Abstract
Free full text

Cytoplasmic m1A reader YTHDF3 inhibits trophoblast invasion by downregulation of m1A-methylated IGF1R
Abstract
N1-methyladenosine (m1A) is one of the important post-transcriptional modifications in RNA and plays an important role in promoting translation or decay of m1A-methylated messenger RNA (mRNA), but the “reader” protein and the exact biological role of m1A remain to be determined. Here, we identified that nine potential m1A “reader” proteins including YTH domain family and heterogeneous nuclear ribonucleoprotein by mass spectrometry, and among them, YTH domain-containing protein 3 (YTHDF3), could bind directly to m1A-carrying RNA. YTHDF3 was then identified to negatively regulate invasion and migration of trophoblast. Mechanistically, we found that the m1A “reader” YTHDF3 bound to certain m1A-methylated transcripts, such as insulin-like growth factor 1 receptor (IGF1R), with the combination of iCLIP-seq (individual-nucleotide resolution ultraviolet crosslinking and immunoprecipitation high-throughput sequencing) and m1A-seq. Furthermore, YTHDF3 could promote IGF1R mRNA degradation and thus inhibit IGF1R protein expression along with its downstream matrix metallopeptidase 9 signaling pathway, consequently decreasing migration and invasion of trophoblast. Thus, we demonstrated that YTHDF3 as an m1A reader decreased invasion and migration of trophoblast by inhibiting IGF1R expression. Our study outlines a new m1A epigenetic way to regulate the trophoblast activity, which suggests a novel therapeutic target for trophoblast-associated pregnancy disorders.
Introduction
Eukaryote RNAs are found to possess more than 100 different types of post-transcriptional modifications, which can regulate RNA splicing, localization, stability, binding, and translation1–3. Methylation at the N6 position of adenosine (m6A) is involved in the epigenetic regulation of gene expression4,5. N1-methyladenosine (m1A) is one frequently occurring modification, which is typically found at many transfer RNAs (tRNAs)6,7. Recent transcriptome-wide mapping also revealed that m1A modification is also present in human messenger RNAs (mRNAs) and suggested the potential roles of m1A in modulating mRNA splicing and translation; meanwhile, m1A modification is dynamically changed upon external heat or starve stimulation8–10. Maternal–fetal interface cells would suffer many external stimuli (hypoxia, lipopolysaccharide (LPS), and hormone), which dynamically changed during the pregnancy, especially the hypoxia environment could affect trophoblast activity11–13. However, whether the trophoblast has dynamic m1A modification upon hypoxia remains unclear. Furthermore, the exact function of dynamic m1A upon different stimuli, especially the m1A modification on specific nucleotide of unique mRNA, has still not been understood and remains to be explored in the future.
Previously, YTH domain-containing proteins were reported to bind directly to m6A and to regulate RNA metabolism as readers14,15. YTH domain-containing protein 3 (YTHDF3) and YTHDF1 promotes protein synthesis by interacting with the translation machinery and YTHDF3 can increase decay of m6A-bearing RNA16,17, but YTHDF2 decreased the stability of m6A-modified mRNA and inhibits the translation of mRNAs18. YTH domain containing 1 (YTHDC1) regulates mRNA splicing by modulating pre-mRNA splicing factors19. Taken together, the YTH domain-containing proteins regulate various biological processes via binding to m6A-modified RNA20. m1A as another important RNA modification has been found to be written by tRNA methyltransferase 6 (TRMT6)/TRMT61A complex (writers)21,22 and erased by a-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3) or ALKBH1 (erasers) in human cells9,23. It has been suggested that YTH domain family can interact with m1A-carrying RNA, potentially functioning as an m1A reader24. However, cellular proteins acted as readers involved in binding to m1A-carrying mRNA remain to be explored, and how the key target transcripts of these m1A readers affect cell activity and which underlying pathways and mechanisms mediate these changes are still far from elucidated.
“Insulin-like growth factor (IGF) axis” plays a key role in regulating cell growth and survival and affecting virtually every organ in the body25–27. In the placenta, the IGF 1 receptor (IGF1R) is expressed in all cell types, including the trophoblast and villous endothelium28. Downregulation of placental IGF1R might be an important factor in pregnancies complicated by intrauterine growth restriction (IUGR)29. Recent report has also shown a significant downregulation of IGF1R protein levels in IUGR pregnancies compared to normal pregnancies30,31. Interestingly, IGF1R protein downregulation in IUGR placentas was accompanied by impaired activation of intracellular signaling molecules of the IGF1R32,33. Moreover, insulin/IGF1R signaling can induce the expression of membrane-type matrix metalloproteinase (MMP), which is the key regulator of cell invasion, migration, and tissue remodeling34,35. However, how to accurately regulate the expression of IGF1R, especially via the RNA epigenetic way, subsequently affecting the downstream signaling of IGF1R, still needs to be illustrated.
Herein, we found that YTHDF3 acted as an m1A “reader” protein, and YTHDF3 expression was significantly downregulated in HTR8/SVneo cells upon hypoxia treatment. YTHDF3 knockdown increased the expression of key molecule IGF1R via enhancing m1A-modified IGF1R mRNA stability, and consequently upregulated MMP9 expression, and finally promoted trophoblast migration and invasion in vitro. Together, our results may add insights to the m1A “reader” YTHDF3 as an important regulator of the IGF signaling pathway in promoting invasion of trophoblast, providing potential targets to control pregnancy-associated diseases induced by hypoxia.
Results
YTHDF3 selectively binds to m1A-containing RNA
To better understand the biological functions of m1A in RNA, we designed a 5′-biotin-labeled m1A-carrying RNA sequence as the probe bait (Fig. (Fig.1a),1a), which was derived from human 28S ribosomal RNA (rRNA) gene and harbored two m1A sites in the stem-loop structure9. First, the m1A and m6A contents of m1A probes were quantified by liquid chromatography with tandem mass spectrometry (LC-MS/MS), we found that the m1A/A ratio is about 30%, and m6A/A ratio is about 1% (Supplementary Fig. S1a), indicating that the m1A probes have no obvious m6A contamination. Furthermore, we verified that m1A antibody can specifically bind to m1A RNA probe, but not to the corresponding unmethylated control bait, and m6A antibody showed a faint nonspecific binding to m1A RNA probe (Fig. (Fig.1b).1b). Using LC-MS/MS to screen systematically for cellular proteins that bound to m1A bearing RNA probe (Supplementary Fig. S1b, c), we identified 155 unique proteins in Raw264.7 cells and 27 unique proteins in HEK293T, respectively (Supplementary Fig. S1d, e). Among nine unique interaction proteins identified in both HEK293T and Raw264.7 cell lines (Supplementary Table S1), we found that YTHDF3 with the highest score in human cells can specificly bind to m1A modification probes (Supplementary Fig. S1f). Using biotin pulldown method, we confirmed that YTHDF3 bound significantly stronger to m1A RNA probes than YTHDF2, but YTHDF1 almost had no interaction (Fig. (Fig.1c);1c); we further confirmed that YTHDF3 could bind to another m1A probe from SOX18 mRNA (Supplementary Fig. S1g). These results indicated that YTHDF3 selectively bound to m1A-methylated RNA and might act as a “reader” protein.
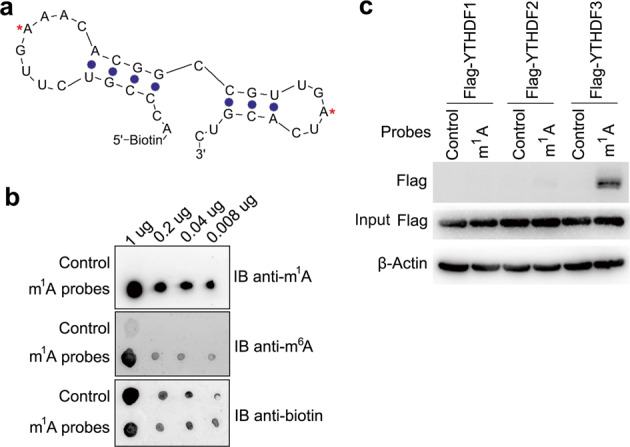
a Illustration of the m1A-modified RNA probe with stem and loop structure derived from the known m1A 1322 site in human 28S rRNA. Red asterisks indicated the m1A-modified sites. b Dot-blot analysis of the m1A probes specificity with m1A or m6A antibody. c IP and immunoblot analyses of the indicated probes in trophoblast HTR8/SVneo transfected with YTHDF1/2/3 plasmid as indicated. Data are representative of three independent experiments.
YTHDF3 inhibited the activity of trophoblast
First, we found that YTHDF3 was significantly downregulated in HTR8/SVneo cells upon hypoxia treatment (Fig. 2a, b), indicating that YTHDF3 may play a potential role in regulating the activity of trophoblast. To investigate the biological significance of YTHDF3, we examined the effects of YTHDF3 knockdown on trophoblast activity. We confirmed that YTHDF3 was stably knocked down by using two different lentiviral short hairpin RNA (shRNA) sequences (Fig. 2c, d). We found that YTHDF3 knockdown significantly promoted the migration, invasion, and proliferation of trophoblast (Fig. 2e–g). In parallel, overexpression of YTHDF3 significantly inhibited the migration, invasion, and proliferation of trophoblast (Fig. 2h–k), suggesting that YTHDF3 could inhibit the activity of trophoblast.
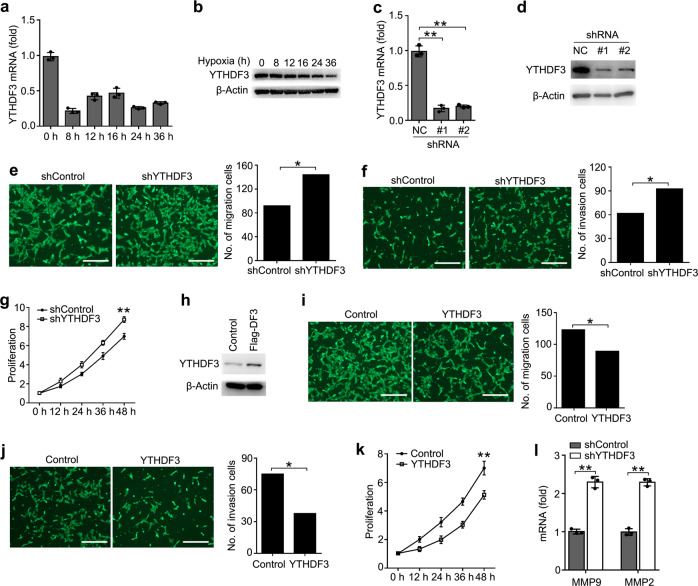
a, b qRT-PCR and immunoblot analysis of YTHDF3 mRNA (a) and protein (b) in trophoblast treated with hypoxia as indicated time. The gene expression was normalized to that of the β-actin internal control in each sample. c, d qRT-PCR and immunoblot analysis of YTHDF3 in trophoblast HTR8/SVneo transfected with lentiviral shControl (NC), YTHDF3 shRNA #1 or #2 as indicated for 72 h. e, f Left: Migration (e) and invasion (f) analysis of HTR8/SVneo cells stably expressing control shRNA (shControl) vs. shRNA targeting YTHDF3 (shYTHDF3) by Transwell assay. Representative images are shown. Right: The number of migration and invasion cells was counted by the ImageJ software. g, k Cell proliferation measured by CCK8 assay of HTR8/SVneo cells stably expressing shYTHDF3 (g) and YTHDF3 (k) as indicated. The value of OD450 was normalized to the value about 24
h. e, f Left: Migration (e) and invasion (f) analysis of HTR8/SVneo cells stably expressing control shRNA (shControl) vs. shRNA targeting YTHDF3 (shYTHDF3) by Transwell assay. Representative images are shown. Right: The number of migration and invasion cells was counted by the ImageJ software. g, k Cell proliferation measured by CCK8 assay of HTR8/SVneo cells stably expressing shYTHDF3 (g) and YTHDF3 (k) as indicated. The value of OD450 was normalized to the value about 24 h after cell seeding. h Immunoblot analysis of HTR8/SVneo cells stably expressing YTHDF3 as indicated. i, j Left: Migration (i) and invasion (j) analysis of HTR8/SVneo cells transfected with YTHDF3 plasmid by Transwell assay. Representative images are shown. Right: The number of migration and invasion cells was counted by the ImageJ software. l qRT-PCR analysis of MMP9 and MMP2 mRNA in HTR8/SVneo stably expressing shControl and shYTHDF3 as indicated. Scale bars, 100
h after cell seeding. h Immunoblot analysis of HTR8/SVneo cells stably expressing YTHDF3 as indicated. i, j Left: Migration (i) and invasion (j) analysis of HTR8/SVneo cells transfected with YTHDF3 plasmid by Transwell assay. Representative images are shown. Right: The number of migration and invasion cells was counted by the ImageJ software. l qRT-PCR analysis of MMP9 and MMP2 mRNA in HTR8/SVneo stably expressing shControl and shYTHDF3 as indicated. Scale bars, 100 μm; *p
μm; *p <
< 0.05, **p
0.05, **p <
< 0.01 (Student’s t test). Data are representative of three independent experiments (mean and s.d. of technical triplicates (a, c, g, k, l)).
0.01 (Student’s t test). Data are representative of three independent experiments (mean and s.d. of technical triplicates (a, c, g, k, l)).
To further determine the role of YTHDF3 in regulating the activity of trophoblast, we examined the downstream genes of YTHDF3 via transcriptome sequencing. The results indicated that 478 genes in HTR8/SVneo were significantly changed upon YTHDF3 stable knockdown (GEO Accession number: GSE135407) (Supplementary Fig. S2). Because extracellular matrix metallopeptidase played an important role in promoting trophoblast invasion36, we further confirmed that knockdown of YTHDF3 enhanced the expression of MMP9 and MMP2 expression in HTR8/SVneo (Fig. (Fig.2l).2l). These results indicated that YTHDF3 inhibited the invasion of trophoblast probably via inhibiting the MMP9 and MMP2 expression.
Identification of YTHDF3-binding RNAs by iCLIP-seq
We further study the underlying mechanisms responsible for YTHDF3-mediated inhibition of trophoblast invasion. As YTHDF3 locates almost exclusively in the cytoplasm of HTR8/SVneo cells treated with hypoxia or LPS (Supplementary Fig. S3a) and it was reported previously that YTHDF3 functions primarily as the RNA-binding protein and m6A reader17, we hypothesized that cytoplasmic YTHDF3 might directly bind a series of RNAs to inhibit trophoblast invasion. To identify YTHDF3-binding RNAs or the direct targeted RNAs, we performed individual-nucleotide resolution ultraviolet (UV) crosslinking and immunoprecipitation (IP) high-throughput sequencing (iCLIP-seq). Immunoblotting of RNA-protein complexes revealed extensive signal for anti-YTHDF3 but not control anti-IgG IPs, suggesting the enrichment for YTHDF3-binding RNAs (Fig. (Fig.3a).3a). Complementary DNA (cDNA libraries were constructed from the purified RNAs and submitted for Illumina deep-sequencing (Supplementary Fig. S3b, c). From four pooled YTHDF3 iCLIP experiments, we obtained 62 and 76 million clean reads from the HTR8/SVneo cells upon normoxia condition, and 72 and 40 million clean reads from hypoxia condition, respectively (GEO Accession number: GSE135642) (Supplementary Table S2). After mapping hg38 human genome and removing PCR duplicates, we yielded 889 and 1076 peaks from the normoxia cells, and 589 and 567 peaks from hypoxia cells (Supplementary Table S2). Interestingly, YTHDF3 peaks were mainly enriched in intergenic (~65%) and introns (~23%), but less (~12%) in coding sequence (CDS), 5′-untranslated regions (5′-UTRs), 3′-UTRs, and noncoding RNA regions (Fig. (Fig.3b),3b), suggesting that YTHDF3 preferentially binds intergenic region of RNA transcripts. Most of the annotated RNA transcripts are mRNA and ribosomal RNAs, and 76 transcripts are shared between 131 ones in normoxia cells and 112 ones in hypoxia cells (Supplementary Fig. S3d, e, f), confirming that the data were highly consistent across different treatments. Hexamer enrichment analysis of iCLIP-seq peaks within all annotated genes identified that YTHDF3 bound to CGGAAGA motif of RNA transcripts in normoxia condition and GAACCGC motif in hypoxia condition (Fig. (Fig.3c).3c). Together, these data showed both location and sequence preference for YTHDF3-binding RNAs.
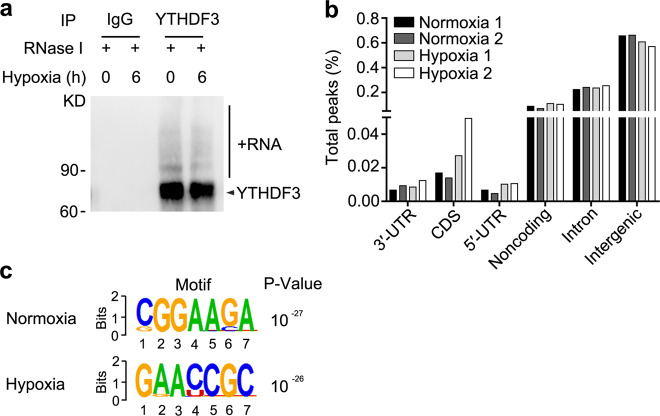
a Immunoblot analysis of immuno-purified and biotin-labeled YTHDF3-RNA complexes transferred to PVDF membrane. IP with IgG as a control shows high specificity of the YTHDF3-RNA signal. b Percentage of total nucleotides under significant iCLIP-seq peaks within refSeq protein-coding genes broken down into transcript feature types extracted from refSeq. c Composite motif logo of the multiple sequence alignment of the most enriched hexamers under significant iCLIP-seq peaks within protein-coding genes as identified by P value, comparing hexamer frequencies to permutations of binding site locations within bound transcripts for normoxia (top) or hypoxia (bottom) cells. Data are representative of three independent experiments with similar results (a).
To understand the functional targets of YTHDF3, we used DAVID (Database for Annotation, Visualization, and Integrated Discovery) to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) terms enriched among protein-coding genes, which were associated with YTHDF3 iCLIP-seq peaks. We observed that YTHDF3 might take part in a set of biological processes and the enrichment for several cellular signaling pathways (Supplementary Fig. S3g, i). Intriguingly, YTHDF3-binding RNAs were enriched in insulin signaling pathway and vascular endothelial growth factor (VEGF) signaling pathway (Supplementary Fig. S3h, j). Thus, the iCLIP-seq peaks further reflect the signaling pathway of YTHDF3 in the regulation of trophoblast invasion.
m1A-seq in trophoblast induced by hypoxia
To explore the mechanisms of m1A “reader” YTHDF3 on trophoblast activity, we intended to identify the transcripts with altered m1A methylation in trophoblast induced by hypoxia. We performed m1A-IP analysis of HTR8/SVneo cells upon hypoxia vs. normoxia (GEO Accession number: GSE135403) (Supplementary Table S3). Trophoblast upon hypoxia exhibited a relatively low total m1A level (Supplementary Fig. S4a). Consistent with previous m1A-seq results and that regions harboring m1A sites have higher GC content8, we identified that the m1A peaks were enriched near the start codons, with the relative conserved GACCCGU and UGAGGXG (X means C, A, U) motifs (Fig. (Fig.4a4a and Supplementary Fig. S4b). In total, we identified about 580 m1A peaks in about 494 transcripts from the normoxia sample and 190 m1A peaks in about 183 transcripts from the hypoxia sample, in which 29 transcripts showed common m1A modifications between the two groups (Fig. 4b, c). In HTR8/SVneo cells treated with hypoxia, 551 peaks in 465 transcripts disappeared and 161 new peaks in 154 transcripts appeared; the 29 peaks in 29 transcripts were found in both normoxia- and hypoxia-treated cells (Fig. 4b, c). We further investigated the m1A distribution patterns within both total and unique peaks. A similar pattern of total and unique m1A distribution in normoxia and hypoxia cells was observed when the RNA species were divided into 5′-UTR, start codon, CDS, stop codon, 3′-UTR regions of mRNAs, and noncoding RNAs (Fig. (Fig.4d).4d). Interestingly, total or unique peaks in hypoxia showed a significant increase of m1A deposit appeared in 5′-UTR regions and a decrease of m1A in CDS regions compared to total peaks in normoxia (Fig. (Fig.4d).4d). The transcripts showing disappeared (normoxia) or new (hypoxia) m1A methylation were enriched in GO terms related to cell migration and cell-matrix adhesion (Supplementary Fig. S4c, e), and KEGG terms related to RNA transport pathway, VEGF signaling pathway, and focal adhesion pathway (Supplementary Fig. S4d, f). These results of GO and KEGG analysis further indicated that m1A modification played important roles in the trophoblast cells mainly via regulating the cell-matrix signaling pathway.
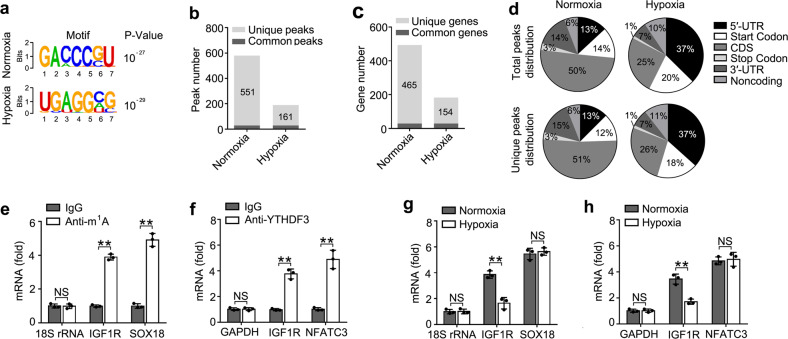
a Composite motif logo of the multiple sequences with significant m1A peaks for normoxia (top) or hypoxia (bottom) HTR8/SVneo cells. b Number of m1A peaks identified in m1A-seq in normoxia and hypoxia HTR8/SVneo cells. c Number of m1A-modified genes identified in m1A-seq. Common m1A genes contain at least one common m1A peak, while unique m1A genes contain no common m1A peak. d Graphs of m1A peak distribution showing the proportion of total m1A peaks in the indicated regions in normoxia and hypoxia cells (top) and loss of existing m1A peaks (unique peaks in normoxia) or the appearance of new m1A peaks (unique peaks in hypoxia) after hypoxia treatment (bottom). e, f Enrichment fold of the indicated RNA transcripts in m1A (e), YTHDF3 (f) IP vs. RNA input control. g, h Enrichment fold of the indicated RNA transcripts in m1A (g), YTHDF3 (h) IP vs. RNA input control in HTR8/SVneo upon hypoxia for 12 h. Each transcript was quantified by qRT-PCR. NS: not significant; **p
h. Each transcript was quantified by qRT-PCR. NS: not significant; **p <
< 0.01 (Student’s t test); NS, not significant. Data are representative of three independent experiments (mean and s.d. of technical triplicates (e–h)).
0.01 (Student’s t test); NS, not significant. Data are representative of three independent experiments (mean and s.d. of technical triplicates (e–h)).
Because the insulin signaling pathway played an important role in maintaining pregnancy37, we hypothesized that YTHDF3 might inhibit the activity of trophoblast through inhibition of the insulin pathway. Taken together with the sequencing results of transcriptome of YTHDF knockdown, iCLIP of YTHDF3 and m1A-IP, transcripts of IGF1R gene were among the most enriched YTHDF3-binding m1A-modified RNAs in insulin signaling pathway. Then, we performed IP of the HTR8/SVneo mRNA with m1A antibody, followed by quantitative reverse transcription PCR (RT-qPCR), and found that, in addition to negative control 18S rRNA, IGF1R mRNAs, and SOX18 (positive control)8 were enriched in the m1A antibody-immunoprecipitated fraction, and found that IGF1R were not enriched in the m6A antibody-immunoprecipitated fraction, confirming the presence of m1A, but no m6A sites on IGF1R transcripts (Fig. (Fig.4e4e and Supplementary Fig. S4g). Meanwhile, unlike glyceraldehyde 3-phosphate dehydrogenase (GAPDH), IGF1R and nuclear factor of activated T cells 3 (NFATC3) (positive control)17 were enriched in the YTHDF3 antibody-immunoprecipitated fraction, confirming that YTHDF3 can bind to IGF1R mRNAs (Fig. (Fig.4f).4f). IGF1R mRNAs, but not SOX18 or NFATC3, showed decreased enrichment in the m1A or YTHDF3 antibody-immunoprecipitated fraction of the hypoxia cells compared to normoxia cells (Fig. 4g, h), thus indicating that m1A sites on IGF1R mRNAs disappeared upon hypoxia treatment and YTHDF3 failed to bind IGF1R mRNA. These results revealed that YTHDF3 regulated the activity of trophoblast probably via targeting IGF1R modification of IGF signaling pathway.
YTHDF3 inhibited IGF1R expression
As IGF1R mRNAs were methylated with m1A, we furtherly investigated the effects of IGF1R m1A modification after up- or downregulation of m1A-associated modification enzyme. We performed IP of the isolated trophoblast RNA with m1A antibody, and found that IGF1R mRNAs, especially positive control SOX188, but not 18S rRNA showed increased enrichment in the m1A antibody-immunoprecipitated fraction when m1A eraser ALKBH3 was knocked down, decreased enrichment when the m1A writer TRMT6 was knocked down, but no effects on enrichment when m6A eraser ALKBH5 knocked down (Fig. (Fig.5a),5a), thus indicating that m1A sites on YTHDF3-binding IGF1R mRNAs are the direct substrates subject to ALKBH3-catalyzed demethylation. Furthermore, the enrichment of IGF1R or SOX18 mRNAs to m1A antibody decreased in trophoblast after overexpression of ALKBH3 (Fig. (Fig.5b).5b). These observations further support the notion that ALKBH3 can erase m1A modification of IGF1R mRNAs.
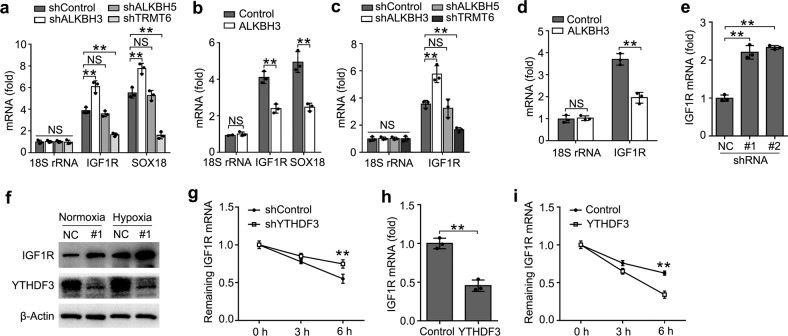
a, b Enrichment fold of the indicated RNA transcripts in m1A IP vs. RNA input control in HTR8/SVneo after transfected with the indicated shRNA for 72 h (a) or plasmids (b) for 48
h (a) or plasmids (b) for 48 h. Each transcript was quantified by qRT-PCR. c, d Enrichment fold of the indicated RNA transcripts in YTHDF3 IP vs. RNA input control in HTR8/SVneo after transfected with the indicated shRNA for 72
h. Each transcript was quantified by qRT-PCR. c, d Enrichment fold of the indicated RNA transcripts in YTHDF3 IP vs. RNA input control in HTR8/SVneo after transfected with the indicated shRNA for 72 h (a) or plasmids (b) for 48
h (a) or plasmids (b) for 48 h. e, f qRT-PCR and immunoblot analysis of the indicated mRNA and protein in HTR8/SVneo transfected with shYTHDF3 for 72
h. e, f qRT-PCR and immunoblot analysis of the indicated mRNA and protein in HTR8/SVneo transfected with shYTHDF3 for 72 h. g, i RNA lifetime for IGF1R in HTR8/SVneo cells transfected with the shYTHDF3 (g) or YTHDF3 plasmids (i) was determined by monitoring transcript abundance after transcription inhibition (TI). h qRT-PCR analysis of the IGF1R in HTR8/SVneo transfected with plasmids as indicated for 48
h. g, i RNA lifetime for IGF1R in HTR8/SVneo cells transfected with the shYTHDF3 (g) or YTHDF3 plasmids (i) was determined by monitoring transcript abundance after transcription inhibition (TI). h qRT-PCR analysis of the IGF1R in HTR8/SVneo transfected with plasmids as indicated for 48 h. NS: not significant; **p
h. NS: not significant; **p <
< 0.01 (Student’s t test); NS, not significant. Data are representative of three independent experiments (mean and s.d. of technical triplicates (a–d, e, g–h)).
0.01 (Student’s t test); NS, not significant. Data are representative of three independent experiments (mean and s.d. of technical triplicates (a–d, e, g–h)).
As YTHDF3 is an m1A reader of RNA methylation, we intended to investigate whether YTHDF3 directly targeted the m1A modification of IGF1R mRNA. We performed IP of the HTR8/SVneo RNA with YTHDF3 antibody, and found that IGF1R showed increased enrichment in the YTHDF3 antibody-immunoprecipitated fraction when ALKBH3 was knocked down, no change when ALKBH5 was knocked down, but decreased enrichment when the m1A “writer” TRMT6 was knocked down (Fig. (Fig.5c).5c). Moreover, we found that IGF1R showed decreased enrichment in the YTHDF3 antibody-immunoprecipitated fraction after ALKBH3 overexpression (Fig. (Fig.5d).5d). All these observations further supported that m1A sites on IGF1R mRNAs were the direct substrates of YTHDF3.
We next explored that whether the expression of IGF1R were affected after binding YTHDF3. We found that knockdown of YTHDF3 increased the mRNA and protein levels of the IGF1R transcript (Fig. 5e, f). Accordingly, the IGF1R mRNA showed decreased RNA decay rates upon YTHDF3 knockdown (Fig. (Fig.5g)5g) and increased RNA decay rates upon ALKBH3 knockdown (Supplementary Fig. S5a), but there was no change on the mRNA and protein level of IGF1R upon knockdown of ALKBH5 (Supplementary Fig. S5b, c). Overexpression of YTHDF3 decreased the mRNA levels of the IGF1R transcript and the transcript showed increased RNA decay rates upon YTHDF3 overexpression (Fig. 5h, i). These results indicated that YTHDF3 inhibited the expression of m1A modified IGF1R via promoting the decay of the transcripts in the cytoplasm.
YTHDF3 regulated the activity of trophoblast via targeting IGF1R
We then investigated the potential roles of IGF1R in regulating the activity of trophoblast. First, we confirmed that IGF1R were efficiently knocked down by two different small interfering RNAs (siRNAs) (Fig. (Fig.6a6a and Supplementary Fig. S6a). We found that knockdown of IGF1R significantly decreased the production of MMP9 and MMP2 (Fig. (Fig.6b6b and Supplementary Fig. S6b), while overexpression of IGF1R could promote MMP9 and MMP2 expression (Fig. (Fig.6c6c and Supplementary Fig. S6c, d). Knockdown of IGF1R inhibited the invasion and migration of trophoblast (Fig. 6d, e) when compared with negative control, which was a phenocopy of our results obtained from overexpression of YTHDF3. Thus, IGF1R and YTHDF3 might act cooperatively in regulating the activity of trophoblast via IGF signaling pathway.
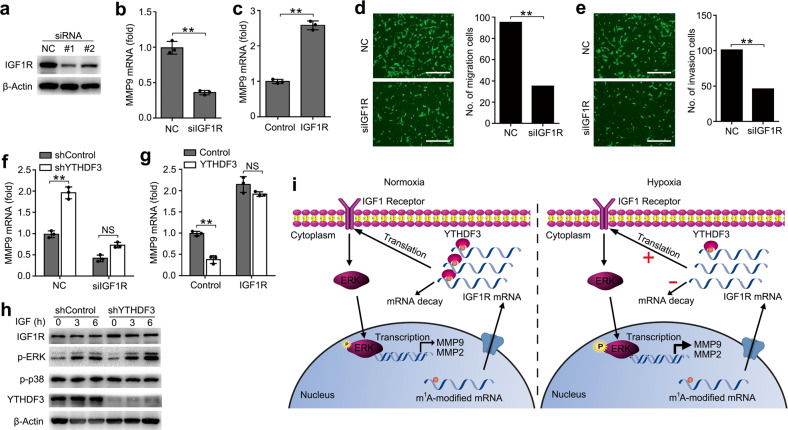
a Immunoblot analysis of the indicated proteins in HTR8/SVneo transfected with IGF1R siRNA #1 or #2 as indicated for 48 h. b, c qRT-PCR analysis of MMP9 mRNA in HTR8/SVneo transfected with the indicated siRNA (b) or plasmid (c). d, e Left: Migration (d) and invasion (e) analysis of HTR8/SVneo cells transfected with NC vs. siIGF1R by Transwell assay. Representative images are shown. Right: The number of migration and invasion cells was counted by the ImageJ software. f qRT-PCR analysis of MMP9 mRNA in stably expressing shYTHDF3 HTR8/SVneo transfected with siIGF1R. g qRT-PCR analysis of MMP9 mRNA in stably expressing YTHDF3 HTR8/SVneo transfected with IGF1R overexpression plasmid. h Immunoblot analysis of the indicated proteins in HTR8/SVneo transfected with shYTHDF3, and stimulated with IGF cytokine for the indicated time. i Working model for the mechanism of YTHDF3 inhibiting trophoblast invasion by decreasing IGF1R expression via promoting m1A-carrying IGF1R decay. P: Phosphorylation. Scale bars, 100
h. b, c qRT-PCR analysis of MMP9 mRNA in HTR8/SVneo transfected with the indicated siRNA (b) or plasmid (c). d, e Left: Migration (d) and invasion (e) analysis of HTR8/SVneo cells transfected with NC vs. siIGF1R by Transwell assay. Representative images are shown. Right: The number of migration and invasion cells was counted by the ImageJ software. f qRT-PCR analysis of MMP9 mRNA in stably expressing shYTHDF3 HTR8/SVneo transfected with siIGF1R. g qRT-PCR analysis of MMP9 mRNA in stably expressing YTHDF3 HTR8/SVneo transfected with IGF1R overexpression plasmid. h Immunoblot analysis of the indicated proteins in HTR8/SVneo transfected with shYTHDF3, and stimulated with IGF cytokine for the indicated time. i Working model for the mechanism of YTHDF3 inhibiting trophoblast invasion by decreasing IGF1R expression via promoting m1A-carrying IGF1R decay. P: Phosphorylation. Scale bars, 100 μm; **p
μm; **p <
< 0.01 (Student’s t test); NS, not significant. Data are representative of three independent experiments (mean and s.d. of technical triplicates (b, c, f, g)).
0.01 (Student’s t test); NS, not significant. Data are representative of three independent experiments (mean and s.d. of technical triplicates (b, c, f, g)).
We next asked whether YTHDF3 regulated the activity of trophoblast mainly through IGF1R signaling. We found that YTHDF3 knockdown-mediated promotion of MMP9 production was rescued by IGF1R knockdown (Fig. (Fig.6f),6f), suggesting that IGF1R knockdown counteracted the effect of YTHDF3 knockdown. Accordingly, overexpression of IGF1R reversed the YTHDF3 overexpression-mediated inhibition of MMP9 production (Fig. (Fig.6g),6g), thus determining that the role of YTHDF3 in inhibiting trophoblast MMP9 production was dependent on IGF1R. We also found that the phosphorylation of extracellular signal-regulated kinase (p-ERK) increased in YTHDF3-knockdown cells and the total IGF1R protein expression also increased, in comparison with the control cells (Fig. (Fig.6h).6h). Taken together, we found that YTHDF3 regulated the activity of trophoblast mainly via targeting IGF1R and affected downstream ERK-MMP9/2 signaling pathway.
On the basis of our findings, we propose the following working model to explain how m1A reader YTHDF3 inhibits the activity of trophoblast (Fig. (Fig.6i).6i). Hypoxia treatment downregulates the m1A reader YTHDF3 expression and demethylates m1A modification of IGF1R, which both decrease the binding between YTHDF3 protein and IGF1R mRNA, and promote IGF1R expression in the cytoplasm, leading to the stronger activation of IGF1R signaling pathway, consequently enhancing MMP9 or MMP2 expression and promoting the invasion and migration of trophoblast cells.
Discussion
We find that YTHDF3 can selectively bind to m1A-carrying RNAs, and hypoxia treatment can downregulate its expression in trophoblast HTR8/SVneo cells. Downregulation of YTHDF3 can enhance trophoblast migration and invasion in vitro. Mechanically, YTHDF3 inhibits the expression of IGF1R via binding m1A modification to enhance the decay of IGF1R mRNA, consequently inhibiting the expression of MMP9, and finally decreasing the invasion of trophoblast. Together, our results indicate that the m1A reader YTHDF3 may be a potential target to control pregnancy-associated diseases induced by hypoxia.
It is well known that YTH domain-containing proteins play as m6A readers and can regulate mRNA metabolism15,38,39, but whether they can act as readers of other RNA modifications is unknown. We use the m1A probes derived from the native present m1A modification site of human 28S rRNA with stem and loop structure40, because they can be more easily and specifically recognized by intracellular reader protein. We found that nine proteins, including mitochondrial ATP synthase subunit alpha (ATP5A1), 78 kDa glucose-regulated protein (HSPA5), transcriptional activator protein Pur-alpha (PURA), heterogeneous nuclear ribonucleoprotein M (HNRNPM), ADP/ATP translocase 2 (SLC25A5), RNA-binding protein 28 (RBM28), poly(rC)-binding protein 1 (PCBP1), YTH domain-containing family protein 3 (YTHDF3), and ruvB-like 2 (RUVBL2), can interact with m1A-carrying RNA. Inconsistent with the report that YTH domain-containing proteins (YTHDF1, YTHDF2, and YTHDF3) all probably play as m1A readers24, our data discriminate the differential binding affinities to m1A-carrying RNAs among them, suggesting different specificities probably endowed by unique protein structure of each one41. The functions of other eight “reader” proteins in regulating m1A-carrying mRNAs are completely unknown and warrant further investigation; nevertheless, our results provide a foundation for further understanding of the biological functions of m1A modification.
kDa glucose-regulated protein (HSPA5), transcriptional activator protein Pur-alpha (PURA), heterogeneous nuclear ribonucleoprotein M (HNRNPM), ADP/ATP translocase 2 (SLC25A5), RNA-binding protein 28 (RBM28), poly(rC)-binding protein 1 (PCBP1), YTH domain-containing family protein 3 (YTHDF3), and ruvB-like 2 (RUVBL2), can interact with m1A-carrying RNA. Inconsistent with the report that YTH domain-containing proteins (YTHDF1, YTHDF2, and YTHDF3) all probably play as m1A readers24, our data discriminate the differential binding affinities to m1A-carrying RNAs among them, suggesting different specificities probably endowed by unique protein structure of each one41. The functions of other eight “reader” proteins in regulating m1A-carrying mRNAs are completely unknown and warrant further investigation; nevertheless, our results provide a foundation for further understanding of the biological functions of m1A modification.
It is known that YTHDF3 can bind to a series of eukaryotic 40S and 60S subunits of RNAs and proteins, and then promote translation of m6A-carrying mRNAs16,17; we also found that most of the noncoding RNAs bound by YTHDF3 are ribosomal RNAs, including different 40S and 60S subunit RNAs, and speculate that YTHDF3 may also recruit 40S and 60S translation machineries to enhance the translation of m1A-carrying RNAs. We found that YTHDF3 peaks are mainly enriched in intergenic (~65%), introns (~23%), and noncoding RNAs (~10%), but merely (~2%) in CDS, 5′-UTR, and 3′-UTR regions, and our iCLIP results in trophoblast do not agree well with previous PAR-CLIP results in human HeLa or HEK293T cells17,42, probably due to our distinct cell line and different CLIP strategies.
Our m1A-seq results from normoxia and hypoxia along with our mechanistic studies in HTR8/SVneo cells reveal regulation of IGF1R expression as an important mediator of trophoblast invasion mediated by YTHDF3. Increased IGF1R expression is probably one of the main mediators of increased invasion in trophoblast with YTHDF3 downregulation, as overexpression of IGF1R is sufficient to rescue the inhibition of MMP9 expression mediated by YTHDF3 overexpression. These findings may be applicable beyond trophoblast invasion to other complicated pregnancy diseases or cancers driven by YTHDF3 overexpression. However, we cannot rule out the involvement of other signaling pathways that could be regulated directly or indirectly by YTHDF3.
We observed a global relative decrease in m1A mRNA methylation upon hypoxia, which could have effects on cellular physiology, especially if the methylation modifications of key transcripts are affected43. We found that many transcripts regarding insulin-like signaling pathway and extracellular matrix are demethylated upon hypoxia compared to normoxia, indicating that the reduced m1A methylation observed in trophoblast cells might affect functions associated with trophoblast invasion. We find that the key transcript IGF1R in IGF signaling pathway is demethylated upon hypoxia. A previous report indicated that m1A modification could enhance the translation of m1A-bearing mRNA, and YTHDF2, but not YTHDF1 or YTHDF3, could promote m1A-carrying mRNA decay and inhibit mRNA translation23,24. However, we found that YTHDF3 can promote m1A-bearing IGF1R decay and significantly inhibit IGF1R expression, and this effect is irrelevant to YTHDF3 functioning as an m6A reader. We speculate whether YTHDF3 targets on unique mRNA depends on its conserved binding motif and its m1A modification status. It was previously reported that YTHDF3 acted as an m6A reader16 and m6A modification plays an important role on physiological changes of trophoblast, and thus we think that YTHDF3 may also regulate the function of trophoblast via m6A modification, but its specific substrate RNA need to be further investigated. Furthermore, we found that m1A site on SOX18 is not affected in hypoxia condition or ALKBH5 overexpression. There might be other specific reader proteins for SOX18, which needs to be further investigated in the future.
Hypoxia has a critical role in the early embryo development and promotes the activity of trophoblast44,45. Abnormal oxygen condition can lead to injury or abortion of fetus (recurrent spontaneous abortion, IUGR)46. Expressions of various genes, especially ones involved in epigenetic modifications, are significantly regulated in hypoxia condition47,48. The hypoxia-inducible molecules have been proposed to be important regulators of first trimester trophoblast behavior49, and we found that YTHDF3 expression is downregulated in trophoblast upon hypoxia condition. These results are consistent with our data that downregulation of YTHDF3 promotes the invasion of trophoblast via upregulation of IGF1R expression, which finally increases MMP9 and MMP2 expression. Our results suggest that regulation of YTHDF3 expression could be a general control mechanism that affects a range of other biological processes, which will be a new direction to be explored in the future.
Materials and methods
Reagents
IGF1 (AFL291) was obtained from R&D System Company (Minneapolis, USA). Antibodies used for IP, immunoblot (IB) analysis and immunofluorescence (IF) were as follows: m1A antibody (D345-3, 1:100 for IP) was from MBL Life Science (Japan). Horse radish peroxidase-coupled secondary antibodies (ab6721, 1:1000 for IB), IGF1R (ab182048, 1:1000 for IB), ALKBH5 (ab69325, 1:1000 for IB), and YTHDF3 (ab83716, 1:1000 for IB) were from Abcam (Cambridge, UK). β-Actin (4970, 1:1000 for IB), p-ERK (4370, 1:1000), and p-p38 (4511, 1:1000) were from Cell Signaling Technology (Danvers, MA, USA). The Flag tags (F7425, 1:1000 for IB) as well as the agarose were used for IP; LPS (L2630) was from Sigma-Aldrich (St. Louis, MO, USA). DAPI (4′,6-diamidino-2-phenylindole) (62247, 1:1000 for IF) Alexa Fluor® 546 (A-11071, 1:500 for IF) and Protein A/G Magnetic Beads (88802) were from Thermo Fisher Scientific.
Cell culture and transfection
HTR8/SVneo cell lines were obtained from American Type Culture Collection (Manassas, VA, USA) and cultured as in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) in normoxia (20% O2) or hypoxia (2% O2). HTR8/SVneo were transfected with siRNAs (final concentration: 20 nM) using INTERFERin (Polyplus-transfection) and plasmids with Lipofectamine™ 2000 (Thermo Fisher, American) according to the manufacturer’s instructions. All siRNAs were obtained from GenePharma (Supplementary Table S4).
nM) using INTERFERin (Polyplus-transfection) and plasmids with Lipofectamine™ 2000 (Thermo Fisher, American) according to the manufacturer’s instructions. All siRNAs were obtained from GenePharma (Supplementary Table S4).
To establish stably transfected HTR8/SVneo cells, G418 was added (1000 μg/mL) 48
μg/mL) 48 h after transfection and maintained at 800
h after transfection and maintained at 800 μg/mL for 3 weeks for positive selection. For stably transfected cells, YTHDF3 expressions were confirmed by Western blot. Lentiviruses harboring YTHDF3 shRNA or non-targeting control sequences were purchased from OBIO Technology (Shanghai, China). HTR8/SVneo cells were infected with the lentivirus at a virus titer of 10:1 to the cell numbers, and with 5
μg/mL for 3 weeks for positive selection. For stably transfected cells, YTHDF3 expressions were confirmed by Western blot. Lentiviruses harboring YTHDF3 shRNA or non-targeting control sequences were purchased from OBIO Technology (Shanghai, China). HTR8/SVneo cells were infected with the lentivirus at a virus titer of 10:1 to the cell numbers, and with 5 μg/mL polybrene (Sigma-Aldrich, USA) to enhance infection efficiency. After HTR8/SVneo cells were transfected for 72
μg/mL polybrene (Sigma-Aldrich, USA) to enhance infection efficiency. After HTR8/SVneo cells were transfected for 72 h, puromycin was added (5
h, puromycin was added (5 μg/mL) for 72
μg/mL) for 72 h and maintained at 2
h and maintained at 2 μg/mL for 3 weeks for positive selection of stably knocked down YTHDF3. All cells were verified to be negative for mycoplasma contamination. The sequences encoding the shRNA are shown in Supplementary Table S5.
μg/mL for 3 weeks for positive selection of stably knocked down YTHDF3. All cells were verified to be negative for mycoplasma contamination. The sequences encoding the shRNA are shown in Supplementary Table S5.
RNA extraction and RT-qPCR
HTR8/SVneo cells were transfected with indicated shRNA, siRNAs, or plasmids. Total RNA was extracted by TRIZOL reagent (Invitrogen) 48 h post transfection. RNA was reversed transcribed using the Reverse Transcription System from TOYOBO (Osaka, Japan). The reverse transcription products from different samples were amplified by real-time PCR and analyzed as described previously50. The primer sequences for qPCR analysis are listed in Supplementary Table S6.
h post transfection. RNA was reversed transcribed using the Reverse Transcription System from TOYOBO (Osaka, Japan). The reverse transcription products from different samples were amplified by real-time PCR and analyzed as described previously50. The primer sequences for qPCR analysis are listed in Supplementary Table S6.
RNA-seq and data analysis
The procedure was adapted from the previous report51. Total RNA was isolated using TRIZOL reagent (Invitrogen). cDNA library was constructed using TruSeq RNA Sample Prep Kit and then sequenced with HiSeq 2000 System (Illumina Inc.). The expression of transcripts was quantified as Reads Per Kilobase of exon model per Million mapped reads.
Affinity purification of m1A-binding proteins and LC-MS/MS
Biotin-labeled oligo ribonucleotides with the sequence of 5′-biotin-ACCCGUCUUGXAACACGGCCGUUGXUCACGUC-3′ (X =
= m1A or A) from human 28S rRNA gene and 5′-biotin-CCGUUCCGCCCXGGCCGCGCCCAGCUGGAAUGCA-3′ (X
m1A or A) from human 28S rRNA gene and 5′-biotin-CCGUUCCGCCCXGGCCGCGCCCAGCUGGAAUGCA-3′ (X =
= m1A or A) from SOX18 mRNA were obtained from Integrated DNA Technologies (IDT). HEK293T or Raw264.7 cells were lysed in cell lysis and binding buffer (containing 10
m1A or A) from SOX18 mRNA were obtained from Integrated DNA Technologies (IDT). HEK293T or Raw264.7 cells were lysed in cell lysis and binding buffer (containing 10 mM Tris-HCl (pH 7.5), 150
mM Tris-HCl (pH 7.5), 150 mM KCl, 1.5
mM KCl, 1.5 mM MgCl2, 0.05% (v/v) NP-40, 10
mM MgCl2, 0.05% (v/v) NP-40, 10 mM NaCl, 0.5
mM NaCl, 0.5 mM DTT, 0.5% (v/v) Triton X-100, 2
mM DTT, 0.5% (v/v) Triton X-100, 2 mM EDTA, and 0.5 units RNase inhibitor) and centrifuged at 13,000
mM EDTA, and 0.5 units RNase inhibitor) and centrifuged at 13,000 r.p.m. at 4
r.p.m. at 4 °C for 15
°C for 15 min. The supernatant was pre-cleared at 4
min. The supernatant was pre-cleared at 4 °C for 1
°C for 1 h by incubation with streptavidin-conjugated agarose beads (Thermo Scientific). The biotinylated m1A and control baits were incubated at 4
h by incubation with streptavidin-conjugated agarose beads (Thermo Scientific). The biotinylated m1A and control baits were incubated at 4 °C with pre-cleared cell lysates for 2
°C with pre-cleared cell lysates for 2 h. Streptavidin-conjugated agarose beads were then added to the mixture and incubated in a shaker at 4
h. Streptavidin-conjugated agarose beads were then added to the mixture and incubated in a shaker at 4 °C for 2
°C for 2 h. The IP samples were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and stained with silver, different bands with intensive signal in m1A probes compared with control probes were cut and digested, and then analyzed by mass spectrometry LC-MS/MS experiments performed as previously described50.
h. The IP samples were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and stained with silver, different bands with intensive signal in m1A probes compared with control probes were cut and digested, and then analyzed by mass spectrometry LC-MS/MS experiments performed as previously described50.
Transwell migration and invasion assay
For invasion assays, cell-culture inserts (0.8 μm, Falcon no. 353097) were pre-coated with Matrigel (40
μm, Falcon no. 353097) were pre-coated with Matrigel (40 μg/insert, Corning) in serum-free medium for 30
μg/insert, Corning) in serum-free medium for 30 min at 37
min at 37 °C. For migration assays, inserts were not pre-coated. Forty thousand cells per insert were seeded in the upper chamber of the insert and cultured in 300
°C. For migration assays, inserts were not pre-coated. Forty thousand cells per insert were seeded in the upper chamber of the insert and cultured in 300 μL RPMI-1640 medium supplemented with 2% FBS. Complete RPMI-1640 medium supplemented with 10% FBS was used in the lower chamber. Following 16–24
μL RPMI-1640 medium supplemented with 2% FBS. Complete RPMI-1640 medium supplemented with 10% FBS was used in the lower chamber. Following 16–24 h of migration or invasion, fluorescent stain using calcein-AM was added to each lower chamber and incubated for 30
h of migration or invasion, fluorescent stain using calcein-AM was added to each lower chamber and incubated for 30 min. Images were collected with a Nikon Eclipse Ti2 with NIS Elements imaging software (version 5.02) and images were analyzed with ImageJ (version 1.51i).
min. Images were collected with a Nikon Eclipse Ti2 with NIS Elements imaging software (version 5.02) and images were analyzed with ImageJ (version 1.51i).
Cell proliferation assay
Five thousand cells were seeded per well in a 96-well plate. The cell proliferation was assessed by assaying the cells at various time points using the Cell Counting Kit-8 (Sigma-Aldrich, 96992) following the manufacturer’s protocols. For each cell line tested, the signal was normalized to the value observed about 24 h after seeding.
h after seeding.
m1A-seq and data analyses
Total RNA was isolated from normoxia or hypoxia HTR8/SVneo cell lines. Polyadenylated RNA was further enriched from total RNA using a Dynabeads mRNA Purification Kit (Invitrogen). RNA fragmentation, m1A-IP, and library preparation were carried out according to previously published protocols9. Sequencing was carried out on an Illumina HiSeq2500 machine in single-read mode with 50 base pairs per read. Significant peaks with false discovery rate <0.05 were annotated to the RefSeq database (hg38). Sequence motifs were identified using Homer. m1A-seq data were analyzed according to the protocol described previously9.
m1A-qRT-PCR
The procedure was adapted from the previous report8. For m1A-qRT-PCR, total RNAs were firstly fragmented to 150 nucleotide using RNA fragmentation buffer (Ambion, AM8740), and then fragmented RNA (as input) was denatured and incubated with m1A antibody for 2 h, and protein A/G magnetic beads were added to the mixture and incubated for additional 2
h, and protein A/G magnetic beads were added to the mixture and incubated for additional 2 h. Beads were washed with buffer and purified, input and immunoprecipitated RNAs were reversed transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific), and then quantified by qPCR using SYBR GREEN mix (TOYOBO). For comparing m1A abundance changes, relative enrichment was first normalized with inputs, and then analyzed by comparing the data from m1A-immunoprecipitated sample. All samples were analyzed in triplicate qPCR.
h. Beads were washed with buffer and purified, input and immunoprecipitated RNAs were reversed transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific), and then quantified by qPCR using SYBR GREEN mix (TOYOBO). For comparing m1A abundance changes, relative enrichment was first normalized with inputs, and then analyzed by comparing the data from m1A-immunoprecipitated sample. All samples were analyzed in triplicate qPCR.
Measurement of RNA lifetime
HTR8/SVneo cells were seeded in 6-well plates at 50% confluency. After the cells were cultured for 24 h, actinomycin D (5
h, actinomycin D (5 mg/mL, A4262, Sigma) was added for 6, 4, 2, and 0
mg/mL, A4262, Sigma) was added for 6, 4, 2, and 0 h before collection. The total RNA was purified using Trizol method and RNA quantities were determined by qRT-PCR. The GAPDH gene was used as a reference gene when carrying out qPCR.
h before collection. The total RNA was purified using Trizol method and RNA quantities were determined by qRT-PCR. The GAPDH gene was used as a reference gene when carrying out qPCR.
Molecular cloning of related genes
Related genes were amplified from human trophoblast by RT-PCR and subsequently cloned into pcDNA vectors. Each construct was confirmed by sequencing. The corresponding primers used in this study are listed in Supplementary Table S7.
Confocal microscopy
HTR8/SVneo cells plated on glass coverslips in six-well plates were treated in hypoxia condition for 12 h or stimulated with LPS for 6
h or stimulated with LPS for 6 h as indicated and fixed in 4% paraformaldehyde. Cells were then stained with YTHDF3 antibody and viewed using a Leica TCS SP2 confocal laser microscope.
h as indicated and fixed in 4% paraformaldehyde. Cells were then stained with YTHDF3 antibody and viewed using a Leica TCS SP2 confocal laser microscope.
Immunoblot
Cells were lysed using Cell Lysis Buffer (Cell Signaling Technology) supplemented with cocktail protease inhibitor (Calbiochem). Protein concentrations of the extracts were measured using a BCA assay (Pierce, USA) and equalized with the extraction reagent. Equivalent amounts of extract were loaded to SDS-PAGE, transferred onto PVDF membranes, and then blotted as described previously52.
Statistical analysis
All experiments were independently repeated at least three times. Comparisons between two groups were performed using Student’s t test. Data were analyzed with the GraphPad Prism 7 Software. Statistical values achieving p <
< 0.05 were considered to be statistically significant. There was no exclusion of data points. No randomization was used.
0.05 were considered to be statistically significant. There was no exclusion of data points. No randomization was used.
Acknowledgements
We thank Zhike Lu for analysis of the sequencing data and Chengqi Yi professor of Peking University for analysis of the m1A and m6A contents of m1A probes. This study was supported by the Key Program of the National Natural Science Foundation of China (81730039), the National Natural Science Foundation of China (81671460, 81871167), the National Key Research and Development Program of China (2017YFC1001401), Shanghai Municipal Medical and Health Discipline Construction Projects (2017ZZ02015), the National Basic Research Program of China (2015CB943300), the Program for Shanghai leaders to L.J. and the Natural Science Foundation of Shanghai (18ZR1430000) to Q.Z.
Author contributions
L.J. and Q.Z. designed the experiments. Q.Z., H.G., F.Y., Y.Y. and F.H. carried out the experiments and analyzed the data and interpreted the findings. L.J. and Q.Z. wrote the manuscript.
Data availability
The source data that support the findings of this study are available from the corresponding author upon request.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qingliang Zheng, Haili Gan
Contributor Information
Qingliang Zheng, Email: nc.ude.ijgnot@2751071.
Liping Jin, Email: moc.361@10plnij.
Supplementary information
Supplementary Information accompanies the paper at (10.1038/s41421-020-0144-4).
References
Articles from Cell Discovery are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/s41421-020-0144-4
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/s41421-020-0144-4.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
RNA modification regulators as promising biomarkers in gynecological cancers.
Cell Biol Toxicol, 40(1):92, 29 Oct 2024
Cited by: 0 articles | PMID: 39472384 | PMCID: PMC11522084
Review Free full text in Europe PMC
Regulations of m6A and other RNA modifications and their roles in cancer.
Front Med, 18(4):622-648, 22 Jun 2024
Cited by: 0 articles | PMID: 38907157
Review
The potential of RNA methylation in the treatment of cardiovascular diseases.
iScience, 27(8):110524, 20 Jul 2024
Cited by: 0 articles | PMID: 39165846 | PMCID: PMC11334793
Review Free full text in Europe PMC
Discovery of a new inhibitor for YTH domain-containing m6A RNA readers.
RSC Chem Biol, 5(9):914-923, 25 Jul 2024
Cited by: 0 articles | PMID: 39211476 | PMCID: PMC11353026
DRAIC mediates hnRNPA2B1 stability and m6A-modified IGF1R instability to inhibit tumor progression.
Oncogene, 43(29):2266-2278, 29 May 2024
Cited by: 0 articles | PMID: 38811846
Go to all (33) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE135403
- (1 citation) GEO - GSE135407
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A functional loop between YTH domain family protein YTHDF3 mediated m6A modification and phosphofructokinase PFKL in glycolysis of hepatocellular carcinoma.
J Exp Clin Cancer Res, 41(1):334, 06 Dec 2022
Cited by: 21 articles | PMID: 36471428 | PMCID: PMC9724358
N6-methyladenosine reader YTHDF3 regulates melanoma metastasis via its 'executor'LOXL3.
Clin Transl Med, 12(11):e1075, 01 Nov 2022
Cited by: 9 articles | PMID: 36324258 | PMCID: PMC9630608
YTHDF3 modulates the progression of breast cancer cells by regulating FGF2 through m<sup>6</sup>A methylation.
Front Cell Dev Biol, 12:1438515, 20 Sep 2024
Cited by: 0 articles | PMID: 39372951 | PMCID: PMC11449838
N1-methyladenosine modification in cancer biology: Current status and future perspectives.
Comput Struct Biotechnol J, 20:6578-6585, 25 Nov 2022
Cited by: 44 articles | PMID: 36467585 | PMCID: PMC9712505
Review Free full text in Europe PMC
Funding
Funders who supported this work.
National Natural Science Foundation of China (National Science Foundation of China) (1)
Grant ID: 81671460,81871167

 #
# 


