Abstract
Free full text

The evolutionarily conserved MAPK/Erk signaling promotes ancestral T-cell immunity in fish via c-Myc–mediated glycolysis
Abstract
The mitogen-activated protein kinase (MAPK) cascade is an ancient and evolutionarily conserved signaling pathway involved in numerous physiological processes. Despite great advances in understanding MAPK-mediated regulation of adaptive immune responses in mammals, its contribution to T-cell immunity in early vertebrates remains unclear. Herein, we used Nile tilapia (Oreochromis niloticus) to investigate the regulatory roles of MAPK/extracellular signal–regulated kinase (Erk) signaling in ancestral T-cell immunity of jawed fish. We found that Nile tilapia possesses an evolutionarily conserved MAPK/Erk axis that is activated through a classical three-tier kinase cascade, involving sequential phosphorylation of RAF proto-oncogene serine/threonine-protein kinase (Raf), MAPK/Erk kinase 1/2 (Mek1/2), and Erk1/2. In Nile tilapia, MAPK/Erk signaling participates in adaptive immune responses during bacterial infection. Upon T-cell activation, the MAPK/Erk axis is robustly activated, and MAPK/Erk blockade by specific inhibitors severely impairs T-cell activation. Furthermore, signals from MAPK/Erk were indispensable for primordial T cells to proliferate and exert their effector functions. Mechanistically, activation of the MAPK/Erk axis promoted glycolysis via induction of the transcriptional regulator proto-oncogene c-Myc (c-Myc), to ensure the proper activation and proliferation of fish T cells. Our results reveal the regulatory mechanisms of MAPK/Erk signaling in T-cell immunity in fish and highlight a close link between immune signals and metabolic programs. We propose that regulation of T-cell immunity by MAPK/Erk is a basic and sophisticated strategy that evolved before the emergence of the tetrapod lineage. These findings shed light on the evolution of the adaptive immune system.
Introduction
Protein kinases comprise one of the largest protein families in the mammalian genome (1, 2). Serine/threonine protein kinase mitogen-activated protein kinase (MAPK)3 and its MAPK kinase (MAPKK) and MAPK kinase (MAPKKK), constitute an evolutionarily conserved MAPK pathway (3, 4). MAPK signaling responds to a broad range of extracellular signals from cytokines, growth factors, and environmental stresses (5, 6). Upon extracellular stimulation, activation of MAPK signaling involves a classical core triple kinase cascade, in which MAPKKK activates MAPKK by phosphorylating two serine residues, and MAPKK in turn phosphorylates and activates MAPK (7). Phosphorylated MAPK can then activate a wide array of downstream molecules, including transcription factors and protein kinases, which facilitates target gene transcription (7,–9). Meanwhile, MAPK signaling is subject to distinct spatiotemporal regulation by cross-talk and feedback mechanisms and can also cooperate with other pathways to control gene transcription (10,–12). Therefore, the MAPK cascade acts as an important integrator of signal transduction, which is crucial for many physiological and pathological processes (13, 14), such as cell proliferation, differentiation, apoptosis, cancer, and almost all aspects of immune responses (3, 14, 16,–18). Three major groups of MAPKs, namely extracellular signal-regulated kinase 1/2 (Erk1/2), P38, and c-Jun N-terminal kinase (JNK), have been identified in mammalian cells (19,–21). Among them, the MAPK/Erk axis, which is composed of Erk1/2 (MAPK), Mek1/2 (MAPKK), and Raf (MAPKKK), is the most evolutionarily ancient and well-characterized MAPK signaling pathway (22).
MAPK/Erk signaling has been implicated in many biological processes related to T-cell immunity (7, 18, 23,–25). Upon engagement of the T-cell receptor (TCR), phosphorylation of lymphocyte-specific protein tyrosine kinase (LCK) and ζ-chain–associated protein kinase 70 (Zap-70) promotes formation of the linker for activation of T cells (LAT) signalosome and activation of phospholipase Cγ1 (PLCγ1). Activated PLCγ1 hydrolyzes the membrane phospholipid phophatidylinositol 4,5-biphosphate into two second messengers, inositol 1,4,5-triphosphate and diacylglycerol (26, 27). Diacylglycerol then allosterically recruits RasGRP1, a guanine nucleotide exchange factor, which in turn acts on Ras to activate the three-tier MAPK cascade, Raf-Mek-Erk (26, 27). Activation of Erk results in transcriptional activation of several important factors, including AP1 (c-Fos/c-Jun), ETS, Elk1, STAT3, and c-Myc, which are crucial for T-cell immunity (18, 23). A study using Erk1-deficient mice demonstrated that MAPK/Erk signaling is required for positive selection of thymocytes (28). Meanwhile, although somewhat controversial, the activity of the MAPK/Erk pathway is believed to be essential for differentiation of both IFN-γ–producing Th1 cells and IL-4/13–producing Th2 cells (29, 30). In addition, the MAPK/Erk pathway regulates several other T-cell processes, such as proliferation, growth, and survival; failure of regulation of these processes caused by a dysfunctional MAPK/Erk signaling is strongly associated with various diseases (4). In recent years, striking advances have been made in our understanding of the regulation of T-cell metabolism by MAPK/Erk signaling. In activated T cells, transcriptional activation of c-Myc and hypoxia-inducible factor-1α (HIF-1α) that act downstream of MAPK/Erk signaling is critical for metabolic reprogramming from oxidative phosphorylation to glycolysis, which ensures the proper activation, proliferation, and effector function of T cells (31,–34). Therefore, the sophisticated regulation of MAPK/Erk signaling is of paramount importance for T-cell immunity in mammals.
Although previous studies have explored the regulatory role of MAPK signaling in T-cell immunity using mammalian models, whether this signaling performs similar functions in early vertebrates remains unclear. From an evolutionary viewpoint, fish were the first group of organisms to evolve primordial T cells, ~450 million years ago; hence, they are ideal species for studying the origin and evolution of the T-cell lineage. Recently, several suspected T-cell subsets, including CD3+, CD8+, CD4-1+, CD4-2+, and TCRγδ+ leukocytes, have been identified in a variety of bony fish (35,–39). Additionally, evidence suggests that CD8+ or CD4+ leukocytes in bony fish perform preliminary immune functions equivalent to cytotoxic or helper T cells in mammals. For example, fish CD8+ leukocytes are cytotoxic and provide efficient protection against secondary infection (35, 36, 40), whereas fish CD4+ leukocytes have potential Th1, Th2, or Th17 cytokine responses during pathogen infection or antigen stimulation (36, 38, 41). However, at present, the regulatory mechanisms of immune response mediated by ancestral T cells in fish remain to be fully elucidated. In previous studies, we found that mTORC1-driven metabolic reprogramming and Ca2+-calcineurin axis–controlled NFAT nuclear translocation are indispensable for teleost T-cell activation, proliferation, and function in a Nile tilapia (Oreochromis niloticus) model (42, 43). These results indicate that ancestral T cells have already developed sophisticated regulatory strategies that are still used in modern T cells. In addition, we identified two components of MAPK/Erk signaling, h-Ras and c-Raf, and revealed their potential involvement in the antibacterial adaptive immune response of teleost (44, 45). Herein, using the same teleost model, we established the complete MAPK/Erk signaling pathway and explored its regulatory roles in the activation, expansion, and effector function of primordial T cells in fish. The findings provide a novel perspective on the regulation of T-cell immunity in an early vertebrate, which is of fundamental importance for understanding evolution of the adaptive immune system.
Results
Nile tilapia possesses an evolutionarily conserved MAPK/Erk signaling pathway
In our previous work, homologues of the h-Ras and the MAPKKK c-Raf were identified from the genomic sequences of Nile tilapia (44, 45). Here, a deep search of the Nile tilapia genome indicated that in addition to h-Ras and c-Raf, the MAPKK Mek1/2 and the MAPK Erk1/2 are encoded in this early vertebrate (Fig. 1A), suggesting the presence of a potentially intact MAPK/Erk signaling pathway in Nile tilapia. Similar to c-Raf, both Mek1/2 and Erk1/2 contain a central serine/threonine protein kinase catalytic (S/TKc) domain (Fig. 1A) that is characteristic of typical serine/threonine protein kinases. Nile tilapia h-Ras and c-Raf are evolutionarily conserved (44, 45), and they share high similarity in domain organization with homologues from mice (Fig. 1, B and C). Accordingly, functional S/TKc domains and activation segments in Nile tilapia Mek1/2 and Erk1/2 proteins are organized in similar ways to their mouse homologues (Fig. 1, D and E).

Evolutionary conservation of MAPK/Erk signaling pathway in Nile tilapia. A, domain prediction of MAPK/Erk pathway elements in Nile tilapia. B–E, comparison of the domain organization of MAPK/Erk pathway elements in Nile tilapia and mice. F, multisequence alignment analysis of the S/TKc domain from Nile tilapia Erk2 with homologs in other animals. Amino acid residues with 100% identity are shown in blue. The four conserved motifs are boxed. 1, the Gly-rich region (GXGXXG) is an ATP-binding loop; 2, the AXK motif could couple with the ATP phosphate group; 3, the HRDXK motif is the catalytic loop; 4, the activation segment (starting with DFG, ending with APE or SPE) of almost all protein kinases. G, prediction of the tertiary structures of Mek1/2 and Erk1/2 from Nile tilapia and mouse by SWISS-MODEL. H, phylogenetic trees constructed with the amino acid sequences of Erk1/2 from the indicated species. The tree was constructed using the neighbor-joining algorithm with the Mega 4.1 program based on multiple sequence alignment by ClustalW. Bootstrap values of 1000 replicates (percentages) are indicated for the branches. The accession numbers of selected sequences are listed in Table S1.
Using multiple-sequence alignment, we revealed that amino acid sequences in Nile tilapia Mek1/2 and Erk1/2, especially in the protein kinase domain, are highly conserved throughout all vertebrates (Fig. S1 and Fig. 1F). Moreover, several vital sites associated with Mek1/2 or Erk1/2 function are present in Nile tilapia, including the ATP-binding loop GXGXXG, the AXK motif, the catalytic K/D/D motif, and two serine residues phosphorylated by upstream kinases (Fig. S1 and Fig. 1F). Furthermore, predicted tertiary structures of Nile tilapia Mek1/2 and Erk1/2, including four residues in the regulatory spine of Mek1/2, are well-conserved with their mouse homologues (Fig. 1G).
To explore the evolutionary relationships of Mek1/2 and Erk1/2, we constructed phylogenetic trees using amino acid sequences. Meks from vertebrates and invertebrates cluster separately, and STE7 (a Mek homologue in yeast) is an outgroup distinct from animal Meks (Fig. S2). Within the cluster comprising vertebrate and invertebrate Meks, Mek1s and Mek2s are well-separated from each other. Both Mek1 and Mek2 from Nile tilapia are clustered with their homologues from other teleosts (Fig. S2), suggesting a close evolutionary distance of Mek1/2 proteins among teleosts. The same is true for Erk1/2; vertebrate Erk1 and Erk2 first form a sister group, which in turn clusters with invertebrate Erk2 (Fig. 1H). However, very few sequences of invertebrate Erk1 are available in the public GenBankTM database. Because Nile tilapia h-Ras and c-Raf are also highly conserved with their mammalian homologues (44, 45), our results suggest that Nile tilapia possesses an intact and evolutionarily conserved MAPK/Erk signaling pathway, which might perform similar functions to that in higher vertebrates.
Nile tilapia MAPK/Erk axis is activated in the primary response of adaptive immunity
To further explore the putative MAPK/Erk signaling pathway in Nile tilapia, we next determined in which tissues/organs this signaling occurs. Using qPCR, Mek1/2 or Erk1/2 mRNA was found to be expressed widely, albeit at varying levels, in all tested tissues, including spleen, head kidney, peripheral blood, trunk kidney, liver, muscle, intestine, and gill (Fig. 2, A–D). Mek1 transcript level was extremely high in muscle, intestine, and gill, but relatively low in spleen (Fig. 2A). Expression of Mek2 was highest in gill and lowest in spleen and intestine (Fig. 2B). Similar to Mek1/2, Erk1/2 mRNA was present at low levels in spleen, but at relatively high levels in gill (Fig. 2, C and D). Interestingly, transcripts of Mek1 and Erk1 were abundant in intestine, but expression of Mek2 and Erk2 was quite low in this tissue (Fig. 2, A–D). The broad expression patterns of Nile tilapia Mek and Erk in tissue were also confirmed at the protein level (Fig. 2, E–G). Interestingly, although mRNA expressions of Mek1/2 and Erk1/2 were quite different in each tissue, there was not much difference in their protein contents among these tissues (Fig. 2, E–G). Together, these results reveal that MAPK/Erk components are broadly expressed in different tissues of Nile tilapia, indicating a potentially wide involvement of this signaling cascade in cellular processes in this bony fish.

MAPK/Erk signaling is involved in the adaptive immune response of Nile tilapia. A–D, Mek1 (A), Mek2 (B), Erk1 (C), and Erk2 (D) expression in the indicated tissues was determined by qPCR (n = 6). E, Mek1/2 and Erk1/2 expression in the indicated tissues was determined by Western blotting. F and G, quantification of Mek1/2 (F) and Erk1/2 (G) expression by calculating the densitometry of target protein/β-actin, n = 3. H–J, Nile tilapia were i.p. injected with A. hydrophila or PBS, and spleen lymphocytes were harvested on the indicated days after infection. H–I, relative mRNA levels of Mek1/2 (H) and Erk1/2 (I) in lymphocytes with or without infection were examined by qPCR (n = 6). J, phosphorylation and total protein level of Mek1/2 or Erk1/2 was examined using the indicated antibodies in lymphocytes with or without A. hydrophila infection. K and L, Nile tilapia were i.p. injected with S. agalactiae or PBS, and spleen lymphocytes were harvested on the indicated days after infection. K, relative mRNA levels of h-Ras, c-Raf, Mek1/2, and Erk1/2 in lymphocytes with or without infection were examined by qPCR (n = 4). L, phosphorylation and total protein levels of Mek1/2 or Erk1/2 were examined using the indicated antibodies in lymphocytes with or without S. agalactiae infection. The experiments in A–D, H and I, and K were repeated two independent times, and experiments in E–G, J, and L were repeated three independent times. *, p < 0.05; **, p < 0.01, determined by two-tailed Student's t test. Error bars, S.E.
To determine whether MAPK/Erk signaling participates in adaptive immune responses in Nile tilapia, we challenged fish with the Gram-negative bacterial pathogen Aeromonas hydrophila. Mek1/2 and Erk1/2 expression in spleen leukocytes was examined during the adaptive immune stage. Although transcripts of Mek1/2 and Erk1/2 were relatively scarce in spleen, their mRNA expression levels were dramatically increased in spleen leukocytes at 5 or 8 days post-infection (DPI; Fig. 2, H and I). The similar temporal expression patterns for Mek1/2 and Erk1/2 imply potentially cooperative roles in regulating adaptive immune responses during bacterial infection in Nile tilapia. Because phosphorylation of Mek1/2 and Erk1/2 is crucial for MAPK/Erk signaling activation and downstream function, we examined protein phosphorylation during bacterial infection in Nile tilapia. Phosphorylation but not total protein of Mek1/2 and Erk1/2 was dramatically enhanced in spleen leukocytes at 3, 5, or 8 DPI compared with uninfected controls (Fig. 2J), indicating the activation of MAPK/Erk signaling during the anti-bacterial immune response. The inducible activation of MAPK/Erk signaling is not specific for Gram-negative infection, because the same was true in fish infected with the Gram-positive bacterium Streptococcus agalactiae. Gram-positive infection resulted in a substantial increase mRNA expression of h-Ras, c-Raf, Mek1/2, and Erk1/2 (Fig. 2K), as well as a dramatic enhancement in phosphorylation but not total protein of Mek1/2 and Erk1/2 in spleen leukocytes during the primary response (Fig. 2L). Overall, these observations suggest that Nile tilapia MAPK/Erk signaling is robustly activated in the primary response of adaptive immunity during bacterial infection.
MAPK/Erk signaling is required for proper T-cell activation in Nile tilapia
Next, we sought to determine how MAPK/Erk signaling participates in adaptive immune responses of Nile tilapia. Activation is a prerequisite to allow lymphocytes to implement adaptive immunity. Because MAPK/Erk signaling was sharply activated in spleen leukocytes during the primary immune response, we first examined the potential involvement of this pathway in lymphocyte activation in vitro. Compared with unstimulated controls, mRNA expression levels of almost all MAPK/Erk components, including h-Ras, c-Raf, Mek1, Erk1, and Erk2, were markedly increased in spleen lymphocytes upon activation by phorbol 12-myristate 13-acetate (PMA) plus ionomycin (Fig. 3A). Furthermore, our previous work (44, 45) and the present results revealed a significant up-regulation of these components when spleen leukocytes were activated by the T cell–specific mitogen PHA (Fig. 3B). These results therefore indicate a tight association between transcriptional up-regulation of MAPK/Erk signaling and proper activation of T cells in Nile tilapia.

MAPK/Erk pathway regulates T-cell activation in Nile tilapia. A and B, Nile tilapia lymphocytes were stimulated with PMA plus ionomycin (A) or PHA (B) in vitro for the indicated time. Relative mRNA levels of the indicated molecules in lymphocytes with or without stimulation were examined by qPCR (n = 4). C–F, Nile tilapia lymphocytes were stimulated or unstimulated with PMA plus ionomycin in vitro. C, immunoblotting analysis showing total protein or phosphorylation levels of the indicated MAPK/Erk pathway elements in lymphocytes with or without stimulation. D, quantification of c-Raf, Mek1/2, and Erk1/2 phosphorylation compared with their total protein, n = 3. E and F, immunofluorescence analysis showing phosphorylation of Mek1/2 (E) and Erk1/2 (F) with or without stimulation. G and H, Nile tilapia individuals were treated with Mek-specific inhibitor trametinib for 2 consecutive days before spleen lymphocytes were harvested for stimulation in vitro. G, lymphocytes were treated with PMA plus ionomycin for examination of phosphorylated Erk1/2 by immunoblotting. H, relative mRNA level of IFN-γ after PHA stimulation in lymphocytes by qPCR (n = 6). I and J, Nile tilapia individuals were treated with Erk-specific inhibitor SCH772984 for 2 consecutive days before spleen lymphocytes were stimulated by PMA plus ionomycin (I) or PHA (J) in vitro. I, Nile tilapia individuals were i.p. injected with Erk-specific inhibitor SCH772984 during S. agalactiae infection, and the spleen lymphocytes were isolated for assay on day 7 after infection. The phosphorylated p90RSK after PMA plus ionomycin stimulation in vitro or S. agalactiae infection in vivo was examined by immunoblotting. J, relative mRNA level of IFN-γ and CD122 after PHA stimulation in lymphocytes by qPCR (n = 6). The experiments in A and B and E–J were repeated two independent times, and experiments in C and D were repeated three independent times. *, p < 0.05; **, p < 0.01; ***, p < 0.001, determined by a two-tailed Student's t test. Error bars, S.E.
At the protein level, we found that expression of c-Raf, Mek1/2, or Erk1/2 was not increased after spleen leukocytes were activated by PMA plus ionomycin (Fig. 3C). However, phosphorylation of all three kinases (c-Raf, Mek1/2, and Erk1/2) was dramatically enhanced upon lymphocyte activation (Fig. 3, C and D). Elevated phosphorylation of Mek1/2 and Erk1/2 was also confirmed by confocal microscopy at 15 min after lymphocytes were stimulated by PMA plus ionomycin (Fig. 3, E and F). To further investigate the regulatory role of MAPK/Erk signaling in T-cell activation, we used the Mek1/2 inhibitor trametinib or the Erk1/2 inhibitor SCH772984 to suppress signaling activity in Nile tilapia. Trametinib effectively blocked MAPK/Erk signaling activation, as revealed by impaired Erk1/2 phosphorylation enhancement upon lymphocyte activation induced by PMA plus ionomycin (Fig. 3G). Additionally, blockade of MAPK/Erk signaling abrogated up-regulation of IFN-γ upon PHA-induced T-cell activation (Fig. 3H). Similarly, SCH772984 suppressed MAPK/Erk signaling activity, as shown by a lack of up-regulation of p90RSK phosphorylation in spleen leukocytes upon PMA plus ionomycin stimulation or S. agalactiae infection (Fig. 3I). SCH772984 also impaired the inducible expression of IFN-γ and CD122 upon T-cell activation (Fig. 3J). In summary, these results suggest that MAPK/Erk signaling is an indispensable regulatory center for T-cell activation in Nile tilapia.
Nile tilapia employs a classical three-tier kinase module to activate MAPK/Erk signaling
Activation of MAPK signaling is achieved through a core triple kinase cascade, MAPKKK-MAPKK-MAPK, which is specific to Raf-Mek1/2-Erk1/2 for MAPK/Erk signaling (5, 9). As stated above, our teleost model demonstrated a concordant phosphorylation of the three kinases in MAPK/Erk signaling upon bacterial infection or lymphocyte activation. To clarify whether this three-tier kinase cascade is absolutely applied for early vertebrates, we treated Nile tilapia spleen leukocytes with inhibitors of Raf, Mek, or Erk before PMA plus ionomycin stimulation was executed. Treatment with the Raf inhibitor dabrafenib did not impair c-Raf phosphorylation during lymphocyte activation but resulted in a decreased Mek1/2 phosphorylation (Fig. 4, A and B), suggesting that c-Raf is a potential upstream kinase of Mek in Nile tilapia. However, dabrafenib had a more subtle effect on Erk1/2 phosphorylation (Fig. 4, A and B). By contrast, the Mek inhibitor trametinib did not suppress the phosphorylation of Mek1/2 but severely impaired Erk1/2 phosphorylation (Figs. 3G and and44 (C and D)), suggesting that Mek functions as a kinase upstream of Erk in Nile tilapia. Interestingly, treatment of Mek inhibitor also reduced the phosphorylation of c-Raf (Fig. 4, C and D). Similarly, the Erk inhibitor SCH772984 not only prevented the phosphorylation of Erk and its downstream p90RSK (Figs. 3I and and44 (E and F)), but also impaired the phosphorylation of c-Raf (Fig. 4, E and F). The defects in Erk phosphorylation induced by Mek or Erk inhibition both caused an impairment in c-Raf phosphorylation (Fig. 4, C–F), indicating a potential feedback regulation of Erk to c-Raf in Nile tilapia. Together, our results suggest that Nile tilapia employs a classical three-tier kinase module to activate MAPK/Erk signaling.

Nile tilapia MAPK/Erk pathway functions by classical tertiary kinase cascade. A, C, and E, Nile tilapia individuals were i.p. injected with Raf-specific inhibitor dabrafenib (A), Mek-specific inhibitor trametinib (C), or Erk-specific inhibitor SCH772984 (E) for 2 consecutive days before spleen lymphocytes were isolated. Lymphocytes from inhibitor-treated or -untreated Nile tilapia were stimulated by PMA plus ionomycin for 15 min, and the phosphorylation levels of c-Raf, Mek1/2, and Erk1/2 were detected by immunoblotting. B, D, and F, quantification of c-Raf (B), Mek1/2 (D), and Erk1/2 (F) phosphorylation compared with their total protein in the indicated groups, n = 3. The experiments in A–F were repeated three independent times. Error bars, S.E.
Nile tilapia MAPK/Erk signaling ensures T-cell expansion during bacterial infection
Following antigen-induced activation, lymphocytes undergo a prompt and robust clonal expansion. Therefore, we sought to establish whether MAPK/Erk signaling is associated with lymphocyte expansion in Nile tilapia. Fish were treated with the Mek inhibitor trametinib during A. hydrophila infection. At 7 DPI, bacterial infection or Mek inhibition did not change the frequency of lymphocytes among spleen leukocytes compared with that in the control group (Fig. 5, A and B). However, the total number of spleen leukocytes was significantly increased following A. hydrophila infection (Fig. 5C) and thus caused a substantial increase of spleen lymphocytes in bacterially infected fish compared with negative controls (Fig. 5, D and E). Once MAPK/Erk signaling was inhibited, the robust expansion of spleen leukocytes or lymphocytes was severely impaired, as revealed by a decrease in the number of spleen leukocytes or lymphocytes compared with the uninhibited group (Fig. 5, C–E). To confirm the regulatory role of MAPK/Erk signaling in lymphocyte proliferation, we injected BrdU to the fish 1 day before scarification. At 5 DPI, the frequency of BrdU+ lymphocytes was dramatically increased in infected fish compared with uninfected controls (Fig. 5F). However, the increase of BrdU+ lymphocytes was sharply impaired once MAPK/Erk activity was blocked (Fig. 5F). The same result was obtained when we examined BrdU incorporation in Nile tilapia during infection by S. agalactiae (Fig. 5G). These observations suggest that MAPK/Erk signaling is crucial for lymphocyte expansion during anti-bacterial adaptive immune responses in Nile tilapia.
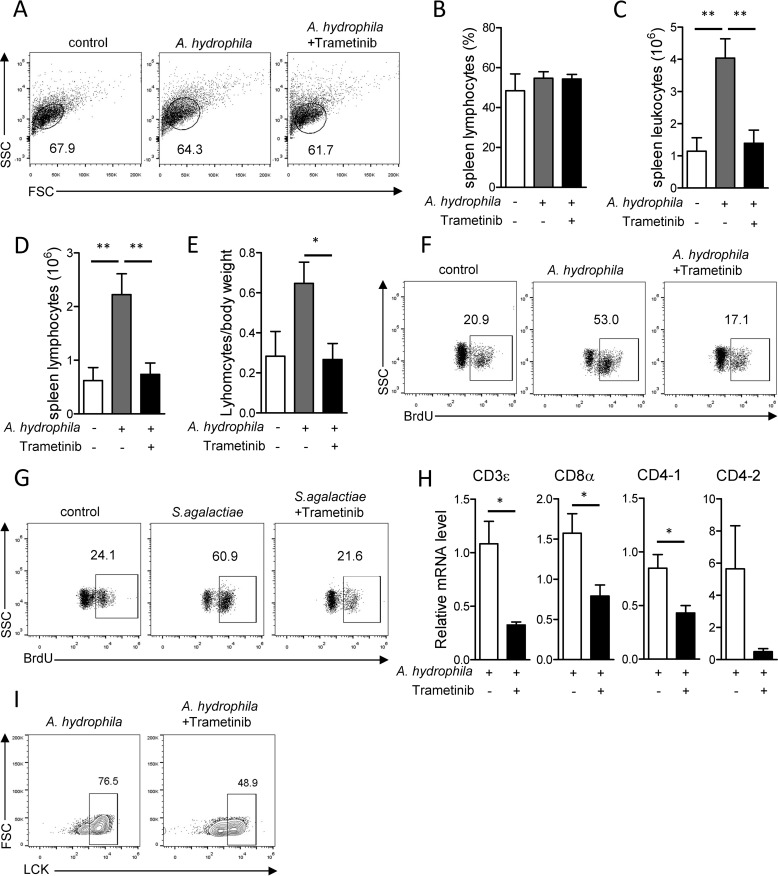
MAPK/Erk signaling is indispensable for pathogen-induced T-cell proliferation in Nile tilapia. Nile tilapia individuals were i.p. injected with Mek-specific inhibitor trametinib on days 2, 3, 4, and 6 after A. hydrophila (A–F, H, and I) or S. agalactiae (G) infection. Spleen lymphocytes were harvested on the indicated days for the assay. A–E, representative dot plots (A), lymphocyte percentage within spleen leukocytes (B), absolute number of spleen leukocytes (C), absolute number of spleen lymphocytes (D), and spleen weight per body weight (E) on day 8 after infection (n = 5–7). F and G, animals were i.p. injected with BrdU 1 day before sacrifice. BrdU incorporation into lymphocytes was examined by flow cytometry on day 5 after A. hydrophila (F) or S. agalactiae (G) infection. H, qPCR revealed the mRNA levels of the indicated genes in lymphocytes with or without trametinib treatment on day 8 after infection (n = 6). I, representative plots show frequencies of LCK+ lymphocytes in spleen on day 8 after infection. The experiments in A–E were repeated three independent times, and experiments in F–I were repeated two independent times. *, p < 0.05; **, p < 0.01, determined by a two-tailed Student's t test. Error bars, S.E.
Bacterial infection activates T cell–mediated immune responses and induces mRNA expression of TCR and co-receptors in Nile tilapia (42). Here, we found that during A. hydrophila infection, the relative expression levels of T-cell surface markers, including CD3ϵ, CD8α, CD4-1, and CD4-2, were substantially decreased in spleen leukocytes upon blockade of MAPK/Erk signaling by trametinib (Fig. 5H); this is suggestive of a potential regulatory role for MAPK/Erk signaling in expansion of the T-cell lineage in Nile tilapia. To confirm the effect of MAPK/Erk activity on T-cell proliferation, we used an LCK antibody that could specifically identify T cells in Nile tilapia (42) to trace the T-cell lineage during A. hydrophila infection. At 7 DPI, blockade of MAPK/Erk signaling markedly reduced the frequency of T cells, as revealed by the LCK+ cells within the lymphocyte population of spleen (Fig. 5I). Therefore, our results suggest that MAPK/Erk signaling is required for proper expansion of T cells during bacterial infection in Nile tilapia.
MAPK/Erk signaling is essential for T cell–mediated elimination of infection
Accompanied by expansion, T cells, especially cytotoxic T cells, are differentiated into effector cells that produce large quantities of cytotoxic cytokines and other molecules to eliminate infected cells. Because Nile tilapia requires MAPK/Erk signaling for proper activation and proliferation of T cells, we wondered whether this pathway plays a regulatory role in effector function during the primary response. Seven days after A. hydrophila infection, mRNA expression levels of the cytotoxic gene granzyme B, the pro-inflammatory cytokines IFN-γ and LT-α, and the pro-apoptotic molecule FasL were up-regulated in spleen leukocytes compared with the control group (Fig. 6, A–C). The inducible expression of these cytotoxic genes was significantly impaired in spleen leukocytes lacking MAPK/Erk signaling activity (Fig. 6, A–C). Moreover, mRNA up-regulation of both KLRG1 and T-bet, an important cell surface marker and transcription factor in effector CD8+ T cells, respectively, was coincidentally suppressed by inhibition of MAPK/Erk activity (Fig. 6, D and E). According to these results, MAPK/Erk signaling is associated with effector T-cell function in Nile tilapia. This finding was further confirmed by the inability of MAPK/Erk-inhibited fish to control bacterial infection, with a higher bacterial burden in spleen relative to uninhibited controls (Fig. 6F). Furthermore, loss of effector function caused by MAPK/Erk signaling inhibition dramatically increased the mortality of fish following infection with the high virulent pathogen S. agalactiae (Fig. 6G). Together, these observations suggest that MAPK/Erk signaling is essential for Nile tilapia effector lymphocytes to be able to eliminate bacterial infection.
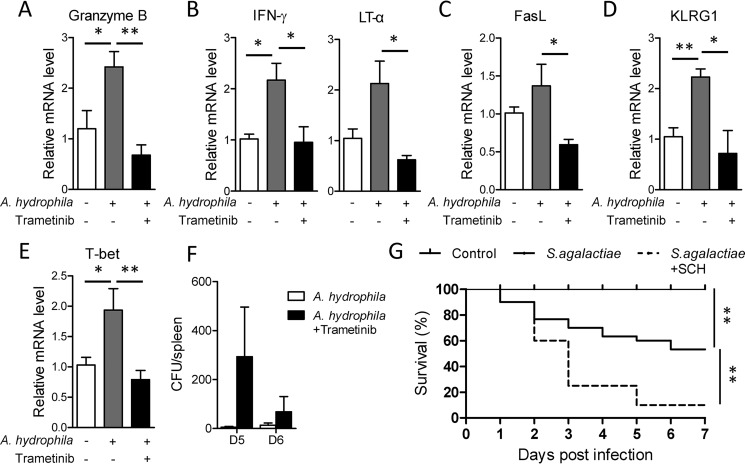
Critical role of MAPK/Erk for Nile tilapia effector cells to eliminate infection. A–F, Nile tilapia individuals infected with A. hydrophila were i.p. injected or not with trametinib on days 2, 3, 4, and 6 post-infection. A–E, relative mRNA levels of the indicated molecules in spleen lymphocytes with or without trametinib treatment on day 8 after infection (n = 6). F, A. hydrophila titers in the liver on day 5 after infection (n = 4). G, Kaplan–Meyer survival plot showing the survival percentage of Nile tilapia with or without SCH772984 treatment after S. agalactiae infection, n = 18. The experiments in A–F were repeated three independent times, and experiments in G were repeated four independent times. *, p < 0.05; **, p < 0.01, determined by a two-tailed Student's t test. Error bars, S.E.
Nile tilapia MAPK/Erk axis promotes T-cell immunity via c-Myc–mediated glycolysis
Upon activation and proliferation, T cells undergo dynamic metabolic switching from oxidative phosphorylation and fatty acid β-oxidation to glycolysis and lipogenesis, to meet the high metabolic requirements of effector T cells (46,–48). In our previous study, we found that such metabolic reprogramming occurs in Nile tilapia T cells during their activation (42). Because the proto-oncogene c-Myc, a transcription factor downstream of MAPK/Erk signaling, is a sophisticated regulator of glycolysis in mammals (31, 49, 50), we are tempted to investigated the potential role of MAPK/Erk activity in T-cell metabolism of fish. In support of our previous finding (42), mRNA expression of glucose transporter 1 (Glut1) was significantly up-regulated upon PHA-induced T-cell activation (Fig. 7A); however, this inducible expression of Glut1 was dramatically impaired in MAPK/Erk-inhibited spleen leukocytes (Fig. 7A). Additionally, impaired Glut1 expression was correlated with defective glucose uptake in spleen effector lymphocytes lacking MAPK/Erk activity (Fig. 7B). Meanwhile, mRNA levels of glycolysis genes, including Glut1, hexokinase 2 (HK2) and phosphofructokinase (pfk), but not pyruvate kinase (pkm), were obviously reduced during primary immune response when MAPK/Erk was inhibited by trametinib (Fig. 7C). Moreover, the enzyme activity of rate-limiting enzyme pfk was concordantly impaired by MAPK/Erk signaling blockade (Fig. 7D). These results collectively reveal that MAPK/Erk signaling coordinates glycolysis programming during T-cell activation in Nile tilapia.
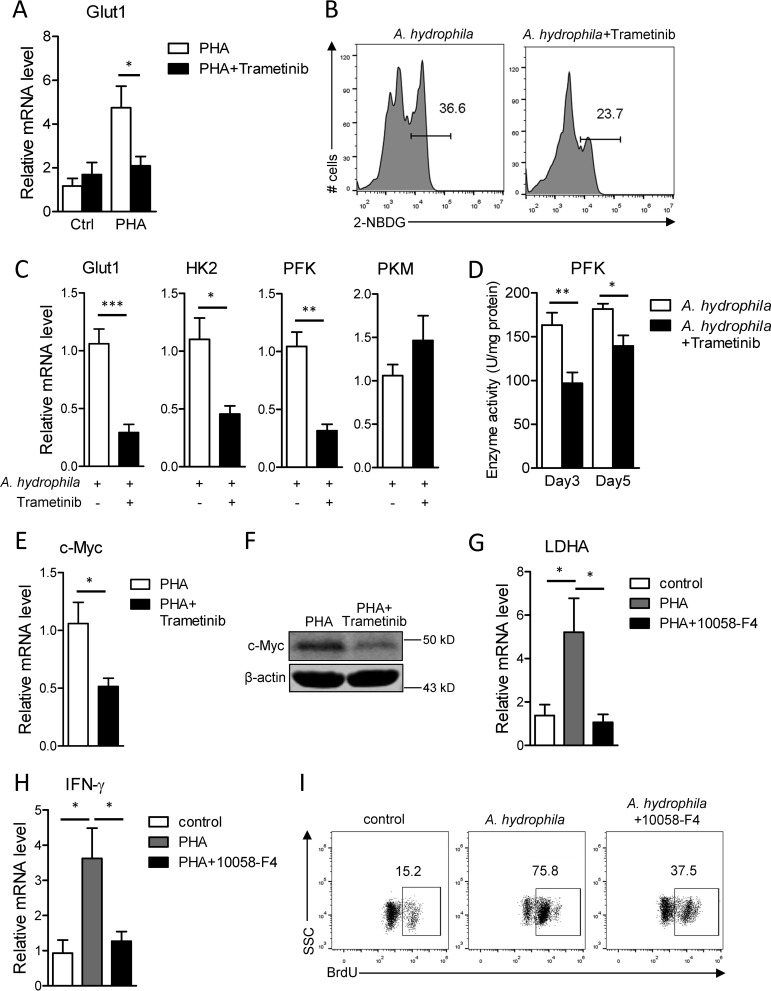
MAPK/Erk pathway regulates T-cell function through c-Myc–controlled glycolysis. A, Nile tilapia individuals were treated with trametinib for 2 consecutive days before spleen lymphocytes were stimulated by PHA for 12 h in vitro. Relative mRNA levels of Glut1 in lymphocytes were examined by qPCR (n = 6). B–D, Nile tilapia individuals infected with A. hydrophila were i.p. injected with trametinib on days 2, 3, 4, and 6 post-infection, and animals were sacrificed on the indicated days for assay. B, glucose uptake, as measured by 2-NBDG labeling in gated spleen lymphocytes on day 8 after infection. C, relative mRNA levels of the indicated genes in lymphocytes were examined by qPCR on day 5 after infection (n = 6–8). D, activities of enzyme pfk in liver with or without trametinib treatment on days 3 and 5 post-infection (n = 6). E and F, Nile tilapia individuals were treated with trametinib for two consecutive days before spleen lymphocytes were stimulated by PHA for 12 h in vitro. Relative mRNA (E) or protein (F) levels of c-Myc in lymphocytes were examined by qPCR (n = 6) or Western blotting. G and H, Nile tilapia individuals were treated with c-Myc–specific inhibitor 10058-F4 for 2 consecutive days before spleen lymphocytes were stimulated by PHA for 12 h in vitro. Relative mRNA levels of LDHA (E) and IFN-γ (F) in lymphocytes were examined by qPCR (n = 6). I, Nile tilapia individuals infected with A. hydrophila were i.p. injected with 10058-F4 on days 2, 3, and 4 post-infection, and animals were i.p. injected with BrdU 1 day before sacrifice. BrdU incorporation in lymphocytes was examined by flow cytometry on day 5 after A. hydrophila infection. The experiments in A–I were repeated two independent times. *, p < 0.05; **, p < 0.01 determined by a two-tailed Student's t test. Error bars, S.E.
Metabolic reprogramming during T-cell activation is controlled by several key transcription factors, among which c-Myc is one of the earliest elevated factors responsible for initiating T-cell activation–induced glycolysis (31, 50). PHA-induced T-cell activation elevates c-Myc expression in Nile tilapia (42). Here, we found that during T-cell activation, both mRNA and protein levels of c-Myc were obviously reduced when MAPK/Erk signaling was blocked (Fig. 7, E and F). This is indicative of a potential association between c-Myc expression and T-cell activation–induced glycolysis. To confirm this association, we treated fish with the c-Myc inhibitor 10058-F4 before spleen leukocytes were activated by PHA. Inhibition of c-Myc by 10058-F4 impaired the elevation of LDHA (Fig. 7G), an important enzyme that catalyzes the conversion of pyruvate into lactate during glycolysis. This finding corroborates that c-Myc plays a regulatory role in glycolysis of Nile tilapia. Moreover, blockade of c-Myc activity strongly inhibited PHA-induced T-cell activation (Fig. 7H) and caused a dramatic defect in lymphocyte expansion during bacterial infection (Fig. 7I). Therefore, our results suggest that MAPK/Erk signaling promotes T-cell immunity via c-Myc–mediated glycolysis programming in Nile tilapia.
Altogether, in a fish model, upon TCR engagement and downstream guanine nucleotide exchange factor activation, Ras is activated and triggers the phosphorylation of the Raf-Mek-Erk cascade. Phosphorylated Erk1/2 then induces the expression of downstream transcription factor c-Myc, which in turn promotes glycolysis in T cells to ensure their proper activation and subsequent expansion. Meanwhile, MAPK/Erk activity is also indispensable for effector T cells to exert their cytotoxic function during infection elimination (Fig. 8).
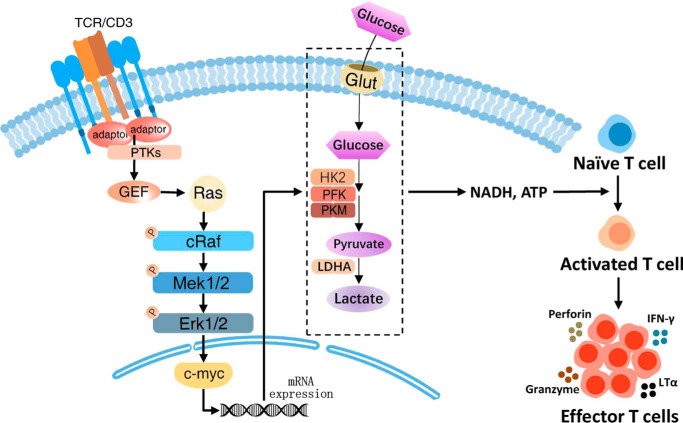
Teleost T cells require MAPK/Erk signaling and c-Myc–mediated metabolic programming for proper activation and function. In Nile tilapia, after native T cells encounter antigen, some adaptors and protein tyrosine kinases recruit guanine nucleotide exchange factors, and once Ras was activated by guanine nucleotide exchange factors, the MAPK/Erk cascade would turn on and phosphorylate the three protein kinases, c-Raf, MEK1/2, and Erk1/2. Then the downstream transcription factor c-Myc coordinates glucose uptake and glycolysis metabolic programs and ensures the proper activation and subsequent expansion of T cell. Meanwhile, Nile tilapia MAPK/Erk signaling is indispensable for effector T cells to exert cytotoxic function and eliminate infection.
Discussion
The MAPK cascade is one of the most ancient signal transduction pathways utilized by diverse biological systems and physiological processes. It is now acknowledged that the MAPK/Erk axis plays a central role in multiple immunological processes involving T cells, including maturation, activation, proliferation, differentiation, and function (18, 23,–25). Although the modulatory roles of MAPK/Erk signaling in adaptive immune responses have been elucidated in mice, little is known about this pathway in T-cell immunity in early vertebrates, especially fish. In the present study, we revealed that the early vertebrate Nile tilapia possesses an intact and evolutionarily conserved MAPK/Erk signaling pathway, which is activated through a classical three-tier kinase module. More importantly, we found that activity of the MAPK/Erk axis is crucial for activation, expansion, and function of ancestral T cells in the bony fish Nile tilapia via regulation of metabolic reprogramming. Given that the regulatory mechanisms of T-cell immunity in early vertebrates are still poorly understood, we anticipated that the present study would provide new insight into evolution of the adaptive immune system.
MAPK signaling is believed to be evolutionarily conserved throughout the plant and animal kingdoms, and many signaling components have been identified in several fish genome-sequencing projects. However, few studies, to date, have systematically analyzed the bioinformatics of MAPK/Erk signaling components in fish species. In our previous and present studies, we identified all key kinase components in the MAPK/Erk axis, including MAPKKK c-Raf, MAPKK Mek1/2, and MAPK Erk1/2, in the genome of bony fish Nile tilapia (45). As expected, primary structures, catalytic sites, functional domains, and tertiary structures of these kinases in the Nile tilapia MAPK/Erk signaling pathway are highly conserved throughout vertebrates. Additionally, the evolutionary conservation of the MAPK/Erk pathway in Nile tilapia was confirmed by phylogenetic trees for both Mek and Erk, in which the different vertebrate MAPKs or MAPKKs cluster with their corresponding orthologues. Moreover, using specific inhibitors, we found that the MAPK/Erk axis in Nile tilapia is activated through a classical three-tier kinase cascade following appropriate stimulation. Therefore, our sequence, structure, phylogenetics, and activation analyses provide strong evidence supporting the evolutionary conservation of the MAPK/Erk axis in vertebrates.
In mammals, MAPK/Erk signaling is usually associated with positive or negative feedback regulation (51, 52). Interestingly, we found that Mek or Erk inhibitor impaired the phosphorylation of Raf, indicating a potential positive feedback regulation of Erk via the Raf-Mek axis in Nile tilapia. In addition to positive activation and feedback regulation, MAPK signaling can be negatively modulated by various mechanisms, such as receptor desensitization, signaling complex dissociation, and kinase dephosphorylation (7). Phosphorylation of threonine residues Thr-286 and Thr-292 in mammalian Mek1 by Erk is reported to negatively regulate the formation of Mek1-Mek2 heterodimers and thus contribute to the inhibition of signaling activity (53,–56). Here, these two sites were found to be different between bony fish and mammal; thus, the negative regulation of MAPK/Erk signaling in Nile tilapia might be quite different from that in higher vertebrates. However, the exact mechanisms underpinning both feedback and negative regulation of the MAPK/Erk axis in bony fish remain a mystery and need to be further elucidated in the future.
It has been acknowledged that MAPK/Erk signaling plays an essential role in the regulation of anti-bacterial immune responses in both invertebrates and vertebrates. Erk-mediated immune signal transduction is of fundamental importance in innate immunity of many invertebrates, including oyster, shrimp, and crab (57,–59). During bacterial infection, phosphorylated oyster Erk interacts with Rel and induces its nuclear translocation, eventually promoting IL-17 and TNF transcription (57). In mice, Erk activation is crucial for phagocytosis by macrophages (60) and the production of TNF and IL-1β following Toll-like receptor stimulation (61, 62). The failure in up-regulation of IL-1β caused by Erk inhibition could increase the susceptibility of mice to Listeria monocytogenes (61). In the adaptive immune system, T cells, including both CD4+ helper T cells and CD8+ cytotoxic T cells, are the main weapons employed during the primary response. The activity of MAPKs differentially controls the differentiation of mammalian T-cell subsets; for example, JNK2 polarizes CD4+ T-cell differentiation into the Th1 lineage (18, 63). In addition, p38 is required for the generation of IFN-γ–producing effector CD8+ T cells (64). However, the regulatory roles of Erk in T-cell differentiation remain controversial. Some studies found that MAPK/Erk signaling contributes to Th2 cell differentiation, and inhibition of signaling activity by either a weak TCR signal, Erk inhibitor, or Mek inhibitor effectively reduces the production of IL-4 or IL-13 (29, 65, 66). By contrast, another study revealed that such signaling is indispensable for opposite-direction Th1 cell differentiation and IFN-γ production (30). In the present study, we showed that mRNA expression and phosphorylation of Mek1/2 and Erk1/2 in spleen lymphocytes were markedly up-regulated during the primary response in Nile tilapia after bacterial infection. Moreover, inhibition of MAPK/Erk signaling reduced levels of pro-inflammatory cytokines and cytotoxic genes, impaired infection clearance, and increased the overall susceptibility of Nile tilapia to infection. These findings suggest that the MAPK/Erk axis plays an indispensable regulatory role in the function of teleost effector T cells. Failed up-regulation of Nile tilapia IFN-γ and its transcription factor T-bet in the absence of MAPK/Erk highlights the potential regulatory role of this signaling in the differentiation of effector CD8 T cells or Th1 cells. However, owing to the lack of effective antibodies for Nile tilapia CD4 or CD8, we cannot currently elucidate the exact contribution of helper or cytotoxic T cells to IFN-γ production in this fish species.
The exquisite activation strategies of modern T cells have been well-illustrated in mammals. Downstream of TCR signals, DAG-RasGRP1-Ras axis–triggered MAPK/Erk signaling and its tightly regulated transcription factor AP-1 are crucial for proper T-cell activation, proliferation, and function (26, 67). Dysregulation or overexpression of MAPK/Erk signaling causes hyperactivation and overproliferation of T cells, which are highly associated with the development of several diseases (4). Herein, using a Nile tilapia model, we found that MAPK/Erk signaling was robustly activated upon T-cell activation, and blockade of the Erk pathway impaired PHA-induced T-cell activation. These results are suggestive of pivotal roles for the MAPK/Erk axis in activation of ancestral T cells in bony fish. Furthermore, this ancient signaling is essential for the proliferation of T-cell lineage in Nile tilapia, because the robust expansion of T cells was severely impaired during bacterial infection, when signaling activity was inhibited. To the best of our knowledge, these results provide the first evidence for the regulation of T-cell activation and proliferation by MAPK/Erk signaling in fish. From an evolutionary viewpoint, we propose that modulation of T-cell immunity by MAPK/Erk signaling might be an ancestral feature that already existed upon the appearance of bony fish 450 million years ago.
Emerging evidence indicates an important role of metabolic control for T-cell function and fate decision (47, 49). Pathways that control T-cell metabolism and function are intimately linked, and changes in cell metabolism have been shown to enhance or suppress specific T-cell functions (48, 49). Accompanied by activation and proliferation, T cells undergo a dynamic metabolic switching from oxidative phosphorylation and fatty acid β-oxidation to glycolysis and lipogenesis, to meet the high metabolic requirements of effect T cells for their rapid growth and division (26, 49, 68). Recent studies using a mouse model highlighted several signals as key regulators for T-cell metabolic reprogramming, such as mTOR, Ca2+-NFAT, and MAPK/Erk (15, 69, 70). Upon T-cell activation, MAPK/Erk signaling is critical for the switch from oxidative phosphorylation to glycolysis, because inhibition of Erk1/2 activity blocks glucose uptake, glycolysis, and the expression and activity of glycolytic enzymes (15). Furthermore, the Erk1/2 pathway promotes glycolysis by inducing downstream transcription factors, among which c-Myc, alone or in cooperation with HIF-1α, regulates several key molecules that contribute to glycolysis, including Glut1, HK2, pkm, and LDHA (31, 33, 34). Our previous study revealed multiple dynamic metabolic reprogramming in ancestral T cells in Nile tilapia (42). Herein, using the same fish model, we demonstrated that MAPK/Erk signaling is essential for glucose uptake and glycolysis during the activation of ancestral T cells. Mechanistically, MAPK/Erk signaling promoted glycolysis-driven T-cell immunity via the downstream transcription factor c-Myc, because inhibition of c-Myc reduced the glycolysis-related gene LDHA and, in turn, impaired the activation and proliferation of T cells in this species. Thus, we conclude that MAPK/Erk signaling-dependent metabolic reprogramming is fundamental to activation and proliferation of ancestral T cells in bony fish.
In summary, we identified an evolutionarily conserved MAPK/Erk pathway in the early vertebrate Nile tilapia and confirmed that it followed a classical three-tier kinase module to activate. In addition, we suggested that activation of MAPK/Erk signaling is of paramount importance for the proper activation, proliferation, and infection elimination of the ancestral T cells in bony fish. Mechanistically, the MAPK/Erk axis promotes glycolysis through the downstream transcription regulator c-Myc, which in turn regulates the activation and proliferation of T cells in this early vertebrate. Our results establish an intimate link between immune signals and metabolic reprogramming in teleost. This study advances our knowledge about the regulatory mechanisms of MAPK/Erk signaling on T-cell immunity in fish species and therefore fills in an important gap regarding evolution of the adaptive immune system.
Experimental procedures
Experimental animals
Healthy Nile tilapia O. niloticus about 6–7 cm in length were purchased from an aquaculture farm in Guangzhou, Guangdong Province, China, and then temporarily cultured in a 90-liter glass tank at 28 °C with continuous aeration and daily feeding. They were used for the experiment at least 2 weeks later. All experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China. The experimental procedures were approved by the East China Normal University Experimental Animal Ethics Committee.
Sequence, structure, and phylogenic analysis
We searched for the related cDNA or amino acid sequences of Nile tilapia in the released whole-genome sequence on NCBI GenBankTM, Ensemble, or KEGG websites and analyzed them using the BLAST algorithm. The potential functional domains of the amino acids were predicted by SMART version 4.0, and domain organization was displayed with DOG version 2.0 software. The analysis of multiple-sequence alignment was performed using ClustalW. The protein tertiary structure was predicted by the SWISS-MODEL server on ExPASy and displayed using PyMOL software. The phylogenetic tree was constructed by MEGA4, using the neighbor-joining method. The corresponding accession numbers are listed in Tables S1 and S2.
Bacterial infection
S. agalactiae or A. hydrophila was cultured in lysogeny broth or brain heart infusion broth to the exponential growth phase and then resuspended in PBS. 100 μl of 2 × 106 CFU of S. agalactiae or 3.6 × 106 CFU of A. hydrophila was intraperitoneally (i.p.) injected into each Nile tilapia for challenge, whereas the control group was injected with 100 μl of sterilized PBS in the same way. At the indicated days post-infection, spleen was harvested, and leukocytes were isolated for assay.
Leukocyte isolation
Spleen leukocytes were isolated using discontinuous Percoll gradient centrifugation according to previous report (42). Tissue was harvested and grinded in precooled L15 medium (Gibco) containing 2% FBS and then filtered with a cell strainer. After 5 min of centrifugation at 1800 rpm at 4 °C, the cells were resuspended with 3 ml of L15 medium. 100% Percoll, configured with Percoll (GE Healthcare) and 10× PBS with a ratio of 9:1, was diluted into 52 or 34% Percoll with L15 medium. 5 ml of 52% Percoll, 5 ml of 34% Percoll, and 3 ml of cell suspension were consecutively added into the 15-ml centrifuge tube in turn and centrifuged at 500 × g at room temperature for 35 min with the lowest-speed acceleration and deceleration. Leukocytes in the layer between 52 and 34% Percoll were collected, washed twice with L15 medium, and resuspended in Dulbecco's modified Eagle's medium (10% FBS) for the assay.
Leukocyte stimulation
For lymphocyte activation, spleen leukocytes resuspended with Dulbecco's modified Eagle's medium containing 10% FBS and 1% penicillin/streptomycin were cultured in a 24-well plate at 28 °C with 5% CO2. The cells were stimulated with 2 μg/ml PHA. At the indicated time after stimulation, leukocytes were collected for corresponding assays. Unstimulated spleen leukocytes were used as control. For lymphocyte signaling activation, spleen leukocytes were resuspended with Dulbecco's PBS and incubated at 30 °C for 30 min to rest the phosphorylated proteins. After that, 50 ng/ml PMA plus 500 ng/ml ionomycin were added into cell suspension. At the indicated time points, the same volume of precooled Dulbecco's PBS was added into cell suspension to stop stimulation. Leukocytes were collected for the assay. The rested cells were used as a control sample.
Mek or Erk inhibition
Nile tilapia was i.p. injected with 0.1 mg/kg/day Mek inhibitor trametinib or 5 mg/kg/day Erk inhibitor SCH772984 (MedChemExpress) for 2 consecutive days before spleen leukocytes were isolated. The isolated leukocytes were stimulated with PHA or PMA plus ionomycin for assay. If the isolated leukocytes were cultured in vitro for more than 4 h, 10 nm trametinib or 1 μg/ml SCH772984 was added to the inhibition group. For the in vivo assay, trametinib or SCH772984 was i.p. injected into Nile tilapia at 2, 3, 4, and 6 days after bacterial infection. At 8 days after infection, spleen leukocytes were isolated for the assay.
Immunofluorescence assay
Spleen leukocytes that were treated or untreated with PMA plus ionomycin for 15 min were spun onto the slide using Cytospin-4 at 1500 rpm for 5 min, and the cells were fixed with methanol at room temperature for 5 min. The prepared slides were blocked in PBS containing 1% BSA at room temperature for 1 h. After washing with PBST three times, the samples were incubated with 1:100 diluted rabbit anti-phospho-Mek1/2 Ser-217/221 or anti-phospho-Erk1/2 Thr-202/Tyr-204 antibody and 1:2400 diluted mouse anti-β-actin antibody (Cell Signaling Technology) at 37 °C for 1 h. After washes, the cells were further incubated in 1:1000 diluted Alexa Fluor 488–conjugated goat anti-rabbit IgG H+L (Abcam) and Alexa Fluor 594-conjugated goat anti-mouse IgG H+L (Abcam) at 37 °C for another 1 h. Another three washes were performed before the slides were sealed by glycerol containing 4′,6-diamidino-2-phenylindole and observed with a Zeiss ApoTome microscope.
Quantitative RT-PCR (qPCR)
Peripheral blood was mixed with an equal volume of precooled anticoagulant (8.17 g/liter NaCl, 5.46 g/liter citric acid, 8.82 g/liter sodium citrate, 19.8 g/liter glucose, 3.72 g/liter EDTA, pH 6.6), and blood cells were collected by centrifugation at 1800 rpm, 4 °C for 5 min. Blood cells, freshly harvested tissues, stimulated or infected cells were put into TRIzol (Invitrogen) for total RNA extraction. After treatment with DNase RQ1 at 37 °C for 30 min, the extracted RNA was used as a template to synthesize the first-strand cDNA with primer oligo(dT)-adaptor and reverse transcriptase Moloney murine leukemia virus (Thermo). The 1:50 diluted cDNA production was then used as template for SYBR Green qPCR on the Bio-Rad CFX96TM real-time system. Melt curves were detected and analyzed to confirm the specificity of each primer. Data are given in terms of mRNA levels relative to β-actin using the 2−ΔΔCt method. The primers used for qPCR are listed in Table S2.
BrdU incorporation
S. agalactiae- or A. hydrophila-infected Nile tilapia was i.p. injected with 0.6 mg of BrdU in 100 μl of PBS on day 4 after infection, and spleen leukocytes were isolated on day 5 for assay. After fixing with BD Cytofix/Cytoperm solution on ice for 30 min, spleen leukocytes were washed twice with BD perm/wash solution. Following a 10-min permeabilization with BD Cytoperm Plus solution on ice, the cells were then refixed with BD Cytofix/Cytoperm solution on ice for another 5 min. Samples were subsequently treated with 300 μg/ml DNase at 37 °C for 1 h before intracellular staining was performed with a 1:100 diluted FITC-anti-BrdU antibody (BD Biosciences) in perm/wash buffer at room temperature for 20 min. After washes, the samples were analyzed by flow cytometry.
Glucose uptake assay
Spleen leukocytes isolated from A. hydrophila-infected Nile tilapia with or without inhibitor treatment were plated in 24-well plates at 1 × 107 cells/well in PBS. After incubating with 100 μm fluorescent 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxyglucose (2-NBDG; Life Technologies, Inc.) at 28 °C for 30 min, the culture medium was removed, and cells were washed twice with prechilled PBS to stop the reaction. Uptake of 2-NBDG was analyzed by a flow cytometer within the lymphocyte population.
Examination of enzyme activity
Enzyme activity of phosphofructokinase (pfk) was performed according to our previous report (42). Briefly, liver harvested from Nile tilapia with or without trametinib treatment on day 3 or 5 after A. hydrophila infection was smashed in extraction buffer using a grinding miller on ice. The samples were then centrifuged at 8000 rpm at 4 °C for 10 min, and the gained supernatant was used for protein concentration and enzyme activity examination. The activity of pfk was measured with commercial assay kits (Nanjing Jiancheng Bioengineering Institute) according to the instructions of the manufacturer. The enzyme activity was calculated per mg of tissue protein.
Flow cytometry
Spleen leukocytes were used for intracellular LCK staining as described previously (42). In brief, leukocytes were fixed with BD Cytofix/Cytoperm solution and washed twice with BD Perm/Wash solution. The fixed cells were then stained with 1:400 diluted Alexa Fluor 647–conjugated LCK antibody (BioLegend) on ice for 30 min, and another two washes were performed before the cells were analyzed. Freshly isolated spleen leukocytes, BrdU-stained cells, and 2-NBDG–stained cells were resuspended in PBS containing 2% FBS for FACS analysis. All of the samples were collected using a BD FACSCalibur flow cytometer. Data were analyzed using FlowJo software.
Western blotting
The infected or stimulated leukocytes were lysed in Nonidet P-40 lysis buffer with phosphatase inhibitor and protease inhibitor, on ice for 30 min. The lysate samples were then centrifuged at 12,000 rpm at 4 °C for 10 min. The supernatants were separated by SDS-PAGE electrophoresis and then transferred onto nitrocellulose filter membrane at 100 V for 2 h. After blocking in PBST containing 4% nonfat powdered milk for 1 h at room temperature, the membrane was incubated with 1:1000 diluted rabbit antibodies against c-Raf (9422), Mek1/2 (8727), Erk1/2 (4695), c-Myc (13987), phospho-c-Raf Ser-338 (9427), phospho-MEK1/2 Ser-217/221 (9154), phospho-ERK1/2 Thr-202/Tyr-204 (4370), phospho-p90RSK Thr-573 (9346), or rabbit anti-β-actin (4967) (Cell Signaling Technology) at 4 °C overnight. The membrane was finally incubated in 1:10,000 diluted goat anti-rabbit IgG H+L Alexa Fluor 790 (Abcam) at room temperature for 1 h. Three washes were performed with PBST following each incubation step. An Odyssey IR fluorescence scanning imaging system was used to observe the membrane.
Statistical analysis
Data are presented as the mean ± S.E., and statistical significance was determined by a two-tailed Student's t test. p values are indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author contributions
X. W. and J. Y. funding acquisition; X. W. and J. Y. writing-original draft; X. W. and J. Y. project administration; Y. Z. software; Y. Z., C. L., K. A., K. L., and H. L. investigation; Y. Z. visualization; J. Y. supervision; J. Y. methodology.
Acknowledgments
We thank the Instrument-sharing Platform of the School of Life Sciences of East China Normal University (ECNU) for instrument sharing, the Flow Cytometry Core Facility of the School of Life Sciences of ECNU for the FACS analysis, and the ECNU Multifunctional Platform for Innovation (011) for animal keeping.
This work was supported by National Natural Science Foundation of China Grants 31972822 and 31872591, Shanghai Pujiang Program Grant 18PJ1402700, and the Fundamental Research Funds for the Central Universities (to J. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1 and S2.
3The abbreviations used are:
- MAPK
- mitogen-activated protein kinase
- MAPKK
- MAPK kinase
- MAPKKK
- MAPKK kinase
- S/TKc
- Ser/Thr kinase core
- Erk
- extracellular signal–regulated kinase; c-Jun N-terminal kinase
- Mek
- MAPK/Erk kinase
- TCR
- T-cell receptor
- LCK
- lymphocyte-specific protein tyrosine kinase
- PLC
- phospholipase C
- NFAT
- nuclear factor of activated T cells
- qPCR
- quantitative PCR
- DPI
- days post-infection
- PHA
- phytohemagglutinin
- IFN
- interferon
- HK2
- hexokinase 2
- LDHA
- lactate dehydrogenase A
- CFU
- colony-forming units
- i.p.
- intraperitoneally
- FBS
- fetal bovine serum
- PMA
- phorbol 12-myristate 13-acetate
- BrdU
- bromodeoxyuridine
- 2-NBDG
- 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxyglucose
- pfk
- phosphofructokinase.
References
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.ra119.012231
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/article/S0021925817495068/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1074/jbc.ra119.012231
Article citations
MEK inhibition prevents CAR-T cell exhaustion and differentiation via downregulation of c-Fos and JunB.
Signal Transduct Target Ther, 9(1):293, 22 Oct 2024
Cited by: 0 articles | PMID: 39438476 | PMCID: PMC11496645
Signal amplification by cyclic extension enables high-sensitivity single-cell mass cytometry.
Nat Biotechnol, 29 Jul 2024
Cited by: 0 articles | PMID: 39075149
Comparative Study on the Mechanism of Macrophage Activation Induced by Polysaccharides from Fresh and Dried Longan.
Nutrients, 16(11):1654, 28 May 2024
Cited by: 0 articles | PMID: 38892587 | PMCID: PMC11174042
CCR7 affects the tumor microenvironment by regulating the activation of naïve CD8+ T cells to promote the proliferation of oral squamous cell carcinoma.
Transl Oncol, 44:101924, 02 Mar 2024
Cited by: 0 articles | PMID: 38430712
<i>E. hellem</i> Ser/Thr protein phosphatase PP1 targets the DC MAPK pathway and impairs immune functions.
Life Sci Alliance, 7(4):e202302375, 10 Jan 2024
Cited by: 0 articles | PMID: 38199846 | PMCID: PMC10781585
Go to all (32) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Akt1/mTORC1 signaling modulates adaptive immune response of Nile tilapia by promoting lymphocyte activation and proliferation.
Dev Comp Immunol, 119:104042, 12 Feb 2021
Cited by: 3 articles | PMID: 33582106
c-Raf participates in adaptive immune response of Nile tilapia via regulating lymphocyte activation.
Fish Shellfish Immunol, 86:507-515, 02 Dec 2018
Cited by: 5 articles | PMID: 30513386
S6K1/S6 axis-regulated lymphocyte activation is important for adaptive immune response of Nile tilapia.
Fish Shellfish Immunol, 106:1120-1130, 21 Sep 2020
Cited by: 1 article | PMID: 32971270
Coordinating ERK/MAPK signalling through scaffolds and inhibitors.
Nat Rev Mol Cell Biol, 6(11):827-837, 01 Nov 2005
Cited by: 669 articles | PMID: 16227978
Review
Funding
Funders who supported this work.
National Natural Science Foundation of China (2)
Grant ID: 31972822
Grant ID: 31872591
Shanghai Pujiang Program (1)
Grant ID: 18PJ1402700





