Abstract
Free full text

Synthesis, Regulation and Degradation of Carotenoids Under Low Level UV-B Radiation in the Filamentous Cyanobacterium Chlorogloeopsis fritschii PCC 6912
Abstract
Carotenoids in cyanobacteria play an important role in protecting against and in repairing damage against low level UV-B radiation. Here we use transcriptomics and metabolomic HPLC pigment analysis to compare carotenoid pathway regulation in the filamentous cyanobacterium Chlorogloeopsis fritschii PCC 6912 exposed to white light and to white light supplemented with low level UV-B. Under UV-B changes in carotenoid transcription regulation were found associated with carotenogenesis (carotenoid synthesis), photoprotection and carotenoid cleavage. Transcriptional regulation was reflected in corresponding pigment signatures. All carotenogenesis pathway genes from geranylgeranyl-diphosphate to lycopene were upregulated. There were significant increases in expression of gene homologs (crtW, crtR, cruF, and cruG) associated with routes to ketolation to produce significant increases in echinenone and canthaxanthin concentrations. There were gene homologs for four β-carotene-ketolases (crtO and crtW) present but only one crtW was upregulated. Putative genes encoding enzymes (CruF, CrtR, and CruG) for the conversion of γ-carotene to myxol 2′-methylpentoside were upregulated. The hydroxylation pathway to nostaxanthin via zeaxanthin and caloxanthin (gene homologs for CrtR and CrtG) were not upregulated, reflected in the unchanged corresponding pigment concentrations in zeaxanthin, caloxanthin and nostaxanthin, Transcripts for the non-photochemical quenching related Orange-Carotenoid-Protein (OCP) and associated Fluoresence-Recovery-Protein (FRP) associated with photoprotection were upregulated, and one carotenoid binding Helical-Carotenoid-Protein (HCP) gene homolog was downregulated. Multiple copies of genes encoding putative apocarotenoid related carotenoid oxygenases responsible for carotenoid cleavage were identified, including an upregulated apo-β-carotenal-oxygenase gene homologous to a retinal producing enzyme. Our study provides holistic insight into the photoregulatory processes that modulate the synthesis, photoprotection and cleavage of carotenoids in cyanobacterial cells exposed to low level UV-B. This is important to understanding how regulation of metabolism responds to a changing environment and how metabolism can be modulated for biotechnological purposes.
Introduction
In contrast to the widely reported detrimental effects that UV has on cyanobacteria we know less about the effects of eustress or low level UV stress. In plants (and in animals) there is increasing evidence that eustress is good. Eustress confers a wide range of adjustment and adaption mechanisms to be invoked enabling and promoting survival (Jansen et al., 2008). Exposure to low levels of UV-B by cells actively promotes survival because it stimulates responses that help to protect against and repair UV-damage (Hideg et al., 2013).
Cyanobacteria are an ancient group of photosynthetic prokaryotes that have had an immense impact on the evolution of other organisms and on the global ecosystem. They are ubiquitous throughout the world and thrive in a wide range of marine and freshwater habitats as well as terrestrial and symbiotic habitats and make an important contribution to global primary production. The ability of cyanobacteria to survive under variable light environments and acclimate to a wide range of spectral conditions has contributed to their ecological and evolutionary success over millions of years (Rastogi et al., 2014; Gan and Bryant, 2015). Despite the importance of cyanobacteria in evolutionary and ecological understanding and more recently in biotechnology applications our understanding on cyanobacterial metabolism, particularly under low level eustress conditions is lacking.
Carotenoids are a group of tetraprenoid metabolites known to play an important role in protecting against photoooxidative damage (Liang et al., 2006). Terrestrial and freshwater cyanobacteria species typically produce a wider range of carotenoids than marine cyanobacteria and contain, in addition to the β-carotene and zeaxanthin found in marine species, keto-xanthophylls such as echinenone, canthaxanthin, and oscillaxanthin and myxol-glycosides (Jeffrey et al., 2011). Freshwater cyanobacteria can also contain the hydroxylated carotenoids caloxanthin and nostoxanthin (Schagerl and Donabaum, 2003), and the dicarboxylated synechoxanthin (Graham and Bryant, 2008).
Unlike in other photosynthetic cells where the majority of light for photosynthesis is harvested by carotenoids, in cyanobacteria (and in rhodophytes), light is additionally harvested for photosynthesis by phycobilin pigments within the phycobilisomes. Phycobilisomes are large light harvesting antenna protein complexes attached externally to the photosystem (Scholes et al., 2011) primarily associated with photosystem II (PSII), although they may also interact with PSI (Ashby and Mullineaux, 1999; Liu et al., 2013). Associated with phycobilisomes is a carotenoid binding protein, the Orange Carotenoid Protein (OCP). The OCP has a photoprotective quenching function protecting the reaction centers within PSI and PSII (Wilson et al., 2008). The protein consists of two domains, with a single keto-carotenoid molecule non-covalently bound between the two domains. OCP acts through non-photochemical quenching (NPQ) to very efficiently quench the excitation energy absorbed by the phycobilisomes (Boulay et al., 2008; Kirilovsky and Kerfeld, 2012; Bao et al., 2017b). This NPQ mechanism is the functional equivalent of the violaxanthin- zeaxanthin xanthophyll cycle present in chlorophytes and plants, and the diadinoxanthin –diatoxanthin xanthophyll cycle present in diatoms. The central carotenoid of the OCP requires a keto group for photoactivity, but the OCP can bind a diversity of these, e.g., the mono keto-carotenoid 3′-hydroxyechinenone, myxol 2′-methylpentosides, echinenone or canthaxanthin (Melnicki et al., 2016; Bao et al., 2017a). Our knowledge of the range of ketocarotenoids involved in this photoprotective mechanism is still emerging.
In addition to carotenoids associated with thylakoid membranes of PSI and PSII, in cyanobacteria, carotenoids can also be associated with cell envelope membranes. These carotenoids are different from, and less diverse than those associated with thylakoid membranes (Murata et al., 1981). Myxoxanthophyll, a monocyclic glycosidic carotenoid is associated with the cytoplasmic membrane and outer membrane of cyanobacteria, and is believed to play a role in the stabilization of cytoplasm and cell wall membranes (Graham and Bryant, 2009). Its glycoside attachment is typically a fucoside, rhamnoside or chinovoside (Takaichi and Mochimaru, 2007) but when the glycoside has not been characterized it is referred to as a myxol-2′-methylpentoside. Myxol-2′-methylpentoside can be both hydroxylated and ketolated to give 2-hydroxymyxol 2′-methylpentoside and 4-ketomyxol 2′-methylpentoside respectively.
Carotenogenesis, the synthesis of carotenoids, can be considered in five stages (Lohr, 2011). The first stage is to produce an isoprene building block unit consisting of isopentenyl diphosphate and its isomer dimethylallyl diphosphate. In cyanobacteria the steps to isoprene are undertaken solely using the methylerythritol phosphate (MEP) pathway (Pattanaik and Lindberg, 2015). The second stage is a stepwise condensation to produce phytoene. Phytoene represents the first committed point to carotenoid biosynthesis. The third stage is sequential desaturation and isomerization to produce lycopene. Lycopene represents the branch point to carotenoid diversity. The fourth stage is cyclization of the linear ends of lycopene to yield cyclic carotenes. The fifth stage is stepwise introduction of hydroxyl and keto groups to produce a diversity of xanthophylls (Figure 3). Although carotenogenesis in plants and, to some extent in green algae, has been well studied, it is only in the last decade or so that genes involved in carotenogenesis in cyanobacteria, algae and land plants have been identified (Chang et al., 2013).
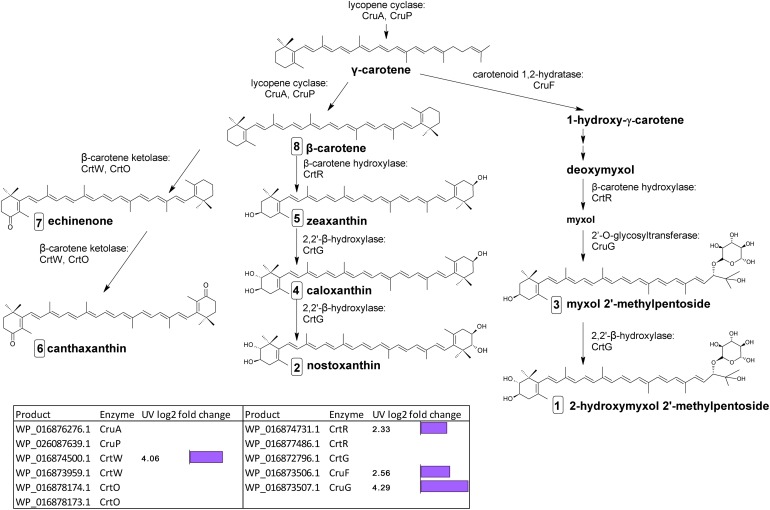
Biosynthetic pathway of carotenoids from γ-carotene in C. fritschii PCC 6912. Genes identified by orthology for enzymes which can catalyze pathway steps are shown together with significant changes in regulation (with –1.5 > log2 fold change > 1.5 and p-adj < 0.05) under UV-B treatment (4h4d samples) illustrated by the purple bars. Numbers in squares indicate the order of elution on the HPLC corresponding to Figure 4. For myxol 2′-methylpentoside and 2-hydroxymyxol 2′-methylpentoside the glycoside is represented as a fucoside.
Whilst most of the enzymes in carotenogenesis have been identified, less is known about carotenoid degradation. The first step in degradation involves enzymatic cleavage of the carotenoid by oxygenases to form a diverse range of bioactive apocarotenoids including volatiles, and photoprotective compounds (Kloer and Schulz, 2006). Carotenoid cleavage dioxygenases (CCDs) generate and modify these biologically important apocarotenoids by the oxidative cleavage of specific carotenoid double bond sites to form aldehyde or ketone products including retinal (Ahrazem et al., 2016). The roles that CCDs play in cyanobacteria are not fully understood.
These recent findings present opportunities to holistically better understand pathways to synthesis of keto-carotenoids, photoprotection and carotenoid turnover and degradation. In particular, the regulation of these carotenoid pathways in cyanobacteria under different conditions is relatively still poorly understood. Specifically, exposure to low level UV irradiation is of importance as it is a common feature of the habitat of many cyanobacterial groups. Thus studies on how carotenoids respond to low level UV-B stress on the cell and on the photosynthesis apparatus are required.
Cyanobacteria are highly diverse in their structure and development and have accordingly been classified into five subgroups or Sections (Castenholz, 2015). Most studies on carotenoids in cyanobacteria have been undertaken on the Section-I unicellular strains Synechocystis sp. PCC 6803 (Ruch et al., 2005; Vajravel et al., 2017) or Synechococcus sp. PCC 7002 (Albrecht et al., 2001; Graham and Bryant, 2008) with fewer studies on the Section-IV filamentous strains Nostoc punctiforme PCC 73102 and Anabaena sp. (also known as Nostoc sp.) PCC 7120 (Takaichi et al., 2005; Marasco et al., 2006). Here we investigate the carotenoid response to low level UV-B in the Section-V filamentous cyanobacterium Chlorogloeopsis fritschii PCC 6912. C. fritschii is a terrestrial species first isolated from the soils of a paddy field in India (Mitra, 1950). C. fritschii has been shown to be a robust thermophilic species with potential to produce a range of useful metabolites including carotenoids (Balasundaram et al., 2012; Kultschar et al., 2019). C. fritschii was the first freshwater cyanobacterial species for which detailed compositional analysis of carotenoids was determined (Evans and Britton, 1983). More recently there has been renewed interest in C. fritschii for its ability to adapt to the far red light (Airs et al., 2014; Ho and Bryant, 2019). Transcription and pigment metabolite profiling were undertaken on cultures exposed to white light and compared to those exposed to white light supplemented with low level UV-B.
Materials and Methods
Experimental Conditions
Chlorogloeopsis fritschii (Mitra) PCC 6912 was inoculated at 1:50 dilution from a master culture and cultivated in 5 L Erlenmeyer flasks containing 2 L BG11 media with 10 mM HEPES buffer at pH 7.5. The culture was perfused with 1% CO2 and was maintained at 38°C under constant white light (410–750 nm: Grolux fluorescent tubes) of 60 μmol photons m–2 s–1. At exponential growth phase, cells were harvested and transferred to nine 500 mL quartz Erlenmeyer flasks containing 200 mL of fresh BG11, and 10 mM HEPES at pH 7.5 to give a concentration of 0.44 g L–1 wet weight (approximating to 0.04 g L–1 dry weight). All 9 cultures were exposed to constant white light (410–750 nm: Grolux fluorescent tubes) of 60 μmol m–2 s–1 photons for 4 days and 4 h (100 h in total). Three flasks were exposed to white light with no UV-B supplementation acting as the control (white), three flasks were exposed to white light supplemented with UV-B for the final 4 h of the experiment (4h) and, three flasks were exposed to white light supplemented with 4 h of UV-B radiation each day for 4 days (4h4d). Flasks exposed to UV-B were placed 10 cm from UV-B tubes supplying 3 μmol m–2 s–1 at wavelength range 300–310 nm. The experiment was set up, guided using prior experiments, where photosynthetic efficiency (Fv/Fm) as previously described (Steele et al., 2018), was measured so that the level of UV-B exposure did not impact detrimentally on photosynthetic efficiency (Supplementary Figure S1). At the end of the experiment samples all 9 flask were placed on ice and were centrifuged (3000 g) at 4°C. The pelleted biomass was snap frozen in liquid nitrogen before storage at −80°C. Transcriptomics was undertaken on the white light (control) and 4h4d samples. Targeted metabolomics to determine pigments was undertaken on the white, 4h and 4h4d samples.
RNA Preparation and Sequencing
For the white light control and the 4h4d samples, RNA was extracted with Trizol followed by terminator exonuclease digestion to enrich for mRNA and subsequently cleaned using a Qiagen RNeasy column. RNA sequencing was conducted at the Centre for Genomic Research, Institute of Integrative Biology at the University of Liverpool, United Kingdom, L69 7ZB, using the Life Technologies SOLiD sequencing platform. For each sample, at least 49,034,856 sequences were obtained (50 bp, min average quality 20 as per manufacturer specifications; per sample average sequence number: 57,516,996.44).
Bioinformatic Analysis
Alignment of reads was carried out using the C. fritschii PCC 6912 genome as reference. The sequences obtained for each sample were aligned on to the reference using bowtie version 0.12.7, using the color space option. Prior the alignment step the sequences required the conversion to a pseudo-fastq file required as input for bowtie. For each of sequence, only the best alignment was reported by bowtie, or one was randomly chosen if many were equally best. The average percentage of unambiguously aligned sequence was 47.67%, with a minimum of 35.9% and the maximum equal to 51.9. Considering the known 6,968 genes, an average of 83.11% of these were identified as expressed across all the samples (ranging between 73.95 and 87.15%). All the obtained alignment files were processed using HTSeq-count (Anders et al., 2015), and reads aligning to the reference genome sequences were counted according to the gene features that they mapped to, as defined in the GTF files.
The differential expression analyses between white and the 4h4d samples were performed using R (version 2.14) and edgeR package. The gene-counts were normalized using “loess smooth” method form the ‘limma’ package. The “GLM” model was applied to the normalized data (with edgeR package) and the dispersion related to each gene (genewise dispersion) and the pairwise group comparisons were performed to identify differentially expressed genes for each of the three possible group comparisons. For each contrast, each gene with a p-value below 0.05 (after adjusting for multiple testing effect using the False Discovery Rate approach, Benjamini and Hochberg, 1995) were selected as differentially expressed for that contrast. Significant changes in regulation were defined as log2 fold change ≤ −1.5 or ≥1.5 and p-adj < 0.05. Data produced in this study can be found within an NCBI BioProject with accession number PRJNA545395.
Pigments
For white, 4h and 4h4d samples, cell pellets (80–170 mg wet weight) were transferred to extraction tubes containing 1 mm glass beads and extracted into 100% methanol using an ultrasonic probe (35 s; 50 W), and 1 mm glass beads. Pellets were repeat extracted with 1 ml methanol five times. Extracts were clarified and analyzed according to the method of Zapata et al. (2000) by injecting onto a Waters Symmetry C8 reverse phase column (150 × 4.6 mm, 3.5 μm particle size) on a Thermo Accela Series HPLC system with chilled autosampler (4°C) and photodiode array detector. The HPLC was calibrated using a suite of standards purchased from DHI (Denmark). The standards were used to generate response factors used for quantification. For pigments where standards were not available (caloxanthin and nostoxanthin) the response factors of the most closely related standard, zeaxanthin, was used. Pigments were identified based on retention time, spectral match using photo-diode array and LC/MS (Supplementary Table S1). Probability (p) of significance between white light control and the 4h4d samples was determined using a Student’s t-test test with a two tailed distribution.
Results
An overview of the log2 fold changes of differential transcriptional expression in carotenoid and carotenoid related gene homologs in C. fritschii PCC 6912 when exposed to supplementary low level UV-B (4h4d) is shown in Figure 1. Further details including levels of expression can be found in Supplementary Table S2. Statistically significant differential transcription regulation changes across carotenogenesis, photoprotection and carotenoid cleavage were observed (Figure 1).
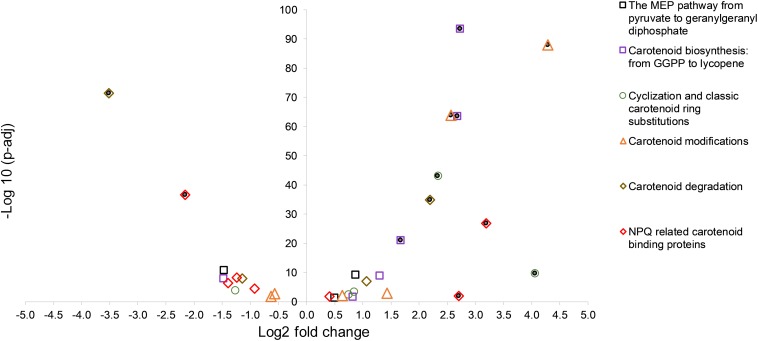
A volcano plot showing significance versus log fold change in expression of carotenoid synthesis and metabolism in C. fritschii PCC 6912 in response to supplementary low level UV-B (4h4d samples). Filled circles indicate significance with −1.5 > log2 fold change > 1.5 p.adj < 0.05.
Carotenogenesis
Focusing on the early stages of biosynthesis of the carotenoids, none of the gene homologs encoding enzymes up to the point of geranylgeranyl diphosphate were differentially upregulated and one putative kinase was slightly differentially downregulated (EC:2.7.1.148: WP_016876009.1: log2 fold change −1.47) in response to the UV treatment (Supplementary Figure S3).
Gene homologs encoding enzymes which catalyze subsequent steps leading to lycopene were differentially upregulated (Figure 2). These were a putative crtB phytoene synthase (WP_016875938.1) with a log2 fold change of 2.68; a putative crtP; 15-cis-phytoene desaturase (WP_016875939.1) with a log2 fold change of 2.73, and a putative crtQ; ζ-carotene desaturase (WP_016872384.1) with a log2 fold change of 1.67. We also found three putative COG1233 phytoene desaturase-related gene homologs (Figure 2).
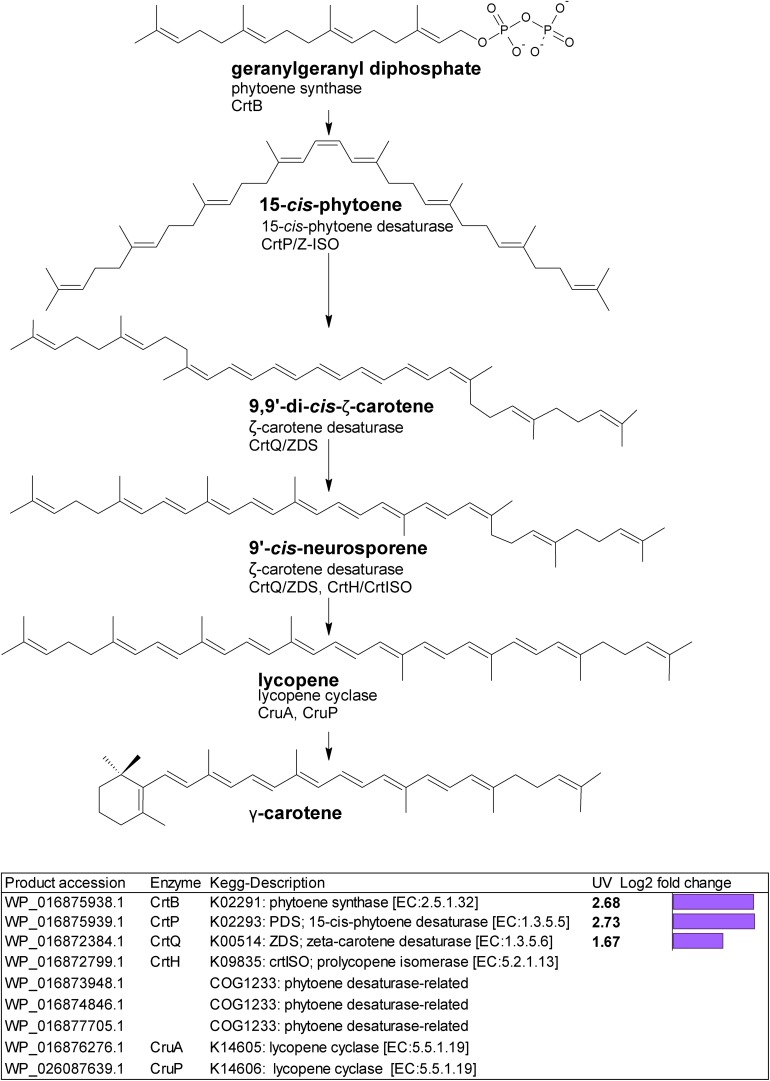
Biosynthetic pathway from geranylgeranyl diphosphate to lycopene. Genes identified by orthology for enzymes which can catalyze pathway steps are shown together with significant changes in regulation (with –1.5 > log2 fold change > 1.5 and p-adj < 0.05) under UV-B treatment (4h4d samples) illustrated by the purple bars.
Lycopene represents a crucial step in carotenoid synthesis, being the precursor to the branch point which generates carotenoid diversity (Lohr, 2011). In cyanobacteria, lycopene is the precursor to γ- and β-carotene. Gene homologs encoding two lycopene cyclases, CruA and CruP which catalyze cyclization from lycopene to produce γ- (one cyclic ring) and β-carotene (two cyclic rings) were not differentially regulated under UV.
The final stages of biosynthesis from γ- and β-carotene leading to the diversity of carotenoids found in C. fritschii involves the stepwise introduction of ketol, hydroxyl, or glycosyl groups using ring-modifying ketolase, hydratase, hydroxylase, and transferase enzymes (Figure 3). Some of these enzymes are promiscuous for carotenoid substrates (Figure 3).
From β-carotene, ketolisation produces echinenone and canthaxanthin. Ketolisation also converts myxol-2′-methylpentoside to 4-ketomyxol 2′-methylpentoside. However, notably we did not observe any 4-ketomyxol 2′-methylpentoside in our HPLC chromatograms (see section Carotenoid Metabolites). We found four β-ketolase homologs involved in catalyzing the conversion of β-carotene to echinenone and canthaxanthin; two of the CrtO type and two of the CrtW type. Expression of one homolog of each type was not detected in this study, and the expressed crtO, WP_016878174.1 was slightly but not significantly downregulated under UV (log2 fold change −1.27). However, the expressed gene encoding a CrtW ketolase homolog (WP_016874500.1) was highly differentially upregulated under UV with a log2 fold change of 4.06 (Figure 3). β-carotene can also be converted by hydroxylation to zeaxanthin, caloxanthin and nostoxanthin (Figure 3). This uses a β-carotene hydroxylase and a 2,2′-β-hydroxylase. There are two genes encoding putative CrtR type beta-carotene hydroxylases, a major (WP_016874731.1) and a minor (WP_016877486.1) according to transcription levels. In this pathway, only the major crtR homolog was upregulated under UV-B (WP_016874731.1) with a log2 fold change of 2.33.
In C. fritschii a number of gene homologs encode enzymes (CruF, CrtR, CruG) to convert γ-carotene to myxol glycosides to produce myxol-2′-methylpentoside (Graham and Bryant, 2009). We found upregulation of all these genes under UV indicating an activation of this pathway (Figure 3). We observed the largest upregulation for a glycosyltransferase cruG (WP_016873507.1: log2 fold 4.29). CruG is required for the conversion of myxol to the myxol 2′-methylpentoside.
We also identified a crtG homolog (WP_016872796.1) encoding for the hydroxylation of zeaxanthin to caloxanthin and nostoxanthin and of myxol 2′-methylpentoside to 2-hydroxymyxol 2′-pentoside. CrtG was not found to be differentially regulated under UV-B.
In terms of the regulation of carotenoid biosynthesis, the global transcription factor NtcA reported to regulate carotenoid biosynthesis in Nostoc PCC 7120 (Sandmann et al., 2016) was not upregulated under UV in C. fritschii PCC 6912. However, we did observe the presence of signature NtcA binding sites in C. fritschii PCC 6912 promoter regions of the crtPB operon, crtO and crtW (Supplementary File S1), which indicates a functional role in the NtcA control of carotenoid transcription in C. fritschii PCC 6912, likely to be modulated by binding of cofactors such as 2-oxoglutarate (Tanigawa et al., 2002).
Photoprotection
Next, we looked for carotenoid binding proteins related transcripts known to be involved in photoprotection. We found a number of orange carotenoid protein (OCP) related transcripts involved in photoprotection of the phycobilisome (PBS) and these showed changes in regulation under UV. One gene homolog encoding a full OCP1 (WP_016875600.1) was found situated next to the gene homolog encoding fluorescence recovery protein (WP_016875601.1); both of these were strongly upregulated in UV (log2 fold change 3.19 and 2.71 respectively) (Table 1).
TABLE 1
Expression changes in OCP and CCD related gene homologs.
 |
Gene homologs encoding N- and C-terminal O, a helical carotenoid protein (HCP) and a C-terminal domain (CTD respectively, were also found. There were four HCPs and one CTD paralog (WP_016872658.1), as reported in Anabaena sp. PCC 7120 (Lopez-Igual et al., 2016). We identified gene homologs for HCP1 (WP_016877404.1), HCP2 (WP_016874882.1), HCP3 (WP_016872653.1) and HCP4 (WP_016872657.1), which is adjacent to the CTD gene homolog. The gene homolog for CTD (WP_016872658.1) and for one HCP (WP_016872653.1) were downregulated under UV (log2 fold changes −1.39 and −2.16 respectively (Table 1).
Carotenoid Cleavage Enzymes
Finally, we looked for gene homologs associated with enzymes responsible for the cleavage of carotenoids. We identified five carotenoid cleavage dioxygenase encoding gene homologues (CCDs); these were associated with the retinal pigment epithelial membrane protein family [COG3670 enzymes (Table 1 and Supplementary Table S2). We compared protein sequences to those of the three genes defined in Nostoc sp. PCC 7120 (Marasco et al., 2006) by multiple sequence alignment. We found WP_016874878.1 was orthologous to NosCCD (all1106), which cleaves C9–C10 double bonds (Scherzinger and Al-Babili, 2008), and WP_016877082.1 to NosDiox2 (all4895) which cleaves apocarotenoids at C13–C14, C13′–C14′, and C15–C15′ double bonds (Heo et al., 2013).
WP_016878062.1 and WP_026087737.1 had protein sequence homology to NosACO (all4284), which cleaves monocyclic or acyclic carotenoids at C15–C15′ double bonds to generate retinal (Ruch et al., 2005; Scherzinger et al., 2006). The fifth gene WP_016873878.1 was for a much longer RPE65 with homology to the lignostilbene-alpha, beta-dioxygenase protein in Nostocales, and unlike the other enzymes identified, this protein contains only two of the four conserved active site histidines characterizing CCD enzymes (Marasco et al., 2006). Only WP_016878062.1 generating retinal showed a change in transcriptional expression in UV light (log2 fold 2.20).
C. fritschii PCC 6912 is one of only 65 cyanobacterial species to possess orthologs for genes for all three enzymes in the putative retinoic pathway (Miles et al., 2019). These are the UV upregulated gene homolog for carotenoid oxygenase (WP_016878062.1), to act on β- carotene to produce retinal, a transcript encoding an aldehyde dehydrogenase (WP_016874813.1 6) to produce retinoic acid from this, which was significantly downregulated under UV (log2 fold −3.94), and an ortholog to the gene for Cyp120 (WP_016874813.1 6) to produce 4-hydroxy-retanoinc acid for which expression was not observed (Table 1 and Supplementary Table S2).
Carotenoid Metabolites
Chlorophyll a concentrations, consistent with the intentional low dose of UV-B, were not significantly different across the nine samples [average 865 ± 128 μg g–1 wet weight (969 nmol g–1 ± 143): p = 0.88 for white and 4h4d]. The total carotenoid pool equated to approximately 28% of that of chl-a (on a molar basis). Likewise the carotenoid pool was determined not to be significantly different between the 9 samples [154 ± 12 μg g–1 (276 nmol g–1 ± 38: p = 0.18 for white and 4h4d)].
The changes observed in levels of carotenoids under UV-B were consistent with the changes observed in transcriptome regulation. The main carotenoids identified in both white and UV supplemented samples were, in order of HPLC elution from polar to non-polar, 2-hydroxymyxol 2′-methylpentoside (tentative), nostoxanthin, myxol 2′-methylpentoside, caloxanthin, zeaxanthin, canthaxanthin, echinenone and β-carotene (Supplementary Figure S2 and Supplementary Table S2). Notably, we did not detect any 4-ketomyxol 2′methylpentoside. β-carotene and echinenone were the two most abundant carotenoids that we detected (Figure 4A). Across all samples these two least polar of the carotenoids were at least double the concentration of other carotenoids (Figure 4A). We found echinenone and β-carotene to increase after 4 h suggesting that the PSI enriched pigments increase whereas the myxol-fucosides, zeaxanthin possibly more associated with PSII decreased in the first 4 h. After 4h4d these more polar carotenoids showed increases whereas echinenone and β-carotene each show an opposite response with echinenone continuing to increase and β-carotene decreasing (Figure 4A).
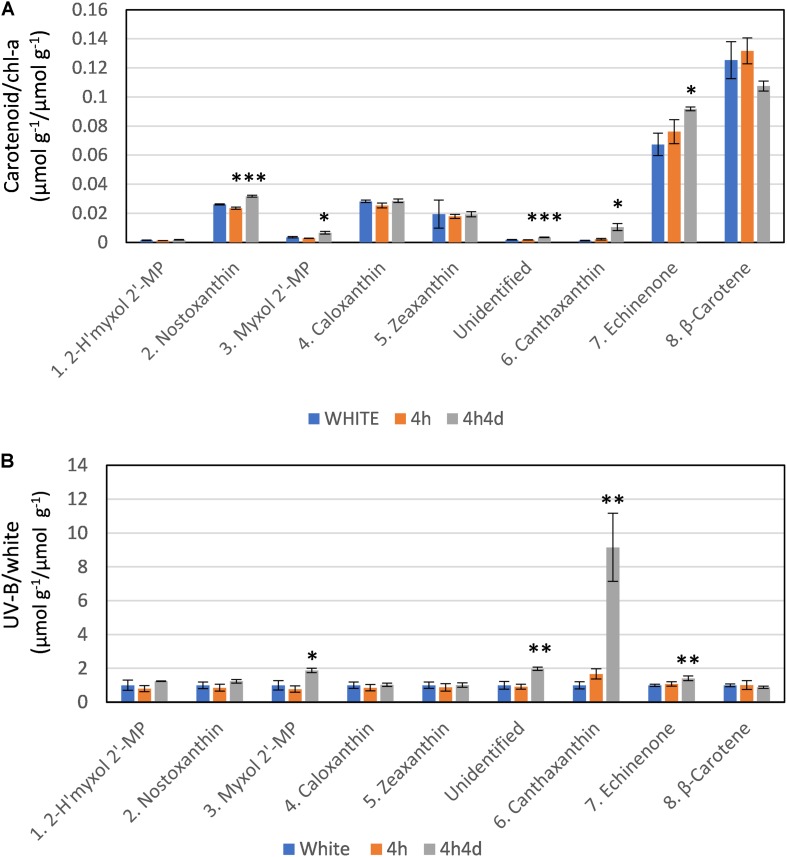
Response of carotenoids to white light and white light supplemented with low level UV-B exposure (4h and 4h4d samples). Carotenoids shown in order of HPLC eluted retention time from polar to non-polar. For HPLC retention time and UV-Vis spectral and LC/MS characterization refer to Supplementary Figure S2 and Supplementary Table S1. (A) Concentration (micromoles per gram of dry weight) of the carotenoid as a ratio of chlorophyll-a concentration (micromoles per gram of dry weight) under the three conditions. (B) Concentration of pigments (micromoles per gram of dry weight) in UV-B exposed samples (4h and 4h4d) as a ratio to the concentration (micromoles per gram of dry weight) in white light. 2- h′myxol 2′-MP, 2-hydroxymyxol 2′-methylpentoside (tentative identification) and myxol 2′-MP, myxol 2′-methylpentoside. Significance difference between white light and 4h4d samples:  = 0.05 > p ≥ 0.01,
= 0.05 > p ≥ 0.01, 
 = 0.01 > p ≥ 0.001, and
= 0.01 > p ≥ 0.001, and 

 = p < 0.001.
= p < 0.001.
Whilst there were some declines in carotenoid concentrations relative to chl-a after 4h of the UV treatment, notably for the more polar carotenoids (Figure 4), we focus here, in correspondence with transcriptome data, on changes observed after 4h4d of UV. For the 4h4d UV samples carotenoids either had similar or increased concentration compared to the white light samples. The only exception was for β-carotene which showed a decrease, although statistically this was not significant (p = 0.14). Three carotenoids showed significant increases measured in μg g–1 wet weight. Echinenone showed a significant increase (p = 0.019) from 36 ± 6 μg g–1 (65 nmol g–1) under white light to 50 ± 24 μg g–1 (92 nmol g–1) under the 4h4d UV exposure conditions. Myxol 2′-methylpentoside and canthaxanthin, whilst both present at relatively low concentration, showed significant (p = 0.016 and 0.019 respectively) increases after 4h4d of UV-B exposure increasing from a mean of 2.5 μg g–1 (3.5 nmol g–1) to 4.8 μg g–1 (6.5 nmol g–1) and from 0.6 μg g–1 (1.1 nmol g–1) to 5.9 μg g–1 (10 nmol g–1) respectively. The most notable difference was for canthaxanthin which showed a nine fold increase in the 4h4d samples when exposed to UV-B (Figure 4B).
Discussion
Photosystem Complexes
Early work found carotenoids distributed differently between the two photosystem complexes (Evans and Britton, 1983): Photosystem I (PSI) of Chlorogloeopsis was found to be preferentially enriched in β-carotene and echinenone whereas PSII was enriched in the more polar carotenoids particularly myxoxanthophyll and zeaxanthin: although notably the location of the majority of echinenone could not be assigned (Evans and Britton, 1983). Association of echinenone with PSI was confirmed more recently in Synechocystis sp. PCC6803 (Vajravel et al., 2017). The increases we observed after 4 h in echinenone and β-carotene suggests that the PSI enriched pigments increase whereas the myxol 2′-methylpentosides and zeaxanthin possibly more associated with PSII decrease. Photosystem II (PSII) is the most light-sensitive complex in the photosynthetic apparatus. Strong light inhibits (photoinhibition) of the activity of PSII. Recent findings suggest that environmental stressors act primarily by inhibiting repair of PSII (Murata et al., 2007). Our results suggest that initially UV-B exposure causes a reduction in carotenoids whereas after 4h4d of exposure, PSII is recovered. This recovery could be activated by the observed increased levels of canthaxanthin.
Carotenoid Transcription Regulation
We found three major types of transcriptional change under UV-B associated with carotenoids; firstly, that associated with carotenoid synthesis (carotenogenesis); secondly photoprotecting-NPQ related, and thirdly, associated with carotenoid cleavage.
Carotenogenesis
The first stage of carotenoid production produces active isoprene; in cyanobacteria this is undertaken solely using the MEP pathway (Lohr et al., 2005; Paniagua-Michel et al., 2012). This is converted into geranylgeranyl diphosphate, the substrate for phytoene synthase. These initial terpenoid biosynthesis steps were not affected by UV-B (Supplementary Figure S3).
Phytoene synthase (CrtB) performs the rate-limiting entry step into carotenoid biosynthesis (Paniagua-Michel et al., 2012), and this together with phytoene desaturase (CrtP), and ζ-carotene desaturase CrtQ, transcripts for all of which we found upregulated under UV, provides the pathway to lycopene (Figure 2). We also observed three putative COG1233 phytoene desaturase-related homologs requiring further investigation. Lycopene represents a crucial step in carotenoid synthesis, generating carotenoid diversity. Upregulation of this pathway opens the gateway to higher carotenoid production, supplying the precursors needed to enable increased photoprotection. It is notable that the cyclases needed to convert lycopene to the diverse carotenoids seen were not transcriptionally upregulated; the pool of enzymes present may be sufficient to allow increased lycopene conversion.
Extending the carotenoid pathway to ketocarotenoids is achieved via carotenoid ketolases; however, these differ in their catalytic properties in different species. For example CrtO in Nostoc PCC 73102 is a diketolase rather than a monoketolase as in Synechocystis PCC 6803 (Schopf et al., 2013). In Anabaena sp. PCC 7120 CrtO and CrtW were functionally characterized to be associated with the conversion of β-carotene to echinenone and canthaxanthin and myxol to 4-keto-myxol respectively (Mochimaru et al., 2005; Takaichi and Mochimaru, 2007). Nostoc PCC 73102 has two carotenoid CrtW ketolases with different properties (CrtW148 and CrtW38) (Steiger and Sandmann, 2004; Takaichi and Mochimaru, 2007; Makino et al., 2008). Enhanced production of canthaxanthin in high light stress in N. punctiforme PCC 73102 has been attributed to the specific upregulation of crtW148 (along with crtB), when it is thought to take over from the other ketolases (Schopf et al., 2013). This contrasts to the light induced upregulation of both crtW and crtO found in Nostoc PCC 7120 (Sandmann et al., 2016).
In C. fritschii PCC 6912 there are two genes encoding putative CrtO ketolases and two genes were also identified encoding putative CrtW ketolases. Only one of these four ketolase gene homologs, crtW (WP_016874500.1), was transcriptionally upregulated under UV with a log2 fold change of 4.06 (Figure 3). Protein sequences for WP_016874500.1 and Nostoc PCC 73102 CrtW148 have a 68.46% identity, and the second crtW (WP_016873959.1) encodes a protein with a 69.29% identity to Nostoc PCC 73102 CrtW38. The pattern of transcription in C. fritschii indicates that upregulation of the CrtW148 -like WP_016874500.1 observed may be the sole transcriptional change responsible for the increase in keto-carotenoids present after UV exposure.
The transcription data indicates an increase in carotenoid synthesis, and a shift toward the production of the ketolated carotenoids echinenone and canthaxanthin under low UV. This was reflected in the significant increases we observed in concentrations of canthaxanthin and echinenone in the HPLC chromatograms. Homologs for all genes (cruF, crtR, cruG) known to contribute to the myxol biosynthesis pathway were upregulated. But notably we did not observe any of the ketolated 4-ketomyxol 2′-methylpentoside in our chromatograms. Echinenone is known to play a key role as the core carotenoid to OCP and red carotenoid protein (RCP), and myxol glycosides are known to provide cell wall photoprotection. However, the role of these carotenoids in photoprotection is still unclear and recent evidence suggests that a wider range of carotenoids including myxol 2′-methylpentosides and canthaxanthin may also play a role in OCP and RCP (Melnicki et al., 2016). In contrast, the hydroxylation pathway to produce zeaxanthin, caloxanthin and nostoxanthin was not affected by UV and again this was reflected in the levels of these pigments which were largely unaltered under UV.
The global transcription factor NtcA regulates carotenoid biosynthesis in Nostoc PCC 7120 through 2-oxoglutarate mediated binding (Sandmann et al., 2016). In C. fritschii PCC 6912 this is encoded by WP_016874320.1, which was not upregulated under UV. Regulation through NtcA via signaling molecules rather than by its increased transcription enables modulation in response to the C/N balance and cellular redox state, and is implicated in regulation in response to high light in Nostoc PCC 7120 (Sandmann et al., 2016). Promotor analysis of C. fritschii PCC 6912 carotenoid pathway gene homologs in comparison to Nostoc PCC 7120 sequences revealed the presence of signature NtcA binding sites giving evidence for similar NtcA regulation of carotenoid biosynthesis in C. fritschii PCC 6912 (For detailed analysis refer to Supplementary File S1).
Photoprotection
We found substantial upregulation of putative genes encoding the NPQ related water soluble photoactive proteins OCP and Fluorescence Recovery Protein (FRP). These proteins together provide the main mechanism by which the PBS is protected from excess light using non-photochemical quenching. The role of Helical Carotenoid Protein (HCP) is still emerging (Bao et al., 2017a). C. fritschii PCC 6912 has a complement of genes analogous to that found in Anabaena: i.e., HCP1-4 and one CTD paralog. In Anabaena, HCP4 containing canthaxanthin bound the PBSs, enabling constitutive fluorescence quenching, and HCP2 and HCP3 exhibited strong singlet oxygen quenching activity, while HCP1 is thought to have an alternate role in stress protection (Lopez-Igual et al., 2016).
We found a putative gene encoding a HCP3, situated nearby the HCP4-CTD gene homologs, and the CTD were downregulated under the UV treatment: Expression of the other HCP transcripts was unchanged. This indicates that, rather than providing additional UV stress protection, HCPs may have alternate roles in C. fritschii. The downregulation of HCP could be interpreted as a requirement to free up bound keto-carotenoids to increase the efficacy of OCP function. The roles of the HCP proteins require further investigation.
Carotenoid Cleavage
Carotenoids are cleaved to form a number of products including retinal. Cyanobacterial genomes have 1-5 CCD gene homologs, with higher numbers in the filamentous cyanobacteria (Cui et al., 2012). Nostoc sp. PCC 7120 has three enzymes, exhibiting different substrate preferences (Marasco et al., 2006). C. fritschii has four corresponding CCD gene homologs including two apocarotenoid oxygenases. Only one of these, which may be involved with retinal synthesis, was upregulated in UV. In cyanobacteria, apocarotenoids act as photoprotective pigments in thylakoid membranes, and retinal acts as the chromophore for rhodopsins, thought to act as photosensory receptors (Jung et al., 2003). However, expression of the C. fritschii rhodopsin (WP_016877934.1) was not modulated by UV light. Cleavage products may also be involved in carotenoid turnover or be involved in signaling or regulation. Induction of an apocarotenoid modulating enzymes has been suggested to allow scavenging of apocarotenoids produced by photodestruction in high light (Ruch et al., 2005). Along with other apocarotenoids, retinal is an industrially valuable metabolite. A greater understanding of the function and regulation of these genes provides useful insights for increasing yields of apocarotenoids including the retinals in industrial biotechnology.
Carotenoid Concentrations
β-carotene and echinenone, the most lipophilic carotenoids we detected, were present in the highest concentrations under white and UV supplemented conditions. Both these pigments act as precursors to canthaxanthin. As a keto-carotenoid canthaxanthin is known to retard the formation of hydroperoxide more efficiently and be more effective as an antioxidant than carotenoids without a keto group (Terao, 1989). Whilst canthaxanthin was present at much lower concentrations under visible and UV, under UV canthaxanthin showed by far the largest change with a ninefold increase in 4h4d exposure. Canthaxanthin has previously been shown to be preferentially formed over zeaxanthin in cells exposed to strong light (500 or 1200 μmol m–2 s–1) with UV-B radiation and, to be a better protectant against solar radiation (Albrecht et al., 2001). An increase in canthaxanthin and a decrease in β-carotene under UV exposure has also been reported in terrestrial cyanobacteria (Lakatos et al., 2001). Also the increases in myxol 2′-methylpentoside and echinenone we observed in the 4h4d UV exposed samples are consistent with other studies where secondary carotenoids have been shown to increase under UV (Ehling-Schulz et al., 1997; Ivanov et al., 2000).
Overall, the changes we found in the carotenoid metabolites in UV exposed cells were consistent with the changes observed in the differential transcription levels, providing strong evidence for pathway activation. The significant increases in both myxol 2′-methylpentoside and canthaxanthin under UV-B were consistent with the significant increases in expression of gene homologs associated with crtW, crtR, cruF, and cruG. However it should be noted that because some genes are involved in multiple reactions and in some cases there are multiple copies then not all genes could be definitively assigned to the production of particular carotenoids.
Conclusion
New insight is provided into the regulatory processes that modulate the synthesis, photoprotection and degradation of carotenoids in the filamentous species of cyanobacteria C. fritschii under low UV-B. Through differential transcript analysis we demonstrated a UV-B induced upregulation of the pathway encoding the initial stages of carotenoid synthesis, producing phytoene and lycopene, but not the cyclases producing β-carotene. Gene homologs encoding enzymes responsible for the synthesis of the keto-carotenoids echinenone and canthaxanthin and of myxol 2′-methylpentoside were upregulated. Metabolite analysis confirmed increases in the keto-carotenoids echinenone and canthaxanthin and of myxol 2′-methylpentoside. This may indicate use of a relatively large β-carotene pool as a mechanism for cellular response to UV via conversion to canthaxanthin via echinenone, with more gradual replacement by lycopene cyclases.
Understanding gene regulation enables optimization of the photochemical performance of cyanobacteria to improve yields of specific carotenoids, such as keto-carotenoids and other valuable biotechnology metabolites such as the apocarotenoid, retinal. Thus our study is relevant to understanding the ecological survival of cyanobacteria under changing climatic conditions and to a society increasingly interested in using cyanobacteria for the production of useful metabolites.
Data Availability Statement
The datasets generated for this study can be found within an NCBI BioProject with accession number PRJNA545395 (data to be released on publication).
Author Contributions
CL secured the funding, conceived the ideas, oversaw the work, and wrote the manuscript. GF secured funding from NBAF and undertook the experiments. CG undertook bioinformatics and contributed to writing the manuscript. RA undertook HPLC of pigment samples, provided the data, and commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Luca Lenzi, Dr. Catharine Sambles, and Dr. Matt Hitchings for their technical support on genome sequencing and transcription profiling. We thank the three reviewers. We are particularly grateful to one of the reviewers who helped us to significantly improve the manuscript providing correct identification for 2-hydroxymyxol 2′-methylpentoside and ensuring that we got essential details correct.
Footnotes
Funding. This work was supported by BBSRC grant BB/E018998/1, Interreg-NWE – EnAlgae and the NERC Biomolecular Analysis Facility at the Centre for Genomic Research, University of Liverpool (NBAF527).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00163/full#supplementary-material
FIGURE S1
Prior experiment to determine level of UV-B at wavelength range 300–310 nm exposure that would not impact detrimentally on photosynthetic efficiency (Fv/Fm). White light, UV 0, 10, and 20 cm represent the control and the distance of the flasks containing C. fritschii PCC 6912 from the UVB source. Average of three replicates with standard deviation. From this, for the main experiment, flasks were placed 10 cm from UV-B tubes measured as supplying 3 μmol m–2 s–1.
FIGURE S3
Gene regulation of terpenoid biosynthesis via the MEP/DOXP pathway to geranylgeranyl diphosphate in C. fritschii WP_016876009.1 was down-regulated (blue shading), other genes were identified but transcription was not observed (light green) or were not differentially regulated (dark green shading).
TABLE S1
Retention time and UV/vis absorption maxima of main peaks detected during HPLC of C. fritschii PCC 6912.
TABLE S2
C. fritschii carotenoid related gene expression in UV supplemented white light.
FILE S1
Promotor signatures of potential NtcA regulation of carotenoid transcription in C. fritschii PCC 6912.
References
- Ahrazem O., Gomez-Gomez L., Rodrigo M. J., Avalos J., Limon M. C. (2016). Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions. Int. J. Mol. Sci. 17 1–38. [Europe PMC free article] [Abstract] [Google Scholar]
- Airs R. L., Temperton B., Sambles C., Farnham G., Skill S. C., Llewellyn C. A. (2014). Chlorophyll f and chlorophyll d are produced in the cyanobacterium Chlorogloeopsis fritschii when cultured under natural light and near-infrared radiation. FEBS Lett. 588 3770–3777. 10.1016/j.febslet.2014.08.026 [Abstract] [CrossRef] [Google Scholar]
- Albrecht M., Steiger S., Sandmann G. (2001). Expression of a ketolase gene mediates the synthesis of canthaxanthin in Synechococcus leading to tolerance against photoinhibition, pigment degradation and UV-B sensitivity of photosynthesis. Photochem. Photobiol. 73 551–555. 10.1562/0031-8655(2001)0730551eoakgm2.0.co2 [Abstract] [CrossRef] [Google Scholar]
- Anders S., Pyl P., Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ashby M. K., Mullineaux C. W. (1999). Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol. Lett. 181 253–260. 10.1111/j.1574-6968.1999.tb08852.x [Abstract] [CrossRef] [Google Scholar]
- Balasundaram B., Skill S. C., Llewellyn C. A. (2012). A low energy process for the recovery of bioproducts from cyanobacteria using a ball mill. Biochem. Eng. J. 69 48–56. 10.1016/j.bej.2012.08.010 [CrossRef] [Google Scholar]
- Bao H., Melnicki M. R., Kerfeld C. A. (2017a). Structure and functions of orange carotenoid protein homologs in cyanobacteria. Curr. Opin. Plant Biol. 37 1–9. 10.1016/j.pbi.2017.03.010 [Abstract] [CrossRef] [Google Scholar]
- Bao H., Melnicki M. R., Pawlowski E. G., Sutter M., Agostoni M., Lechno-Yossef S., et al. (2017b). Additional families of orange carotenoid proteins in the photoprotective system of cyanobacteria. Nat. Plants 3:17089. 10.1038/nplants.2017.89 [Abstract] [CrossRef] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x [CrossRef] [Google Scholar]
- Boulay C., Abasova L., Six C., Vass I., Kirilovsky D. (2008). Occurrence and function of the orange carotenoid protein in photoprotective mechanisms in various cyanobacteria. Biochim. Biophys. Acta Bioenerg. 1777 1344–1354. 10.1016/j.bbabio.2008.07.002 [Abstract] [CrossRef] [Google Scholar]
- Castenholz R. W. (2015). “Cyanobacteria,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. Whitman W. B. (Hoboken, NJ: John Wiley & Sons, Inc; ). [Google Scholar]
- Chang W. C., Song H., Liu H. W., Liu P. (2013). Current development in isoprenoid precursor biosynthesis and regulation. Curr. Opin. Chem. Biol. 17 571–579. 10.1016/j.cbpa.2013.06.020 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cui H., Wang Y., Qin S. (2012). Genomewide analysis of carotenoid cleavage dioxygenases in unicellular and filamentous cyanobacteria. Comp. Funct. Genom. 2012:164690. 10.1155/2012/164690 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ehling-Schulz M., Bilger W., Scherer S. (1997). UV-B-induced synthesis of photoprotective pigments and extracellular polysaccharides in the terrestrial cyanobacterium Nostoc commune. J. Bacteriol. 179 1940–1945. 10.1128/jb.179.6.1940-1945.1997 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Evans E. H., Britton G. (1983). Relationship between growth-conditions, cell morphology and carotenoid composition of the cyanobacterium Chlorogloeopsis (Chlorogloea) fritschii, and carotenoid compositions of Photosystem-1 and Photosystem-2 preparations. Arch. Microbiol. 135 284–286. 10.1007/bf00413482 [CrossRef] [Google Scholar]
- Gan F., Bryant D. A. (2015). Adaptive and acclimative responses of cyanobacteria to far-red light. Environ. Microbiol. 17 3450–3465. 10.1111/1462-2920.12992 [Abstract] [CrossRef] [Google Scholar]
- Graham J. E., Bryant D. A. (2008). The biosynthetic pathway for synechoxanthin, an aromatic carotenoid synthesized by the euryhaline, unicellular cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 190 7966–7974. 10.1128/JB.00985-08 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Graham J. E., Bryant D. A. (2009). The biosynthetic pathway for myxol-2′ fucoside (myxoxanthophyll) in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 191 3292–3300. 10.1128/JB.00050-09 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Heo J., Kim S. H., Lee P. C. (2013). New insight into the cleavage reaction of Nostoc sp. strain PCC 7120 carotenoid cleavage dioxygenase in natural and nonnatural carotenoids. Appl. Environ. Microbiol. 79 3336–3345. 10.1128/AEM.00071-13 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hideg E., Jansen M. A., Strid A. (2013). UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 18 107–115. 10.1016/j.tplants.2012.09.003 [Abstract] [CrossRef] [Google Scholar]
- Ho M. Y., Bryant D. A. (2019). Global transcriptional profiling of the cyanobacterium Chlorogloeopsis fritschii PCC 9212 in far-red light: insights into the regulation of chlorophyll d synthesis. Front. Microbiol. 10:465. 10.3389/fmicb.2019.00465 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ivanov A. G., Miskiewicz E., Clarke A. K., Greenberg B. M., Huner N. P. A. (2000). Protection of photosystem II against UV-A and UV-B radiation in the cyanobacterium Plectonema boryanum: the role of growth temperature and growth irradiance. Photochem. Photobiol. 72 772–779. 10.1562/0031-8655(2000)0720772popiau2.0.co2 [Abstract] [CrossRef] [Google Scholar]
- Jansen M. A. K., Hectors K., O’brien N. M., Guisez Y., Potters G. (2008). Plant stress and human health: do human consumers benefit from UV-B acclimated crops? Plant Sci. 175 449–458. 10.1016/j.plantsci.2008.04.010 [CrossRef] [Google Scholar]
- Jeffrey S. W., Wright S. W., Zapata M. (2011). “Microalgal classes and their pigment signatures,” in Phytoplankton Pigments, eds Roy S., Llewellyn C. A., Egeland E. S., Johnsen G. (Cambridge, MA: Cambridge Press; ), 3–77. [Google Scholar]
- Jung K. H., Trivedi V. D., Spudich J. L. (2003). Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 47 1513–1522. 10.1046/j.1365-2958.2003.03395.x [Abstract] [CrossRef] [Google Scholar]
- Kirilovsky D., Kerfeld C. A. (2012). The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim. Biophys. Acta Bioenerg. 1817 158–166. 10.1016/j.bbabio.2011.04.013 [Abstract] [CrossRef] [Google Scholar]
- Kloer D. P., Schulz G. E. (2006). Structural and biological aspects of carotenoid cleavage. Cell. Mol. Life Sci. 63 2291–2303. 10.1007/s00018-006-6176-6 [Abstract] [CrossRef] [Google Scholar]
- Kultschar B., Dudley E., Wilson S., Llewellyn C. A. (2019). Intracellular and extracellular metabolites from the cyanobacterium Chlorogloeopsis fritschii PCC 6912 during 48 Hours of UV-B exposure. Metabolites 9 1–15. 10.3390/metabo9040074 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lakatos M., Bilger W., Budel B. (2001). Carotenoid composition of terrestrial cyanobacteria: response to natural light conditions in open rock habitats in Venezuela. Eur. J. Phycol. 36 367–375. 10.1080/09670260110001735518 [CrossRef] [Google Scholar]
- Liang C., Zhao F., Wei W., Wen Z., Qin S. (2006). Carotenoid biosynthesis in cyanobacteria: structural and evolutionary scenarios based on comparative genomics. Int. J. Biol. Sci. 2 197–207. 10.7150/ijbs.2.197 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Liu H., Zhang H., Niedzwiedzki D. M., Prado M., He G., Gross M. L., et al. (2013). Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 342 1104–1107. 10.1126/science.1242321 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lohr M. (2011). “Carotenoid metabolism in phytoplankton,” in Phytoplankton Pigments, eds Roy S., Llewellyn C. A., Egeland E., Johnsen S. G. (Cambridge, MA: Cambridge University Press; ), 113–164. [Google Scholar]
- Lohr M., Im C. S., Grossman A. R. (2005). Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii. Plant Physiol. 138 490–515. 10.1104/pp.104.056069 [Abstract] [CrossRef] [Google Scholar]
- Lopez-Igual R., Wilson A., Leverenz R. L., Melnicki M. R., Bourcier De Carbon C., Sutter M., et al. (2016). Different functions of the paralogs to the N-terminal domain of the orange carotenoid protein in the cyanobacterium Anabaena sp. PCC 7120. Plant Physiol. 171 1852–1866. 10.1104/pp.16.00502 [Abstract] [CrossRef] [Google Scholar]
- Makino T., Harada H., Ikenaga H., Matsuda S., Takaichi S., Shindo K., et al. (2008). Characterization of cyanobacterial carotenoid ketolase CrtW and hydroxylase CrtR by complementation analysis in Escherichia coli. Plant Cell Physiol. 49 1867–1878. 10.1093/pcp/pcn169 [Abstract] [CrossRef] [Google Scholar]
- Marasco E. K., Vay K., Schmidt-Dannert C. (2006). Identification of carotenoid cleavage dioxygenases from Nostoc sp. PCC 7120 with different cleavage activities. J. Biol. Chem. 281 31583–31593. 10.1074/jbc.m606299200 [Abstract] [CrossRef] [Google Scholar]
- Melnicki M. R., Leverenz R. L., Sutter M., Lopez-Igual R., Wilson A., Pawlowski E. G., et al. (2016). Structure, diversity, and evolution of a new family of soluble carotenoid-binding proteins in cyanobacteria. Mol. Plant 9 1379–1394. 10.1016/j.molp.2016.06.009 [Abstract] [CrossRef] [Google Scholar]
- Miles J. A., Machattou P., Nevin-Jones D., Webb M. E., Millard A., Scanlan D. J., et al. (2019). Identification of a cyanobacterial aldehyde dehydrogenase that produces retinoic acid in vitro. Biochem. Biophys. Res. Commun. 510 27–34. 10.1016/j.bbrc.2018.12.171 [Abstract] [CrossRef] [Google Scholar]
- Mitra A. K. (1950). Two new algae from Indian soils. Ann. Bot. 14 457–464. 10.1016/j.jphotobiol.2019.111638 [Abstract] [CrossRef] [Google Scholar]
- Mochimaru M., Masukawa H., Takaichi S. (2005). The cyanobacterium Anabaena sp. PCC 7120 has two distinct beta-carotene ketolases: CrtO for echinenone and CrtW for ketomyxol synthesis. FEBS Lett. 579 6111–6114. 10.1016/j.febslet.2005.09.081 [Abstract] [CrossRef] [Google Scholar]
- Murata N., Sato N., Omata T., Kuwabara T. (1981). Separation and characterization of thylakoid and cell-envelope of the blue-green-alga (Cyanobacterium) Anacystis nidulans. Plant Cell Physiol. 22 855–866. [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S. I. (2007). Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta Bioenerg. 1767 414–421. 10.1016/j.bbabio.2006.11.019 [Abstract] [CrossRef] [Google Scholar]
- Paniagua-Michel J., Olmos-Soto J., Ruiz M. A. (2012). Pathways of carotenoid biosynthesis in bacteria and microalgae. Methods Mol. Biol. 892 1–12. 10.1007/978-1-61779-879-5_1 [Abstract] [CrossRef] [Google Scholar]
- Pattanaik B., Lindberg P. (2015). Terpenoids and their biosynthesis in cyanobacteria. Life 5 269–293. 10.3390/life5010269 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rastogi R. P., Sinha R. P., Moh S. H., Lee T. K., Kottuparambil S., Kim Y. J., et al. (2014). Ultraviolet radiation and cyanobacteria. J. Photochem. Photobiol. B Biol. 141 154–169. 10.1016/j.jphotobiol.2014.09.020 [Abstract] [CrossRef] [Google Scholar]
- Ruch S., Beyer P., Ernst H., Al-Babili S. (2005). Retinal biosynthesis in eubacteria: in vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Mol. Microbiol. 55 1015–1024. 10.1111/j.1365-2958.2004.04460.x [Abstract] [CrossRef] [Google Scholar]
- Sandmann G., Mautz J., Breitenbach J. (2016). Control of light-dependent keto carotenoid biosynthesis in Nostoc 7120 by the transcription factor NtcA. Z. Naturforsch. C 71 303–311. 10.1515/znc-2016-0117 [Abstract] [CrossRef] [Google Scholar]
- Schagerl M., Donabaum K. (2003). Patterns of major photosynthetic pigments in freshwater algae. 1. Cyanoprokaryota, Rhodophyta and Cryptophyta. Ann. Limnol. Int. J. Limnol. 39 35–47. 10.1065/espr2007.01.379 [Abstract] [CrossRef] [Google Scholar]
- Scherzinger D., Al-Babili S. (2008). In vitro characterization of a carotenoid cleavage dioxygenase from Nostoc sp. PCC 7120 reveals a novel cleavage pattern, cytosolic localization and induction by highlight. Mol. Microbiol. 69 231–244. 10.1111/j.1365-2958.2008.06282.x [Abstract] [CrossRef] [Google Scholar]
- Scherzinger D., Ruch S., Kloer D. P., Wilde A., Al-Babili S. (2006). Retinal is formed from apo-carotenoids in Nostoc sp. PCC7120: in vitro characterization of an apo-carotenoid oxygenase. Biochem. J. 398 361–369. 10.1042/bj20060592 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Scholes G. D., Fleming G. R., Olaya-Castro A., Van Grondelle R. (2011). Lessons from nature about solar light harvesting. Nat. Chem. 3 763–774. 10.1038/nchem.1145 [Abstract] [CrossRef] [Google Scholar]
- Schopf L., Mautz J., Sandmann G. (2013). Multiple ketolases involved in light regulation of canthaxanthin biosynthesis in Nostoc punctiforme PCC 73102. Planta 237 1279–1285. 10.1007/s00425-013-1846-8 [Abstract] [CrossRef] [Google Scholar]
- Steele D. J., Kimmance S. A., Franklin D. J., Airs R. L. (2018). Occurrence of chlorophyll allomers during virus-induced mortality and population decline in the ubiquitous picoeukaryote Ostreococcus tauri. Environ. Microbiol. 20 588–601. 10.1111/1462-2920.13980 [Abstract] [CrossRef] [Google Scholar]
- Steiger S., Sandmann G. (2004). Cloning of two carotenoid ketolase genes from Nostoc punctiforme for the heterologous production of canthaxanthin and astaxanthin. Biotechnol. Lett. 26 813–817. 10.1023/b:bile.0000025880.91269.cf [Abstract] [CrossRef] [Google Scholar]
- Takaichi S., Mochimaru M. (2007). Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides. Cell. Mol. Life Sci. 64 2607–2619. 10.1007/s00018-007-7190-z [Abstract] [CrossRef] [Google Scholar]
- Takaichi S., Mochimaru M., Maoka T., Katoh H. (2005). Myxol and 4-ketomyxol 2′-fucosides, not rhamnosides, from Anabaena sp. PCC 7120 and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol. 46 497–504. 10.1093/pcp/pci049 [Abstract] [CrossRef] [Google Scholar]
- Tanigawa R., Shirokane M., Maeda S., Omata T., Tanaka K., Takahashi H., et al. (2002). Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. U.S.A. 99 4251–4255. 10.1073/pnas.072587199 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Terao J. (1989). Antioxidant activity of beta-carotene-related carotenoids in solution. Lipids 24 659–661. 10.1007/bf02535085 [Abstract] [CrossRef] [Google Scholar]
- Vajravel S., Kis M., Klodawska K., Laczko-Dobos H., Malec P., Kovacs L., et al. (2017). Zeaxanthin and echinenone modify the structure of photosystem I trimer in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta Bioenerg. 1858 510–518. 10.1016/j.bbabio.2017.05.001 [Abstract] [CrossRef] [Google Scholar]
- Wilson A., Punginelli C., Gall A., Bonetti C., Alexandre M., Routaboul J. M., et al. (2008). A photoactive carotenoid protein acting as light intensity sensor. Proc. Natl. Acad. Sci. U.S.A. 105 12075–12080. 10.1073/pnas.0804636105 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zapata M., Rodriguez F., Garrido J. L. (2000). Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C-8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 195 29–45. 10.3354/meps195029 [CrossRef] [Google Scholar]
Articles from Frontiers in Microbiology are provided here courtesy of Frontiers Media SA
Full text links
Read article at publisher's site: https://doi.org/10.3389/fmicb.2020.00163
Read article for free, from open access legal sources, via Unpaywall:
https://www.frontiersin.org/articles/10.3389/fmicb.2020.00163/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.3389/fmicb.2020.00163
Article citations
Extensive Genomic Rearrangement of Catalase-Less Cyanobloom-Forming Microcystis aeruginosa in Freshwater Ecosystems.
J Microbiol, 08 Oct 2024
Cited by: 0 articles | PMID: 39377859
Review
Microbial canthaxanthin: an orange-red keto carotenoid with potential pharmaceutical applications.
BioTechnologia (Pozn), 104(3):315-328, 25 Sep 2023
Cited by: 2 articles | PMID: 37850112 | PMCID: PMC10578118
Review Free full text in Europe PMC
Protection and Damage Repair Mechanisms Contributed To the Survival of Chroococcidiopsis sp. Exposed To a Mars-Like Near Space Environment.
Microbiol Spectr, 10(6):e0344022, 01 Dec 2022
Cited by: 2 articles | PMID: 36453906 | PMCID: PMC9769825
Metabolomics of Chlorophylls and Carotenoids: Analytical Methods and Metabolome-Based Studies.
Antioxidants (Basel), 10(10):1622, 15 Oct 2021
Cited by: 4 articles | PMID: 34679756 | PMCID: PMC8533378
Review Free full text in Europe PMC
Metabolic engineering of Synechocystis sp. PCC 6803 for the photoproduction of the sesquiterpene valencene.
Metab Eng Commun, 13:e00178, 13 Aug 2021
Cited by: 8 articles | PMID: 34466381 | PMCID: PMC8382996
Go to all (10) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (2 citations) BioProject - PRJNA545395
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of cyanobacterial carotenoid ketolase CrtW and hydroxylase CrtR by complementation analysis in Escherichia coli.
Plant Cell Physiol, 49(12):1867-1878, 05 Nov 2008
Cited by: 18 articles | PMID: 18987067
Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides.
Cell Mol Life Sci, 64(19-20):2607-2619, 01 Oct 2007
Cited by: 79 articles | PMID: 17643187 | PMCID: PMC11136355
Review Free full text in Europe PMC
2,2'-beta-hydroxylase (CrtG) is involved in carotenogenesis of both nostoxanthin and 2-hydroxymyxol 2'-fucoside in Thermosynechococcus elongatus strain BP-1.
Plant Cell Physiol, 49(11):1678-1687, 15 Sep 2008
Cited by: 25 articles | PMID: 18794175
Carotenoids, versatile components of oxygenic photosynthesis.
Prog Lipid Res, 52(4):539-561, 26 Jul 2013
Cited by: 91 articles | PMID: 23896007
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (1)
Optimising yield of antioxidants and sunscreens in microalgae for sustainable biosynthesis of ingredients for health and beauty products
Dr Llewellyn, Plymouth Marine Laboratory
Grant ID: BB/E018998/1
Interreg
NERC Environmental Bioinformatics Centre
Natural Environment Research Council (7)
Grant ID: NBAF010004
Grant ID: NE/R015953/1
Grant ID: NBAF010002
Grant ID: NE/C510016/1
Grant ID: NE/N001974/1
Grant ID: NE/P006434/1
Grant ID: NE/P021409/1





