Abstract
Free full text

An HIV Envelope gp120-Fc Fusion Protein Elicits Effector Antibody Responses in Rhesus Macaques
Associated Data
ABSTRACT
A goal for HIV prevention programs is to develop safe, effective vaccines that elicit durable and broadly protective antibodies. Many vaccine programs focus on the immune responses to critical epitopes in the gp120 portion of HIV envelope glycoprotein (Env) and seek to improve the quality and quantity of antibodies by altering the sequence, conformation, oligomerization, or glycosylation of gp120 to activate appropriate germ line B cells and mimic the subsequent maturation pathways seen in infected individuals. As a complement to these strategies, we developed dimeric fusion protein immunogens consisting of HIVBaL gp120 monomer attached to a Gly/Ser linker that is, in turn, fused to one half of the dimeric Fc domain from rhesus macaque IgG1 (Env-rFc). We envisioned that Env-rFc may mimic some aspects of immune complexes by binding Fc gamma receptors (FcγRs) on immune cells to increase the strength, breadth, and durability of Env-specific antibody responses. The Env-rFc retained a capacity to bind both cell surface CD4 and FcγRs. In a rhesus macaque immunization study, Env-rFc elicited higher gp120 binding antibody titers than Env and elicited antibodies that recognize CD4-induced epitopes. Env-rFc also induced antibodies capable of neutralizing tier 1A HIV pseudotyped viruses and mediating antibody-dependent cellular cytotoxicity, outcomes not observed with monomeric gp120 in our study. Serum antibodies produced in Env-rFc-immunized macaques had increased durability compared to that of Env monomer immunization. Our work suggests that adding IgG1 Fc to Env-based immunogens may stimulate increased effector capacity in the immune sera and improve the protective serum antibody response.
INTRODUCTION
Developing a preventive HIV vaccine remains an unmet global public health goal. The RV144 Thai trial consisted of a prime-boost strategy that combined a recombinant canarypox vaccine with monomeric gp120 envelope glycoprotein (Env) and demonstrated modest efficacy in reducing the risk of HIV infection (1). This vaccine strategy showed that improvements are needed to increase the quantity, quality, and durability of immune responses. Current research on HIV vaccines focuses on designing Env immunogens that induce the activation and maturation of broadly neutralizing antibody (bNAb) germ line B cells (2). Strategies include engineering well-ordered Env trimers mimicking those found during natural infection (e.g., SOSIP trimers) or trimers designed to bind with high affinity to bNAb germ line B cells (e.g., eOD-GT8) (3). Other strategies include modifying the conformation or glycosylation patterns of Env to expose conserved sites or multimerizing critical epitopes using scaffold proteins in order to enhance the immune response (reviewed in reference 4).
Previous immunization studies in mice tested recombinant proteins where HIV proteins were fused to IgG Fc and showed improved immune responses. When p55 Gag or Env V3 loop was fused to murine Fc from the IgG2a subclass, which binds with the highest affinity to activating Fc gamma receptors (FcγRs) in mice, immune responses were stronger than when Fc from the weakly binding subclass IgG1 Fc was used (5). Mice immunized with gp41 prehairpin fusion intermediate attached to Fc developed neutralizing antibody responses against HIV IIIb (6). Immunizing mice with HIVCN54 gp120 fused to IgG Fc (gp120-Fc), but not gp120, elicited Env-specific humoral responses in the absence of adjuvant, suggesting that the Fc sequence acted as an adjuvant (7). Env-specific antibody titers were significantly lower in mice immunized with a mutated gp120-Fc that did not bind Fc receptors, implying that the increased immunogenicity of Fc fusion protein was likely due to engagement of Fc receptors on immune effector cells. A key question not addressed in this earlier study is whether the gp120-Fc immunogen generated an antibody response that was quantitatively and qualitatively different from that generated by the use of gp120 plus soluble adjuvant.
Fusing an Fc segment to Env immunogens might be a strategy to increase their potency. Our group constructed fusion proteins containing HIV-1 gp120 (BaL strain) and the Fc region from rhesus IgG1 (Env-rFc) to test whether immune responses against the relatively weak immunogen, gp120, could be improved in rhesus macaques by fusing it to the Fc region. Our findings show that Env-rFc fusion proteins were more potent than monomeric Env alone and highlight the potential for improving the quantity and quality of antibody responses by incorporating an Fc region within a suitable protein immunogen.
RESULTS
Env-rFc design, expression, and functional characterization.
We engineered an Env-rFc construct containing gp120 (Env) fused to rhesus IgG1 Fc fragment (rFc) through a 14-amino-acid Gly/Ser linker. The IgG1 Fc binds activating FcγRs with higher affinity than does the Fc of other IgG subclasses. In our constructs, the gp120 molecule (plus the G/S linker) is fused to each chain of a dimeric Fc fragment including the hinge region (Fig. 1A). Env-rFc DNA expression constructs were transfected into CHO cells, and Fc fusion protein expression was verified by Western blotting (Fig. 1B).
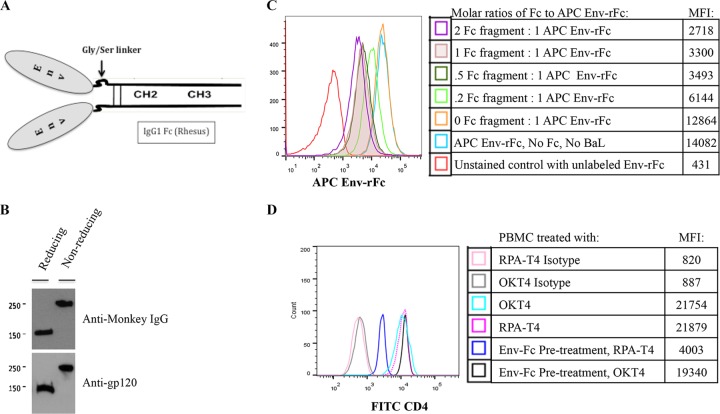
Design, expression, and functional characterization of Env-rFc. (A) Schematic of Env-rFc fusion protein. An HIVBaL envelope glycoprotein gp120 (Env) is fused to each arm of the rhesus IgG1 Fc fragment (including the hinge) through a Gly/Ser linker. (B) A Western blot illustrates the expression of Env-rFc when probed with anti-rhesus IgG1 antibody or anti-gp120. Env-rFc formed monomers under reducing conditions and disulfide-linked homodimers under nonreducing conditions. (C) Env-rFc binding to FcγRs on THP-1 cells. THP-1 cells were pretreated with HIVBaL gp120 to block Env-rFc binding to CD4 receptors. THP-1 cells were then combined with various molar concentrations of unlabeled IgG Fc fragments, washed, and stained with APC-labeled Env-rFc (molar ratios of Fc fragments to APC Env-rFc were 0:1, 0.2:1, 0.5:1, 1:1, and 2:1). The isotype control was THP-1 cells treated with unlabeled Env-rFc. MFI, mean fluorescence intensity. (D) Env-rFc binding to CD4 on PBMCs. FcγRs on PBMCs were blocked with IgG Fc fragments. PBMCs were stained with unlabeled Env-rFc and either FITC-labeled RPA-T4, which competes with Env for binding CD4, or FITC-labeled OKT-4, which binds CD4 at a distinct epitope from Env. RPA-T4 and OKT-4 binding to PBMCs without prior addition of Env-rFc is also shown.
We assessed the functional integrity of Env-rFc by testing its binding to cell surface FcγRs or CD4 receptors by use of flow cytometry. Env-rFc was conjugated to the fluorescent label allophycocyanin (APC). An inhibition assay was used to measure Env-rFc binding to FcγRs on THP-1, a human leukemic monocytic/macrophage cell line (8). We preincubated THP-1 cells with unlabeled HIVBaL gp120 to block any Env-rFc binding to cell surface CD4. THP-1 cells treated with gp120 were incubated with various concentrations of unlabeled IgG Fc fragment, washed, and stained with APC-labeled Env-rFc. As expected, the highest mean fluorescence intensity (MFI) signal from labeled Env-rFc was achieved when no competing Fc fragments were present. When cells were treated with increasing amounts of Fc fragment, we observed a dose-dependent inhibition of Env-rFc binding (Fig. 1C).
We next tested whether Env-rFc binds to cell surface CD4. Peripheral blood mononuclear cells (PBMCs) were incubated with saturating concentrations (determined previously) of Fc fragment before Env-rFc was added. We tested for Env binding to CD4 by staining with either an RPA-T4 antibody that does not bind when Env glycoprotein is bound to CD4 or an OKT-4 antibody that binds in the presence or absence of Env glycoprotein. After the addition of Env-rFc, RPA-T4 binding was decreased, showing that our fusion protein was binding to CD4 on PBMCs (Fig. 1D). Judging by FcγRs and CD4 binding, our Env-rFc appeared to be intact, with normal ligand activities.
Env-rFc elicits IgG titers higher in magnitude and breadth than Env does in rhesus macaques.
Rhesus macaques were immunized with either Env-rFc or Env by the intramuscular (i.m.) route to test whether Env-rFc is more immunogenic than Env. The six animals in the Env-rFc group and five animals in the Env group each received 100 μg of immunogen plus 50 μg of CpG adjuvant by the i.m. route at week 0 and again at week 4. Sera were collected at weeks 0, 4, and 7, and an enzyme-linked immunosorbent assay (ELISA) was used to test for total IgG levels (Fig. 2A). Immune responses were minimal at week 4. Our studies focused on week 7 when peak immune responses were achieved. ELISA measurements were corrected by subtracting preimmunization background levels for each animal.
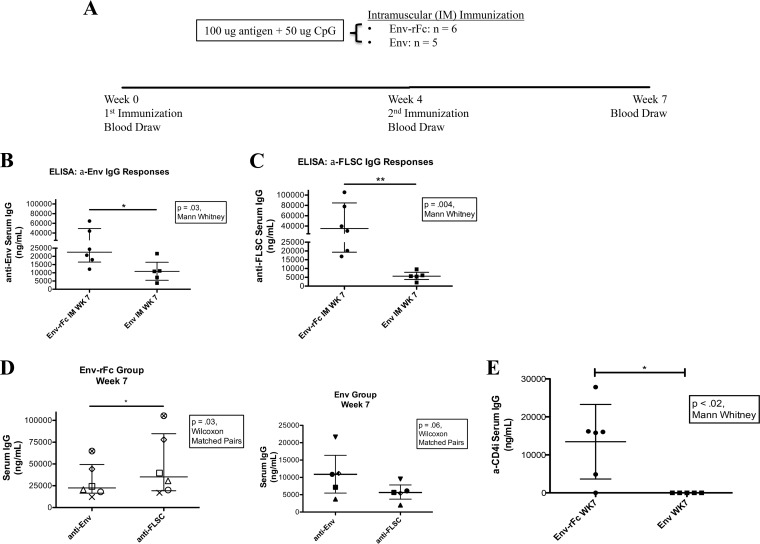
Env-rFc elicits higher-magnitude titers and a broader immune response than Env. (A) Immunization schedule. Rhesus macaques were immunized by the i.m. route with 100 μg of immunogen plus 50 μg of CpG at week 0 and then a second time at week 4. Blood was drawn at weeks 0 (preimmunization), 4, and 7 (peak immune responses). The Env-rFc group consisted of 6 animals, and the Env group consisted of 5 animals. (B and C) Anti-Env (B) and anti-FLSC (C) week 7 serum IgG levels were measured by ELISA. IgG levels were calculated relative to the standard VRC01 antibody. P values were calculated using the Mann-Whitney test. (D) Env-specific versus FLSC-specific IgG responses at week 7 comparing each animal in a group. IgG levels were calculated relative to the standard VRC01 antibody. P values were calculated using the Wilcoxon matched-pairs signed-rank test. (E) Anti-CD4i responses were measured by ELISA. Plates were coated with FLSC. Prior to addition, animal sera were preadsorbed with HIVBaL gp120 such that only conformationally induced CD4i-specific IgG was detected. IgG levels were calculated relative to the standard VRC01 antibody. The P value was calculated using the Mann-Whitney test.
Serum anti-Env IgG levels were significantly higher for the Env-rFc group than the Env group at 7 weeks after immunization (P = 0.03) (Fig. 2B). These differences in week 7 sera between the Env-rFc and Env groups became further pronounced (P = 0.004) when IgG levels specific for the full-length single-chain protein (FLSC) were measured (9) (Fig. 2C). FLSC contains gp120 fused to a flexible linker plus domains 1 and 2 of CD4 such that the CD4 binding site is always occupied and gp120 is always in the open, or CD4-induced (CD4i), conformation (9). Thus, FLSC exposes epitopes that are less accessible in the native, or closed, conformation of Env. We assessed the breadth of antibody responses by comparing binding levels of serum antibodies to Env or FLSC. In the Env-rFc animals, there were significant increases in anti-FLSC IgG levels compared to their anti-Env levels, suggesting a broadening of the immune response in monkeys receiving the fusion protein. Anti-FLSC IgG responses in the Env group were below the anti-Env IgG levels (approaching significance, P = 0.06) (Fig. 2D). Increased breadth of serum responses in the Env-rFc group was further demonstrated by the presence of serum IgG reacting to CD4i epitopes in five of six animals from the Env-rFc group and in none of the animals in the Env group (Fig. 2E). The anti-CD4i IgG response comprised approximately 20% of the total IgG FLSC-specific response (see Table S1 in the supplemental material). These results suggest that targeting the Env immunogen to FcγR by fusing it to an IgG Fc moiety both augments peak immune responses and broadens the response.
Env-rFc immunization elicits a neutralizing antibody and antibody-dependent cellular cytotoxicity (ADCC) response.
We tested for the presence of neutralizing antibodies at week 7 by use of the TZM-bl cell assay (10). Six of six animals in the Env-rFc group had modest levels of neutralizing antibodies against tier 1A MN.3 virus (range of titer for 50% neutralization was 30 to 341 dilution of serum; average neutralization titer, 143), but neutralizing activity was not detected in sera from the Env immunization group (50% infective dose [ID50] values were below the minimum serum dilution of 20) (Table 1). We did not detect neutralization of the more neutralization-resistant tier 1B or tier 2 viruses in any of the sera tested.
TABLE 1
Week 7 neutralization titers
| Env group and animal | ID50 in TZM-bl cellsa | |||
|---|---|---|---|---|
| MN.3 virus, clade B, tier 1A | BaL.26 virus, clade B, tier 1B | Bx08.16 virus, clade B, tier 1B | TRO.11 virus, clade B, tier 2 | |
| Env-rFc | ||||
    811 811 | 341 | <20 | <20 | <20 |
    814 814 | 32 | <20 | <20 | <20 |
    815 815 | 45 | <20 | <20 | <20 |
    819 819 | 305 | <20 | <20 | <20 |
    821 821 | 107 | <20 | <20 | <20 |
    822 822 | 30 | <20 | <20 | <20 |
| Env | ||||
    802 802 | <20 | <20 | <20 | <20 |
    809 809 | <20 | <20 | <20 | <20 |
    816 816 | <20 | <20 | <20 | <20 |
    817 817 | <20 | <20 | <20 | <20 |
    820 820 | <20 | <20 | <20 | <20 |
We measured serum ADCC activity using a standard flow cytometry-based ADCC-GranToxiLux (ADCC-GTL) assay (described in reference 11). ADCC was detected in four of six Env-rFc-immunized animals but in none of the Env-immunized animals (Fig. 3). In Fig. 3, we report serum ADCC activity as the percentage of target cells that received a lethal hit of granzyme B (peak percent granzyme B activity ranged between 8.82% and 20.83%). The titer of dilution sera at which this peak activity was observed ranged from 132 to 330 (Table S2).
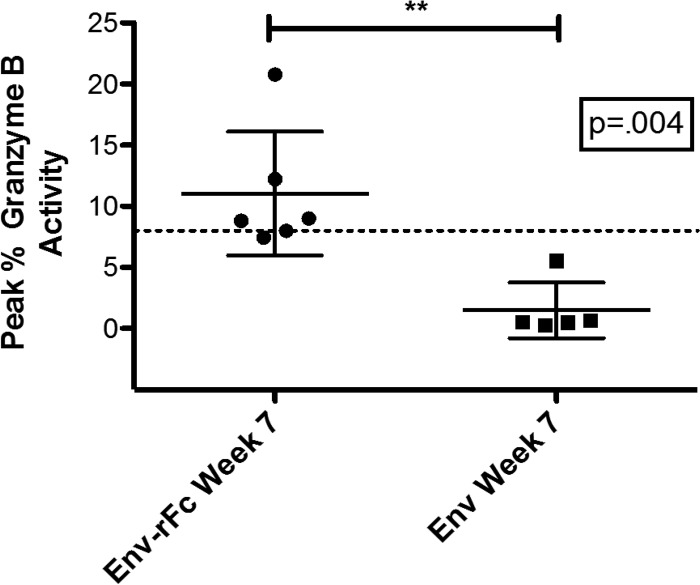
Env-rFc immunization elicits ADCC responses. A fluorescence-based ADCC-GTL assay was used to detect ADCC activity in the sera of Env-rFc- or Env-immunized animals. CEM.NKRCCR5 target cells were coated with HIVBaL gp120, which was the immunogen used to immunize animals. Effector cell-derived granzyme B (GzB) is delivered into target cells as a result of antigen-specific antibody-FcγR interactions. Shown here is the percentage of target cells with GzB activity as a result of GzB hydrolysis of a fluorogenic peptide substrate. The cutoff for positivity for GzB activity is 8%. The P value was calculated using the Mann-Whitney test.
Immunization with Env-rFc yields a more durable immune response.
The decay rates for Env or FLSC binding antibodies were measured for 17 weeks after immunization. Antibody levels in animals immunized with Env-rFc declined over a 10-week period from the peak at week 7 until the last measured time point at week 17, while the initial phase of decay in the Env-immunized group ended by 5 weeks after the peak at week 12 (Fig. 4; Table S3). The average levels of antibody at week 17 in the Env-rFc group were 5 times higher than the levels seen in the Env group at week 12 (Fig. 4; Table S3).
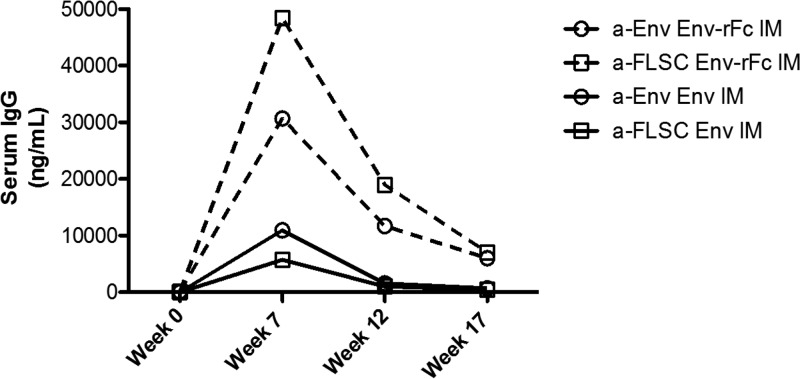
Env-rFc immunization elicits a more durable immune response than Env immunization. Anti-Env (circles) or anti-FLSC (squares) IgG responses in the Env (solid line) and Env-rFc (dashed line) groups were measured with ELISA. Responses are shown from week 0 (preimmunization) through week 17. IgG levels were calculated relative to the standard VRC01 antibody.
In animals immunized with Env-rFc, the portion of antibodies binding specifically to FLSC declined more rapidly than Env binding titers. By week 7 or 12, FLSC binding titers were 1.6 times higher than Env binding titers, but by week 17 they were nearly at the same level. Thus, CD4i-specific responses were detectable for only about 10 weeks after the peak response time point.
DISCUSSION
We designed a fusion protein containing the gp120 portion of HIV envelope glycoprotein from the BaL strain joined at the C terminus to a poly-serine/glycine linker which, in turn, was joined at the C terminus to the Fc region of rhesus macaque IgG1. The dimeric fusion protein was highly expressed in CHO cells. Env-rFc was functionally intact, based on its binding to CD4 or Fcγ cell surface receptors.
Our immunization study evaluated the magnitude and quality of antibody responses against this fusion protein (Env-rFc) in rhesus macaques. Compared to monomeric gp120, the Env-rFc protein showed increased potency and greater breadth of antibody responses, including both neutralizing and antibody-dependent cellular cytotoxicity activities, and serum antibody responses were more durable.
Immunization studies with Env-rFc employed the soluble adjuvant, CpG, and the i.m. route of delivery. After two immunizations with Env-rFc, macaques showed higher peak titers for gp120-binding antibodies than animals immunized with gp120 alone. Env-rFc was more potent for inducing antibody responses to the CD4-induced epitopes on gp120. While we were able to measure antibodies against these conformational epitopes in sera from Env-rFc-immunized macaques, these responses were of short duration and decayed more rapidly than the overall gp120-binding antibodies. The gp120-binding antibodies appeared to be more durable in the Env-rFc-immunized macaques than in the Env-immunized macaques. Further, a modest neutralizing activity against tier 1A virus was detected in animals immunized with Env-rFc but not in animals immunized with gp120 alone. Similar results were observed when immune sera were tested for ADCC. Again, only the Env-rFc animals manifested serum antibodies capable of directing the killing of cellular targets in a standard ADCC-GTL assay.
The peak titers of gp120 binding antibody responses among Env-rFc-immunized macaques were similar or greater than levels observed by using gp120 alone (12) or by following a DNA priming-recombinant viral vector boosting strategy (13, 14). Gp120-binding antibody titers greater than what we attained have been reported by others using more extensive immunization regimens and/or incorporating more potent adjuvants (15). Broadening the antibody response to increase the recognition of CD4-induced epitopes has been achieved using the full-length single-chain version of gp120 (16), and the Fc fusion protein studied here also produced CD4i targeting antibodies.
Neutralizing antibody responses obtained in our Env-rFc-immunized macaques are probably insufficient to protect animals from homologous or heterologous virus challenge (17, 18); for this reason, we did not challenge macaques in this study. However, responses to Env-rFc were substantially improved over what has been achieved using monomeric gp120 (12, 16, 19). Of critical importance to our work is that the Env-rFc immunogen was superior to gp120 for inducing the modest neutralizing antibody responses reported here, suggesting that the addition of an Fc segment altered both the quantity and quality of the resulting serum antibodies. Inasmuch as we have not tested a broad range of adjuvants and did not conduct extensive tests to optimize the dose and schedule for immunization, it is notable that Env-rFc was capable of improving the immune response.
Monomeric gp120 alone has not elicited strong protective immunity in macaque models (12) or human clinical trials (1, 20, 21). Here, we asked whether immune responses to gp120 could be improved by building an Env-rFc fusion protein that would carry two molecules of gp120 in close proximity and bind to cell surface Fc receptors to mimic some properties of IgG immune complexes (ICs). Env-rFc might bind the low-affinity gp120-specific germ line BCR with higher avidity, lowering the activation threshold of naive B cells and resulting in the formation of bNAb. ICs engage FcγRs and augment cell-mediated and humoral responses to immunogens bound by the antibody Fab regions (reviewed in reference 22). Multiple mechanisms have been proposed for IC effects on immunity (reviewed in references 22 and 23). IC binding to activating FcγRs on dendritic cells (DCs) induces maturation, increasing major histocompatibility complex and costimulatory molecule expression, and leads to enhanced antigen presentation (24, 25, 26). Further, ICs are deposited on follicular dendritic cells within lymphoid follicles, where they promote immunoglobulin somatic hypermutation and affinity maturation of antigen-specific B cells (27, 28, 29, 30).
We also know that the capacity for binding to the neonatal Fc receptor (FcRn) is important for increasing the half-life of IgG molecules compared to other subtypes that do not bind FcRn (31). Since Env-rFc contains an FcRn binding site, it is expected to recycle through acidic cytoplasmic vesicles (such as endosomes) and be released back into the extracellular milieu without degradation, similar to the trafficking of noncomplexed IgG. This recycling mechanism increases the IgG half-life by allowing temporary protection from catabolism in the harsh circulatory environment (32) and should also increase the half-life of Env-rFc and influence the serum antibody response to immunization.
Our study was designed to make a preliminary assessment of the effect of adding Fc to gp120 in terms of altering the quantity or quality of the resulting antibody responses. We did not attempt to optimize the immunogen by changing the formulation, adjuvant composition, or immunization schedule, including a prime-boost component or giving repeat doses. We reasoned that gp120 was unlikely to suffice as a component of preventive HIV vaccine and that the current status of the field was focused on well-ordered envelope glycoprotein immunogens that may be found as trimers or other conformationally constrained molecules (3, 33). Based on our results, we argue that adding Fc segments to any of the high-performing envelope glycoprotein immunogens may improve their capacity to elicit durable antibody and that such modifications should be considered part of the vaccine development pathway. Mutations in the IgG1 Fc can increase binding to FcγRs (34). Env-rFc immunogens with gain-of-function mutations can be tested in future studies to optimize the fusion protein and further improve immune responses. We note the previous publication from our group showing that a fusion protein containing HIV p24 Gag linked to mouse Fc (35) was extraordinarily potent for eliciting cellular or antibody responses. Perhaps Fc is a generally useful modification for immunogens and may bridge the gap between current products and truly effective HIV vaccines.
MATERIALS AND METHODS
Cells and reagents.
Chinese hamster ovary (CHO-K) cells and THP-1 human monocytic cells were purchased from the American Tissue Culture Collection (ATCC TIB-202; ATCC, Manassas, VA). CHO cell lines were grown in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 mM HEPES (N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid), 10% fetal calf serum (Sigma-Aldrich, Inc., Darmstadt, Germany), 1% l-glutamine, nonessential amino acids, and 1% penicillin-streptomycin. THP-1 cells were grown in complete RPMI 1640 (Thermo Fisher Scientific Inc., Waltham, MA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific Inc., Waltham, MA), 1% l-glutamine, 0.05 mM 2-mercaptoethanol, and 1% penicillin-streptomycin (ATCC TIB-202, ATCC, Manassas, VA). Cells were grown in 5% CO2 at 37°C. Peripheral blood mononuclear cells (PBMCs) were purified from whole blood that was obtained from commercial suppliers, and PBMCs were cultured in the same RPMI-based medium.
PG9 antibody (catalog no. 12149) and VRC01 antibody (catalog no. 12033) were acquired from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH.
Manufacturing of Env-rFc and Env glycoproteins was performed by the Viral and Cell Core Facility at the Institute of Human Virology, University of Maryland School of Medicine. HIVBaL gp120 was expressed in 293FS cells from an expression construct purchased from Aldevron (Fargo, ND).
Constructing the Env-rFc fusion protein expression vector.
The cDNA encoding gp120 from the HIV-1 isolate BaL was amplified using PCR primers (5′-CTCTGAATTCACCGCCATGGGGTCTCTGCAACCG-3′, 5′-AGATCCCGAGCCACCTCCTCCGGACCCACCACCGCCTGATCCGGCCACGCCCAGGGGCTC-3′). The antisense primer was extended to comprise 14 codons for glycine and serine (GSGGGGSGGGGSGS) that formed a linker between the gp120 and Fc sequences. The Fc fragment of monkey IgG1 containing hinge, CH2, and CH3 domains was amplified by reverse transcription PCR (RT-PCR) from the monkey PBMC cDNA using primer pairs 5′-GGATCAGGCGGTGGTGGGTCCGGAGGAGGTGGCTCGGGATCTCCTCCCACGTGCCCACCG-3′ and 5′-TATACTCGAGTTATTTACCCGGAGACAGGGA-3′. The forward primer for IgG1 Fc is complementary to the G/S linker region. PCR amplification reaction products were annealed and digested with restriction endonucleases EcoRI and XhoI before ligation into a pCDNA3 vector containing a gp120 secretion signal sequence. Each construct was verified by DNA sequencing.
Plasmids containing the Env-rFc were transfected into CHO cells using Effectene (Qiagen, Valencia, CA). G418-resistant clones were selected and tested for secretion of Env-rFc fusion proteins. SDS-PAGE and Western blotting were performed to assess the recombinant fusion proteins using mouse anti-gp120 or anti-monkey IgG antibody. The highest-expressing clones were preserved in cell banks for Env-rFc production. Env-rFc fusion proteins secreted in CHO cell supernatants were first cleaned by ultrafiltration and further purified by affinity chromatography using protein A Sepharose 4 fast flow (Amersham Pharmacia, Piscataway, NJ). Protein concentrations were measured with Bradford protein assay kits (Thermo Fisher Scientific, Inc., Waltham, MA), using mouse IgG2a as a standard.
Biochemical characterization of Env-rFc.
Env-rFc was labeled with allophycocyanin (APC) fluorophore using the Lightning Link APC-XL conjugation kit (Innova Biosciences Ltd., Cambridge, England) according to the manufacturer's instructions.
Inhibition assay: Env-rFc binding to FcγRs.
THP-1 cells were pretreated with HIVBaL gp120 (to block Env-rFc binding to cell surface CD4) and with various molar concentrations of unlabeled IgG Fc fragments for 1 h at 4°C. Treated THP-1 cells were washed and stained with 0.5 μg of APC-labeled Env-rFc (this amount produced maximum signal intensity in the absence of competing Fc fragments or HIVBaL gp120). Molar ratios of Fc fragments to APC-labeled Env-rFc were 0:1, 0.2:1, 0.5:1, 1:1, and 2:1. The isotype control was THP-1 cells treated with unlabeled Env-rFc. Binding of labeled proteins was analyzed with an Accuri C6 flow cytometer (BD Biosciences, San Jose, CA).
Competition assay: Env-rFc binding to CD4.
PBMCs were preincubated with 2 μg of IgG Fc fragments for 30 min on ice. Cells were treated with Env-rFc for 30 min on ice and then washed and stained with a fluorescein isothiocyanate (FITC) anti-CD4 antibody that does not compete with Env for binding to CD4 (OKT4; BioLegend, San Diego, CA) or with an FITC anti-CD4 antibody that competes with Env for the CD4 binding site (RPA-T4; BioLegend, San Diego, CA). Stained PBMCs were analyzed using an Accuri C6 flow cytometer (BD Biosciences, San Jose, CA).
Animals and immunizations. (i) Rhesus macaques.
Juvenile rhesus macaques with equal numbers of males and females were obtained from commercial suppliers. Animal housing and experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Maryland School of Medicine (IACUC protocol no. 0115004).
(ii) Immunizations.
Rhesus macaques were immunized with 100 μg of recombinant HIVBaL envelope gp120 (Env) glycoprotein (n = 5) or 100 μg of Env-rFc fusion protein (n = 6) per dose. Proteins were formulated in 0.5 ml of sterile saline plus 50 μg of CpG ODN (InvivoGen, San Diego, CA). Immunizations were performed at week 0 and week 4 via the i.m. route into the thigh. Blood samples were collected from animals that had received ketamine restraint anesthesia. Venous blood samples were obtained at weeks 0 (preimmunization), 4, and 7. Immune responses were analyzed at week 7 (3 weeks after the second immunization). Subsequent blood samples were obtained to evaluate the durability and specificity of antibody responses.
HIVBaL Env gp120, full-length single-chain protein (FLSC) ELISA.
Ninety-six-well microtiter ELISA plates were coated with 5 μg/ml antigen (HIVBaL gp120 or FLSC) in sodium bicarbonate solution, pH 9.6 (Sigma-Aldrich, Inc., Darmstadt, Germany), and incubated overnight at 4°C. Plates were washed with phosphate-buffered saline (PBS)–0.05% Tween 20 (PBS-T) and blocked with 5% bovine serum albumin (BSA) for 1.5 h at 37°C. After washing 4 times, 100 μl of animal serum or VRC01 monoclonal antibody standard was added to duplicate wells, and antibody concentrations were determined in control tests. Experimental serum samples added to coated plates were incubated for 1.5 h at 37°C and washed 6 times. One hundred microliters of horseradish peroxidase (HRP)-conjugated sheep anti-human IgG (GE Healthcare, Chicago, IL) was added at a 1:1,000 dilution and incubated for 1 h at 37°C. Plates were washed 6 times again, and 100 μl of TMB (3,3′,5,5′-tetramethylbenzidine) Ultra solution (Thermo Fisher Scientific, Inc., Waltham, MA) was added for 15 min. The reaction was quenched with 100 μl of 0.16 M sulfuric acid stop solution (Thermo Fisher Scientific, Inc., Waltham, MA). The absorbance was measured at a wavelength of 450 nm.
CD4-induced (CD4i)-specific IgG levels were measured in ELISAs using FLSC bound to the plate. Animal sera were preadsorbed with 10 μg HIVBaL gp120 for 30 min at 4°C to block gp120-specific antibodies and reveal only the fraction of antibodies specific for the conformationally constrained FLSC.
Calculations.
All samples were run in duplicate. Average optical density (OD) values were adjusted by subtracting the background OD values (blank wells with zero added standard). Preimmunization (week 0) serum IgG levels were subtracted from the week 7 serum concentrations. The concentration of HIV-1 IgG antibody was calculated relative to the standard curve using a four-parameter fit curve that was generated using the GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA).
HIV neutralization assay.
Serum antibody neutralization titers were detected by measuring a reduction in virus infection using TZM-bl cells as the target for HIV (36). Briefly, heat-inactivated sera (56°C, 1 h) were combined with tier 1A, tier 1B, or tier 2 clade B Env-pseudotyped viruses and overlaid on virus-permissive TZM-bl cells expressing a Tat-regulated firefly luciferase (Luc) reporter gene. Luciferase signal, measured in relative luminescence units (RLU), is directly proportional to infectious virus particles. Neutralization titers are reported as the dilution at which the RLU declined to 50% of the maximum RLU from virus control wells (no test sample) after subtraction of background luminescence in cell control wells. The lower limit of detection was a neutralizing titer of 20.
ADCC.
A flow-based ADCC-GranToxiLux (ADCC-GTL) assay (described in reference 11) was used to measure ADCC activity in macaque sera. Recombinant HIVBaL gp120 was used to coat target CEM.NKRCCR5 cells in the GTL assay. Preimmunization or week 7 macaque sera were used to decorate the gp120-coated cells. PBMCs obtained from an HIV-seronegative healthy donor were used as effectors. Antibody-dependent cellular cytotoxicity (ADCC) is measured as the percentage of target cells with granzyme B activity. Effector cell-derived granzyme B hydrolyzes a fluorogenic peptide substrate, generating a fluorescent signal that allows individual target cells that have received a lethal hit to be identified by flow cytometry. Target and effector cell populations, in the absence of serum, were used to calculate background. The cutoff for positivity in the GTL assay was >8% of granzyme B activity. The recombinant gp120 representing the HIVBaL isolate was chosen because it matched the immunogen used in the vaccine study.
Statistical analysis.
Results from experiments are presented as the median with interquartile range. Data were analyzed with GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA), and P values of <0.05 were considered to be statistically significant.
ACKNOWLEDGMENTS
Research reported in this study was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI102680 (C. D. Pauza and X. Zhu, principal investigators).
The funders had no role in the study design, data collection and interpretation, or decision to submit the work for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare that they have no conflicting interests.
Z.S. collected, analyzed, and interpreted the data for the biochemical studies (except the expression of protein with Western blotting) and immunization studies and wrote the manuscript. W.L. designed and engineered the Env-rFc immunogen, confirmed its expression with Western blotting, and contributed to the editing of the manuscript. B.P. participated in the interpretation of data and provided intellectual insight. G.F. oversaw the collection of data, analysis, and interpretation for the ADCC-GTL assay and contributed to the editing of the manuscript. C.L. and D.M. oversaw the collection of data, analysis, and interpretation for the neutralization assay. D.M. contributed to the editing of the manuscript. X.Z. was a major contributor to the study design, oversaw the production of the Env-rFc immunogen, and was a major contributor to the editing of the manuscript. C.D.P. was the main contributor to the design and direction of the study, and he edited the manuscript.
We acknowledge the following people at the Institute of Human Virology, University of Maryland at Baltimore: staff at the animal facility for caring for the rhesus macaques in our study and for performing the anesthesia and immunizations, Rex Lin for providing FLSC, and Felisa Diaz-Mendez for her technical and organizational support throughout the study. We appreciate technical support from Liming Wu at VA-MD College of Veterinary Medicine, University of Maryland College Park.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CVI.00028-17.
REFERENCES
Articles from Clinical and Vaccine Immunology : CVI are provided here courtesy of American Society for Microbiology (ASM)
Citations & impact
Impact metrics
Article citations
Preclinical assessment of a recombinant RBD-Fc fusion protein as SARS-CoV-2 candidate vaccine.
Eur J Microbiol Immunol (Bp), 14(3):228-242, 16 May 2024
Cited by: 0 articles | PMID: 38753442 | PMCID: PMC11393645
Antigen-dependent modulation of immune responses to antigen-Fc fusion proteins by Fc-effector functions.
Front Immunol, 14:1275193, 05 Oct 2023
Cited by: 1 article | PMID: 37868961 | PMCID: PMC10585040
Multimeric Epitope-Scaffold HIV Vaccines Target V1V2 and Differentially Tune Polyfunctional Antibody Responses.
Cell Rep, 28(4):877-895.e6, 01 Jul 2019
Cited by: 27 articles | PMID: 31340151 | PMCID: PMC6666430
Improving the Breadth of the Host's Immune Response to Lassa Virus.
Pathogens, 7(4):E84, 28 Oct 2018
Cited by: 14 articles | PMID: 30373278 | PMCID: PMC6313495
Review Free full text in Europe PMC
Incomplete Downregulation of CD4 Expression Affects HIV-1 Env Conformation and Antibody-Dependent Cellular Cytotoxicity Responses.
J Virol, 92(13):e00484-18, 13 Jun 2018
Cited by: 50 articles | PMID: 29669829 | PMCID: PMC6002730
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A Trimeric HIV-1 Envelope gp120 Immunogen Induces Potent and Broad Anti-V1V2 Loop Antibodies against HIV-1 in Rabbits and Rhesus Macaques.
J Virol, 92(5):e01796-17, 12 Feb 2018
Cited by: 20 articles | PMID: 29237847 | PMCID: PMC5809733
HIV-1 gp120-CD4-Induced Antibody Complex Elicits CD4 Binding Site-Specific Antibody Response in Mice.
J Immunol, 204(6):1543-1561, 17 Feb 2020
Cited by: 3 articles | PMID: 32066595 | PMCID: PMC7065964
Cross-Linking of a CD4-Mimetic Miniprotein with HIV-1 Env gp140 Alters Kinetics and Specificities of Antibody Responses against HIV-1 Env in Macaques.
J Virol, 91(19):e00401-17, 12 Sep 2017
Cited by: 5 articles | PMID: 28490585 | PMCID: PMC5599731
Designing immunogens to elicit broadly neutralizing antibodies to the HIV-1 envelope glycoprotein.
Curr HIV Res, 5(6):514-541, 01 Nov 2007
Cited by: 24 articles | PMID: 18045109
Review
 a,*
a,*



