Abstract
Free full text

Isolation and characterization of the bacteriophage T4 tail-associated lysozyme.
Abstract
Direct evidence has been obtained that the tail-associated lysozyme of bacteriophage T4 (tail-lysozyme) is gp5, which is a protein component of the hub of the baseplate. Tails were treated with 3 M guanidine hydrochloride containing 1% Triton X-100, and the tail-lysozyme was separated from other tail components by preparative isoelectric focusing electrophoresis as a peak with a pI of 8.4. The molecular weight as determined from sodium dodecyl sulfate electrophoresis was 42,000. The tail-lysozyme was unambiguously identified as gp5 when the position of the lysozyme was compared with that of gp5 of tube-baseplates from 5ts1/23amH11/eL1ainfected Escherichia coli cells by two-dimensional gel electrophoresis. The tail-lysozyme has N-acetylmuramidase activity and the same substrate specificity as gene e lysozyme; the optimum pH is around 5.8, about 1 pH unit lower than for the e lysozyme. We assume that the tail-lysozyme plays an essential role in locally digesting the peptidoglycan layer to let the tube penetrate into the periplasmic space. The tail-lysozyme is presumably also responsible for "lysis from without."
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
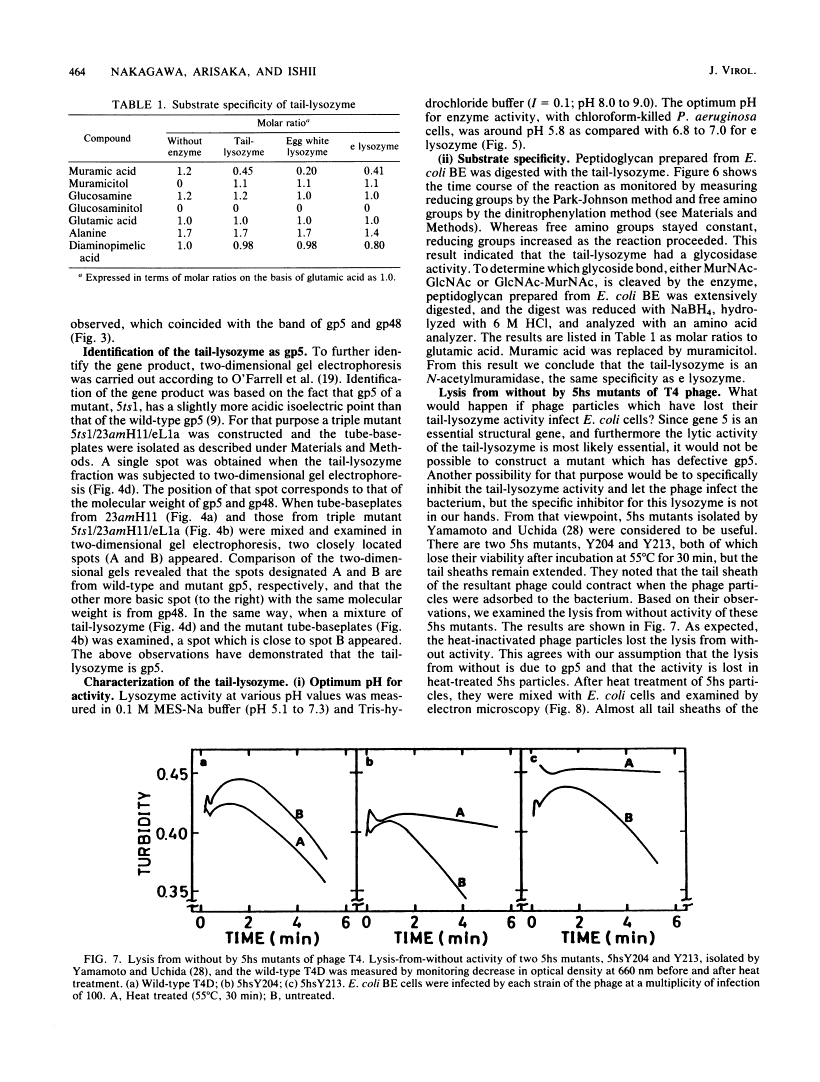
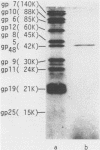
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black LW, Hogness DS. The lysozyme of bacteriophage lambda. I. Purification and molecular weight. J Biol Chem. 1969 Apr 25;244(8):1968–1975. [Abstract] [Google Scholar]
- Crowther RA, Lenk EV, Kikuchi Y, King J. Molecular reorganization in the hexagon to star transition of the baseplate of bacteriophage T4. J Mol Biol. 1977 Nov 5;116(3):489–523. [Abstract] [Google Scholar]
- Duda RL, Eiserling FA. Evidence for an internal component of the bacteriophage T4D tail core: a possible length-determining template. J Virol. 1982 Aug;43(2):714–720. [Europe PMC free article] [Abstract] [Google Scholar]
- Fairbanks G, Steck TL, Wallach DF. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. [Abstract] [Google Scholar]
- Furukawa H, Kuroiwa T, Mizushima S. DNA injection during bacteriophage T4 infection of Escherichia coli. J Bacteriol. 1983 May;154(2):938–945. [Europe PMC free article] [Abstract] [Google Scholar]
- Iba H, Nanno M, Okada Y. Identification and partial purification of a lytic enzyme in the bacteriophage phi 6 virion. FEBS Lett. 1979 Jul 15;103(2):234–237. [Abstract] [Google Scholar]
- Iwashita S, Kanegasaki S. Smooth specific phage adsorption: endorhamnosidase activity of tail parts of P22. Biochem Biophys Res Commun. 1973 Nov 16;55(2):403–409. [Abstract] [Google Scholar]
- Kao SH, McClain WH. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J Virol. 1980 Apr;34(1):95–103. [Europe PMC free article] [Abstract] [Google Scholar]
- KATZ W, WEIDEL W. [Isolation and characterization of the lysozyme bound to T2 phage]. Z Naturforsch B. 1961 Jun;16B:363–368. [Abstract] [Google Scholar]
- Kells SS, Haselkorn R. Bacteriophage T4 short tail fibers are the product of gene 12. J Mol Biol. 1974 Mar 15;83(4):473–485. [Abstract] [Google Scholar]
- Labedan B, Goldberg EB. Requirement for membrane potential in injection of phage T4 DNA. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4669–4673. [Europe PMC free article] [Abstract] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. [Abstract] [Google Scholar]
- Loeb MR. Bacteriophage T4-mediated release of envelope components from Escherichia coli. J Virol. 1974 Mar;13(3):631–641. [Europe PMC free article] [Abstract] [Google Scholar]
- Moody MF. Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol. 1973 Nov 15;80(4):613–635. [Abstract] [Google Scholar]
- Oakley BR, Kirsch DR, Morris NR. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. [Abstract] [Google Scholar]
- O'Farrell PZ, Goodman HM, O'Farrell PH. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. [Abstract] [Google Scholar]
- PARK JT, JOHNSON MJ. A submicrodetermination of glucose. J Biol Chem. 1949 Nov;181(1):149–151. [Abstract] [Google Scholar]
- Simon LD, Anderson TF. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. II. Structure and function of the baseplate. Virology. 1967 Jun;32(2):298–305. [Abstract] [Google Scholar]
- Szewczyk B, Skórko R. Lysozyme activity of bacteriophage T4 ghosts. Biochim Biophys Acta. 1981 Nov 13;662(1):131–137. [Abstract] [Google Scholar]
- Szewczyk B, Skórko R. Purification and some properties of bacteriophage T4 particle-associated lysozyme. Eur J Biochem. 1983 Jul 1;133(3):717–722. [Abstract] [Google Scholar]
- Tschopp J, Arisaka F, van Driel R, Engel J. Purification, characterization and reassembly of the bacteriophage T4D tail sheath protein P18. J Mol Biol. 1979 Feb 25;128(2):247–258. [Abstract] [Google Scholar]
- Tsugita A, Inouye M. Purification of bacteriophage T4 lysozyme. J Biol Chem. 1968 Jan 25;243(2):391–397. [Abstract] [Google Scholar]
- Tsukagoshi N, Schäfer R, Franklin RM. Structure and synthesis of a lipid-containing bacteriophage. An endolysin activity associated with bacteriophage PM2. Eur J Biochem. 1977 Aug 1;77(3):585–588. [Abstract] [Google Scholar]
- Wyckoff M, Rodbard D, Chrambach A. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate-containing buffers using multiphasic buffer systems: properties of the stack, valid Rf- measurement, and optimized procedure. Anal Biochem. 1977 Apr;78(2):459–482. [Abstract] [Google Scholar]
- Yamamoto M, Uchida H. Organization and function of bacteriophage T4 tail. I. Isolation of heat-sensitive T4 tail mutants. Virology. 1973 Mar;52(1):234–245. [Abstract] [Google Scholar]
- Zorzopulos J, Kozloff LM, Chapman V, DeLong S. Bacteriophage T4D receptors and the Escherichia coli cell wall structure: role of spherical particles and protein b of the cell wall in bacteriophage infection. J Bacteriol. 1979 Jan;137(1):545–555. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.54.2.460-466.1985
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc254817?pdf=render
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/reprint/54/2/460
Free to read at jvi.asm.org
http://jvi.asm.org/cgi/content/abstract/54/2/460
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.54.2.460-466.1985
Article citations
Genome analysis of vB_SupP_AX, a novel N4-like phage infecting Sulfitobacter.
Int Microbiol, 27(4):1297-1306, 08 Jan 2024
Cited by: 0 articles | PMID: 38190086
Vibrio Phage VMJ710 Can Prevent and Treat Disease Caused by Pathogenic MDR V. cholerae O1 in an Infant Mouse Model.
Antibiotics (Basel), 12(6):1046, 14 Jun 2023
Cited by: 2 articles | PMID: 37370365 | PMCID: PMC10295236
Pseudomonas Phage ZCPS1 Endolysin as a Potential Therapeutic Agent.
Viruses, 15(2):520, 13 Feb 2023
Cited by: 4 articles | PMID: 36851734 | PMCID: PMC9961711
Evolutionary Dynamics between Phages and Bacteria as a Possible Approach for Designing Effective Phage Therapies against Antibiotic-Resistant Bacteria.
Antibiotics (Basel), 11(7):915, 07 Jul 2022
Cited by: 21 articles | PMID: 35884169 | PMCID: PMC9311878
Review Free full text in Europe PMC
Major tail proteins of bacteriophages of the order Caudovirales.
J Biol Chem, 298(1):101472, 08 Dec 2021
Cited by: 27 articles | PMID: 34890646 | PMCID: PMC8718954
Review Free full text in Europe PMC
Go to all (53) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification of T4 gene 25 product, a component of the tail baseplate, as a 15K lysozyme.
Mol Gen Genet, 202(3):363-367, 01 Mar 1986
Cited by: 11 articles | PMID: 3520236
Mapping of functional sites on the primary structure of the tail lysozyme of bacteriophage T4 by mutational analysis.
Biochim Biophys Acta, 1384(2):243-252, 01 May 1998
Cited by: 16 articles | PMID: 9659385
Purification and some properties of bacteriophage T4 particle-associated lysozyme.
Eur J Biochem, 133(3):717-722, 01 Jul 1983
Cited by: 3 articles | PMID: 6345157
The tail lysozyme complex of bacteriophage T4.
Int J Biochem Cell Biol, 35(1):16-21, 01 Jan 2003
Cited by: 43 articles | PMID: 12467643
Review