Abstract
Free full text

Metabolic and epigenetic coordination of T cell and Macrophage immunity
Abstract
Recognition of pathogens by innate and adaptive immune cells instructs rapid alterations of cellular processes to promote effective resolution of infection. To accommodate increased bioenergetic and biosynthetic demands, metabolic pathways are harnessed to maximize proliferation and effector molecule production. In parallel, activation initiates context-specific gene-expression programs that drive effector functions and cell fates that correlate with changes in epigenetic landscapes. Many chromatin- and DNA-modifying enzymes make use of substrates and cofactors that are intermediates of metabolic pathways, providing potential cross talk between metabolism and epigenetic regulation of gene expression. In this review, we discuss recent studies of T cells and macrophages supporting a role for metabolic activity in integrating environmental signals with activation-induced gene-expression programs through modulation of the epigenome and speculate as to how this may influence context-specific macrophage and T cell responses to infection.
Introduction
Activation of immune cells in response to infection is the result of the summation of antigen-induced gene-expression programs integrated with environmental signals. Both innate and adaptive immune cells increase their metabolic throughput upon stimulation, promoting energy generation and biosynthesis, while shifting the relative usage of metabolic pathways to support proliferation, effector molecule production, and differentiation (O’Neill and Pearce 2016; MacIver, Michalek, and Rathmell 2013; Pollizzi and Powell 2014; Buck, O’Sullivan, and Pearce 2015). While the rapid transitions in cellular metabolism have long been described, few studies focused on determining the intrinsic cellular impacts metabolic activity can have on gene expression, effector molecule production, and, ultimately, the fate and function of a given cell. Recent interest in understanding the impact of these fundamental metabolic changes on immune cell differentiation and function has yielded numerous studies examining the pathways and metabolites involved in driving protective immune responses. A key question that has emerged is whether differential metabolic activity (i.e. use of glycolysis vs mitochondrial respiration) is coincident with differentiation, a consequence of changes in phenotype or localization, or a direct driver of alterations in immune cell differentiation and function. The complex interplay metabolism can have on nearly all cellular processes and the link between metabolites and epigenetic modifiers have been extensively reviewed (Kinnaird et al. 2016; Kaelin and McKnight 2013; Etchegaray and Mostoslavsky 2016). Here we focus on studies that lay the ground work for a direct link between metabolic intermediates and regulation of epigenetic landscapes of two key immune cells, T cells and macrophages.
The discovery of numerous epigenetic modifications and the enzymes responsible for them have revealed another extensive network of potential regulators of immune cell function. Traditionally, attempts to identify factors critical for the differentiation and function of immune cell populations have relied on comparisons of global gene-expression changes following stimuli. The regulation of individual epigenetic modifications, such as histone methylation and acetylation, as well as DNA methylation, are a recent topic of interest. Progress toward characterization of the global epigenetic landscapes of many immune cell populations in distinct differentiation states allow the linkage of epigenetic modifications to alterations in differentiation and function. Further, numerous signals (i.e. T cell receptor (TCR), Toll-like receptors (TLR), inhibitory receptors, and cytokines) drive changes in the epigenome that result in downstream modulation of immune responses.
As studies of immunometabolism and epigenetics continue to reveal mechanisms that regulate immune cell differentiation and function, the exciting possibility has emerged that alterations in availability of metabolic intermediates may directly impact the epigenome of immune cells as well. This fits well with studies in cancer cells and stem cells suggesting a link between cellular metabolism and alterations in epigenetic landscapes (Kinnaird et al. 2016; Etchegaray and Mostoslavsky 2016). The enzymes required for epigenetic modifications, for example DNA demethylases (Ten-eleven Translocation (Tet) enzymes) and histone demethylases (HMTs), often require cofactors and substrates that are also critical intermediate metabolites. For example, 2-oxoglutarate is a cofactor for Tet enzymes, suggesting a link between cellular metabolism and regulation of gene expression through alterations in the epigenetic landscape of immune cells. While this link has been hypothesized and explored in non-immune cell types (Kinnaird et al. 2016; Kaelin and McKnight 2013; Etchegaray and Mostoslavsky 2016), this premise remains a nascent area of study in the context of immunity. In this review, we highlight recent studies that emphasize the impact stimulation-induced metabolism has on macrophage and T cell phenotype and effector function. Further, we discuss recent discoveries in the unique epigenetic profiles of macrophage and T cell subsets as differentiation is induced following activation. Finally, we address the exciting possibility that alterations in cellular metabolism may provide an additional mechanistic link between environmental signals and downstream gene-expression changes by immune cells critical for the specification of immune cell function due to recent studies that suggest coordination of epigenetic modifications by metabolic activity.
Epigenetic landscapes
The program of gene expression within each cell type is controlled by interactions of sequence-specific transcription factors (TFs) with promoters and distal enhancers. While promoters represent the obligatory sites of initiation of mRNAs, enhancers are major determinants of cell-specific patterns of expression and physiologic responses to internal and external signals (Heinz et al. 2015; Levine 2010). Promoters and enhancers exhibit distinct chromatin signatures defined by modifications of both DNA and histones. This topic has recently been reviewed (Tessarz and Kouzarides 2014; Allis and Jenuwein 2016) and for the purposes of this discussion we consider promoters to exhibit a histone modification signature of trimethylation of histone H3 lysine 4 (H3K4me3), whereas enhancers are typically characterized by monomethylation of histone H3 lysine 4 (H3K4me1) (Heintzman et al. 2009; Heintzman et al. 2007) (Figure 1A). Active enhancers and promoters are further distinguished by histone acetylation, for example at histone H3 lysine 27 (H3K27ac) (Creyghton et al. 2010). Intriguingly, recent studies of macrophages indicate that the histone acetyltransferase p300 can use Crotonyl-CoA as an alternative substrate to Acetyl-CoA for histone tail modification as an activating mark (Sabari et al. 2015). These modifications are recognized by ‘reader’ proteins that play various roles in transcriptional activation or repression. For example, histone acetylation is recognized by proteins that contain bromo domains, such as Brd4, which interacts with the positive transcription elongation factor (pTEFb) and stimulates the transition of RNA Pol II from a paused to elongating form (Bres, Yoh, and Jones 2008). Many other DNA and histone modifications are associated with distinct chromatin functions. For example, DNA methylation of cytosine at CpG and histone methylation of H3K9 or H3K27 are associated with active repression (Blattler and Farnham 2013; Wiles and Selker 2016; Wang, Jia, and Jia 2016), whereas CpG methylation within gene bodies prevents aberrant intragenic transcriptional initiation (Neri et al. 2017).
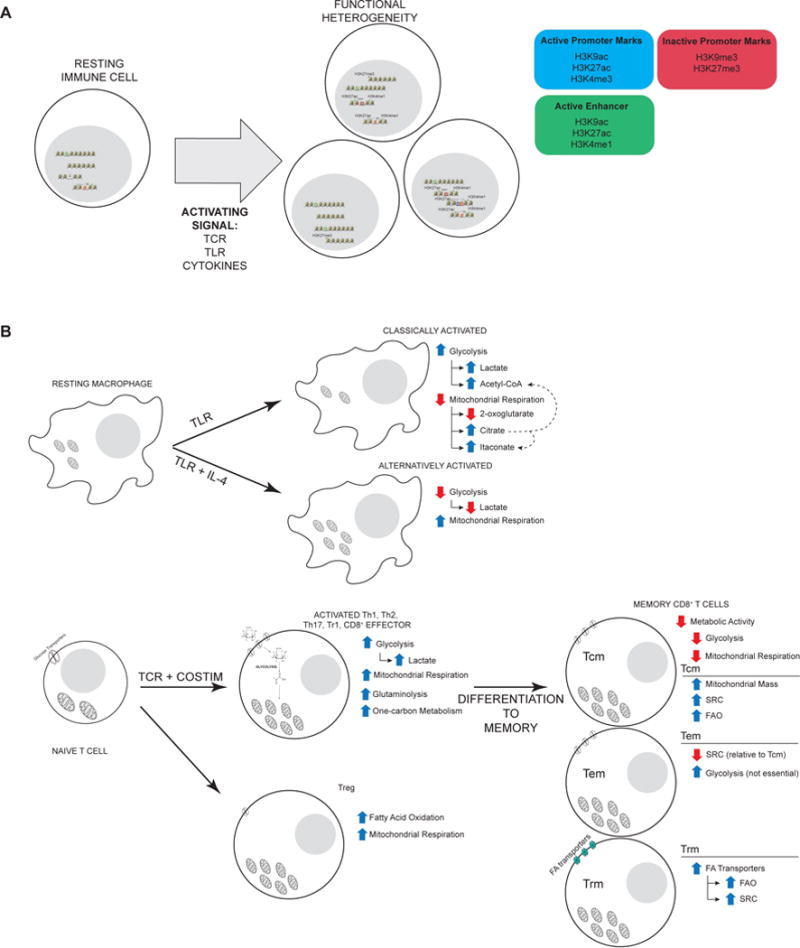
A Activation of immune cells results in substantial remodeling of epigenetic landscapes. Genes associated with the naive state are repressed and effector genes become active through deposition of repressive marks (i.e. H3K9me3 and H3K27me3) and activating marks at promoters and enhancers (i.e. H3K27ac and H3K4me1). Diversity in epigenetic landscapes correlates with diversity in responses and plays a role in specifying differentiation. B Classical activation of macrophages elicits increased glycolytic metabolism and reduced reliance on mitochondrial respiration. These gross alterations in metabolic pathway usage underlie specific changes in the abundance of several metabolites. In particular, increased glycolytic metabolism drives production of lactate and supports a pool of acetyl-CoA by slowing the TCA cycle resulting in a buildup of citrate that can be converted to acetyl-CoA as well. Reduced flux through the TCA cycle, also reduces the production of 2-oxoglutarate and increases the production of itaconate. Alternatively, TLR stimulation of macrophages along with IL-4 reduces glycolytic metabolism and drives increased mitochondrial respiration and oxidative phosphorylation. C TCR and co-receptor stimulation of Th1, Th2, Th17, Tr1, and CD8+ T cells drives a general increase in both glycolytic metabolism and mitochondrial metabolism as measured by extracellular flux analysis. Glucose transport is increased substantially to facilitate glycolytic metabolism and mitochondrial mass increases as well. Tregs on the other hand do not exhibit as dramatic an increase in glucose uptake and glycolytic metabolism, but increase reliance on oxidative phosphorylation and FAO. Biosynthetic pathways are engaged to fuel proliferation, in particular serine-driven folate metabolism, a branch of one-carbon metabolism, that promotes nucleotide synthesis. Extensive characterization of metabolic changes that occur during memory CD8+ T cell differentiation show that in general memory CD8+ T cells exhibit reduced metabolic activity, glycolytic and mitochondrial, with metabolic heterogeneity exhibited within the memory pool. Tcm maintain high mitochondrial mass, substantially increased SRC fueled by increased FAO. Tem maintain lower levels of SRC and can be supported by high levels of glycolytic metabolism. Examination of Trm metabolism show that skin Trm uniquely rely on high levels of fatty acid import to fuel FAO and SRC.
The recent application of Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) has facilitated the global identification of open chromatin corresponding to sites of TF binding with high genomic resolution and minimal cellular input, which allows for feasible ex vivo analyses (Buenrostro et al. 2013; Amit, Winter, and Jung 2016). TF binding motifs within accessible chromatin regions can be used to infer the binding of known TFs and identify potential gene targets of these TFs. When combined with chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq) that identifies active promoters and enhancers, ATAC-seq proves especially powerful to pinpoint TF binding sites inside the regulatory elements. For instance, studies examining open chromatin identified by ATAC-seq with corresponding chromatin modifications have successfully revealed tissue-specific TF and enhancers responsible for the specific identity of tissue-resident macrophages, lineage-determining TF (LDTF) in hematopoiesis (Lavin et al. 2014; Lara-Astiaso et al. 2014), and more recently TFs regulating T cell differentiation to infection (Sen et al. 2016; Yu et al. 2017).
Cofactor requirements for epigenetic modifications
Gene-expression within cell types is tightly regulated by TFs and their interactions with promoters and distal enhancers. Therefore, regulation of TF binding and activity by the epigenome is a crucial step in modulating differentiation programs and, in immune cells can be essential for permitting or inhibiting the execution of gene-expression downstream of immune stimuli (Kanno et al. 2012). For example, Ezh2, the catalytic subunit of polycomb repressive complex 2 (PRC2), is a histone methyltransferase (HMT) that mediates gene repression by facilitating deposition of H3K27 trimethylation (H3K27me3) and has been implicated in repressing cytokine production and limiting CD4+ T helper (Th) cell subset plasticity (Tumes et al. 2013). Specifically, analysis of Ezh2 binding and H3K27me3 of in vitro differentiated Th1 and Th2 cells demonstrated that Ezh2 activity downstream of polarizing cytokine conditions was necessary to repress expression of alternative Th lineage TFs and cytokines, maintaining the differentiation state of CD4+ Th cell subsets (Tumes et al. 2013). Thus, understanding the regulation of enzymes that add chemical groups to histones or DNA (“writers”, histone acetyltransferases (HATs), HMTs, and DNA methyltransferases (DNMTs)) and enzymes that remove those same modifications (“erasers”, histone deacetylases (HDACs) and Tet enzymes) is essential for building a model of how immune cells modulate chromatin to regulate function and cell fate.
Several families of “writer” and “eraser” enzymes that shape the epigenetic landscape have been identified of which several have been shown to be critical regulators of immune cell activity (Chang, Wherry, and Goldrath 2014; Wherry and Kurachi 2015; Link, Gosselin, and Glass 2015). These enzymes are often grouped based on the substrate and the mark they regulate: acetylation of histones (HATs and HDACs) and methylation of histones (HMTs and HDMs) and DNA (DNMTs and Tet) (Allis and Jenuwein 2016). As our mechanistic understanding of these “writer” and “eraser” proteins grows, an appreciation for the importance of cofactors that take part in the establishment of epigenetic marks as well as those necessary for their removal has emerged. While there is an extensive array of cofactors and substrates necessary for the epigenetic modification of the genome, (reviewed extensively in (Kinnaird et al. 2016; Kaelin and McKnight 2013; Etchegaray and Mostoslavsky 2016)) we will focus on several key metabolites that have demonstrated roles in deposition of epigenetic marks that have been implicated in regulation of T cell and macrophage activation.
Acetyl-CoA
Histone subunits contain multiple positively charged lysine residues that attract negatively charged DNA. Acetylation of these residues not only creates docking sites for Bromodomain-containing readers, but also neutralizes the positive charge of the lysine residue, resulting in loosening of the interaction between DNA and histone (Huang et al. 2015). This results in a more accessible chromatin structure permissive to transcriptional machinery. Histone acetylation is performed by HATs and requires the availability of acetyl-CoA, which is the sole source of acetyl groups in eukaryotic cells (Choudhary et al. 2009). Thus, regulation of the production of acetyl-CoA, which can be produced from cytoplasmic acetate, citrate, or pyruvate by distinct enzymes, may serve as a critical sensing step for HAT activity within cells (Kinnaird et al. 2016). This sensing mechanism is supported in studies of Saccharomyces cerevisiae where reduction of cytosolic acetyl-CoA production was sufficient to drive a rapid reduction in histone acetylation within the nucleus (Takahashi et al. 2006). A seminal study in colon cell carcinoma cells also showed that modulation of acetyl-CoA levels produced by ATP citrate lyase (ACLY) was sufficient to regulate HAT activity (Wellen et al. 2009). Building on those findings, the culture of mammalian cells in nutrient limiting conditions initiates protein kinase B (also known as AKT)-driven uptake and metabolism of glucose and phosphorylation of ACLY, that sustains high ACLY activity despite reduced citrate levels, increasing the pool of acetyl-CoA, and promoting HAT activity (Lee et al. 2014). Notably, a recent study of CD4+ T cells draws a link between glycolytic metabolism and acetyl-CoA levels; deletion of the glycolytic enzyme lactate dehydrogenase a (LDHA) inhibited glycolytic metabolism, reduced acetyl-CoA levels, and lowered H3K9ac of the Ifng enhancer (Peng et al. 2016). This study argues the reduction in H3K9ac is a direct result of reduced acetyl-CoA levels in LDHA-deficient CD4+ T cells. However, it is unclear how LDHA deficiency directly modulates acetyl-CoA levels, whether by reducing levels of citrate available for conversion to acetyl-CoA or through inhibition of acetyl-CoA synthetase (Peng et al. 2016). Furthermore, manipulation of any single metabolic pathway may result in disruption of other metabolic and signaling pathways as energy or biosynthetic processes are perturbed, requiring significant care in interpreting data in complex scenarios such as immune cell activation and the downstream impacts on epigenetic enzymes.
In contrast to promotion of acetylation by increasing acetyl-CoA levels, CoA-SH, a product of HAT catalyzed histone acetylation, can serve as a negative regulator of HAT activity. This negative feedback regulates HAT activity in tumor cells and may serve as a critical upper limit for histone acetylation in non-transformed cells and suggest that clearance of CoA-SH is an equally important regulator of HAT activity as acetyl-CoA levels (Lee et al. 2014). Given the central role acetyl-CoA plays in energy production, many other pathways may indirectly regulate acetyl-CoA levels and subsequently histone acetylation as well. Whether regulation of histone acetylation on such a global level can drive cell fate decisions remains an open question.
Nicotinamide adenine dinucleotide (NAD+)
NAD+ and NADH are central coenzymes necessary to accept or carry electrons in numerous cellular processes. Reflective of their importance, numerous processes regulate their abundance. The discovery of SIR2, the first identified Sirtuin (SIRT) NAD-dependent deacetylase, as the enzyme responsible for mediating the effects of caloric restriction on yeast lifespan linked energy balance to histone acetylation through levels of NAD+ (Imai et al. 2000). Specifically, in yeast and mice, SIR2 required cleavage of NAD+ to deacetylate histones and yeast mutants with defects in NAD+ metabolism showed reduced SIR2-dependent repression of gene expression (Imai et al. 2000; Tanner et al. 2000; Smith et al. 2000). Expanding on these findings nutrient-deficient conditions activated AMPK, increased NAD+ levels, and supported SIRT activity in skeletal muscle cells, resulting in deacetylation of peroxisome proliferator-activated receptor γ coactivator 1α (PGC1a) (Cantó et al. 2010; Cantó et al. 2009). These studies detail how cellular energy balance, read out by NAD+ and NADH levels, can regulate protein acetylation including histone acetylation.
The complex regulation of NAD+ availability and the impact on SIRTs have been reviewed previously, highlighting that regulation of NAD+ levels are dependent on synthesis, compartmentalization, and usage by metabolic and non-metabolic pathways (Houtkooper, Pirinen, and Auwerx 2012). For example, poly ADP-ribose polymerases (PARPs) have been shown to be NAD+-consuming enzymes implicated in the regulation of NAD+ levels and Sirt activity (Schreiber et al. 2006). Specifically, deletion of Parp1 in mice increased intracellular NAD+ levels in brown adipose tissue and muscle resulting in increased SIRT1 activity (Bai et al. 2011). Particularly relevant to immune cell responses, the compound resveratrol indirectly activates SIRTs, driving histone deacetylation, by increasing the availability of NAD+ in an AMPK-dependent fashion (Um et al. 2010).
S-adenosylmethionine (SAM)
DNMTs and HMTs transfer methyl groups to DNA and histones via the same molecular mechanism, utilizing a methyl group from SAM to generate methylated DNA/histones and a molecule of S-adenosyl homocysteine (SAH). SAM is produced via one-carbon metabolism from methionine by the enzyme methionine adenosyltransferase (Sakata et al. 1993). One-carbon metabolism is a critical component of cellular biosynthesis in immune cells and can regulate proliferation, as evidenced by a recent study that restricted serine, a methyl donor for the conversion of tetrahydrofolate (THF) to 5,10-metheylene-THF in the folate cycle of one-carbon metabolism, suppressing T cell proliferation by abrogating purine nucleotide biosynthesis without affecting energy production (Ma et al. 2017). Multiple metabolic inputs affect one-carbon metabolism and the regulation of this pathway is complex making it difficult to predict outcomes given changes in any branch. In mouse embryonic stem cells, catabolism of threonine (one branch of one-carbon metabolism) was important for maintaining high levels SAM as removal of threonine from culture media resulted in reduced SAM levels and diminished H3K4me3 (Shyh-Chang et al. 2013). At the simplest level, high levels of SAH can be equated with low nutrient conditions and high SAM concentrations with nutrient rich conditions, highlighting a role for the overall nutritional state of a cell as a modulator of DNMT and HMT activity.
2-oxoglutarate
A citric acid cycle intermediate, 2-oxoglutarate, also called α-ketoglutarate, is typically generated in the mitochondria from isocitrate by isocitrate dehydrogenase (IDH). 2-oxoglutarate functions as a critical cofactor for the function of 2-oxoglutarate-dependent dioxygenases that hydroxylate proteins (prolylhydroxylases), demethylate histones (JmjC domain-containing Histone Demethylases (JHDMs)), or de-methylate DNA (Tet enzymes). In the case of JHDMs, 2-oxoglutarate is decarboxylated to succinate during demethylation of the substrate (Chervona and Costa 2012). The importance of 2-oxoglutarate on JHDM and Tet function has primarily been examined in cancer cell lines where tumor-associated mutations in IDH result in the production of the R-enantiomer of 2-hydroxyglutarate (2-HG), which along with succinate and fumarate can inhibit 2-oxoglutarate-dependent dioxygenases (Kaelin and McKnight 2013). This raises the possibility that regulation of 2-oxoglutarate or competitors (2-HG) can regulate the epigenome via modulation of 2-oxoglutarate-dependent dioxygenase activity. Little is currently known about how physiologic fluctuations in 2-oxoglutarate levels, compartmentalization, or competition between 2-oxoglutarate-consuming enzymes play into the regulation of JHDM or Tet enzyme activity.
The metabolites discussed above by no means constitute the only metabolic intermediates that exhibit rapid alterations in concentration or use following macrophage and T cell stimulation and can modulate “writer” and “eraser” activity. For example, lactate, which can play a role in modulating T cell function in the tumor microenvironment, was demonstrated to be a weak inhibitor of HDACs and may impact immune cell epigenomes (Latham et al. 2012; Brand et al. 2016). Furthermore, many epigenetic modifying enzymes (i.e. HATs and SIRTs) have substrates beyond histones and DNA, which are beyond the scope of this review, but must be considered when evaluating the activity of these enzymes. In the following sections, we focus the discussion on central metabolic molecules that are dynamically regulated or have been shown to influence epigenetic enzymes in T cells or macrophages directly.
Epigenetic landscapes in macrophages
Genome-wide analysis of open chromatin and histone modifications associated with active enhancers in many cell types and tissues indicate that mammalian genomes contain on the order of a million potential regulatory elements (Roadmap Epigenomics et al. 2015). Each cell type typically selects ~30,000 such elements that act as primary drivers of that cell’s program of gene expression and specific pattern of responses to internal and external signals. How each cell acquires its unique set of enhancers remains a major question. As in other cell types, promoters and enhancers in macrophages are selected from the genome by specific combinations of lineage determining transcription factors (LDTFs) and more broadly expressed TFs (Link, Gosselin, and Glass 2015). Cell type-specific transcriptional responses are primarily driven by enhancers, which are the most numerous sites of DNA binding of LDTFs. In peritoneal and bone marrow derived macrophages, PU.1, AP-1, CCAAT/Enhancer binding protein (C/EBP), and interferon regulatory factors (IRF) are major LDTFs and ChIP-seq experiments demonstrate that they are found alone or in combination at most macrophage enhancers (Ostuni et al. 2013; Ghisletti et al. 2010; Heinz et al. 2010; Kaikkonen et al. 2013). These factors play important pioneering roles in establishing open regions of chromatin that become sites of action for signal dependent TFs, such as NFκB and nuclear hormone receptors (Glass and Natoli 2016). Specific combinations of TFs recruit co-activator and/or corepressor complexes that in turn control the transcriptional functions of the corresponding regulatory elements. Many of these complexes contain enzymatic activities that modify chromatin through effects on DNA methylation, histone tail methylation, acetylation, etc. Studies of nuclear hormone receptors established a paradigm by which signaling could induce a switch in the activity of a regulatory element by inducing the exchange of co-repressor complexes for co-activator complexes (Rosenfeld, Lunyak, and Glass 2006). For example, unliganded retinoic acid receptors bind NCoR and SMRT co-repressor complexes that contain HDACs and repressive HMTs that exert active repression functions. Upon binding retinoic acid, NCoR/SMRT complexes are exchanged for SRC co-activator complexes that contain HATs such as CBP and p300 and activating HMTs such as CARM1, leading to gene activation. Thus, enhancers and promoters occupied by LDTFs and other sequence-specific TFs can range from being strongly repressed to maximally activated depending on the specific combinations of factors and their activity states (Figure 1A).
Treatment of macrophages with lipopolysaccharides (LPS) in the presence or absence of interferon-γ (IFNγ) to induce a ‘classically’ activated phenotype or with interleukin-4 (IL-4) to induce an ‘alternatively’ activated phenotype have been extensively used as in vitro paradigms for investigation of signal-dependent gene expression and metabolic reprogramming (O’Neill and Pearce 2016; Baardman et al. 2015; Pearce and Pearce 2013; Glass and Natoli 2016; Wynn, Chawla, and Pollard 2013). The LPS response in macrophages is initially driven by latent TFs that include NFκB, AP-1 and IRFs that subsequently result in secondary transcriptional responses to type I interferons and other cytokines. Hundreds of genes are activated or repressed and are associated with corresponding changes in the surrounding epigenetic landscapes (Kaikkonen et al. 2013; Ostuni et al. 2013). Although most changes in epigenetic features occur at pre-existing regulatory regions, there are also inactive genomic loci that acquire features of active enhancers (Kaikkonen et al. 2013; Ostuni et al. 2013), so-called ‘latent’ or ‘de novo’ enhancers. Some of these features are retained following resolution of the initial LPS response, and are associated with more rapid responses of higher magnitude to subsequent stimuli, suggesting a form of epigenetic ‘memory’ (Ostuni et al. 2013).
The transcriptional response to IL-4 in macrophages is driven by a STAT-6-PPARγ-PGC1α signaling axis (Vats et al. 2006), which play key roles in regulating mitochondrial function and oxidative metabolism. In addition, metabolic reprogramming has also recently been proposed to involve a mammalian target of rapamycin complex 2 (mTORC2)-IRF4 signaling axis (Huang et al. 2016). As with LPS treatment, responses to IL-4 are associated with extensive alterations of the epigenetic landscapes (Piccolo et al. 2017). Intriguingly, IFNγ and IL-4 were found to mutually inhibit the epigenomic and transcriptional changes induced by each cytokine alone (Piccolo et al. 2017).
Metabolic reprogramming is also a notable aspect of the phenomenon of trained immunity, in which an initial response of an innate immune cell to a particular stimulus results in altered responses to a subsequent stimulus by mechanisms that are thought to be epigenetic in nature (Netea et al. 2016; Arts, Joosten, and Netea 2016). Immune tolerance of macrophages is a well-studied example in which treatment with an initial dose of LPS renders them less responsive to a second dose for a subset of inducible genes (distinct from those associated with latent enhancers noted above). This phenomenon may contribute to the immune deficiency of sepsis (Netea et al. 2016). Treatment of monocytes with the Candida albicans cell wall constituent β-glucan provides an additional model of trained immunity, in which trained (i.e., β-glucan-treated) monocytes exhibit an increased response to LPS treatment 7 days later relative to naive monocytes (Cheng et al. 2014). In this model, β-glucan treatment is associated with immediate changes in expression of genes associated with immune responses and metabolism, and corresponding changes in the epigenetic landscape. While the initial transcriptional response resolves, epigenetic changes persist (Cheng et al. 2014), suggesting a mechanistic basis for training, analogous to findings made in LPS-treated macrophages. Intriguingly, β-glucan treatment has recently been found to partially reverse LPS tolerance in vitro through mechanisms suggested to involve reprogramming the epigenetic landscape (Novakovic et al. 2016). The two most significant functional annotations associated with β-glucan-induced genes were oxidative reduction and metabolism, suggesting roles of these processes in training.
Metabolic reprogramming in macrophages
A conventional view of signal-dependent gene expression places metabolic intermediates downstream of TFs and their targets that play direct roles in regulation of metabolic processes (e.g., PPARγ and enzymes involved in fatty acid oxidation). This view is challenged, however, by numerous recent findings indicating that metabolic intermediates are required for aspects of gene expression that are specific for distinct polarization states. There are potentially a broad range of mechanisms that could account for these findings, acting at both epigenetic and non-epigenetic levels.
LPS ligation of TLR4, while exerting direct effects on gene expression by activating signal-dependent TFs such as NFκB, AP-1 and IRFs, also results in a shift to glycolytic metabolism and impaired mitochondrial respiration (Rodriguez-Prados et al. 2010). The shift to glycolysis in response to LPS is thought to be important in the context of acute bacterial infection because glycolysis, although being less efficient in generating adenosine triphosphate (ATP), can be up-regulated many fold and therefore results in a faster production of ATP compared to oxidative phosphorylation (Arts, Joosten, and Netea 2016). Glycolysis results in an increase in lactate, which is a weak inhibitor of Class II histone de-acetylases (HDACs) (Latham et al. 2012). TLR signaling also results in marked shifts in NAD+-NADH ratios, which influence the activities of the Class III HDACs SIRT1 and SIRT6, potentially altering de-acetylation of histone and non-histone substrates (Figure 1B) (Liu et al. 2012).
The response to LPS plus IFNγ also results in alterations in the tricarboxylic acid (TCA) cycle (Lampropoulou et al. 2016; Jha et al. 2015). Down-regulation of isocitrate dehydrogenase, which converts isocitrate to 2-oxoglutarate, leads to accumulation of citrate, required for fatty acid biosynthesis and itaconic acid formation. Citrate can be converted to acetyl-CoA by ATP-citrate lyase, which is used as a substrate for histone acetylation in cancer cells (Wellen et al. 2009). Thus, one consequence of the metabolic switch of classical macrophage activation could be generation of acetyl-CoA for histone acetylation and reduced levels of 2-oxoglutarate, needed for JHDMs and Tet proteins (Figure 1B). These possibilities have yet to be examined directly. The break in the TCA cycle induced by LPS and IFNγ is compensated for by a variant of the aspartate-arginosuccinate shunt. Notably, inhibition of aspartate-aminotransferase, a key enzyme of the shunt, inhibited nitric oxide and IL-6 production in M1 macrophages, indicating a role of shunt intermediates in establishing the classically activated phenotype (Jha et al. 2015). The underlying mechanisms have not been established.
Succinate represents another TCA cycle intermediate with regulatory functions. Succinate has been shown to activate HIF-1α and promote inflammatory gene expression (Kelly and O’Neill 2015). Succinate levels were recently shown to be modulated by itaconate, which is one of the most highly induced metabolites in classically activated macrophages (Figure 1B) (Lampropoulou et al. 2016). Itaconate modulates macrophage metabolism and effector functions by inhibiting succinate dehydrogenase-mediated oxidation of succinate. Bone marrow macrophages from mice lacking Irg1, required for production of Itaconate, exhibit marked reductions in succinate levels and reductions in levels of some inflammatory cytokines following treatment with LPS (Lampropoulou et al. 2016).
In contrast to classical activation of macrophages, treatment of macrophages with IL-4 in addition to TLR signaling results in a shift to oxidative metabolism (Vats et al. 2006; Jha et al. 2015). Overall, the program of macrophage alternative activation is coupled to changes in polyamine synthesis and fatty acid oxidation (Figure 1B). In addition, enhanced use of glucose for UDP-GlcNAc synthesis has been suggested as a metabolic signature of alternatively activated macrophages (Jha et al. 2015).
Epigenetic landscapes in T cell activation
Activation of T cells initiates dramatic changes in gene expression within hours and over the course of days the T cell response evolves with acquisition of heterogeneous effector fates distinguished by phenotype, effector capacity, and potential for establishing long-lived protective immunity. Many conditions influence the composition and outcome of T cell responses including the duration of antigen exposure, availability of costimulation, and cytokine milieu. Naive CD4+ T cells give rise to effector populations with helper and regulatory functions distinguished on the basis of cytokine production and expression of ‘lineage defining’ TF i.e. Th1 (IFNγ and T-bet), Th2 (IL-4 and GATA3), Th17 (IL-17 and RORγt), follicular helper T cells (Tfh)(IL-21 and Bcl-6), regulatory T cells (Treg)(IL-10 and FoxP3)(Bonelli et al. 2014). Notably, significant complexity within each Th population including overlapping TF expression and cytokine production as well as plasticity among fates are observed (Bonelli et al. 2014), particularly when studied ex vivo. In contrast, activation of naive CD8+ T cells leads to an effector population that broadly produces IFNγ and TNFα and possesses cytotoxic activity but with phenotypic complexity and TF dependence that can indicate potential for differentiation to long-lived memory cells, i.e. IL-7R is expressed by memory-precursor cells that depend on TCF-1, Eomes, Id3, and FOXO1, while KLRG1 is expressed by shorter-lived effector and effector-memory cells that depend on T-bet, Blimp-1, Id2, IRF4 and Zeb2 (Chang, Wherry, and Goldrath 2014). In contexts where antigen exposure is sustained beyond the initial expansion and effector phases, such as in chronic infections or tumors, responding T cells often exhibit a progressive loss of function or “exhaustion” that can modulate immunopathology as well as lead to a permissive dysfunctional state and escape of virus or tumors. Exhaustion of CD8+ T cells is regulated by expression of inhibitory receptors such as PD-1, the TFs T-bet and Eomes, and is accompanied by metabolic and epigenetic states distinct from both effector and memory T cell subsets (Wherry and Kurachi 2015). Recent studies make clear that LDTF provide only a portion of the information needed to establish stable regulatory networks that drive T cell effector function and memory cell fates; the epigenetic landscapes accompanying T cell activation and effector cell polarization show dynamic remodeling of DNA and histone modifications that mediate access of key TFs to their targets and provide the framework for the relative stability or flexibility between cell fates.
Comparisons of global DNA methylation among naive and effector CD8+ T cell populations in the response to acute viral infection reveal differentially methylated regions at promoters and enhancers consistent with the dynamic changes in gene expression following T cell activation, and emphasize that gains in methylation in effector cells compared to naive T cells are consistent with repression of the naive state in addition to demethylation of necessary effector loci to drive differentiation and effector function (Scharer et al. 2013). More recent studies using ATAC-seq to capture dynamic changes in chromatin accessibility as antigen-specific polyclonal or TCR transgenic CD8+ T cells differentiated in the response to acute or chronic LCMV infection allowed the identification of stable changes in chromatin accessibility by effector, memory, and exhausted populations (Scott-Browne et al. 2016; Scharer et al. 2017). More than 70,000 regions that were “open” in at least one differentiation state could be discerned and approximately half of those were stable in all cell types examined, indicating a core pattern of chromatin organization common to the CD8+ lineage regardless of activation state (Scott-Browne et al. 2016). Integration of chromatin accessibility with known TF binding motifs allow the prediction of TF that promote gene expression at each differentiation state (Scott-Browne et al. 2016; Scharer et al. 2017). Memory CD8+ T cells maintained an open configuration at many regulatory sites providing evidence for “memory primed” genes that can be more rapidly induced compared to naive populations. The ATAC-seq analysis of CD8+ T cells in chronic LCMV infection of mouse or HIV-specific cells in humans revealed a unique chromatin state associated with exhaustion, where many accessible sites were shared with effector populations in acute infection, but additional regions were uniquely lost or gained which could mediate expression of genes that drive exhaustion such as PD-1 (Scott-Browne et al. 2016; Pauken et al. 2016; Sen et al. 2016). Notably, blockade of PD-1 initiated the re-engagement of genes associated with effector function but could not reverse the exhausted configuration of chromatin, further emphasizing the central role that epigenetic landscapes play in differentiation and functional state of T cells during infection.
Genome-wide analysis of H3K4me3 and H3K27me3 histone methylation for polyclonal naive and endogenous memory subpopulations at the steady state and antigen-specific naive, effector, and memory CD8+ T cells responding to influenza A virus revealed dynamic changes in promoter methylation correlating with gene expression (with H3K4me3 associated with expressed genes and H3K27me3 associated with reduced expression) (Araki, Wang, et al. 2009; Russ et al. 2014). Notably, naive T cells showed co-deposition of H3K4me3 and H3K27me3 at promoter regions of genes important for cellular differentiation whereas genes associated with immune effector function lacked the permissive H3K4me3 modification, suggesting epigenetic mechanisms underlying CD8+ T cell differentiation and acquisition of effector function. Interestingly, a subset of effector-associated genes are rapidly, and in some cases, transiently induced after T cell activation (Best et al. 2013; Kakaradov et al. 2017), and may be marked by H3K4me2 modification in naive T cells (Russ et al. 2014). Treatment of effector or memory T cells subsets with histone acetylase or deacetylase inhibitors modulates expression of effector-associated molecules (Araki, Wang, et al. 2009) and affects memory function and potential (Northrop, Wells, and Shen 2008), further emphasizing a role for epigenetic changes in regulating gene expression at different stages of the T cell response (Figure 1A). Recent studies of antigen-specific CD8+ T cells expand these analyses, including genome-wide assessment of histone modification of H3K4me1 and H3K27ac marks (He et al. 2016) as well as ATAC-seq (Yu et al. 2017), to show extensive remodeling of enhancers as the effector and memory populations differentiate and, along with improved computational approaches, predict TF binding activity at each differentiation state with greater resolution. Of more than 50,000 enhancers identified, approximately 50% were unchanged over the course of the immune response. However, of enhancers that showed dynamic regulation, as many or more were lost as gained at each stage, in agreement with the idea that repression of alternative fates is key to differentiation (Yu et al. 2017). Furthermore, single-cell gene expression analysis at the first division following infection reveals an early burst of transcriptional activity by daughter cells destined for terminal differentiation compared to memory-precursor cells at the same stage (Kakaradov et al. 2017). This burst was followed by epigenetic silencing of genes associated with long-lived memory formation with deposition of H3K27me3 and a requirement of Ezh2 (Kakaradov et al. 2017; Gray et al. 2017). These recent studies further emphasize a role for epigenetic regulation at each stage of the CD8+ T cell response to infection and show that accessibility of specific regulatory regions by differentiation intermediates provide the basis for TF that are ubiquitously expressed to drive one cell fate over another.
Similar studies providing a comparative epigenetic landscape for CD4+ Th subsets, generated by in vitro polarization and directly ex vivo from a range of disease states, reveal dynamic changes in DNA and histone modifications as Th cells differentiate and have recently been reviewed (Tripathi and Lahesmaa 2014; Bonelli et al. 2014; Vahedi et al. 2013). Notably, the presence of H3K4me3 and H3K27me3 modifications which indicate a potential for both activation and repression at key regulatory sites of LDTF in differentiated Th cells provides insight into the substantial plasticity observed between distinct Th subsets (Figure 1A). Observations that many of the Th subset-distinct enhancer landscapes established in response to external stimuli such as cytokines are regulated by STAT family TFs and not LDTFs emphasize that differentiation is regulated not only by expression of key TF but also by accessibility of regulatory elements for which the landscape may be remodeled in response to triggering of environmental sensors (Bonelli et al. 2014; Tripathi and Lahesmaa 2014; Vahedi et al. 2013).
Metabolic reprogramming in T cells
A number of reviews have detailed the changes in T cell metabolism that occur following activation (MacIver, Michalek, and Rathmell 2013; Pollizzi and Powell 2014) and here we will focus on highlighting studies that suggest metabolic links between T cell receptor stimulation and “writers” and “erasers” of the epigenetic code discussed above. Activation of T cells initiates a molecular program that rapidly alters gene expression and metabolic activity in T cells. Numerous signaling pathways induce increases in metabolic capacity and the engagement of biosynthetic pathways following TCR engagement and costimulation (reviewed extensively (MacIver, Michalek, and Rathmell 2013; Pollizzi and Powell 2014)), promoting the activity of TFs such as, c-MYC, mTOR, PI3K and AKT, Hypoxia inducible factor 1 alpha (HIF1α), and AMP-activated protein kinase (AMPK).
Initial reports suggested that both CD4+ and CD8+ T cells engaged the same metabolic program upon activation emphasizing the use of glycolytic metabolism to fuel energy production and preserve non-glucose carbon sources for biosynthesis (Wang et al. 2011; Bental and Deutsch 1993). However, recent studies have revealed complexity in both the usage of TFs as well as differential reliance on glycolysis versus mitochondrial respiration following activation reflective of the differences between CD4+ Th cell subsets and CD8+ T cells (Wang et al. 2011; Cao et al. 2014). For example, studies of mTORC1 versus mTORC2 usage revealed distinct roles for each pathway in Th subsets and CD8+ T cell differentiation. Specifically, loss of mTORC1 or mTORC2 signaling in CD4+ T cells resulted in failure of CD4+ T cells to differentiate into Th1 and Th17 or Th2 cells, respectively (Figure 1C) (Delgoffe et al. 2011). Notably, mTORC1 and mTORC2 signaling in CD4+ T cells contributed to Tfh differentiation, while conversely loss of mTORC signaling in CD4+ T cells promoted Foxp3 expression, but reduced suppressive ability (Delgoffe et al. 2011; Zeng et al. 2016; Zeng et al. 2013). In contrast for CD8+ T cells, loss of mTORC1 significantly perturbed the generation of effector responses; while mTORC2 loss did not impact effector responses but appeared to promote memory CD8+ T cell generation (Pollizzi et al. 2015). In each of these scenarios, mTORC1 and mTORC2 signaling is thought to promote glycolytic metabolism, but critically, it remains to be determined whether the alterations in differentiation following their loss is primarily a function of changes in energy production or biogenesis due to reduced glycolytic metabolism or loss of regulation of non-metabolic target genes. These questions (and many others discussed further in PEARCE AND JONES) are areas of active investigation and a number of studies that propose some direct effects on differentiation and gene expression are discussed below.
Recent studies examining the energetic profile of CD4+ and CD8+ T cells following activation supported glycolytic metabolism as a crucial contributor to fueling effector responses and proliferation (Peng et al. 2016; Doedens et al. 2013; Chang et al. 2013). It is also clear that mitochondrial metabolism contributes to effector CD8+ T cell metabolism beyond the established paradigm of supporting memory T cells (Figure 1C) (Cao et al. 2014; Scharping et al. 2016; Bengsch et al. 2016). Following in vitro activation, CD4+ and CD8+ T cell proliferation was inhibited by pharmacological blockade of either glycolysis, via 2-deoxyglucose treatment, or mitochondrial respiration, via oligomycin treatment (Cao et al. 2014; Phan et al. 2016). Examination of exhausted antigen-specific CD8+ T cells responding in chronic viral infection or tumors treated with anti-PD-1 suggested that in some inflammatory contexts mitochondrial function is critical for reinvigorating T cell responses (Bengsch et al. 2016; Scharping et al. 2016).
Given the substantial increase in the use of glycolytic metabolism by T cells following activation, most studies have focused on elucidating the impact of energy producing pathways on T cell biology (Blagih et al. 2015; O’Sullivan et al. 2014; Chang et al. 2013; van der Windt et al. 2013; van der Windt et al. 2012; Phan et al. 2016; Okoye et al. 2015; Wang et al. 2011; Pearce et al. 2009; Sukumar et al. 2013). This focus has yielded a number of critical insights, including how energy producing pathways support production of cytokines by effector CD8+ T cells and CD4+ Th subsets (Chang et al. 2013; Peng et al. 2016; Ho et al. 2015). Specifically, the regulation of IFNγ production by glycolytic metabolism has revealed two potential mechanisms of metabolism-dependent cytokine production (Chang et al. 2013; Peng et al. 2016). One centers on sustained glycolytic throughput occupying glyceraldehyde 3-phosphate dehydrogenase (GAPDH) thereby preventing inhibition of IFNγ mRNA translation, while the other links glycolytic metabolism to deposition of epigenetic marks that drive IFNγ expression (Chang et al. 2013; Peng et al. 2016). While the exact mechanism by which glycolytic metabolism facilitates production of IFNγ remains controversial, the correlative changes in T cell metabolism following activation clearly have functional ramifications and support survival of long-lived memory T cells as well as set the stage for rapid secondary responses by memory T cells (Chang, Wherry, and Goldrath 2014; Pollizzi and Powell 2014). Importantly, this encompasses biosynthetic pathways as well, such as serine-fueled one carbon metabolism, essential for fueling efficient proliferation (Ma et al. 2017).
Impact of metabolic switching on T cell function and differentiation
A strategy by which T cells integrate the substantial array of in situ signals that initiate, sustain, and mediate effector responses may be reflected in the marked shifts in energy production and biosynthetic pathways following activation (Pollizzi and Powell 2014; Chang, Wherry, and Goldrath 2014). At present, the fundamental question of whether metabolic reprogramming or switching during the course of T cell responses drives fate decisions remains unanswered primarily due to the difficulty in distinguishing defects in survival versus alterations in differentiation. However, there is strong data for both CD4+ and CD8+ T cells demonstrating usage of glycolysis, mitochondrial respiration, biosynthetic pathways, and the factors that regulate them facilitate distinct aspects of effector functionality (Bengsch et al. 2016; Chang et al. 2013; Doedens et al. 2013; Scharping et al. 2016; Peng et al. 2016; Ma et al. 2017; Ho et al. 2015; Shi et al. 2011; Zeng et al. 2013; Gerriets et al. 2015). Moreover, recent studies support the hypothesis that microenvironmental nutrient availability (discussed below) can directly impact T cell metabolism and result in modulation of gene-expression, function, and T cell fate (Ho et al. 2015; Ananieva et al. 2014).
Studies examining the level of metabolic heterogeneity in CD4+ T cells and Th subsets show that Th1, Th2, Th17, and Tfh cell subsets exhibit increased expression of Glut1 and glycolytic rate in comparison to Treg cells following activation (Figure 1C) (Zeng et al. 2016; Shi et al. 2011; Michalek et al. 2011). In particular, HIF1α-driven glycolytic metabolism can support differentiation of Th17 cells, and pharmacologic inhibition of glycolytic metabolism in activated CD4+ T cells skewed them towards iTreg differentiation and reduced disease severity in a mouse model of multiple sclerosis (Shi et al. 2011). In line with the initial observations that Th1, Th2, and Th17 cells relied on glycolytic metabolism, abrogated mTOR signaling in CD4+ T cells increased Foxp3 expression (Zeng et al. 2013). As mentioned above, a nuance to the increased generation of Foxp3+ cells is the reduced capacity of those cells to suppress. A number of hypotheses could explain this loss of function, including a requirement for mTOR in effector Treg differentiation, but perhaps the more intriguing question is whether glycolytic metabolism serves to facilitate cytokine production by suppressive Tregs. The hypothesis that glycolysis also maintains suppressive cytokine production in Tregs is supported by the observation that Type 1 regulatory (Tr1) cells, an IL-10 producing CD4+ T cell subset that does not express Foxp3, exhibited a metabolite profile associated with glycolytic metabolism similar to Th17 cells (Mascanfroni et al. 2015).
In contrast to the effect metabolic activity has on effector function and proliferation of T cells, the causative impacts on differentiation of CD4+ and CD8+ T cell subsets remains to unclear. Two key studies renewed interest in T cell metabolism as a modulator of cell-fate decisions (Araki, Turner, et al. 2009; Pearce et al. 2009). Inhibition of mTOR by treatment with rapamycin or activation of AMPK by treatment with metformin potentiated the formation of memory CD8+ T cells following infection by inhibiting glycolytic metabolism and supporting fatty acid oxidation (FAO) (Araki, Turner, et al. 2009; Pearce et al. 2009). These studies suggested that a critical step in the specification of the memory T cell fate is a transition away from ‘effector’ metabolism, (reliance on glycolysis) to ‘memory’ metabolism (reliance on fatty acid oxidation and mitochondrial respiration). Memory CD8+ T cells exhibited greater oxygen consumption, a surrogate for mitochondria-driven energy production, than that of naive and effector CD8+ T cells and most importantly, maintained a greater maximal capacity for oxygen consumption dubbed mitochondrial spare respiratory capacity (SRC) (van der Windt et al. 2012). Furthermore, this increased SRC was shown to be a product of increased mitochondrial mass and fueled by T cell-intrinsic lipolysis that supplied fatty acids for use within the mitochondria (O’Sullivan et al. 2014; van der Windt et al. 2012).
These studies established a paradigm in which T cells require the transition from glycolytic metabolism to mitochondria-dependent oxidative phosphorylation (OxPhos) for the generation of memory and it has been argued the shift in T cell metabolism itself drives the formation of memory CD8+ T cells (O’Sullivan et al. 2014; van der Windt et al. 2012). Deletion or inhibition of molecules critical for various metabolic processes provide complementary evidence showing metabolic transitions and differential usage of glycolysis and OxPhos occur during T cell responses and can impact the generation of effector and memory cell function and survival (Pollizzi et al. 2015; Chaoul et al. 2015; Okoye et al. 2015; Rolf et al. 2013; Sena et al. 2013; Rao et al. 2010; Shrestha et al. 2014; Balmer et al. 2016). For example, inhibition of glycolysis with 2-deoxyglucose during activation can promote a memory-like gene-expression program and longer-term survival of CD8+ T cells responsive to infection or tumors supporting the necessity of a metabolic transition in the specification of a memory cell fate (Sukumar et al. 2013). Additionally, IL-7 driven expression of glycerol channel aquaporin 9 (AQP9) was essential for the production of triacylglcerides, necessary for FAO-driven ATP production and survival of memory T cells (Cui et al. 2015). Similarly, increased intracellular levels of L-Arginine improved survival of human and mouse T cells (Geiger et al. 2016). While these studies certainly demonstrate that T cell metabolism supports the differentiation of memory T cells and that the shift between reliance on glycolytic metabolism and FAO-driven mitochondrial respiration correlate with the memory T cell fate, it remains to be determined whether metabolic pathways function as a driving force behind cell-fate decisions such as memory differentiation. More specifically, parsing out the difference between inducing differentiation and enhancing the likelihood of survival by improving energy production will need to be considered in future studies.
The importance of discriminating improvements in survival versus driving memory differentiation is shown in studies of von Hippel Lindau tumor suppressor (VHL)-deficient CD8+ T cells which sustain high levels of glycolytic metabolism while actively suppressing mitochondrial respiration due to constitutive stabilization of HIFα subunits (Phan et al. 2016). Following acute viral infection, VHL-deficient CD8+ T cells form protective long-lived memory T cell populations despite sustained reliance on glycolytic metabolism, reduced SRC, and downregulated mitochondrial respiration deemed essential for memory differentiation by previous studies (Phan et al. 2016). Intriguingly, the sustained usage of glycolytic metabolism by VHL-deficient CD8+ T cells did not result in a bioenergetic disadvantage as cellular ATP levels during the contraction or memory phases of the response to infection were similar to wildtype CD8+ T cells. These data strongly argue that the memory CD8+ T cell fate in general is independent of the usage of either glycolytic or mitochondrial metabolism and that the specification of memory relies on the provision of adequate cellular energy for survival allowing other mechanisms (i.e. LDTFs) to delineate T cell fate (Phan et al. 2016). Furthermore, examination of metabolic activity of memory CD8+ T cell subsets, central memory (Tcm) and effector memory (Tem) T cells, revealed a previously unappreciated level of metabolic heterogeneity within the memory CD8+ T cell pool that may reflect the distinct functional and migratory characteristics of each subset (Figure 1C) (Phan et al. 2016). Whether this metabolic heterogeneity drives the differentiation of Tcm or Tem cells is unknown, however, a recent study of tissue-resident memory CD8+ T (Trm) cell metabolism showed that skin Trm utilize distinct strategies relative to circulating memory cells for acquiring fatty acids for energy production and survival, potentially reflective of their localization, which supports the premise that T cells exhibit substantial metabolic flexibility (Figure 1C) (Pan et al. 2017).
Similarly, metabolic heterogeneity has been observed in Th subsets which express LDTFs, as discussed above, while differential metabolic pathway usage affects functionality it remains to be determined whether the metabolic heterogeneity observed contributes mechanistically to Th subset differentiation (Michalek et al. 2011; Shi et al. 2011; Mascanfroni et al. 2015; Zeng et al. 2016). These studies raise a number of questions about when, where, and how fluctuations in T cell metabolism impact T cell function and support a model in which cellular metabolism serves as a context-specific integrator of extracellular cues such as cytokines and nutrients (discussed more thoroughly in this issue by Vander Heiden, Olenchock and Rathmell).
Impacts of metabolism on gene expression and links to epigenetics
One of the difficulties in studying the impact of metabolism on T cell function and differentiation lies in the limited ability to manipulate metabolic pathways in vivo where the full complement of signals governing the immune response are present. Many studies have relied on in vitro models that at times translate well to T cell responses in situ, but some conclusions drawn directly about T cell metabolism require further examination in physiological contexts as the availability of nutrients, cytokines, and cellular interactions are critical and support a model where shifts in cellular metabolism are a likely means for integration of multiple exogenous and endogenous signals downstream of pathogen recognition. In particular, two recent studies demonstrate in CD4+ T cells that levels of phosphoenolpyruvate (PEP) and leucine feedback into well-studied signaling pathways altering T cell gene expression (Ho et al. 2015; Ananieva et al. 2014). PEP, a glycolytic intermediate, inhibited SERCA-mediated calcium uptake and thus supported a model where PEP accumulation is critical for sustaining Ca2+-NFAT signaling linking the upregulation of glycolytic metabolism in T cells to maintenance of TCR signaling (Ho et al. 2015). Functionally, this is particularly relevant in the context of tumors where glucose can be limiting due to competition for uptake by tumor cells as overexpression of phosphoenolypyruvate carboxykinase 1, which converts oxaloacetate into PEP, improved Ca2+-NFAT signaling in tumor infiltrating CD4+ T cells, bolstering their tumoricidal ability. Similar effects were also seen for CD8+ T cells activated in vitro and cultured in glucose-poor conditions (Ho et al. 2015). Cytosolic branched chain aminotransferase (BCATc) is induced upon T cell activation and catalyzes the transamination of cytosolic leucine (Ananieva et al. 2014). Loss of BCATc in CD4+ T cells resulted in an accumulation of cytosolic leucine, a known activator of mTORC1, following activation and promoted increased mTORC1 activation, measured by phosphorylation of S6 and 4EBP-1, further increasing glycolytic metabolism and demonstrating another metabolic layer of regulation of TCR-driven signaling (Ananieva et al. 2014). These data highlight the direct impact alterations in metabolite levels can have on gene expression in T cells and support the possible regulation of T cell function via epigenetic enzymes.
Two recent studies directly raised the possibility that metabolites may modulate the epigenome of T cells following activation. As discussed above, glycolytic metabolism supports effector function and, in the case of IFNγ production, one proposed mechanism is that engagement of glycolytic metabolism by T cells can facilitate the production of IFNγ through maintenance of high acetyl-CoA levels that facilitated deposition of H3K9Ac at the IFNγ promoter driving IFNγ expression (Figure 2) (Peng et al. 2016). It remains unclear what other loci exhibit increased acetylation following LDHA deletion and whether this can be modulated on a gene-specific level remains unexplored. The second study that shows direct modulation of the epigenome in T cells centers around the oncometabolite 2-hydroxyglutarate (Tyrakis et al. 2016). In vitro activated CD8+ T cells treated with S-enantiomer of 2-hydroxyglutarate (S-2-HG) acquired a Tcm-like phenotype that was sustained following transfer into wildtype host mice. Furthermore, S-2-HG treated CD8+ T cells exhibited enhanced proliferation upon stimulation in vivo, persistence, and anti-tumor capacity following adoptive transfer. These changes coincided with alterations in global DNA and histone methylation which may be a direct effect of S-2-HG inhibition of Tet2 and Utx3 (Figure 2) (Tyrakis et al. 2016). Critically, while it was observed that T cells do produce S-2-HG at physiologically significant levels following in vitro activation and culture, it remains to be determined whether S-2-HG is produced in situ at levels necessary to have similar impacts (Tyrakis et al. 2016). Furthermore, whether global DNA and histone methylation changes occur and at what level of specificity in physiological conditions remain to be defined. Clearly the wealth of data regarding the impact of metabolic pathways on T cells supports modulation of enzymes that modify epigenetic landscapes, however the small number of studies directly examining this connection highlight that further exploration of molecular mechanisms is needed.
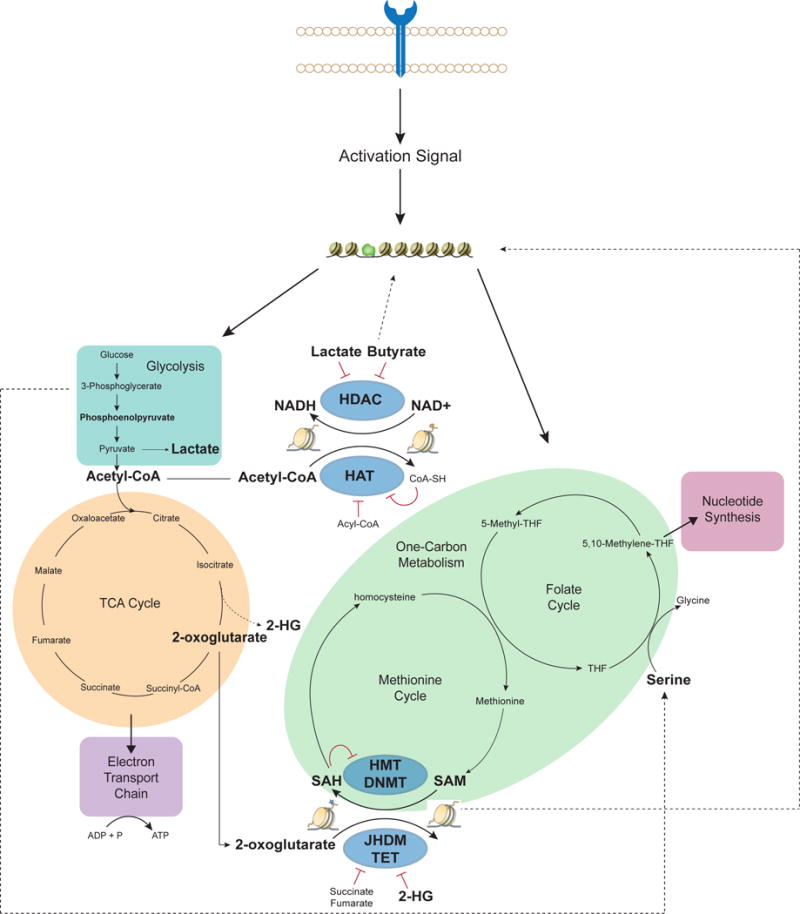
Recognition of pathogen by immune cells initiates a genetic program that promotes use of numerous metabolic pathways for biogenesis (i.e. nucleotide synthesis) and energy production (i.e. glycolysis, TCA cycle). Metabolites involved in glycolysis (teal), TCA cycle (orange), and one-carbon metabolism (light-green) can play critical roles as substrates such as acetyl-CoA and SAM, for histone acetylation and methylation respectively, or cofactors such as 2-oxoglutarate and NAD+, for demethylation of DNA and histone deacetylation respectively. Epigenetic enzymes (blue) catalyze the addition (HAT, HMT or DNMT) and removal (HDAC, JHDM, or TET) of epigenetic marks and can be impacted by multiple metabolic pathways. Metabolites shown to be controlled by pathways relevant in T cells or macrophages (bold) are critical for energy production (i.e. lactate, acetyl-CoA) and proliferation (i.e. serine). Flexibility in metabolic pathway usage is reflected by the small network depicted here; for example, 3-phosphoglcerate can be converted to serine for use in one-carbon metabolism. Clear links between activity of epigenome modifying enzymes and metabolites relevant in T cells and macrophages that will impact the epigenetic landscape of activated cells can be drawn.
Influence of tissue environment on gene expression and epigenetic landscapes
These studies raise a number of exciting questions about how metabolites regulate macrophage and T cell biology and whether physiological conditions such as diverse tissue microenvironments, inflammation, or diet may significantly modulate their function and differentiation. Nearly all studies connecting signaldependent gene expression, metabolism and epigenetic consequences have been performed in vitro. However, in vivo, these interactions occur in the context of specific tissue environments, in which infection and disease can radically alter both the populations of immune cells within tissues as well as gene-expression patterns with a given cell type.
In the case of macrophages, the steady state populations are derived from primitive erythro-myeloid progenitor cells during fetal development and/or monocyte-derived macrophages that originate from hematopoietic stem cells (Ginhoux et al. 2016; Schulz et al. 2012). In addition to playing general roles in innate immunity, each tissue-resident macrophage is specialized to support tissue-specific homeostatic functions (Wynn, Chawla, and Pollard 2013). For example, microglia, the main resident macrophage population of the central nervous system, secrete factors that modulate neurogenesis and participate in the refinement and elimination of synapses, while resident peritoneal macrophages play roles in gut immunity. Consistent with this functional specialization, different tissue resident macrophages express tissue-specific genes in addition to genes that enable core macrophage functions, such as phagocytosis and pathogen surveillance (Gautier et al. 2012). Comparing microglia to peritoneal macrophages, for example, nearly 2000 mRNAs are more than 16-fold differentially expressed that are enriched for functional annotations related to brain homeostasis and gut immunity, respectively (Gosselin et al. 2014). These differences in gene expression are associated with corresponding differences in enhancer landscapes (Gosselin et al. 2014; Lavin et al. 2014). While tissue macrophages normally serve adaptive functions, dysregulation of macrophage activities contributes to a diverse range of human diseases, including cardiovascular, metabolic, neurodegenerative and neoplastic diseases. Therefore, dissecting the full complement of signals that modulate and maintain tissue-resident macrophage gene-expression may yield insight into novel stimuli that drive human diseases.
Similar to tissue-resident macrophages, recent studies have identified Trm cells that reside in peripheral tissues without recirculation that are critical contributors to host defense upon reinfection (Jiang et al. 2012; Schenkel et al. 2013; Schenkel et al. 2014; Iijima and Iwasaki 2014; Ariotti et al. 2014). Both CD8+ and CD4+ Trm cells have been identified and exhibit important sentinel functions that drive recruitment of circulating adaptive immune cells as well as unique niche-driven roles in the case of CD4+ Trm cells in the female reproductive tract (FRT) (Schenkel et al. 2014; Iijima and Iwasaki 2014; Ariotti et al. 2014). Due to the recent identification of Trm, only a small number of Trm populations have been profiled for gene-expression from different tissues. However similar to tissue-resident macrophages, these studies show tissue-specific gene expression in addition to the core Trm gene signature necessary for long-term survival and function in non-lymphoid tissues (Mackay et al. 2013). In the case of T cells, it remains to be seen whether there are essential tissue-specific gene programs beyond the core Trm signature currently identified. Further studies characterizing populations from other tissues where Trm have been identified coupled with analysis of enhancer landscapes and chromatin dynamics will provide clarity as to whether there are additional tissue-specific functions of Trm similar to the CD4+ Trm in the FRT and hint at tissue-specific signals that may regulate these programs. Furthermore, clarifying our understanding of Trm biology may contribute to our understanding of a diverse subset of human diseases in which memory T cells are thought to participate.
Tissue environment has emerged as an important determinant of tissue-specific macrophage gene expression programs. Primitive macrophages acquire tissue-specific gene signatures soon after entering developing organs in the mouse embryo (Matcovitch-Natan et al. 2016; Mass et al. 2016). Plasticity in the tissue-specific program of adult macrophage gene expression was demonstrated by transfer of peritoneal macrophages to the alveolar air spaces of the lung, which resulted in substantial reprogramming of gene expression to an alveolar macrophage-like pattern (Lavin et al. 2014). Conversely, transfer of microglia or peritoneal macrophages to an in vitro environment resulted in loss of expression of a large fraction of the genes that are specific for each cell type (Gosselin et al. 2014). Collectively, these observations imply that each tissue environment provides signals that instruct entering macrophages to acquire tissue-specific programs of gene expression (Figure 3). However, the identities of the signaling molecules that play these instructive roles are for the most part unknown.

Activated immune cells initiate rapid metabolic and epigenetic changes that are responsive to additional stimuli encountered in peripheral tissues. Migration to peripheral tissues submit immune cells to additional challenges such as changes in nutrient availability, tissue-specific signals, as well as alter the metabolic pathways that are utilized by the cell in situ. Cells that enter the gut microenvironment must contend with microbiome derived metabolites, such as butyrate, that has been shown to inhibit HDACs and modulate macrophage and T cell differentiation and function. Similarly, immune cells that infiltrate tumors encounter a challenging environment with increased competition for glucose, higher concentrations of lactate, and reduced oxygen levels. Reduced glucose availability could impair glycolytic metabolism reduce acetyl-CoA levels and inhibit the acetylation of histones necessary for gene expression. Increased lactate concentrations may inhibit HDAC activity further dysregulating the epigenetic landscapes of tumor-infiltrating immune cells. These examples highlight the possible effects of extrinsic signals on epigenetic enzymes and support a role for metabolites as integrators of cell-intrinsic and –extrinsic signals for the modulation of gene expression.
In the case of T cells, it remains to be determined whether tissue environment directly defines the residency characteristics of each Trm population and whether any level of plasticity exists in Trm populations. However, a number of studies provide a hint that local tissue environment provides essential cues that regulate T cell responses. For example, T cell-intrinsic expression of prolyl hydroxylases (PHD), oxygen-sensing negative regulators of HIF TFs, limited the induction of Th1 responses, promoted Treg induction, and suppressed CD8+ effector function specifically in the lung suggesting that oxygen levels regulate T cell responses (Clever et al. 2016). Furthermore, PHD deletion in T cells abrogated the number of lung tumors in mice, but did not alter the size of the subcutaneous tumor in a B16 model of secondary tumor colonization indicating that oxygen-sensing specifically restrained pulmonary T cell responses (Clever et al. 2016). In line with these observations, the gut microenvironment, which contains diet-derived antigens, commensal microbes and their metabolic byproducts, and local inflammatory signals, induces Treg differentiation through several mechanisms and modulates immune responses (Mucida et al. 2005; Sun et al. 2007; Arpaia et al. 2013; Atarashi et al. 2011). As the field moves to interrogate signals critical for T cell responses to understand Trm ontogeny and function, it is likely that microenvironmental cues may regulate or play a role in T cell metabolism and epigenetics (Figure 3).
A reasonable assumption is that many of the factors that regulate tissue-specific macrophage phenotypes are classical signaling molecules that bind to specific receptors that in turn regulate gene expression. For example, local retinoic acid has been suggested to be a key regulator of peritoneal macrophage phenotypes by inducing expression of GATA6 in a retinoic acid-dependent manner (Okabe and Medzhitov 2014). Conversely, TGFβ has been established to play important roles in maintaining Trm and microglia-specific phenotypes by regulating the activities of SMAD proteins (Butovsky et al. 2014; Chang, Wherry, and Goldrath 2014). However, several lines of evidence suggest that small molecules involved in various aspects of metabolism could also play important regulatory roles. For example, lactic acid produced by tumor cells, as a by-product of aerobic or anaerobic glycolysis, dramatically alters gene expression and functional polarization of tumor-associated macrophages as well as infiltrating T cells (Figure 3) (Colegio et al. 2014; Brand et al. 2016). Remarkably, recent studies indicate that β-hydroxyl butyrate, a short chain organic acid that accumulates during starvation and provides an alternative source of energy to glucose, covalently modifies histone lysine residues in the liver and serves as an activation mark analogous to histone acetylation (Xie et al. 2016). Also, in CD4+ T cells, microbiome-derived butyrate has been shown to modulate Foxp3 stability and histone acetylation at Foxp3 regulatory elements in vitro (Figure 3) (Arvey et al. 2014). By extension, it is conceivable that other biochemical intermediates present in the diet and/or produced by the microbiome could have analogous effects on epigenetic landscapes of various cell types, including immune cells.
Concluding remarks
The study of immunometabolism and epigenetic landscapes of macrophages and T cells has produced significant advances in recent years. As the independent roles of metabolic pathways and epigenetics expand in macrophage and T cell biology, a link between metabolites and the epigenetic modifying enzymes that require them, previously explored in cancer cells and other cell types, is beginning to emerge. Intriguing reports suggest a significant role for intermediate metabolites and the pathways that generate them in modulating the epigenomes of activated macrophages and T cells. Many questions remain to be explored. Studies to date have focused analyses on particular loci of interest, but have yet to define the broad effects metabolite levels have on global epigenetic landscapes. Further, the extent to which global changes in metabolic intermediates impact the epigenome in a gene-specific manner remains undetermined. It is likely that modulation of global metabolite levels drive broad effects that require additional mechanisms to target epigenetic modifications to specific loci. Moreover, defining the compartmentalization of particular metabolites in relation to epigenetic “writers” and “erasers” may clarify the impact of macrophage and T cell metabolism on epigenetic landscapes following activation. In sum, there is an enticing link between cell-intrinsic and –extrinsic metabolites and gene-expression with sparse experimental evidence of molecular mechanisms resulting in functional consequences in immune cells that warrant further exploration.
Acknowledgments
Funding of this work provided by the National Institutes of Health (AI072117, AI067545) for A.W.G. and DK091183, NS096170) for C.K.G. C.K.G. was also supported by a Leducq Transatlantic Network Grant and the Ben and Wanda Hildyard Chair in Hereditary Diseases. We thank Nathan Spann and Kyla Omilusik for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. [Abstract] [Google Scholar]
- Amit Ido, Winter Deborah R, Jung Steffen. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. 2016;17:18–25. [Abstract] [Google Scholar]
- Ananieva Elitsa A, Patel Chirag H, Drake Charles H, Powell Jonathan D, Hutson Susan M. Cytosolic branched chain aminotransferase (BCATc) regulates mTORC1 signaling and glycolytic metabolism in CD4+ T cells. The Journal of biological chemistry. 2014;289:18793–804. [Europe PMC free article] [Abstract] [Google Scholar]
- Araki Koichi, Turner Alexandra P, Shaffer Virginia Oliva, Gangappa Shivaprakash, Keller Susanne A, Bachmann Martin F, Larsen Christian P, Ahmed Rafi. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–12. [Europe PMC free article] [Abstract] [Google Scholar]
- Araki Yasuto, Wang Zhibin, Zang Chongzhi, Wood William H, Schones Dustin, Cui Kairong, Roh Tae-Young, Lhotsky Brad, Wersto Robert P, Peng Weiqun, Becker Kevin G, Zhao Keji, Weng Nan-ping. Genome-wide Analysis of Histone Methylation Reveals Chromatin State-Based Regulation of Gene Transcription and Function of Memory CD8+ T Cells. Immunity. 2009;30:912–25. [Europe PMC free article] [Abstract] [Google Scholar]
- Ariotti Silvia, Hogenbirk Marc A, Dijkgraaf Feline E, Visser Lindy L, Hoekstra Mirjam E, Song Ji-Ying, Jacobs Heinz, Haanen John B, Schumacher Ton N. Skin-resident memory CD8<sup>+</sup> T cells trigger a state of tissue-wide pathogen alert. Science. 2014;346:101. [Abstract] [Google Scholar]
- Arpaia Nicholas, Campbell Clarissa, Fan Xiying, Dikiy Stanislav, van der Veeken Joris, deRoos Paul, Liu Hui, Cross Justin R, Pfeffer Klaus, Coffer Paul J, Rudensky Alexander Y. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–55. [Europe PMC free article] [Abstract] [Google Scholar]
- Arts RJ, Joosten LA, Netea MG. Immunometabolic circuits in trained immunity. Semin Immunol. 2016;28:425–30. [Abstract] [Google Scholar]
- Arvey Aaron, van der Veeken Joris, Samstein Robert M, Feng Yongqiang, Stamatoyannopoulos John A, Rudensky Alexander Y. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nature Immunology. 2014;15:580–87. [Europe PMC free article] [Abstract] [Google Scholar]
- Atarashi Koji, Tanoue Takeshi, Shima Tatsuichiro, Imaoka Akemi, Kuwahara Tomomi, Momose Yoshika, Cheng Genhong, Yamasaki Sho, Saito Takashi, Ohba Yusuke, Taniguchi Tadatsugu, Takeda Kiyoshi, Hori Shohei, Ivanov Ivaylo I, Umesaki Yoshinori, Itoh Kikuji, Honda Kenya. Induction of Colonic Regulatory T Cells by Indigenous <em>Clostridium</em> Species. Science. 2011;331:337. [Europe PMC free article] [Abstract] [Google Scholar]
- Baardman J, Licht I, de Winther MP, Van den Bossche J. Metabolic-epigenetic crosstalk in macrophage activation. Epigenomics. 2015;7:1155–64. [Abstract] [Google Scholar]
- Bai Péter, Cantó Carles, Oudart Hugues, Brunyánszki Attila, Cen Yana, Thomas Charles, Yamamoto Hiroyasu, Huber Aline, Kiss Borbála, Houtkooper Riekelt H, Schoonjans Kristina, Schreiber Valérie, Sauve Anthony A, Murcia Josiane Menissier-de, Auwerx Johan. PARP-1 Inhibition Increases Mitochondrial Metabolism through SIRT1 Activation. Cell Metabolism. 2011;13:461–68. [Europe PMC free article] [Abstract] [Google Scholar]
- Balmer Maria L, Ma Eric H, Bantug Glenn R, Grählert Jasmin, Pfister Simona, Glatter Timo, Jauch Annaïse, Dimeloe Sarah, Slack Emma, Dehio Philippe, Krzyzaniak Magdalena A, King Carolyn G, Burgener Anne-Valérie, Fischer Marco, Develioglu Leyla, Belle Réka, Recher Mike, Bonilla Weldy V, Macpherson Andrew J, Hapfelmeier Siegfried, Jones Russell G, Hess Christoph. Memory CD8+ T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity. 2016;44:1312–24. [Abstract] [Google Scholar]
- Bengsch Bertram, Johnson Andy L, Kurachi Makoto, Odorizzi Pamela M, Pauken Kristen E, Attanasio John, Stelekati Erietta, McLane Laura M, Paley Michael A, Delgoffe Greg M, Wherry E John. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8+ T Cell Exhaustion. Immunity. 2016;45:358–73. [Europe PMC free article] [Abstract] [Google Scholar]
- Bental Michal, Deutsch Carol. Metabolic changes in activated T cells: An NMR study of human peripheral blood lymphocytes. Magnetic Resonance in Medicine. 1993;29:317–26. [Abstract] [Google Scholar]
- Best J Adam, Blair David A, Knell Jamie, Yang Edward, Mayya Viveka, Doedens Andrew, Dustin Michael L, Goldrath Ananda W. Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–12. [Europe PMC free article] [Abstract] [Google Scholar]
- Blagih Julianna, Coulombe François, Vincent Emma EE, Dupuy Fanny, Galicia-Vázquez Gabriela, Yurchenko Ekaterina, Raissi Thomas CC, van der Windt Gerritje JW, Viollet Benoit, Pearce Erika LL, Pelletier Jerry, Piccirillo Ciriaco AA, Krawczyk Connie MM, Divangahi Maziar, Jones Russell GG, van der Windt Gerritje JW, Viollet Benoit, Pearce Erika LL, Pelletier Jerry, Piccirillo Ciriaco AA, Krawczyk Connie MM, Divangahi Maziar, Jones Russell GG. The Energy Sensor AMPK Regulates T Cell Metabolic Adaptation and Effector Responses In Vivo. Immunity. 2015;42:41–54. [Abstract] [Google Scholar]
- Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288:34287–94. [Europe PMC free article] [Abstract] [Google Scholar]
- Bonelli Michael, Shih Han-Yu, Hirahara Kiyoshi, Singelton Kentner, Laurence Arian, Poholek Amanda, Hand Tim, Mikami Yohei, Vahedi Golnaz, Kanno Yuka, O’Shea John J. Helper T Cell Plasticity: Impact of Extrinsic and Intrinsic Signals on Transcriptomes and Epigenomes. In: Ellmeier Wilfried, Taniuchi Ichiro., editors. Transcriptional Control of Lineage Differentiation in Immune Cells. Springer International Publishing; Cham.: 2014. [Europe PMC free article] [Abstract] [Google Scholar]
- Brand Almut, Singer Katrin, Koehl Gudrun E, Kolitzus Marlene, Schoenhammer Gabriele, Thiel Annette, Matos Carina, Bruss Christina, Klobuch Sebastian, Peter Katrin, Kastenberger Michael, Bogdan Christian, Schleicher Ulrike, Mackensen Andreas, Ullrich Evelyn, Fichtner-Feigl Stefan, Kesselring Rebecca, Mack Matthias, Ritter Uwe, Schmid Maximilian, Blank Christian, Dettmer Katja, Oefner Peter J, Hoffmann Petra, Walenta Stefan, Geissler Edward K, Pouyssegur Jacques, Villunger Andreas, Steven André, Seliger Barbara, Schreml Stephan, Haferkamp Sebastian, Kohl Elisabeth, Karrer Sigrid, Berneburg Mark, Herr Wolfgang, Mueller-Klieser Wolfgang, Renner Kathrin, Kreutz Marina. LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metabolism. 2016;24:657–71. [Abstract] [Google Scholar]
- Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–40. [Europe PMC free article] [Abstract] [Google Scholar]
- Buck Michael D, O’Sullivan David, Pearce Erika L. T cell metabolism drives immunity. The Journal of Experimental Medicine. 2015;212:1345. [Europe PMC free article] [Abstract] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–43. [Europe PMC free article] [Abstract] [Google Scholar]
- Cantó Carles, Gerhart-Hines Zachary, Feige Jerome N, Lagouge Marie, Noriega Lilia, Milne Jill C, Elliott Peter J, Puigserver Pere, Auwerx Johan. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. [Europe PMC free article] [Abstract] [Google Scholar]
- Cantó Carles, Jiang Lake Q, Deshmukh Atul S, Mataki Chikage, Coste Agnes, Lagouge Marie, Zierath Juleen R, Auwerx Johan. Interdependence of AMPK and SIRT1 for Metabolic Adaptation to Fasting and Exercise in Skeletal Muscle. Cell Metabolism. 2010;11:213–19. [Europe PMC free article] [Abstract] [Google Scholar]
- Cao Yilin, Rathmell Jeffrey C, Macintyre Andrew N, Moran R, Tamagnine J. Metabolic Reprogramming towards Aerobic Glycolysis Correlates with Greater Proliferative Ability and Resistance to Metabolic Inhibition in CD8 versus CD4 T Cells. PLoS ONE. 2014;9:e104104–e04. [Europe PMC free article] [Abstract] [Google Scholar]
- Chang Chih-Hao, Curtis Jonathan D, Maggi Leonard B, Faubert Brandon, Villarino Alejandro V, O’Sullivan David, Huang Stanley Ching-Cheng, van der Windt Gerritje JW, Blagih Julianna, Qiu Jing, Weber Jason D, Pearce Edward J, Jones Russell G, Pearce Erika L. Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell. 2013;153:1239–51. [Europe PMC free article] [Abstract] [Google Scholar]
- Chang John T, Wherry E John, Goldrath Ananda W. Molecular regulation of effector and memory T cell differentiation. Nature Immunology. 2014;15:1104–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Chaoul Nada, Fayolle Catherine, Desrues Belinda, Oberkampf Marine, Tang Alexandre, Ladant Daniel, Leclerc Claude. Rapamycin Impairs Antitumor CD8+ T-cell Responses and Vaccine-Induced Tumor Eradication. Cancer Research. 2015;75:3279–91. [Abstract] [Google Scholar]
- Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, Manjeri GR, Li Y, Ifrim DC, Arts RJ, van der Veer BM, Deen PM, Logie C, O’Neill LA, Willems P, van de Veerdonk FL, van der Meer JW, Ng A, Joosten LA, Wijmenga C, Stunnenberg HG, Xavier RJ, Netea MG. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. [Europe PMC free article] [Abstract] [Google Scholar]
- Chervona Yana, Costa Max. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radical Biology and Medicine. 2012;53:1041–47. [Europe PMC free article] [Abstract] [Google Scholar]
- Choudhary Chunaram, Kumar Chanchal, Gnad Florian, Nielsen Michael L, Rehman Michael, Walther Tobias C, Olsen Jesper V, Mann Matthias. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science. 2009;325:834–40. [Abstract] [Google Scholar]
- Clever David, Roychoudhuri Rahul, Constantinides Michael G, Askenase Michael H, Sukumar Madhusudhanan, Klebanoff Christopher A, Eil Robert L, Hickman Heather D, Yu Zhiya, Pan Jenny H, Palmer Douglas C, Phan Anthony T, Goulding John, Gattinoni Luca, Goldrath Ananda W, Belkaid Yasmine, Restifo Nicholas P. Oxygen Sensing by T Cells Establishes an Immunologically Tolerant Metastatic Niche. Cell. 2016;166:1117–31.e14. [Europe PMC free article] [Abstract] [Google Scholar]
- Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, Cline GW, Phillips AJ, Medzhitov R. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–63. [Europe PMC free article] [Abstract] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Cui Guoliang, Staron Matthew M, Gray Simon M, Ho Ping-Chih, Amezquita Robert A, Wu Jingxia, Kaech Susan M. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell. 2015;161:750–61. [Europe PMC free article] [Abstract] [Google Scholar]
- Delgoffe Greg M, Pollizzi Kristen N, Waickman Adam T, Heikamp Emily, Meyers David J, Horton Maureen R, Xiao Bo, Worley Paul F, Powell Jonathan D. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nature Immunology. 2011;12:295–303. [Europe PMC free article] [Abstract] [Google Scholar]
- Doedens Andrew L, Phan Anthony T, Stradner Martin H, Fujimoto Jessica K, Nguyen Jessica V, Yang Edward, Johnson Randall S, Goldrath Ananda W. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nature Immunology. 2013;14:1173–82. [Europe PMC free article] [Abstract] [Google Scholar]
- Etchegaray Jean-Pierre, Mostoslavsky Raul. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol Cell. 2016;62:695–711. [Europe PMC free article] [Abstract] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ, Consortium Immunological Genome Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–28. [Europe PMC free article] [Abstract] [Google Scholar]
- Geiger Roger, Rieckmann Jan C, Wolf Tobias, Basso Camilla, Feng Yuehan, Fuhrer Tobias, Kogadeeva Maria, Picotti Paola, Meissner Felix, Mann Matthias, Zamboni Nicola, Sallusto Federica, Lanzavecchia Antonio. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell. 2016;167:829–42.e13. [Europe PMC free article] [Abstract] [Google Scholar]
- Gerriets Valerie A, Kishton Rigel J, Nichols Amanda G, Macintyre Andrew N, Inoue Makoto, Ilkayeva Olga, Winter Peter S, Liu Xiaojing, Priyadharshini Bhavana, Slawinska Marta E, Haeberli Lea, Huck Catherine, Turka Laurence A, Wood Kris C, Hale Laura P, Smith Paul A, Schneider Martin A, MacIver Nancie J, Locasale Jason W, Newgard Christopher B, Shinohara Mari L, Rathmell Jeffrey C. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. Journal of Clinical Investigation. 2015;125:194–207. [Europe PMC free article] [Abstract] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–28. [Abstract] [Google Scholar]
- Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17:34–40. [Abstract] [Google Scholar]
- Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2016;17:26–33. [Europe PMC free article] [Abstract] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–40. [Europe PMC free article] [Abstract] [Google Scholar]
- Gray Simon M, Amezquita Robert A, Guan Tianxia, Kleinstein Steven H, Kaech Susan M. Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8+ T Cell Terminal Differentiation and Loss of Multipotency. Immunity. 2017;46:596–608. [Europe PMC free article] [Abstract] [Google Scholar]
- He Bing, Xing Shaojun, Chen Changya, Gao Peng, Teng Li, Shan Qiang, Gullicksrud Jodi A, Martin Matthew D, Yu Shuyang, Harty John T, Badovinac Vladimir P, Tan Kai, Xue Hai-Hui. CD8+ T Cells Utilize Highly Dynamic Enhancer Repertoires and Regulatory Circuitry in Response to Infections. Immunity. 2016;45:1341–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. [Europe PMC free article] [Abstract] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. [Abstract] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. [Europe PMC free article] [Abstract] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Ho Ping-Chih, Bihuniak Jessica Dauz, Macintyre Andrew N, Staron Matthew, Liu Xiaojing, Amezquita Robert, Tsui Yao-Chen, Cui Guoliang, Micevic Goran, Perales Jose C, Kleinstein Steven H, Abel E Dale, Insogna Karl L, Feske Stefan, Locasale Jason W, Bosenberg Marcus W, Rathmell Jeffrey C, Kaech Susan M. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–28. [Europe PMC free article] [Abstract] [Google Scholar]
- Houtkooper Riekelt H, Pirinen Eija, Auwerx Johan. Sirtuins as regulators of metabolism and healthspan. Nature Reviews Molecular Cell Biology. 2012;13:225–25. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang He, Lin Shu, Garcia Benjamin A, Zhao Yingming. Quantitative Proteomic Analysis of Histone Modifications. Chemical Reviews. 2015;115:2376–418. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang SC, Smith AM, Everts B, Colonna M, Pearce EL, Schilling JD, Pearce EJ. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity. 2016;45:817–30. [Europe PMC free article] [Abstract] [Google Scholar]
- Iijima Norifumi, Akiko Iwasaki. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 2014;346:93. [Europe PMC free article] [Abstract] [Google Scholar]
- Imai Shin-ichiro, Armstrong Christopher M, Kaeberlein Matt, Guarente Leonard. Transcriptional silencing and longevity protein Sir2 is an NAD-dependenthistone deacetylase. Nature. 2000;403:795–800. [Abstract] [Google Scholar]
- Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–30. [Abstract] [Google Scholar]
- Jiang Xiaodong, Clark Rachael A, Liu Luzheng, Wagers Amy J, Fuhlbrigge Robert C, Kupper Thomas S. Skin infection generates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature. 2012;483:227–31. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaelin William G, Jr, McKnight Steven L. Influence of Metabolism on Epigenetics and Disease. Cell. 2013;153:56–69. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–25. [Europe PMC free article] [Abstract] [Google Scholar]
- Kakaradov Boyko, Arsenio Janilyn, Widjaja Christella E, He Zhaoren, Aigner Stefan, Metz Patrick J, Yu Bingfei, Wehrens Ellen J, Lopez Justine, Kim Stephanie H, Zuniga Elina I, Goldrath Ananda W, Chang John T, Yeo Gene W. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nature Immunology 2017 [Europe PMC free article] [Abstract] [Google Scholar]
- Kanno Yuka, Vahedi Golnaz, Hirahara Kiyoshi, Singleton Kentner, O’Shea John J. Transcriptional and Epigenetic Control of T Helper Cell Specification: Molecular Mechanisms Underlying Commitment and Plasticity. Annu Rev Immunol. 2012;30:707–31. [Europe PMC free article] [Abstract] [Google Scholar]
- Kelly B, O’Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25:771–84. [Europe PMC free article] [Abstract] [Google Scholar]
- Kinnaird Adam, Zhao Steven, Wellen Kathryn E, Michelakis Evangelos D. Metabolic control of epigenetics in cancer. Nature reviews Cancer. 2016;16:694–707. [Abstract] [Google Scholar]
- Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC, Griss T, Weinheimer CJ, Khader S, Randolph GJ, Pearce EJ, Jones RG, Diwan A, Diamond MS, Artyomov MN. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016;24:158–66. [Europe PMC free article] [Abstract] [Google Scholar]
- Lara-Astiaso David, Weiner Assaf, Lorenzo-Vivas Erika, Zaretsky Irina, Jaitin Diego Adhemar, David Eyal, Keren-Shaul Hadas, Mildner Alexander, Winter Deborah, Jung Steffen, Friedman Nir, Amit Ido. Chromatin state dynamics during blood formation. Science. 2014;345:943. [Europe PMC free article] [Abstract] [Google Scholar]
- Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, Hayward L, Langridge-Smith P, Gilbert N, Ramsahoye BH. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012;40:4794–803. [Europe PMC free article] [Abstract] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–26. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee Joyce V, Carrer Alessandro, Shah Supriya, Snyder Nathaniel W, Wei Shuanzeng, Venneti Sriram, Worth Andrew J, Yuan Zuo-Fei, Lim Hee-Woong, Liu Shichong, Jackson Ellen, Aiello Nicole M, Haas Naomi B, Rebbeck Timothy R, Judkins Alexander, Won Kyoung-Jae, Chodosh Lewis A, Garcia Benjamin A, Stanger Ben Z, Feldman Michael D, Blair Ian A, Wellen Kathryn E. Akt-Dependent Metabolic Reprogramming Regulates Tumor Cell Histone Acetylation. Cell Metabolism. 2014;20:306–19. [Europe PMC free article] [Abstract] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–63. [Europe PMC free article] [Abstract] [Google Scholar]
- Link VM, Gosselin D, Glass CK. Mechanisms Underlying the Selection and Function of Macrophage-Specific Enhancers. Cold Spring Harb Symp Quant Biol. 2015;80:213–21. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu TF, Vachharajani VT, Yoza BK, McCall CE. NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J Biol Chem. 2012;287:25758–69. [Europe PMC free article] [Abstract] [Google Scholar]
- Ma Eric H, Bantug Glenn, Griss Takla, Condotta Stephanie, Johnson Radia M, Samborska Bozena, Mainolfi Nello, Suri Vipin, Guak Hannah, Balmer Maria L, Verway Mark J, Raissi Thomas C, Tsui Harmony, Boukhaled Giselle, Costa Sofia Henriques da, Frezza Christian, Krawczyk Connie M, Friedman Adam, Manfredi Mark, Richer Martin J, Hess Christoph, Jones Russell G. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metabolism 2017 [Abstract] [Google Scholar]
- MacIver Nancie J, Michalek Ryan D, Rathmell Jeffrey C. Metabolic Regulation of T Lymphocytes. Annu Rev Immunol. 2013;31:259–83. [Europe PMC free article] [Abstract] [Google Scholar]
- Mackay Laura K, Rahimpour Azad, Ma Joel Z, Collins Nicholas, Stock Angus T, Hafon Ming-Li, Vega-Ramos Javier, Lauzurica Pilar, Mueller Scott N, Stefanovic Tijana, Tscharke David C, Heath William R, Inouye Michael, Carbone Francis R, Gebhardt Thomas. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;14:1294–301. [Abstract] [Google Scholar]
- Mascanfroni Ivan D, Takenaka Maisa C, Yeste Ada, Patel Bonny, Wu Yan, Kenison Jessica E, Siddiqui Shafiuddin, Basso Alexandre S, Otterbein Leo E, Pardoll Drew M, Pan Fan, Priel Avner, Clish Clary B, Robson Simon C, Quintana Francisco J. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α Nature Medicine, advance on 2015 [Europe PMC free article] [Abstract] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, Crozet L, Jacome-Galarza CE, Handler K, Klughammer J, Kobayashi Y, Gomez-Perdiguero E, Schultze JL, Beyer M, Bock C, Geissmann F. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353 [Europe PMC free article] [Abstract] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Aguilar S Vargas, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Gonzalez F Zelada, Perrin P, Keren-Shaul H, Gury M, Lara-Astaiso D, Thaiss CA, Cohen M, Halpern K Bahar, Baruch K, Deczkowska A, Lorenzo-Vivas E, Itzkovitz S, Elinav E, Sieweke MH, Schwartz M, Amit I. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. [Abstract] [Google Scholar]
- Michalek Ryan D, Gerriets Valerie A, Jacobs Sarah R, Macintyre Andrew N, MacIver Nancie J, Mason Emily F, Sullivan Sarah A, Nichols Amanda G, Rathmell Jeffrey C. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4+ T Cell Subsets. The Journal of Immunology. 2011;186:3299–303. [Europe PMC free article] [Abstract] [Google Scholar]
- Mucida Daniel, Kutchukhidze Nino, Erazo Agustin, Russo Momtchilo, Lafaille Juan J, de Lafaille Maria A Curotto. Oral tolerance in the absence of naturally occurring Tregs. The Journal of Clinical Investigation. 2005;115:1923–33. [Abstract] [Google Scholar]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. [Abstract] [Google Scholar]
- Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, O’Neill LA, Xavier RJ. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. [Europe PMC free article] [Abstract] [Google Scholar]
- Northrop John K, Wells Andrew D, Shen Hao. Cutting Edge: Chromatin Remodeling as a Molecular Basis for the Enhanced Functionality of Memory CD8 T Cells. The Journal of Immunology. 2008;181:865. [Abstract] [Google Scholar]
- Novakovic B, Habibi E, Wang SY, Arts RJ, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J, Matarese F, van Heeringen SJ, Janssen–Megens EM, Sharifi N, Wang C, Keramati F, Schoonenberg V, Flicek P, Clarke L, Pickkers P, Heath S, Gut I, Netea MG, Martens JH, Logie C, Stunnenberg HG. beta-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell. 2016;167:1354–68.e14. [Europe PMC free article] [Abstract] [Google Scholar]
- O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. [Europe PMC free article] [Abstract] [Google Scholar]
- O’Sullivan David, van der Windt Gerritje JW, Huang Stanley Ching-Cheng, Curtis Jonathan D, Chang Chih-Hao, Buck Michael D, Qiu Jing, Smith Amber M, Lam Wing Y, DiPlato Lisa M, Hsu Fong-Fu, Birnbaum Morris J, Pearce Edward J, Pearce Erika L. Memory CD8+ T Cells Use Cell-Intrinsic Lipolysis to Support the Metabolic Programming Necessary for Development. Immunity. 2014;41:75–88. [Europe PMC free article] [Abstract] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–44. [Europe PMC free article] [Abstract] [Google Scholar]
- Okoye Isobel, Wang Lihui, Pallmer Katharina, Richter Kirsten, Ichimura Takahuru, Haas Robert, Crouse Josh, Choi Onjee, Heathcote Dean, Lovo Elena, Mauro Claudio, Abdi Reza, Oxenius Annette, Rutschmann Sophie, Ashton-Rickardt Philip G. The protein LEM promotes CD8+ T cell immunity through effects on mitochondrial respiration. Science. 2015;348:995–1001. [Abstract] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–71. [Abstract] [Google Scholar]
- Pan Youdong, Tian Tian, Park Chang Ook, Lofftus Serena Y, Mei Shenglin, Liu Xing, Luo Chi, O’Malley John T, Gehad Ahmed, Teague Jessica E, Divito Sherrie J, Fuhlbrigge Robert, Puigserver Pere, Krueger James G, Hotamisligil Gökhan S, Clark Rachael A, Kupper Thomas S. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 2017;543:252–56. [Europe PMC free article] [Abstract] [Google Scholar]
- Pauken Kristen E, Sammons Morgan A, Odorizzi Pamela M, Manne Sasikanth, Godec Jernej, Khan Omar, Drake Adam M, Chen Zeyu, Sen Debattama, Kurachi Makoto, Barnitz R Anthony, Bartman Caroline, Bengsch Bertram, Huang Alexander C, Schenkel Jason M, Vahedi Golnaz, Haining W Nicholas, Berger Shelley L, Wherry E John. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science (New York, NY) 2016;354:1160–65. [Europe PMC free article] [Abstract] [Google Scholar]
- Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–43. [Europe PMC free article] [Abstract] [Google Scholar]
- Pearce Erika L, Walsh Matthew C, Cejas Pedro J, Harms Gretchen M, Shen Hao, Wang Li-San, Jones Russell G, Choi Yongwon. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–07. [Europe PMC free article] [Abstract] [Google Scholar]
- Peng Min, Yin Na, Chhangawala Sagar, Xu Ke, Leslie Christina S, Li Ming O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–84. [Europe PMC free article] [Abstract] [Google Scholar]
- Phan Anthony T, Doedens Andrew L, Palazon Asis, Tyrakis Petros A, Cheung Kitty P, Johnson Randall S, Goldrath Ananda W. Constitutive Glycolytic Metabolism Supports CD8+ T Cell Effector Memory Differentiation during Viral Infection. Immunity. 2016;45:1024–37. [Europe PMC free article] [Abstract] [Google Scholar]
- Piccolo V, Curina A, Genua M, Ghisletti S, Simonatto M, Sabo A, Amati B, Ostuni R, Natoli G. Opposing macrophage polarization programs show extensive epigenomic and transcriptional crosstalk. Nat Immunol 2017 [Europe PMC free article] [Abstract] [Google Scholar]
- Pollizzi Kristen N, Patel Chirag H, Sun Im-Hong, Oh Min-Hee, Waickman Adam T, Wen Jiayu, Delgoffe Greg M, Powell Jonathan D. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. Journal of Clinical Investigation. 2015;125:2090–108. [Europe PMC free article] [Abstract] [Google Scholar]
- Pollizzi Kristen N, Powell Jonathan D. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–46. [Europe PMC free article] [Abstract] [Google Scholar]
- Rao Rajesh R, Li Qingsheng, Odunsi Kunle, Shrikant Protul A. The mTOR Kinase Determines Effector versus Memory CD8+ T Cell Fate by Regulating the Expression of Transcription Factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. [Europe PMC free article] [Abstract] [Google Scholar]
- Roadmap Epigenomics, Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [Europe PMC free article] [Abstract] [Google Scholar]
- Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, Cascante M, Bosca L. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185:605–14. [Abstract] [Google Scholar]
- Rolf Julia, Zarrouk Marouan, Finlay David K, Foretz Marc, Viollet Benoit, Cantrell Doreen A. AMPKα1: A glucose sensor that controls CD8 T-cell memory. European Journal of Immunology. 2013;43:889–96. [Europe PMC free article] [Abstract] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–28. [Abstract] [Google Scholar]
- Russ Brendan E, Olshanksy Moshe, Smallwood Heather S, Li Jasmine, Denton Alice E, Prier Julia E, Stock Angus T, Croom Hayley A, Cullen Jolie G, Nguyen Michelle LT, Rowe Stephanie, Olson Matthew R, Finkelstein David B, Kelso Anne, Thomas Paul G, Speed Terry P, Rao Sudha, Turner Stephen J. Distinct Epigenetic Signatures Delineate Transcriptional Programs during Virus-Specific CD8+ T Cell Differentiation. Immunity. 2014;41:853–65. [Europe PMC free article] [Abstract] [Google Scholar]
- Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, Roeder RG, Allis CD. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell. 2015;58:203–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Sakata SF, Shelly LL, Ruppert S, Schutz G, Chou JY. Cloning and expression of murine S-adenosylmethionine synthetase. The Journal of biological chemistry. 1993;268:13978–86. [Abstract] [Google Scholar]
- Scharer Christopher D, Bally Alexander PR, Gandham Bhanu, Boss Jeremy M. Cutting Edge: Chromatin Accessibility Programs CD8 T Cell Memory. The Journal of Immunology. 2017;198:2238. [Europe PMC free article] [Abstract] [Google Scholar]
- Scharer Christopher D, Barwick Benjamin G, Youngblood Benjamin A, Ahmed Rafi, Boss Jeremy M. Global DNA Methylation Remodeling Accompanies CD8 T Cell Effector Function. The Journal of Immunology. 2013;191:3419. [Europe PMC free article] [Abstract] [Google Scholar]
- Scharping Nicole E, Menk Ashley V, Moreci Rebecca S, Whetstone Ryan D, Dadey Rebekah E, Watkins Simon C, Ferris Robert L, Delgoffe Greg M. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity. 2016;45:374–88. [Europe PMC free article] [Abstract] [Google Scholar]
- Schenkel Jason M, Fraser Kathryn A, Beura Lalit K, Pauken Kristen E, Vezys Vaiva, Masopust David. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346:98. [Europe PMC free article] [Abstract] [Google Scholar]
- Schenkel Jason M, Fraser Kathryn A, Vezys Vaiva, Masopust David. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. 2013;14:509–13. [Europe PMC free article] [Abstract] [Google Scholar]
- Schreiber Valérie, Dantzer Françoise, Ame Jean-Christophe, de Murcia Gilbert. Poly(ADP-ribose): novel functions for an old molecule. Nature Reviews Molecular Cell Biology. 2006;7:517–28. [Abstract] [Google Scholar]
- Schulz C, Perdiguero E Gomez, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, Frampton J, Liu KJ, Geissmann F. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. [Abstract] [Google Scholar]
- Scott-Browne James P, López-Moyado Isaac F, Trifari Sara, Wong Victor, Chavez Lukas, Rao Anjana, Pereira Renata M. Dynamic Changes in Chromatin Accessibility Occur in CD8+ T Cells Responding to Viral Infection. Immunity 2016 [Europe PMC free article] [Abstract] [Google Scholar]
- Sen Debattama R, Kaminski James, Barnitz R Anthony, Kurachi Makoto, Gerdemann Ulrike, Yates Kathleen B, Tsao Hsiao-Wei, Godec Jernej, LaFleur Martin W, Brown Flavian D, Tonnerre Pierre, Chung Raymond T, Tully Damien C, Allen Todd M, Frahm Nicole, Lauer Georg M, Wherry E John, Yosef Nir, Haining W Nicholas. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165. [Europe PMC free article] [Abstract] [Google Scholar]
- Sena Laura A, Li Sha, Jairaman Amit, Prakriya Murali, Ezponda Teresa, Hildeman David A, Wang Chyung-Ru, Schumacker Paul T, Licht Jonathan D, Perlman Harris, Bryce Paul J, Chandel Navdeep S. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–36. [Europe PMC free article] [Abstract] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. Journal of Experimental Medicine. 2011;208:1367–76. [Europe PMC free article] [Abstract] [Google Scholar]
- Shrestha Sharad, Yang Kai, Wei Jun, Karmaus Peer WF, Neale Geoffrey, Chi Hongbo. Tsc1 promotes the differentiation of memory CD8+ T cells via orchestrating the transcriptional and metabolic programs. Proceedings of the National Academy of Sciences. 2014;111:14858–63. [Europe PMC free article] [Abstract] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, Daley GQ, Cantley LC. Influence of Threonine Metabolism on S-Adenosylmethionine and Histone Methylation. Science. 2013;339:222–26. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith Jeffrey S, Brachmann Carrie Baker, Celic Ivana, Kenna Margaret A, Muhammad Shabazz, Starai Vincent J, Avalos Jose L, Escalante-Semerena Jorge C, Grubmeyer Charles, Wolberger Cynthia, Boeke Jef D. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proceedings of the National Academy of Sciences. 2000;97:6658–63. [Europe PMC free article] [Abstract] [Google Scholar]
- Sukumar Madhusudhanan, Liu Jie, Ji Yun, Subramanian Murugan, Crompton Joseph G, Yu Zhiya, Roychoudhuri Rahul, Palmer Douglas C, Muranski Pawel, Karoly Edward D, Mohney Robert P, Klebanoff Christopher A, Lal Ashish, Finkel Toren, Restifo Nicholas P, Gattinoni Luca. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. The Journal of Clinical Investigation. 2013;123:4479–88. [Europe PMC free article] [Abstract] [Google Scholar]
- Sun Cheng-Ming, Hall Jason A, Blank Rebecca B, Bouladoux Nicolas, Oukka Mohamed, Mora J Rodrigo, Belkaid Yasmine. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. The Journal of Experimental Medicine. 2007;204:1775. [Europe PMC free article] [Abstract] [Google Scholar]
- Takahashi Hidekazu, McCaffery J Michael, Irizarry Rafael A, Boeke Jef D. Nucleocytosolic Acetyl-Coenzyme A Synthetase Is Required for Histone Acetylation and Global Transcription. Mol Cell. 2006;23:207–17. [Abstract] [Google Scholar]
- Tanner Kirk G, Landry Joseph, Sternglanz Rolf, Denu John M. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proceedings of the National Academy of Sciences. 2000;97:14178–82. [Europe PMC free article] [Abstract] [Google Scholar]
- Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–8. [Abstract] [Google Scholar]
- Tripathi Subhash K, Lahesmaa Riitta. Transcriptional and epigenetic regulation of T-helper lineage specification. Immunological Reviews. 2014;261:62–83. [Europe PMC free article] [Abstract] [Google Scholar]
- Tumes Damon J, Onodera Atsushi, Suzuki Akane, Shinoda Kenta, Endo Yusuke, Iwamura Chiaki, Hosokawa Hiroyuki, Koseki Haruhiko, Tokoyoda Koji, Suzuki Yutaka, Motohashi Shinichiro, Nakayama Toshinori. The Polycomb Protein Ezh2 Regulates Differentiation and Plasticity of CD4+ T Helper Type 1 and Type 2 Cells. Immunity. 2013;39:819–32. [Abstract] [Google Scholar]
- Tyrakis Petros A, Palazon Asis, Macias David, Lee Kian L, Phan Anthony T, Veliça Pedro, You Jia, Chia Grace S, Sim Jingwei, Doedens Andrew, Abelanet Alice, Evans Colin E, Griffiths John R, Poellinger Lorenz, Goldrath Ananda W, Johnson Randall S. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature. 2016;540:236–41. [Europe PMC free article] [Abstract] [Google Scholar]
- Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. AMP-Activated Protein Kinase-Deficient Mice Are Resistant to the Metabolic Effects of Resveratrol. Diabetes. 2010;59:554–63. [Europe PMC free article] [Abstract] [Google Scholar]
- Vahedi Golnaz, Kanno Yuka, Sartorelli Vittorio, O’Shea John J. Transcription factors and CD4 T cells seeking identity: masters, minions, setters and spikers. Immunology. 2013;139:294–98. [Abstract] [Google Scholar]
- van der Windt GJW, W GJ, O’Sullivan D, Everts B, Huang SCC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences. 2013;110:14336–41. [Europe PMC free article] [Abstract] [Google Scholar]
- van der Windt Gerritje JW, Bart Everts, Chih-Hao Chang, Jonathan DD Curtis, Tori CC Freitas, Eyal Amiel, Edward J Pearce, Erika L Pearce. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity. 2012;36:68–78. [Europe PMC free article] [Abstract] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang J, Jia ST, Jia S. New Insights into the Regulation of Heterochromatin. Trends Genet. 2016;32:284–94. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Ruoning, Dillon Christopher P, Shi Lewis Zhichang, Milasta Sandra, Carter Robert, Finkelstein David, McCormick Laura L, Fitzgerald Patrick, Chi Hongbo, Munger Joshua, Green Douglas R. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. [Europe PMC free article] [Abstract] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. [Europe PMC free article] [Abstract] [Google Scholar]
- Wherry E John, Kurachi Makoto. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–99. [Europe PMC free article] [Abstract] [Google Scholar]
- Wiles ET, Selker EU. H3K27 methylation: a promiscuous repressive chromatin mark. Curr Opin Genet Dev. 2016;43:31–37. [Europe PMC free article] [Abstract] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–55. [Europe PMC free article] [Abstract] [Google Scholar]
- Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y, Xia C, Peng C, Ruan H, Kirkey M, Wang D, Jensen LM, Kwon OK, Lee S, Pletcher SD, Tan M, Lombard DB, White KP, Zhao H, Li J, Roeder RG, Yang X, Zhao Y. Metabolic Regulation of Gene Expression by Histone Lysine beta-Hydroxybutyrylation. Mol Cell. 2016;62:194–206. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu Bingfei, Zhang Kai, Milner J Justin, Toma Clara, Chen Runqiang, Scott-Browne James P, Pereira Renata M, Crotty Shane, Chang John T, Pipkin Matthew E, Wang Wei, Goldrath Ananda W. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat Immunol. 2017 advance online publication. [Abstract] [Google Scholar]
- Zeng Hu, Cohen Sivan, Guy Cliff, Shrestha Sharad, Neale Geoffrey, Brown Scott A, Cloer Caryn, Kishton Rigel J, Gao Xia, Youngblood Ben, Do Mytrang, Li Ming O, Locasale Jason W, Rathmell Jeffrey C, Chi Hongbo. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity. 2016;45:540–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Zeng Hu, Yang Kai, Cloer Caryn, Neale Geoffrey, Vogel Peter, Chi Hongbo. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature. 2013;499:485–90. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.immuni.2017.04.016
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1074761317301814/pdf
Citations & impact
Impact metrics
Article citations
The significance of CD16+ monocytes in the occurrence and development of chronic thromboembolic pulmonary hypertension: insights from single-cell RNA sequencing.
Front Immunol, 15:1446710, 13 Aug 2024
Cited by: 0 articles | PMID: 39192976 | PMCID: PMC11347785
Metabolic flux in macrophages in obesity and type-2 diabetes.
J Pharm Pharm Sci, 27:13210, 26 Jun 2024
Cited by: 1 article | PMID: 38988822 | PMCID: PMC11233469
Review Free full text in Europe PMC
A new era of cancer immunotherapy: combining revolutionary technologies for enhanced CAR-M therapy.
Mol Cancer, 23(1):117, 01 Jun 2024
Cited by: 5 articles | PMID: 38824567 | PMCID: PMC11143597
Review Free full text in Europe PMC
Effects of Iron Status on Adaptive Immunity and Vaccine Efficacy: A Review.
Adv Nutr, 15(6):100238, 08 May 2024
Cited by: 1 article | PMID: 38729263 | PMCID: PMC11251406
Review Free full text in Europe PMC
Fasting hyperglycaemia and fatty liver drive colorectal cancer: a retrospective analysis in 1145 patients.
Intern Emerg Med, 19(5):1267-1277, 26 Apr 2024
Cited by: 3 articles | PMID: 38668822 | PMCID: PMC11364717
Go to all (160) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Macrophage metabolism: a wound-healing perspective.
Immunol Cell Biol, 97(3):268-278, 21 Feb 2019
Cited by: 19 articles | PMID: 30779212
Review
Modulation of macrophage activation and programming in immunity.
J Cell Physiol, 228(3):502-512, 01 Mar 2013
Cited by: 86 articles | PMID: 22777800
Review
Transcriptional regulation of T cell metabolism.
Mol Immunol, 68(2 pt c):520-526, 19 Aug 2015
Cited by: 9 articles | PMID: 26298576 | PMCID: PMC4679508
Review Free full text in Europe PMC
Unraveling the Complex Interplay Between T Cell Metabolism and Function.
Annu Rev Immunol, 36:461-488, 01 Apr 2018
Cited by: 379 articles | PMID: 29677474 | PMCID: PMC6323527
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Ben and Wanda Hildyard Chair in Hereditary Diseases
Leducq Transatlantic Network Grant
NHLBI NIH HHS (1)
Grant ID: P01 HL088093
NIAID NIH HHS (4)
Grant ID: R01 AI096852
Grant ID: R01 AI072117
Grant ID: R37 AI067545
Grant ID: R01 AI067545
NIDDK NIH HHS (1)
Grant ID: R01 DK091183
NIGMS NIH HHS (1)
Grant ID: T32 GM007240
NIH (4)
Grant ID: AI067545
Grant ID: NS096170
Grant ID: DK091183
Grant ID: AI072117
NINDS NIH HHS (1)
Grant ID: R01 NS096170





