cancer cells upon nutrient deprivation, mut-p53, but not WT-p53, binds to one of the AMPK subunits, a major energy sensor, which in turn inhibits its function, leading to anabolic metabolism (Zhou et al. 2014). In addition, it was reported that in lung and breast carcinoma, mut-p53 stimulates glucose intake by up-regulating the RhoA-ROCK pathway that results in translocation of GLUT1 to the plasma membrane (Zhang et al. 2013). mut-p53 was also shown to modulate the mevalonate pathway by binding to the TF SREBP (Freed-Pastor et al. 2012). One explanation for the Warburg effect is that proliferating tumor cells generate hypoxic conditions, which confer an advantage for cells with decreased dependence on aerobic respiration (Hsu and Sabatini 2008). In addition, to overcome the shortage in oxygen supply, cancer cells induce angiogenesis (Hanahan and Weinberg 2011), which WT-p53 was shown to suppress (Dameron et al. 1994; Van Meir et al. 1994; Mukhopadhyay et al. 1995). In contrast, mut-p53 acts to enrich the nutrients and oxygen supply to the tumor through cobinding with E2F1 to ID4 promoter leading to neoangiogenesis (Fontemaggi et al. 2009). In agreement with these observations, we found that mut-p53 elevates ROS levels by attenuating the expression of phase 2 detoxifying enzymes, NQO1 and HO-1 (Kalo et al. 2012). High ROS levels lead to an increase of HIF1, which is responsible for up-regulation of VEGF-A, a pivotal angiogenesis signal (Khromova et al. 2009). In summary, mut-p53 mediates a tumorigenic metabolic shift and angiogenesis aiming to provide nutrients, building blocks, and oxygen supply to support the developing tumor.
mut-p53 INTERFERES WITH DNA-REPAIR MECHANISMS AND MEDIATES GENOMIC INSTABILITY
Tumorigenesis is associated with compromised DNA-repair pathways. This perturbation results in reduced DNA-repair capacity and increased genetic instability in tumor cells (Helleday et al. 2008). In response to DNA damage and according to the type of perturbation and the cell-cycle state, WT-p53 mediates the proper DNA-repair response, including nucleotide-excision repair (NER), base-excision repair (BER), DNA-mismatch repair (MMR), nonhomologous end-joining (NHEJ), and homologous recombination (HR) (Offer et al. 1999; Zurer et al. 2004; Sengupta and Harris 2005). Thus, in the absence of WT-p53, genomic instability arises. HUPKI mice that harbor mut-p53 showed interchromosomal translocations rarely seen in p53-null mice (Song et al. 2007), indicating mut-p53 GOF. This observation of increased genomic instability was apparent in other mut-p53 mice models as well (Murphy et al. 2000; Hingorani et al. 2005; Caulin et al. 2007). Several mechanisms were suggested for mut-p53 GOF in modifying DNA-repair pathways. It was shown that mut-p53 inhibits the pathway downstream from ataxia telangiectasia mutated (ATM) by suppressing the establishment of Mre11-Rad50-NBS1 complex, which is essential for DNA double-stranded break repair. Alternatively, it was shown that mut-p53 cooperates with E2F4 in binding to BRCA1 and RAD17 promoters that result in their down-regulation (Valenti et al. 2015). Interestingly, a recent study showed that mut-p53 does not only affect transcription but also localization of the proteins. In breast cancer cells, DNA-repair genes, PARP1 and PCNA, were shown to be associated with the chromatin and absent in the cytosol only in the presence of mut-p53 (Polotskaia et al. 2015). Another mechanism that controls genomic stability in human cancer is the accumulation of ROS that can be mediated by mut-p53 (Kalo et al. 2012). It is well accepted that DNA perturbations that promote tumorigenic processes are acquired with time (Stratton et al. 2009). However, in certain cancer cases a short single catastrophic event, termed chromothripsis, was evident (Stephens et al. 2011). It was suggested that chromothripsis is formed by shattering of vast areas of chromosome(s), followed by an error-prone, NHEJ, reconstruction mechanism (Rausch et al. 2012). Despite being a rare event, it was found to be prevalent in specific tumor types, such as SHH (sonic-hedgehog-driven)-medulloblastoma of LFS patients and AML (acute myeloid leukemia), both expressing mut-p53 (Rausch et al. 2012). In all, these data suggest that mut-p53 actively contributes to a genome instability phenotype, mostly through modifications in DNA-repair pathways and elevated ROS levels.
mut-p53 PROTECTS FROM CELL DEATH AND MEDIATES DRUG RESISTANCE
WT-p53 serves as a central inducer of programmed cell death following anticancer therapy (Yonish-Rouach et al. 1991; Lowe et al. 1993). Perhaps the most devastating mut-p53 GOF is its ability to confer drug resistance (broadly described in Shetzer et al. 2014b), which is one of the reasons for the mut-p53 association with a poor prognosis (Olivier et al. 2005; Petitjean et al. 2007). mut-p53-mediated drug resistance was shown in many tumor types following treatment with different agents (Li et al. 1998; Blandino et al. 1999; Matas et al. 2001; Pugacheva et al. 2002; Capponcelli et al. 2005; Tsang et al. 2005; Bossi et al. 2006; Buganim et al. 2006; Kawamata et al. 2007; Wong et al. 2007; Do et al. 2012; Wang et al. 2014). Mechanistically, mut-p53 protects against apoptosis by affecting many proteins involved in the apoptotic pathway, both at the transcriptional level (Li et al. 1998) and by protein–protein interaction (Chee et al. 2013). Among these apoptotic proteins are Fas/Apo-1 (Gurova et al. 2003; Zalcenstein et al. 2003), caspase 9 (Chee et al. 2013), caspase 3 (Pohl et al. 1999; Wong et al. 2007), and Bcl-xL (Huang et al. 2013). Ample data indicate that tumors acquire various drug resistance mechanisms (Holohan et al. 2013). For example, one of the canonical drug resistance mechanisms adopted by the cancer cells is the efflux of chemotherapeutics agents out of the cell (Gottesman 2002). In this respect, it was shown that mut-p53 enhances the expression of MDR1, ATP-binding cassette (ABC) transporters, notorious for coffering drug-resistance against xenobiotic compounds with broad substrate specificity (Chin et al. 1992; Dittmer et al. 1993). Other mechanisms of drug resistance used by cancer cells include activation of alternative signaling pathways and evasion of cell death (Holohan et al. 2013). Vitamin D3 can also induce apoptosis and possess antiproliferative activities (Colston et al. 1992) and therefore is being extensively explored as a cancer-preventive and even a cancer-therapeutic agent. Strikingly, we showed that mut-p53 modulates the antiproliferative effects of vitamin D3 by physical interaction with the vitamin D3 receptor, thereby converting vitamin D3 into an antiapoptotic agent (Stambolsky et al. 2010). A number of interesting studies reported that mut-p53 mediates drug resistance via alteration of miRs expression. It was shown that in lung cancer cells, mut-p53 inhibits apoptosis and confers increased chemoresistance to multiple agents by induction of miR-128-2 expression that targets E2F5 (Donzelli et al. 2012). Similarly, Masciarelli and colleagues have shown that down-regulation of miR-223 expression by mut-p53 and the consequent up-regulation of stathmin-1 in breast and colon cancer cell lines sensitized these cells to treatment with DNA damaging agents (Masciarelli et al. 2014). Therefore, targeting the mechanisms underlying mut-p53-mediated drug resistance may be seen as a prime aim for overcoming cancer recurrence following chemotherapy.
mut-p53 GOF IN CSCs
Heterogeneity of cancer development at large can be explained by at least two main theories: the “stochastic” or “clonal evolution” model and the “hierarchical” model (Reya et al. 2001). The stochastic model postulates that every cancer cell within the tumor has the same potential to proliferate and to propagate into a tumor. However, the hierarchical model suggests that only a minor subset of cells within the tumor has the potential to generate new tumors that recapitulate the original one (Visvader and Lindeman 2008). This minor population is regarded as the tumor-initiating cell population that has the ability to self-renew and to differentiate into heterogeneous lineages (Vermeulen et al. 2008). In all, the latter theory served as the basis for coining the term “cancer stem cells” (or CSCs) (Lapidot et al. 1994; Bonnet and Dick 1997).
It is now well accepted that CSCs represent an important target population for anticancer therapeutics, as their survival following therapy is likely to result in disease relapse (Holohan et al. 2013). CSCs are characterized as quiescent cells within the tumors. Notably, cytotoxic agents are primarily effective against proliferative cells; therefore, these quiescent cells show a degree of drug insensitivity relative to cycling cells and persist following chemotherapy (Agarwal and Kaye 2003). Moreover, CSCs have the ability to efflux cytotoxic compounds, as well as to display high activity of aldehyde dehydrogenase (ALDH) detoxifying enzymes. Additional features of CSCs include the capacity to form spheres in soft agar and the expression of typical surface markers such as CD44, CD133, and many others (Magee et al. 2012).
Apparently, mut-p53 GOF and characteristics of CSCs seem to coincide (broadly described in Shetzer et al. 2014b). CSCs display tolerance to chemotherapy and play a crucial role in cancer recurrence (Visvader and Lindeman 2008); likewise p53 mutants show GOF in conferring drug resistance in numerous tumor types as elaborated above. mut-p53 shows GOF by up-regulating MDR1 (Dittmer et al. 1993), although these very same efflux pumps are considered to be pivotal means to detect and isolate CSCs. Additionally, mut-p53 confers apoptosis resistance by affecting Bcl-2 family members (Brosh and Rotter 2009; Huang et al. 2013). Similarly, CSCs show abundant expression of prosurvival proteins of the Bcl-2 family members compared with normal adult stem cells (ASCs) and somatic cells, allowing the former cells to sustain cellular stress (Merritt et al. 1995; Mandal et al. 2011). Another pathway that is shared by CSCs and mut-p53 GOF is the ability to induce angiogenesis by its main regulator VEGF (Bao et al. 2006; Calabrese et al. 2007).
CSCs may originate from malignant transformation of normal ASCs or progenitor cells that underwent oncogenic genetic alterations or following dedifferentiation of somatic cells that already harbor precancer genetic defects (Sugihara and Saya 2013; Aloni-Grinstein et al. 2014). Ample data suggest that the emergence of CSCs occurs in part as a result of EMT. Transformed mammary epithelial cells that were induced to undergo EMT gave rise to cells with breast CSCs markers and features such as the increased capacity to form mammospheres, soft agar colonies, and tumors (Mani et al. 2008). Recently, a landmark report stated that there is a significant correlation between lifetime risk to develop a specific type of cancer and the number of lifetime stem cell (SC) divisions in the host tissue. Using meta-analysis, this study showed correlative evidence that SCs are the origin of two-thirds of the human cancer types examined (Tomasetti and Vogelstein 2015).
Importantly, WT-p53 blocks the formation of CSCs regardless of their origin. In fact, it was found that WT-p53 governs embryonic and ASCs properties. It ensures the genomic stability of SCs following genotoxic insults and also controls their differentiation and proliferation (Aloni-Grinstein et al. 1993; Lin et al. 2005; Molchadsky et al. 2008, 2010, 2013; Solozobova and Blattner 2011; Rivlin et al. 2014b). In contrast, p53 mutations in SCs seem to equip them with accentuated oncogenic activity. The initial evidence that linked mut-p53 GOF and dedifferentiation was the association of p53 mutations and poorly differentiated tumors such as thyroid carcinomas (Donghi et al. 1993; Fagin et al. 1993), gastric cancer (Han et al. 1993), chondrosarcomas (Yamaguchi et al. 1996), skin tumors (Kemp et al. 1993), adenoid cystic carcinomas (Nagao et al. 2003), and prostate cancer (Matsushima et al. 1998). These studies showed that accumulation of mut-p53 was restricted to high-grade/poorly differentiated tumors. Moreover, one tumor showed two distinct areas of differentiated and undifferentiated thyroid carcinoma, yet mut-p53 was detected only in the undifferentiated regions (Donghi et al. 1993). Nevertheless, mut-p53 GOF was not acknowledged as the driving force behind dedifferentiation, rather this data was interpreted as an association between mut-p53 and aggressiveness of the disease. As of today, CSCs with p53 perturbations were successfully isolated by different combination of CSC markers from various cancer types such as gliomas (Zheng et al. 2008; Wang et al. 2009; Friedmann-Morvinski et al. 2012), breast cancer (Vadakkan et al. 2014), and ovarian cancer (Motohara et al. 2011; Flesken-Nikitin et al. 2013).
In recent years, the development of reprogramming technology allowed the generation of iPSCs by dedifferentiation of somatic cells (Takahashi and Yamanaka 2006) and opened a new platform to study the potential contribution of various factors required for SC formation. Because reprogramming and tumorigenesis share overlapping mechanisms (Semi et al. 2013), the reprogramming technology may be used to mimic the process of CSC formation via dedifferentiation of somatic cells bearing oncogenic genetic aberrations. We and others have revealed that p53 functions as a reprogramming barrier (Krizhanovsky and Lowe 2009). This activity of WT-p53 is manifested by attenuation of cell proliferation (Hanna et al. 2009; Yi et al. 2012) and by inhibition of Klf4-induced mesenchymal–epithelial transition, essential in the early stages of iPSC generation (Brosh et al. 2013). In contrast to the suppressive actions of WT-p53, various mutations in p53 confer an opposite effect by promoting the reprogramming process and concomitantly displaying an oncogenic GOF (Sarig et al. 2010; Yi et al. 2012; Shetzer et al. 2014a). Indeed, we have shown that mouse embryonic fibroblasts carrying mut-p53R172H undergo the reprogramming process with shorter latency and higher efficiency compared with their p53-deficient counterparts (Sarig et al. 2010). Importantly, although these mut-p53-expressing iPSCs were able to differentiate into the three germ layers in vitro, displaying the features of normal iPSCs, upon injection into immunocompromised mice, mut-p53 iPSCs formed malignant and invasive tumors instead of the benign teratomas generated by WT-p53 iPSCs. This illustrates the oncogenic GOF of mut-p53 that alters both the quantity and quality of the reprogramming process, permitting generation of CSCs. Interestingly, we showed that heterozygous mut-p53 iPSCs (WT/mut-p53R172H) were comparable to WT-p53 iPSCs as manifested by similar reprogramming kinetics and formation of benign teratomas. Most of the clones retained heterozygosity for prolonged time in culture; however, the small percentage of clones that underwent p53 LOH formed malignant tumors in vivo. This intriguing observation suggests that, during the reprograming process of untransformed cells harboring endogenous mut-p53, WT-p53 dominates over the mut-p53 and only upon p53 LOH were these cells able to induce malignant tumors in mice (Shetzer et al. 2014a). Although iPSCs are known to show features that are comparable to authentic embryonic stem cells (ESCs) (Takahashi et al. 2007), we observed that murine ESCs heterozygous for mut-p53 do not undergo LOH in vitro and in vivo. Moreover, both mut-p53 heterozygous and homozygous ESCs generate benign teratomas following injection into immunosuppressed mice. This interesting phenomenon, reflecting the unique mechanism existing in ESCs that functions to protect against CSC formation, is mediated by the proteomic stabilization of mut-p53 and the conversion to WT conformation (Rivlin et al. 2014a).
Examination of humanized mouse models harboring mut-p53 that closely mimic LFS patients indicated an augmented self-renewal potential that is reflected by a higher number of mesenchymal and hematopoietic SCs compared with p53-deficient mice (Hanel et al. 2013). Mesenchymal stem cells (MSCs), which are known to be less genomically stable than ESCs and iPSCs (Krtolica 2005; Shetzer et al. 2014a), were proposed as the cell of origin of soft tissue and bone sarcoma in adult life (Li et al. 2009; Mohseny and Hogendoorn 2011; Rodriguez et al. 2012). Although sarcomas comprise <1.5% of human tumor burden (Zahm and Fraumeni 1997; Virtanen et al. 2006), in LFS patients the incidence of soft-tissue and bone sarcoma is the highest (Petitjean et al. 2007). We found that MSCs heterozygous for mut-p53 are prone to undergo p53 LOH, which may lead to sarcomagenesis (Shetzer et al. 2014a). Likewise, it was reported that, in the human osteosarcoma cell line, mut-p53 GOF resulted in facilitated CSC formation, promoted proliferation, invasiveness, and resistance to apoptosis (Di Fiore et al. 2014). Recently, another study showed the involvement of mut-p53 GOF in the development of osteosarcoma. LFS patients' fibroblasts (heterozygous for mut-p53G245D) were reprogrammed into iPSCs and further induced to differentiate into MSCs and osteoblasts. Only osteoblasts were able to form tumors and recapitulate osteosarcoma of LFS patients characterized with defective osteoblastic differentiation and expression patterns (Lee et al. 2015). This discrepancy in the cell of origin of sarcoma between studies may result from either the different p53 mutation type (mut-p53R172H vs. mut-p53G245D) or from differences between human and mouse models.
Importantly, mutations in p53 are not a marker for CSCs. Nevertheless, p53 mutations augment the probability to generate CSCs by either malignant transformation of normal SCs or dedifferentiation of somatic cells. The observation that CSCs and mut-p53 share common features makes it tempting to speculate that the ability to form CSCs comprises the essence of mutant p53 GOF features (Fig. 2).
mut-p53-BASED THERAPEUTICS
The notion that p53 is the most frequently mutated gene in human cancer makes it an attractive target for cancer therapy. Numerous approaches and drugs directed to restore WT activity in mut-p53-bearing tumors are currently in different stages of preclinical and clinical trials (broadly discussed in Muller and Vousden 2014). However, today, there is no p53-based approved therapy. One of the approaches aiming to restore WT activity is based on reverting mut-p53 into WT conformation. Potentially, this strategy has two major advantages. On one hand, it will permit discarding of mut-p53 oncogenic GOF; on the other hand, it will allow reacquiring WT-p53 tumor-suppressor capabilities. This approach uses small molecules that change the structure of mut-p53 protein and enable it to partially retain transactivation activity. High-throughput screening led to identification of a group of small synthetic molecules such as APR-017 (PRIMA-1) and its methylated form APR-246 (PRIMA-1MET). These first-in-class drugs are able to interact with the DNA-binding domain of multiple p53 mutant proteins, promoting their folding into WT conformation and thereby inducing apoptosis and activating several p53 target genes in human tumor cells carrying mut-p53 (Bykov et al. 2002; Lehmann et al. 2012). Yet, the exact mechanism of action remains to be established (Lambert et al. 2009). Notably, PRIMA-1MET is the first drug of this class that has reached a clinical phase (Cheok et al. 2011). Other small molecules that interact specifically with mut-p53Y220C and restore WT functionality are PhiKan083 and PK7088 (Boeckler et al. 2008; Liu et al. 2013). These molecules represent an example of structure-based drug design that can identify small molecules that stabilize oncogenic p53 mutants. We have attempted a different approach harnessing peptide-based therapy through a large phage display screening (P Tal, S Eizenberger, E Cohen et al., unpubl.). Additional strategies are based on induction of mut-p53 degradation. One option is proteasomal degradation by the ubiquitin ligase MDM2. It was shown that inhibiting HDAC by SAHA disrupts HSP90 and HSP70 protection, which leads to mut-p53 degradation (Li et al. 2011). However, one should bear in mind that HDAC inhibitors affect WT-p53 transcription as well (Murphy et al. 1999) with possible deleterious consequences. Another possibility is targeting mut-p53-interacting proteins, such as family members. One example for this strategy is RETRA, which induces the release of mut-p53-p73 interaction, therefore enabling p73 proper function and preventing mut-p53 oncogenic GOF (Kravchenko et al. 2008). Surprisingly, RETRA was shown to exert anticancer properties independent of p53 status (Sonnemann et al. 2015). Other approaches use degradation of mut-p53 through autophagy (Vakifahmetoglu-Norberg et al. 2013) and inhibition of mut-p53 downstream pathways such as receptor tyrosine kinase signaling (Muller et al. 2009, 2013) or cholesterol synthesis (Freed-Pastor et al. 2012).
CONCLUDING REMARKS
The concept that a single amino acid substitution in p53 leads to formation of an oncogenic protein that was first evident more than 30 years ago is now widely accepted. Oncogenic mut-p53 GOF is manifested in many fundamental aspects of the malignant transformation, the most pivotal of which are shown in Figure 2. The powers gained by p53 mutants facilitate enhanced proliferation and avoidance of cell death. They confer genome instability, as well as promote metabolic shift and angiogenesis that provide the tumor with its essential nutrients. Cells with mutations in p53 acquire plasticity that permits migration, invasion, and metastasis. Moreover, p53 mutants are able to modify the microenvironment and support chronic inflammation that further contributes to tumor development. We have addressed the various accepted mechanisms underlying mut-p53 GOF, including interactions with its family members and other TFs. The ample data accumulated in the field of mut-p53 GOF pertaining to the above-mentioned mechanisms mostly originate from the examination of tumor bulk population. Importantly, however, it is seems that mut-p53 enables the evolvement of CSCs, which serve as the cornerstone of initiation and progression of tumorigenesis. Furthermore, the notion that the CSCs are the drug resistance entity in tumors makes it tempting to suggest that, in addition to the conventional chemotherapeutic agents that eliminate the bulk of proliferating tumor cells, a second line of treatment that targets CSCs might prove beneficial. Our findings that ESCs expressing mut-p53 induce a shift toward a WT-p53 conformation and that ESCs heterozygous for mut-p53 do not undergo LOH, thus avoiding its oncogenic activity, suggest that ESCs have unique mechanisms that may suppress malignant transformation. The identification of these physiological mechanisms of preventing the onset of a mut-p53-dependent oncogenic process in ESCs may pave the way to novel cancer therapeutic approaches.
Finally, one could speculate that therapy aimed at mut-p53 conversion into its WT conformation could target a major driving force in the formation of CSCs. Hence, future efforts should be invested in this direction. Our growing understanding and experience gained in preclinical and clinical trials will permit future development of more sophisticated and efficient cancer therapies, at large, and p53-targeted cancer therapy, in particular.
ACKNOWLEDGMENTS
This study is supported by the Center of Excellence of the Flight-Attendant Medical Research Institute (FAMRI), Israel Science Foundation ISF-MOKED center from the Israeli Academy of Science, the Israel Cancer Research Funds (ICRF), and the German–Israeli Helmholtz Research School in Cancer Biology (to Y.S.). V.R. is the incumbent of the Norman and Helen Asher Professorial Chair for Cancer Research at the Weizmann Institute. We are grateful to Tamar Shetzer for assistance with graphic design.
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
Articles from Cold Spring Harbor Perspectives in Medicine are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/cshperspect.a026203
Read article for free, from open access legal sources, via Unpaywall:
http://perspectivesinmedicine.cshlp.org/content/6/10/a026203.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/112043721
Article citations
From regulation to deregulation of p53 in hematologic malignancies: implications for diagnosis, prognosis and therapy.
Biomark Res, 12(1):137, 14 Nov 2024
Cited by: 0 articles | PMID: 39538363
Review
Modeling lung adenocarcinoma metastases using patient-derived organoids.
Cell Rep Med, 5(10):101777, 01 Oct 2024
Cited by: 0 articles | PMID: 39413736 | PMCID: PMC11513837
Unmet Horizons: Assessing the Challenges in the Treatment of TP53-Mutated Acute Myeloid Leukemia.
J Clin Med, 13(4):1082, 14 Feb 2024
Cited by: 0 articles | PMID: 38398394 | PMCID: PMC10889132
Review Free full text in Europe PMC
Cell fate regulation governed by p53: Friends or reversible foes in cancer therapy.
Cancer Commun (Lond), 44(3):297-360, 04 Feb 2024
Cited by: 7 articles | PMID: 38311377 | PMCID: PMC10958678
Review Free full text in Europe PMC
TP53 mutations correlate with the non-coding RNA content of small extracellular vesicles in melanoma.
J Extracell Biol, 2(8):e105, 31 Jul 2023
Cited by: 0 articles | PMID: 38939511
Go to all (32) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers.
Oncogene, 37(12):1669-1684, 18 Jan 2018
Cited by: 65 articles | PMID: 29343849 | PMCID: PMC6448595
A Mutant p53-Dependent Embryonic Stem Cell Gene Signature Is Associated with Augmented Tumorigenesis of Stem Cells.
Cancer Res, 78(20):5833-5847, 28 Aug 2018
Cited by: 17 articles | PMID: 30154152
A Gain-of-Function p53-Mutant Oncogene Promotes Cell Fate Plasticity and Myeloid Leukemia through the Pluripotency Factor FOXH1.
Cancer Discov, 9(7):962-979, 08 May 2019
Cited by: 48 articles | PMID: 31068365 | PMCID: PMC6606372
Gain-of-Function Mutant p53: All the Roads Lead to Tumorigenesis.
Int J Mol Sci, 20(24):E6197, 08 Dec 2019
Cited by: 88 articles | PMID: 31817996 | PMCID: PMC6940767
Review Free full text in Europe PMC




