Abstract
Free full text

Modulating Heparanase Activity: Tuning Sulfation Pattern and Glycosidic Linkage of Oligosaccharides
Abstract
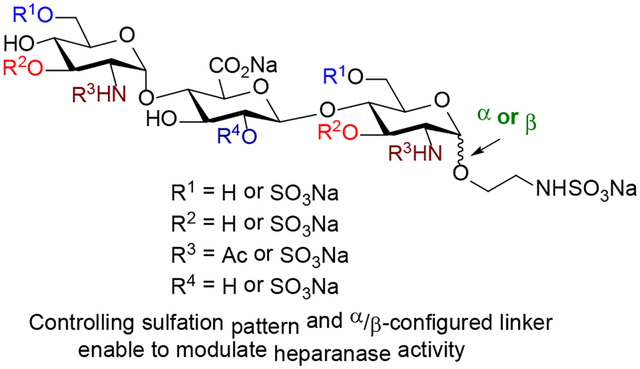
Heparanase cleaves polymeric heparan sulfate (HS) molecules into smaller oligosaccharides, allowing for release of angiogenic growth factors promoting tumor development and autoreactive immune cells to reach the insulin-producing β cells. Interaction of heparanase with HS chains is regulated by specific substrate sulfation sequences. We have synthesized eleven trisaccharides that are highly tunable in structure and sulfation pattern, allowing us to determine how heparanase recognizes HS substrate and selects a favorable cleavage site. Our study shows that (1) N-SO3− at +1 subsite and 6-O-SO3− at −2 subsite of trisaccharides are critical for heparanase recognition; (2) addition of 2-O-SO3− at the −1 subsite and of 3-O-SO3− to GlcN unit is not advantageous; and (3) the anomeric configuration (α or β) at the reducing end is crucial in controlling heparanase activity. Our study also illustrates that the α-trisaccharide having N- and 6-O-SO3− at −2 and +1 subsites inhibited heparanase and was resistant toward hydrolysis.
INTRODUCTION
Heparanase is an endolytic enzyme that hydrolyzes the internal β-(1,4)-linkage between glucuronic acid (GlcA) and N-sulfated glucosamine (GlcNS) along heparan sulfate (HS) polysaccharide chains of proteoglycans.1 HS chains are ubiquitously found on the cell surface, extracellular matrix (ECM), and basement membranes.1–2 Cleavage of HS chains releases, among other biologically important molecules, HS-bound proangiogenic and protumorigenic growth factors, cytokines and chemokines.3–5 Pre-clinical and clinical studies illustrate that high levels of heparanase expression correlate with increased tumor growth, angiogenesis, enhanced metastasis and poor patient prognosis for hematological and solid malignancies.6–8 Pancreatic β cells contains high levels of HS, essential for their survivial, and it has been demonstrated that β cells rapidly lost their HS in people with type I diabetes (T1D).9–10 Heparanase, which degrades HS and therefore assists migrating immune cells to enter the pancreas and kidney tissues, is induced by metabolic inflammation under the diabetic condition. During T1D, the immune system produces heparanase to destroy HS chains within β cells and cause their death.9–10 Increased heparanase activity aids autoreactive immune cells to enter and attack the insulin-producing pancreatic β cells and functional kidney cells. These studies attest to the importance of targeting heparanase for cancer and diabetic therapy.
Heparanase is similar to the rest of the GH79 family of carbohydrate-processing enzymes, which are featured by a conserved (β/α)8-TIM barrel fold where the catalytic residues reside.11 The proenzyme is secreted into the extracellular environment from the Golgi in its inactive 65 kDa form. Conversion to its active form, a non-covalently linked 50 + 8 kDa heterodimer, is achieved by proteolytic removal of a 6 kDa linker peptide by cathepsin L.1, 11 A major breakthrough was the report of the crystal structure of human heparanase alone and when bound to ligands.11 A sulfated HS trisaccharide spanning −2, −1, and +1 subsites (Figure 1) in the binding cleft of heparanase is a recognition sequence.11–12 Analysis also showed that the GlcAβ(1,4)GlcNS fragment of HS trisaccharide is located in the +1 and −1 subsites,13 next to the catalytic residues Glu225 and Glu343, which are responsible for cleavage of the scissile bond between a −1 GlcA unit and a +1 GlcNS unit (Figure 1). The binding cleft is flanked by positively charged residues, termed heparin-binding domains (HBD-1 and HBD-2).
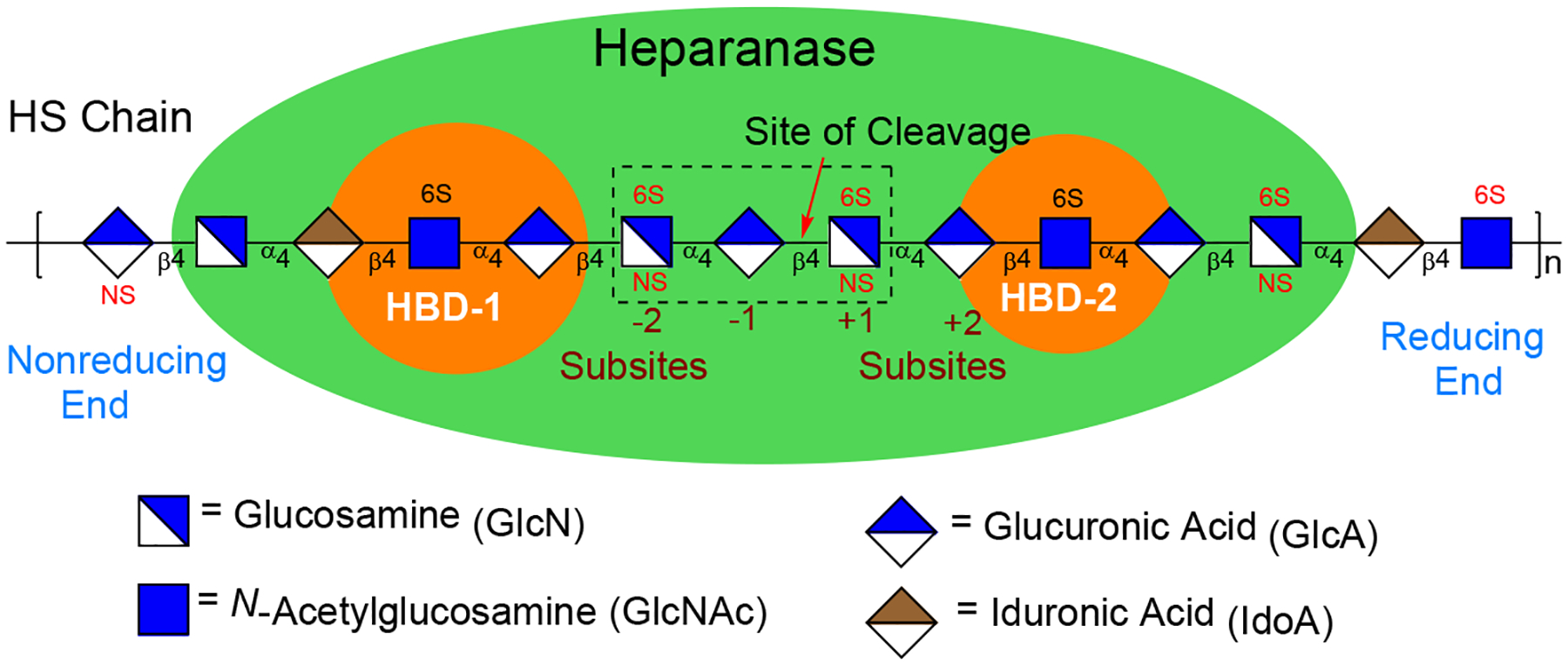
Interactions between heparanase and heparan sulfate. HS trisaccharide fragment across the −2, −1, and +1 subsites of the enzyme’s binding cleft is a recognition sequence.
Interaction of heparanase with HS is regulated by substrate sulfation sequences, and only HS chains with specific sulfation patterns are cleaved by heparanase.1, 11 Studies using HS oligomers have suggested that heparanase cleaves the β(1,4) linkage between a GlcA unit and a GlcNS unit, bearing a 6-O-SO3− and/or a 3-O-SO3−, at the −1 and +1 binding subsites.14–15 The crystal structure of heparanase-HS ligand further indicates that a GlcN unit carrying −2 N-SO3− and +1 6-O-SO3− is key for recognition due to their contact with the enzyme through a hydrogen bonding network.11 However, inspection of the GlcN unit at the −2 binding subsite revealed that the electron density for 6-O-SO3− is significantly weaker than that for N-SO3−, indicating that this subsite was occupied by a mixture of GlcNS6S and GlcNS units.11 Although these studies provide insight into how heparanase “reads” the sulfation pattern of HS substrates, some ambiguity remains: (1) whether a GlcN unit, carrying N-SO3− at +1 subsite and 6-O-SO3− at −2 subsite is important for heparanase recognition and cleavage; (2) which 6-O-SO3− at the −2 or +1 position has a higher binding affinity; (3) what effects does addition of a 3-O-SO3− at the −2 or +1 position have on the recognition of heparanase; (4) whether addition of a 2-O-SO3− at the −1 site could impact the binding affinity; and (5) does the glycosidic linkage configuration (α or β) at the reducing end of a +1 GlcN unit plays a role in controlling heparanase activity. These questions remain unsolved as selective sulfation at specific position cannot be distinguished through enzymatic oligosaccharide synthesis or the use of isolated and depolymerized heparin mixtures.12, 14
To resolve the aforementioned questions, we report herein a modular chemical approach for the creation of well-defined trisaccharides from several strategically protected building blocks. This strategy installs 6-O-, 3-O, and N-sulfate as well as N-acetyl at each sugar unit independently. To demonstrate the significance of this parallel combinatorial approach, eleven sulfated trisaccharides which are equipped with α- or β-anomeric configured aminoethyl linker at the reducing end have been prepared. These trisaccharides will allow us to systematically determine how heparanase recognizes HS’s sulfation pattern and selects a favorable cleavage site.16 Importantly, this study demonstrates the importance of the precise sequence of sulfation in promoting heparanase recognition and of the anomeric configuration of the reducing end linker of trisaccharides in modulating heparanase activity. Trisaccharide bearing the β-configured linker activated heparanase and displayed hydrolysis by heparanase; in contrast, trisaccharide bearing the α-configured linker inhibited heparanase and was resistant toward hydrolysis. Overall, this study provides a clear picture of substrate specificity and affinity and may be of substantial use in identifying structurally-defined and specific small molecule carbohydrate inhibitors of heparanase.
RESULTS AND DISCUSSION
Rational Design of Sulfated Trisaccharides.
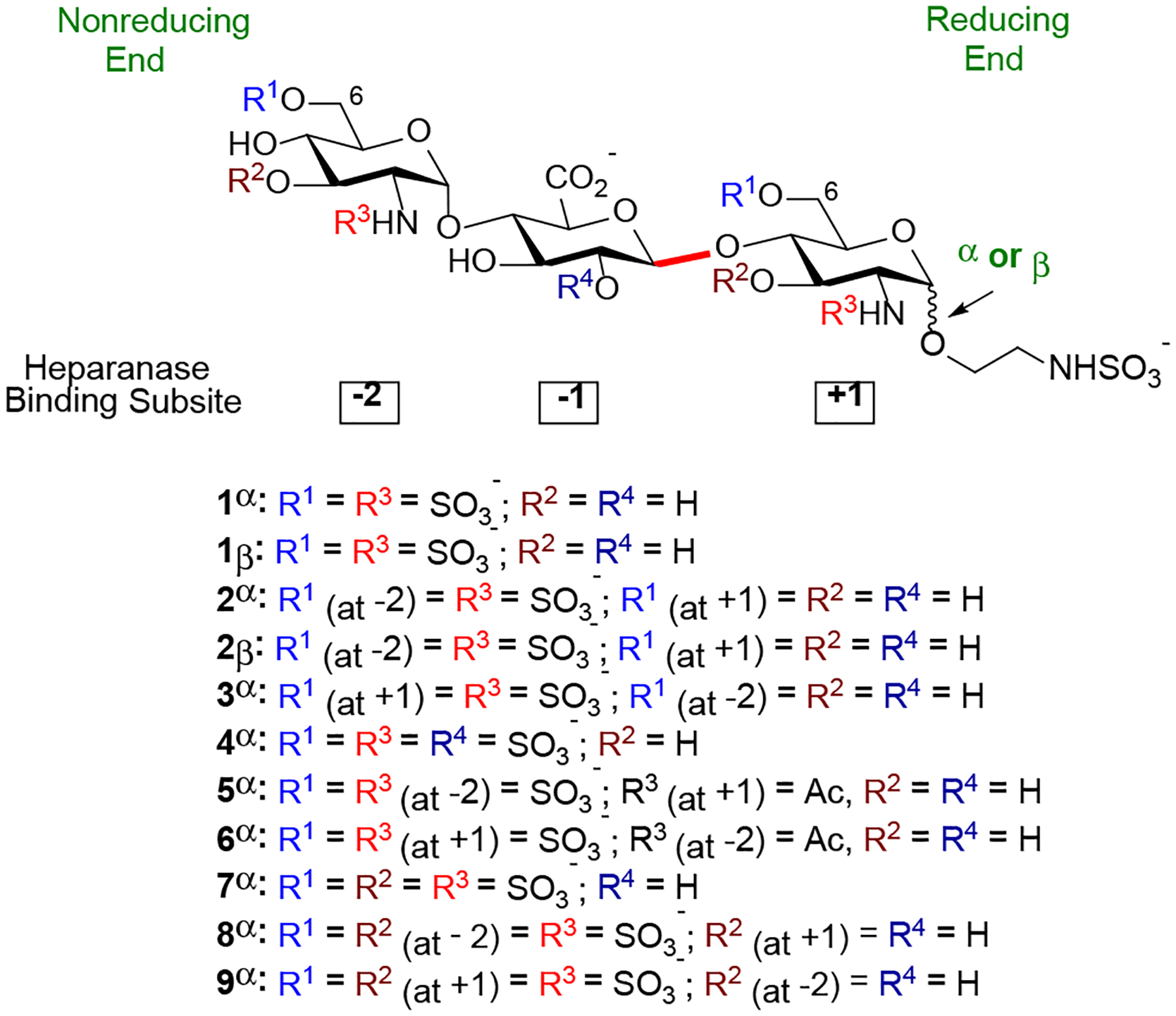
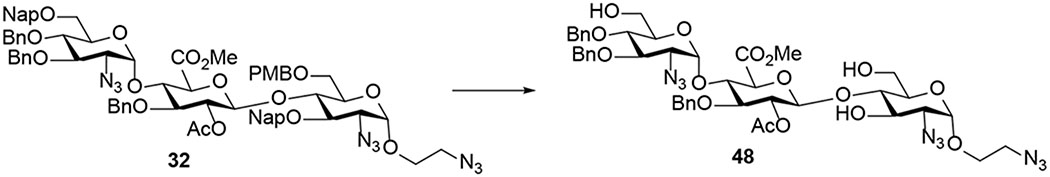
Our goal is to design the synthesis of a library of sulfated trisaccharides (Figure 2), representing all possible N-acetyl as well as 6-O-, 3-O-, 2-O- and N-sulfate motifs. Our approach allows us to dissect the contribution of an individual sulfate. We hypothesize that any additional sulfate or change in the pattern could disrupt the positioning of the saccharide, reducing ionic salt bridges and hydrogen bond interactions. Since trisaccharides 1α and 1β are structurally similar to the natural HS substrate (Figure 1),11–12 they are used as model substrates for comparison and determination of the role of the anomeric configuration at the reducing end in modulating heparanase activity. On the other hand, trisaccharides 2α and 2β determine whether 6-O-SO3− located at the +1 binding subsite is critical for recognition. Trisaccharide 3α validates that preference at the −2 subsite is likely to be GlcNS6S >>> GlcNS due to potential electrostatic and hydrogen-bonding interactions between 6-O-SO3− and the side chain of Lys159 near HBD-2.11 While 2α and 3α examine whether 6-O-SO3− located at either −2 or +1 subsite is more important for heparanase recognition, 5α and 6α determine the relative importance of N-SO3− located at the −2 or +1 subsite. Furthermore, 8α and 9α elucidate the effects of 3-O-SO3− at the −2 or +1 subsite on HS substrate-heparanase interactions. Trisaccharide 7α determines whether a highly sulfated substrate has a negative or positive impact on HS-heparanase interactions. Finally, trisaccharide 4α determines whether the sulfate group located at C2 position of the GlcA unit could affect the binding affinity as it was reported that the presence of 2-O-SO3− on GlcA or IdoA (iduronic acid) unit located at the +1 subsite could not be tolerated by heparanase due to steric interactions between the sulfate group and residue Asn224 in the binding cleft.11
To develop a modular strategy for the preparation of eleven sulfated trisaccharides (Figure 2), a set of properly protected monosaccharide building blocks that resemble the different GlcN and GlcA units found in HS at −2, −1, and +1 binding subsites were employed. A key issue in this approach is the proper selection of a set of conditions and protecting groups that satisfy the following criteria: (1) the C2-amino group of the glucosamine (GlcN) units needs to be functionalized in such a way that α−1,2-cis linkage can be preferentially formed between a −2 GlcN unit and a −1 GlcA unit and between a +1 GlcN unit and a reducing-end aminoethyl linker; (2) the C2-amino group needs to be differentially functionalized in such a way that either N-sulfate or N-acetyl group can be selectively introduced at −2 and +1 subsites; (3) the C2-hydroxyl protecting group of the GlcA unit allows for the stereoselective β−1,2-trans-linkage between a −1 GlcA unit and a +1 GlcN unit and for selective sulfation; (4) the C2-amino protecting group of the GlcN unit allows for the stereoselective β−1,2-trans-linkage between a +1 GlcN unit and an aminoethyl spacer; (5) protecting groups can be selectively removed to reveal hydroxyls that need to be selectively sulfated at C6 or C3 of the GlcN units; and (6) a standard set of reaction conditions for oligosaccharide formation, selective sulfation, and global deprotection.
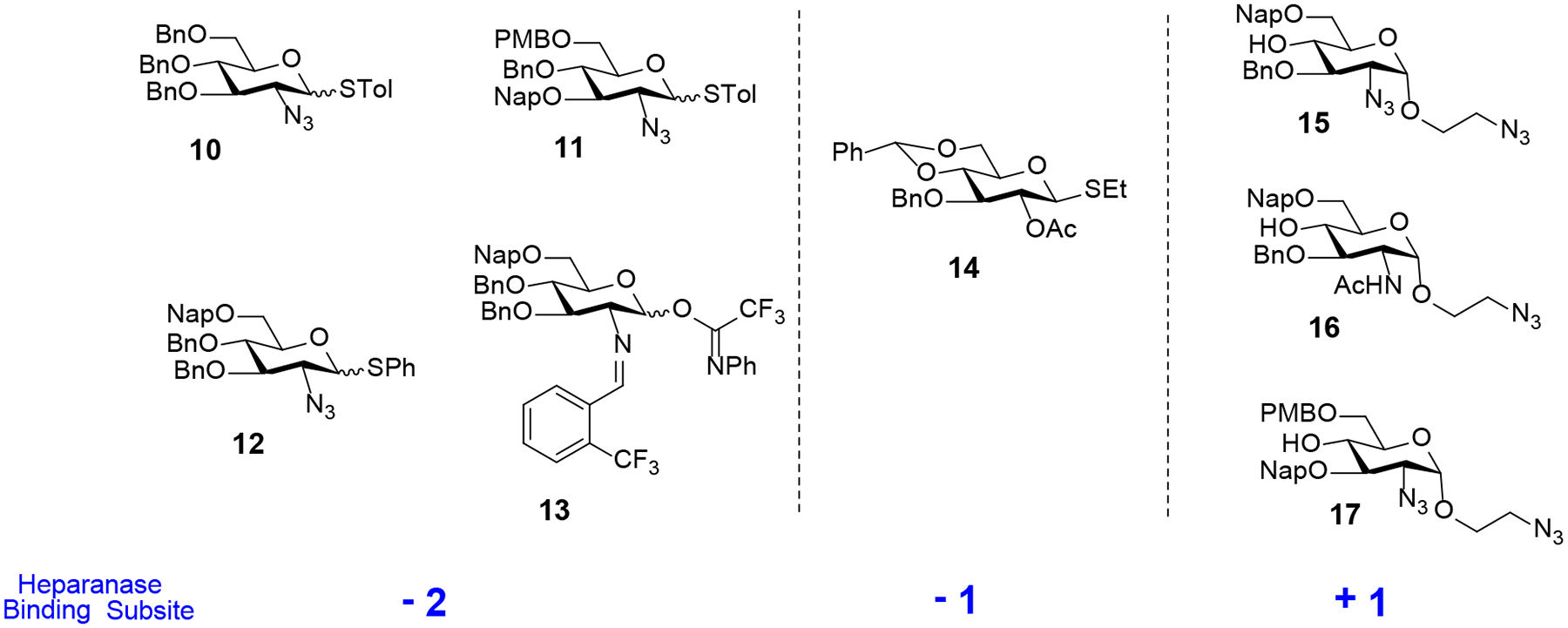
The monosaccharide building blocks (10 to 17) that we developed for the assembly of sulfated trisaccharides are illustrated in Figure 3. The napthylmethyl (Nap) and p-methoxybenzyl (PMB) protecting groups will be utilized for those hydroxyls at C6 and/or C3 that need sulfation. An important feature of the Nap and PMB groups is that they can chemoselectively be removed by 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) in the presence of the benzyl (Bn) group. Both azido and benzylideneamino groups will be utilized as a C2-amino masking functionality. Additionally, they allow the installation of α−1,2-cis linkage and can be converted into an amine for selective sulfation and acetylation. The benzylidene acetal on monosaccharide 14 can be selectively removed to reveal a diol whose C6-hydroxyl can be selectively oxidized to generate a GlcA unit at the −1 binding site. The C2-acetyl group on 14 is able to direct β−1,2-trans glycoside formation by neighboring group participation strategies and can be selectively removed for selective sulfation.17 The C6-acetyl protecting group of commercially available trisaccharide 34 (Scheme 4, vide infra),18 can be selectively removed to reveal hydroxyls at −2 and +1 subsites for selective sulfation. Finally, benzyl (Bn) groups are used as permanent protecting groups for other hydroxyls that would be free in the final compounds.
Synthesis of Disaccharide and Trisaccharide Building Blocks.
With the aforementioned requirements in mind, monosaccharide units 10 - 17 were prepared according to the standard reaction conditions (see Supporting Information 2.1). Having electrophilic donors 10 – 14 and nucleophilic acceptors 15 – 17 in hand, we next focused on the parallel synthesis of disaccharide building blocks incorporating 1,2-trans relative stereochemistry (Scheme 1). A combination of N-iodosuccinimide and silver triflate (NIS/AgOTf)-mediated glycosylation of each of the acceptors 15-17 with thioglycoside donor 14 afforded three different disaccharides (18, 19, and 24) bearing an aminoethyl spacer at the reducing end. In each coupling, only a β−1,2-trans glycoside was observed due to neighboring group participation by the C2-acetyl of the donor 14, affording the corresponding disaccharides in excellent yields (85% to 96%). Next, the coupling products 18, 19, and 24 were transformed into disaccharide acceptors 22, 23, and 26 bearing a −1 GlcA derivative and a +1 GlcN derivative, utilizing a standard set of reaction conditions (Scheme 1). Accordingly, the benzylidene acetals of 18, 19, and 24 were removed by treatment with 80% acetic acid (AcOH) at 80 °C to generate the desired diols 20, 21, and 25, respectively, whose C6-hydroxyls were selectively oxidized with substoichiometric amount of 2,2,6,6-teteramethyl-1-piperidinyloxy (TEMPO) in the presence of excess iodobenzene diacetate (BAIB) as a co-oxidant.19 The resulting carboxylic acids were then protected as methyl esters by treatment with methyl iodide (MeI) to afford disaccharide acceptors 22, 23, and 26 ranging from 78% to 83% over two steps.
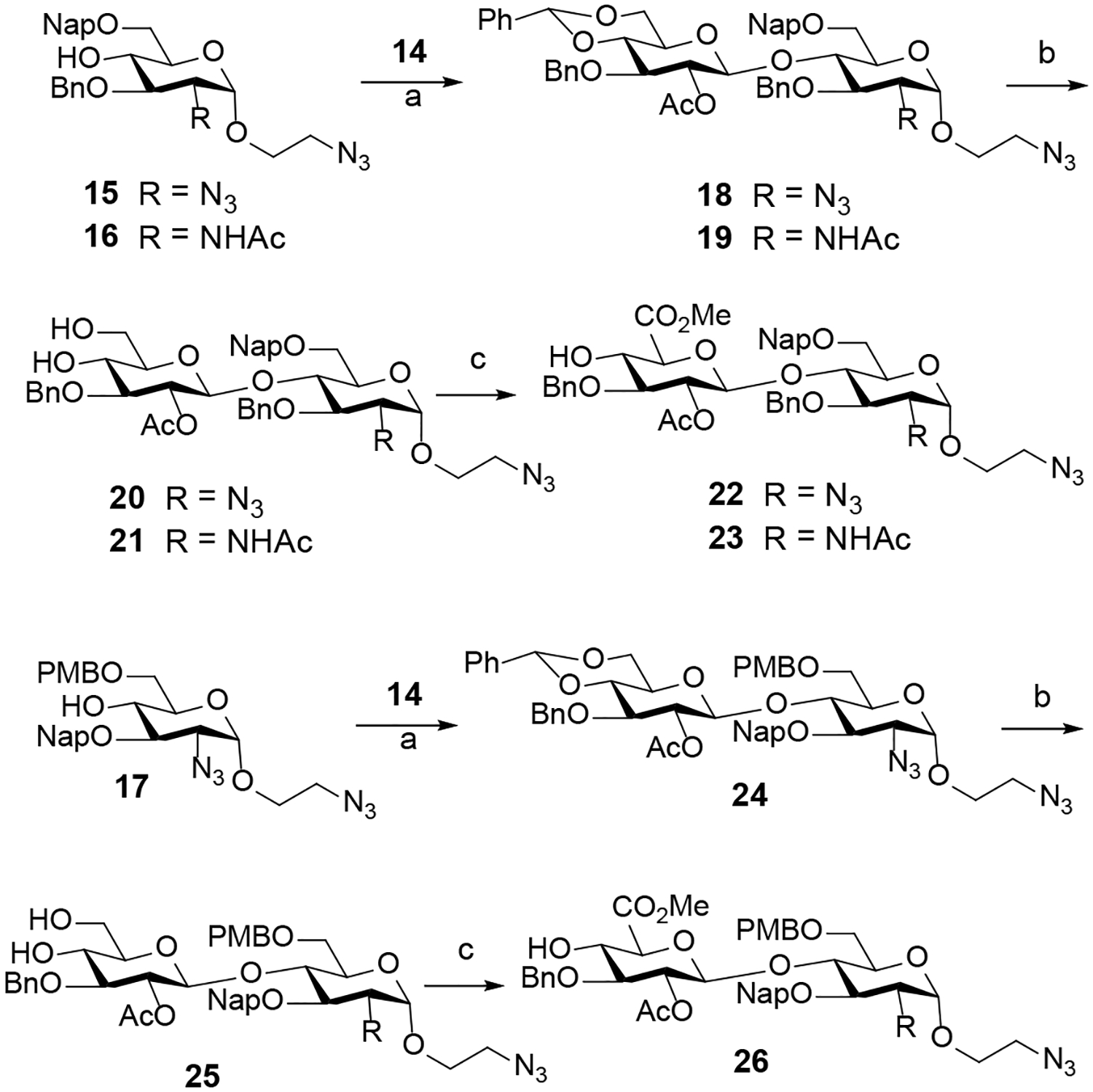
Synthesis of disaccharides with GlcA-GlcN backbonea
a Reagents and conditions: (a) NIS, AgOTf, DCM, 0 °C (18, 85%; 19, 96%; 24, 87%); (b) 80% AcOH, 80 °C (20, 72%; 21, 82%; 25, 75%); (c) BAIB, TEMPO, DCM/H2O; then MeI, KHCO3, DMF (22, 78%; 23, 80%; 26, 83%).
To illustrate that the spacer-containing disaccharide acceptors 22, 23, and 26 (Scheme 1) and −2 GlcN-derived donors 10 – 13 (Figure 3) can be employed for the stereoselective construction of α−1,4-linkages of trisaccharides 27 – 33, we first investigated a NIS/AgOTf-mediated coupling of disaccharide acceptor 23 with thioglycoside donor 12 (Table 1). The desired trisaccharide 27 was not observed at 0 °C or room temperature (entries 1 and 2) due to relatively low reactivity of C4-hydroxyl of the GlcA unit. Increasing the reaction temperature (25°C → 35°C) and longer reaction time (2 h → 12 h) afforded 27 in 56% yield (entry 4) as a mixture of anomers (α:β = 3:1). Following the seminal work of Mong and co-workers,20 we investigated N,N-dimethylforamide (DMF) as an α-modulating additive to direct the stereochemical course of glycosylation. As illustrated in entry 5, the stereoselectivity of the reaction with DMF (6 equiv.) was significantly much better (α:β >20:1). This optimal set of glycosylation conditions were then applied to the coupling of disaccharide acceptors 22 and 26 with thioglycoside donors 10, 11 and 12 to furnish the trisaccharides 28 – 32 (57% to 65%) with exclusive α-anomeric configuration at the newly-formed glycosidic bond (Scheme 2).
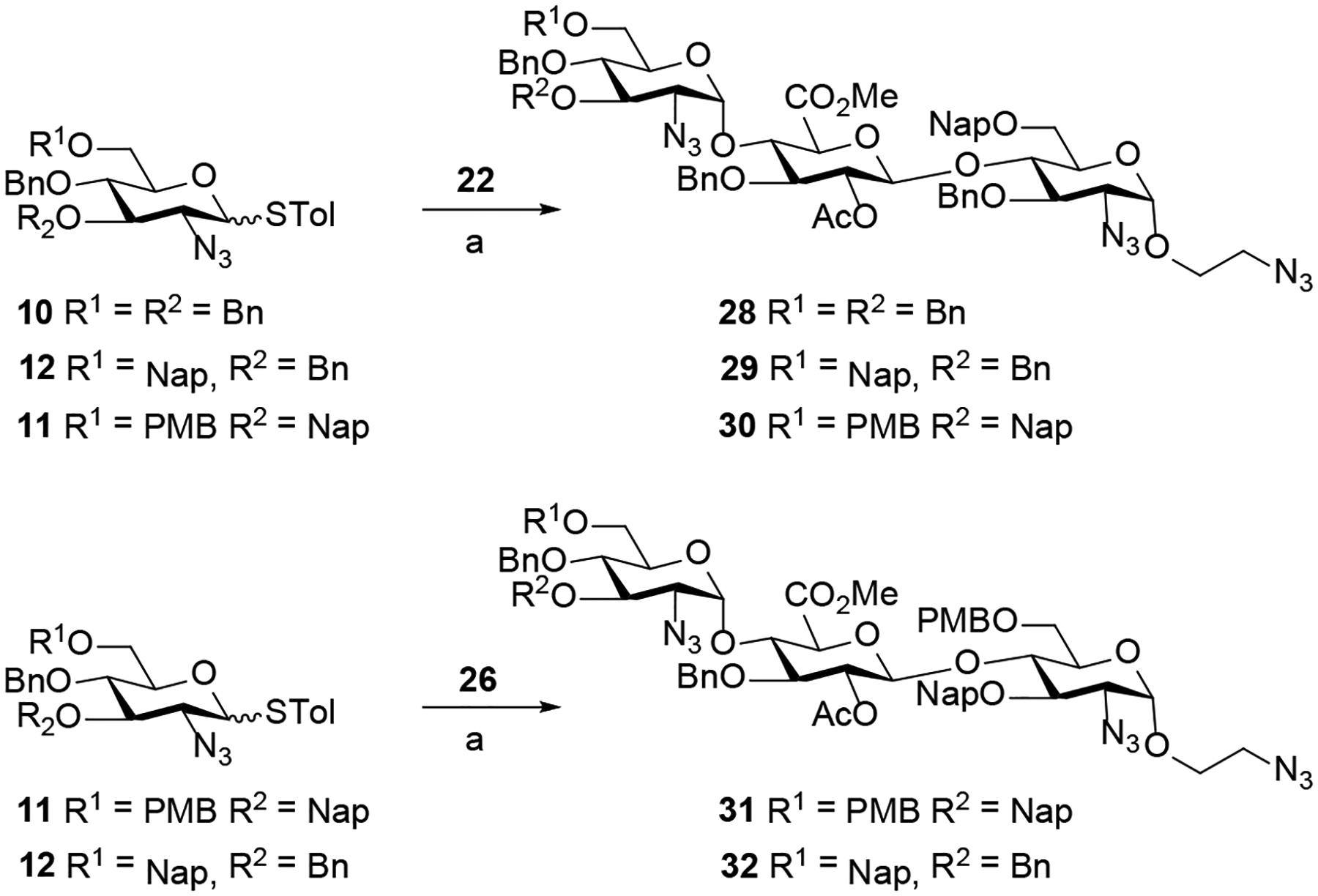
Stereoselective construction of trisaccharides 28-32a
a Reagents and conditions: (a) NIS, AgOTf, DMF (6 equiv.), DCM, 35 °C, 5 h (28, 63%; 29, 65%; 30, 62%; 31, 57%; 32, 60%; α:β >20:1).
Table 1.
Stereoselective glycosylation of disaccharide acceptor 23with C2-azido thioglycoside donor 12a
 | ||||||
|---|---|---|---|---|---|---|
| entry | AgOTf (equiv) | additive | Temp. | time (h) | yield (%) | α:β |
| 1 | 0.1 | none | 0 °C | 1 | NR | |
| 2 | 0.1 | none | 25 °C | 1 | NR | |
| 3 | 0.1 | none | 35 °C | 2 | 45 | 3:1 |
| 4 | 0.1 | none | 35 °C | 12 | 56 | 3:1 |
| 5 | 0.1 | DMF (6 equiv) | 35 °C | 12 | NR | |
| 6 | 1 | DMF (6 equiv) | 35 °C | 12 | 51 | >20:1 |
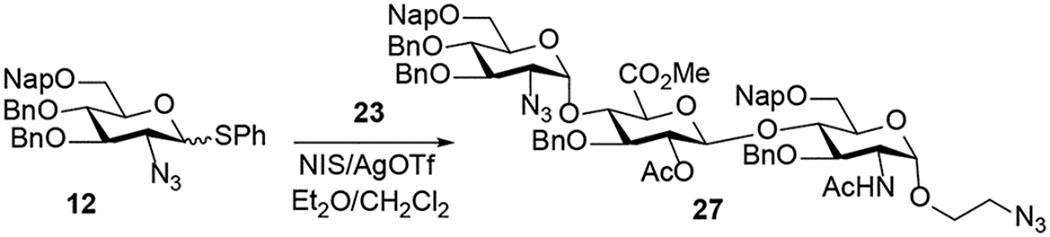
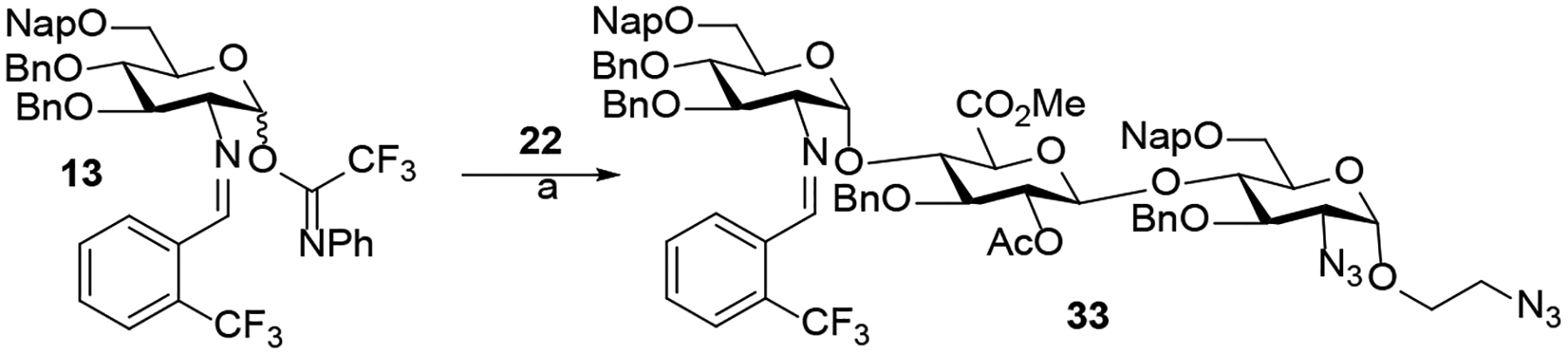
After establishing the effectiveness of glycosylation conditions for the synthesis of trisaccharides 27 – 32, we examined the coupling of disaccharide acceptor 22 with C2-N-benzylidene donor 13 (Scheme 3). The N-benzylidene group can be selectively removed under acidic conditions in the presence of the azido moiety. The N-benzylidene and azido offer a set of orthogonal amino protecting groups that allows selective functionalization of each function, further increasing the structural diversity of trisaccharides. We have recently discovered that the release of triflic acid from nickel triflate in combination with the unique stereoelectronic nature of the C(2)-N-benzylidene group on glycosyl N-phenyl trifluoroacetimidates donor provided the highly favorable formation of 1,2-cis-amino- glycosides.21–24 We found that the reaction of donor 13 with disaccharide acceptor 22 (Scheme 2) using triflic acid (TfOH) provided trisaccharide 33 with moderate α-selectivity (α:β = 5:1). Addition of DMF as an α-modulating additive20 significantly improved α-selectivity (α:β > 20:1) of 33 (Scheme 3). In this reaction, it is worth noting that stoichiometric amount of triflic acid was required because the DMF additive can quench the acid. In addition, room temperature and long reaction time (8 h) were neccesaary for the coupling to proceed to completion while degradation was observed at 35 °C.
Preparation of trisaccharide 33 containing C2-N-benzylidenea
a Reagents and conditions: (a) TfOH (1 equiv.), DMF (6 equiv.), 25 °C, 8 h, DCM, 57%, α:β > 20:1.
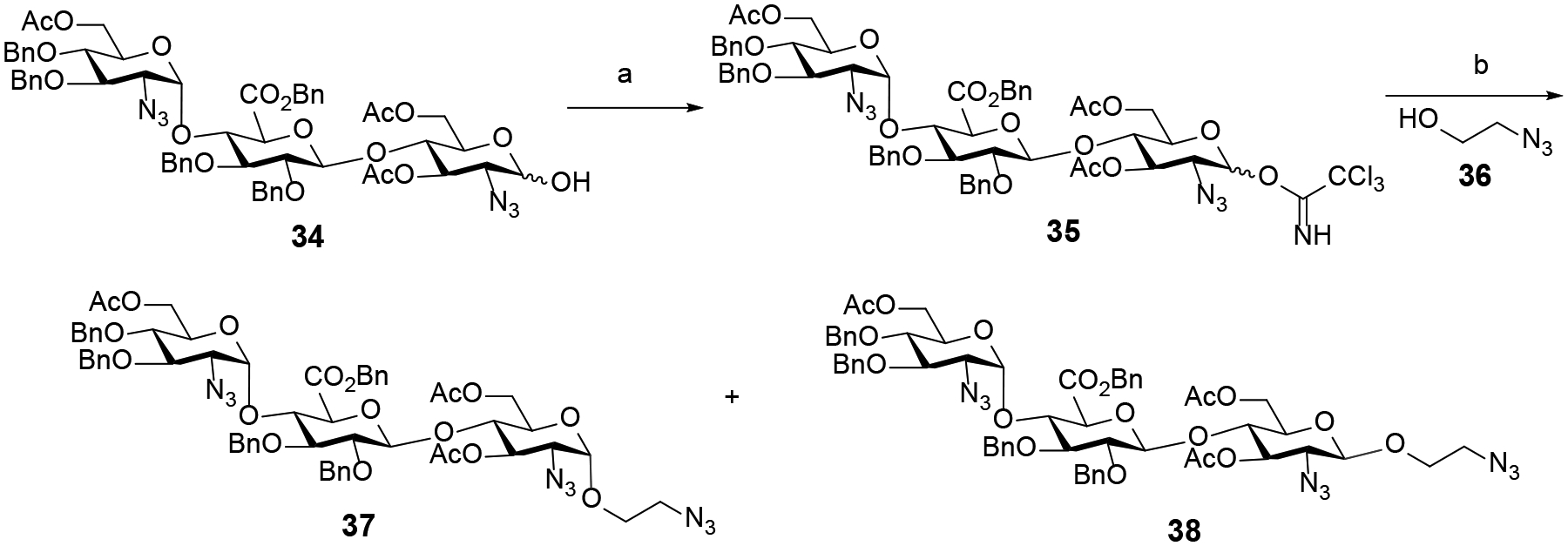
We next focused on the synthesis of trisaccharides having the β-anomeric configuration at the reducing end so that we can compare their ability to modulate heparanase activity with their α-counterparts. Accordingly, we embarked on the synthesis of α- and β-adducts 37 and 38 (Scheme 4) starting from commercially available trisaccharide 34.18 The hemiacetal 34 was modified at the reducing end to furnish trichloroacetimidate 35, ready for coupling to the aminoethyl spacer 36. Since the C2-azido moiety of the reducing end glucosamine unit of 35 lacks traditional neighboring participation, we anticipated that a mixture of α-and β-anomers would be generated. The stereochemical outcome of this reaction is necessary for us to access both α- and β-trisaccharides for subsequent studies of heparanase activity. Accordingly, glycosylation of the acceptor 36 with trisaccharide trichloroacetimidate 35 promoted by trimethylsilyl trifluoromethanesulfonate (TMSOTf) afforded α-adduct 37 (51%) and β-adduct 38 (30%), which could be separated by traditional silica column chromatography (Scheme 4).
Synthesis of Sulfated Trisaccharides.
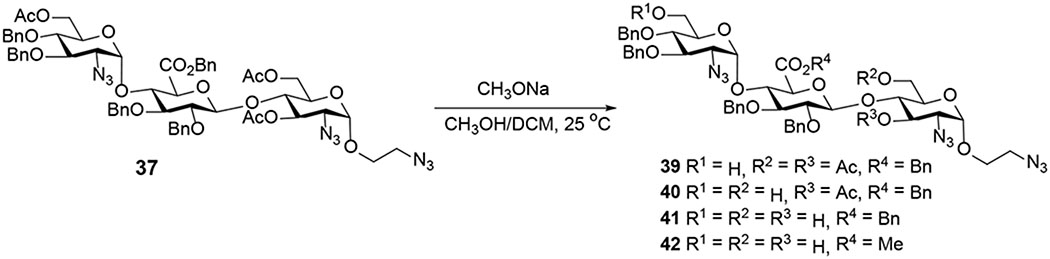
After constructing the trisaccharide frameworks, functional group transformations were next conducted to achieve the desired products incorporating different sulfation patterns at 6-O, 3-O and 2-N of glucosamine unit at −2 and +1 binding subsites as well as 2-O of glucuronic acid unit at the −1 binding subsite. First, selective deacetylation at C6 of trisaccharide 37 were investigated (Table 2) using sodium methoxide (NaOMe) in a 1:1 mixture of methanol (MeOH) and dichloromethane (DCM). Employing substoichiometric amount (0.2 equiv.) of NaOMe at low reaction concentration (0.02 M) for 5 h (entry 3) afforded a separable mixture of 39 (removal of −2 C6-acetyl group of GlcN unit) and 40 (removal of +1 C6-acetyl group of GlcN unit), further increasing the structural diversity of synthetic targets. On the other hand, using equimolar sodium methoxide at high concentration (0.1 M) predominantly yielded 40 (entry 6). In all cases, an inseparable mixture of the byproducts 41 and 42 which were tentatively identified by Mass spectrometry was also detected. A higher amount of 41 and 42 was observed at higher concentration and longer reaction time. The optimal conditions developed for the selective hydrolysis were then exploited with trisaccharide 38 incorporating the reducing end β-linkage (Scheme 5) to deliver the separable products 43 (41%) and 44 (35%).
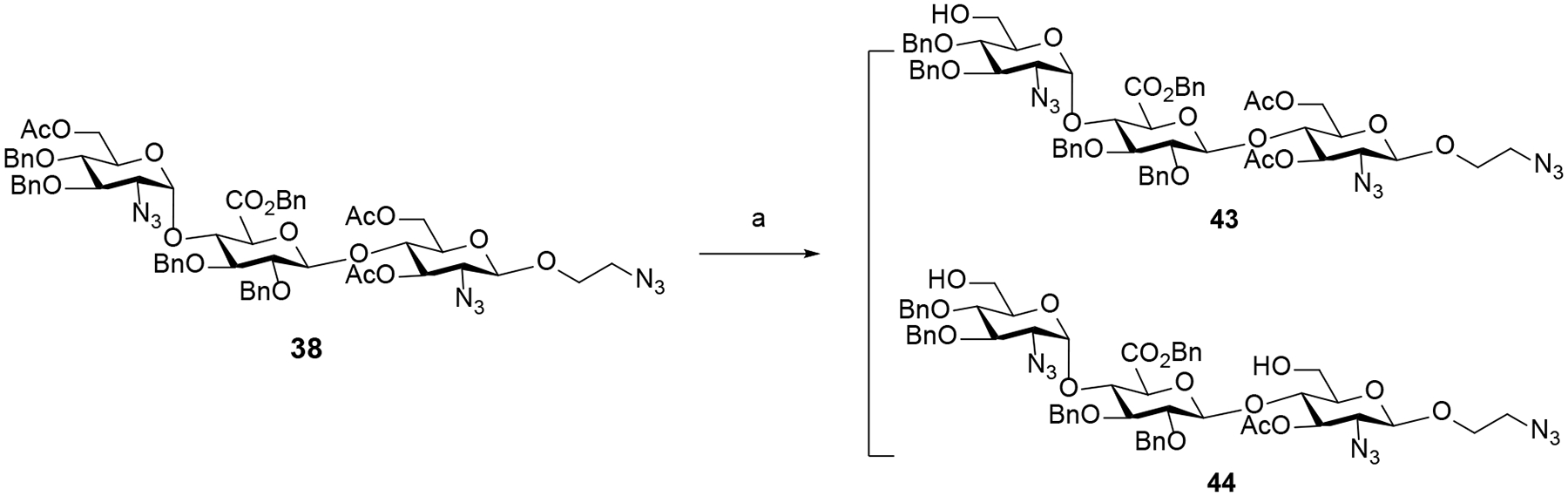
Selective C6-deacetylation of −2 and +1 glucosamine units of β-linked trisaccharide 38a
a Reagents and conditions: (a) CH3ONa (0.2 equiv.), 1:1 mixture of MeOH and DCM (0.02 M), 25 °C, 5 h (43, 41%; 44, 35%).
Table 2.
Selective C6-deacetylation of −2 and +1 glucosamine units of α-linked trisaccharide 37a
 | ||||||
|---|---|---|---|---|---|---|
| entry | CH3ONa (equiv.) | concentration (M) | time (h) | 39: yield (%) | 40: yield (%) | 41+42: yield (%) |
| 1 | 0.1 | 0.02 | 8 | 23 | 8 | none |
| 2 | 0.2 | 0.02 | 8 | 35 | 40 | 17 |
| 3 | 0.2 | 0.02 | 5 | 42 | 33 | 11 |
| 4 | 0.5 | 0.02 | 5 | 24 | 48 | 22 |
| 5 | 1.0 | 0.1 | 3 | 7 | 57 | 21 |
| 6 | 1.0 | 0.1 | 5 | 5 | 51 | 32 |
Prior to examining the conditions for selective removal of the Nap and PMB groups, the C2-acetyl group of −1 glucuronic acid (GlcA) derivative of trisaccharide 29 was first hydrolyzed under mild conditions to reveal secondary alcohol 45 (Scheme 6). Next, the C2-N-benzylidene of −2 GlcN moiety of trisaccharide 33 was removed in less than 20 min under acidic conditions. Subsequent acetylation of the resulting amine salt intermediate 46 afforded trisaccharide 47 (Scheme 6) having a −2 GlcNAc moiety. The stage was set for chemoselective cleavage of the Nap and PMB groups from trisaccharides 27, 28, 30-32, 45, and 47.
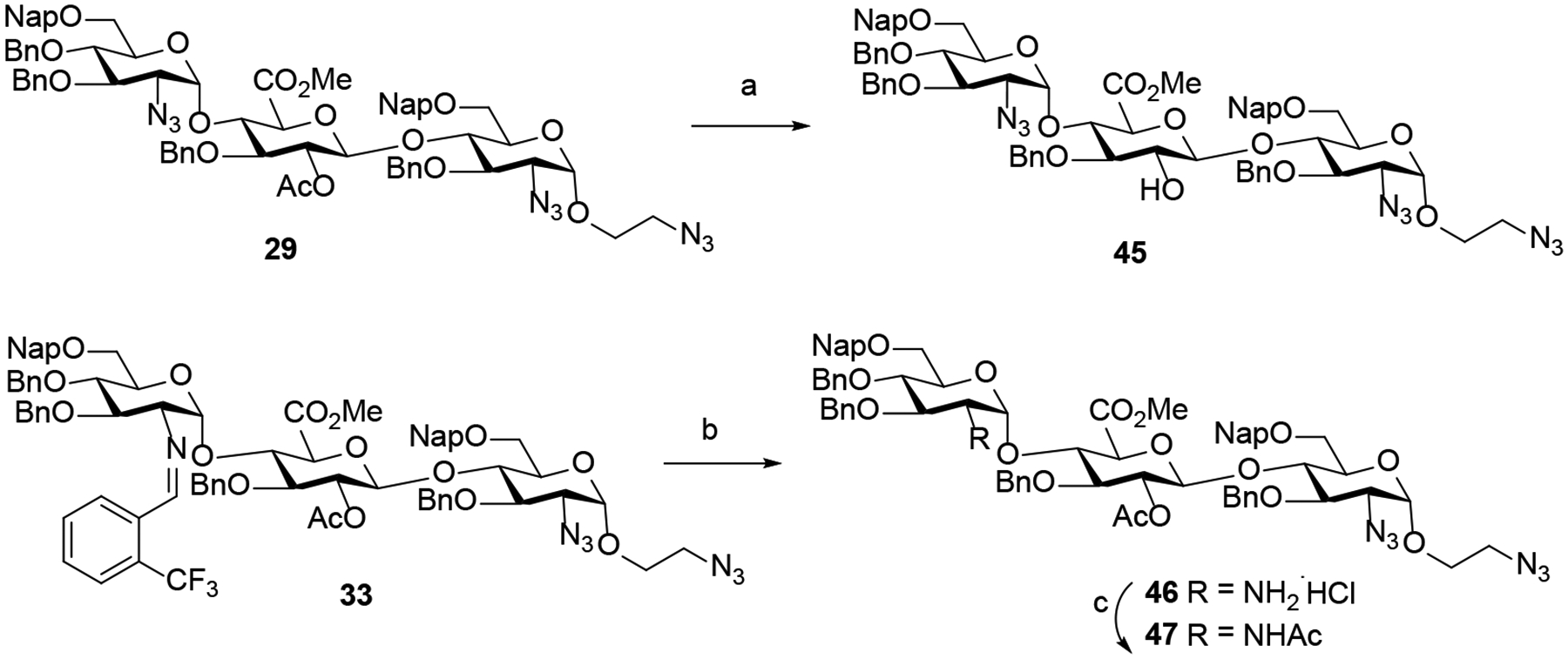
Removal of C2-acetyl group of −1 GlcA unit and C2-N-benzylidene group of −2 GlcN unita
a Reagents and conditions: (a) CH3ONa (4.0 equiv.), CH3OH/DCM (45, 85%); (b) 6N HCl, acetone, 25 °C, 20 min (46, 78%); (c) Ac2O, Et3N, DCM/CH3OH (47, 81%).
Proper Nap and PMB deprotection procedure is necessary to reveal key hydroxyls for subsequent O-sulfonations. As such, we first examined the conditions with the most available trisaccharide 32 as it contains the C6-Nap group at −2 GlcN unit as well as the C6-PMB and C3-Nap groups at +1 GlcN unit (Table 3). To this end, we studied the reaction of 32 under acidic conditions using a 8:1 mixture of TFA and DCM (entries 1 and 2), affording the corresponding triol 48 in poor yield (26% to 35%). Reasoning that the acidic conditions were causing product decomposition, we next examined the reaction of 32 with DDQ in the presence of DCM and water.25 In the absence of an acid scavenger of the acidic 2,3-dichloro-5,6-dicyanohydroquinone byproduct resulting from the DDQ deprotection, the desired adduct 48 was isolated in moderate yield (entries 3–4). Following the seminal work of Bennett and co-workers,26 we investigated the strained olefin, β-pinene, as an acid scavenger in combination with DDQ, leading to the formation of the desired product 48 in a significantly higher yield (73%, entry 6). The optimal conditions were applied to other trisaccharides to give compounds 49 – 54 in an overall yield ranging from 65% to 82% (Scheme 7).
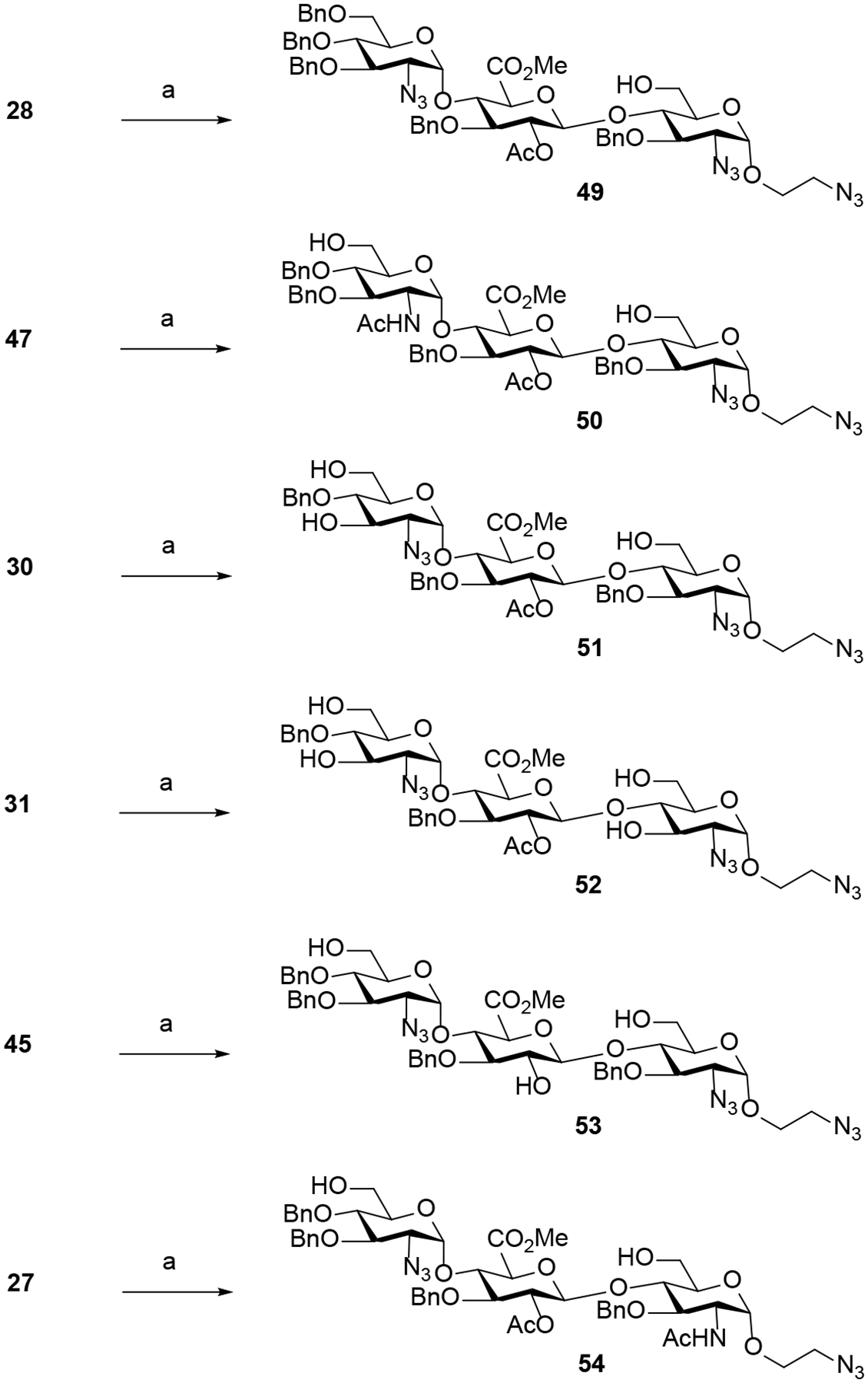
Removal of the Nap and PMB protecting groups.
a Reagents and conditions: (a) DDQ (9 equiv.), β-pinene (9 equiv.), 25 °C, DCM/H2O (10/1) (49, 82%; 50, 75%; 51, 71%; 52, 76%; 53, 71%; 54, 65%).
Table 3.
Chemoselective cleavage of the Nap and PMB protecting groups of trisaccharide 32a
 | ||||||
|---|---|---|---|---|---|---|
| entry | solvent | DDQ | β-pinene | temperature | time (h) | yield (%) |
| 1 | TFA/toluene (8/1) | none | none | 0 °C | 3 | 26 |
| 2 | TFA/toluene (8/1) | none | none | 0 °C→25 °C | 3 | 35 |
| 3 | DCM/H2O (10/1) | 9 equiv. | none | 25 °C | 4 | 36 |
| 4 | DCM/H2O (10/1) | 9 equiv. | none | 25 °C | 16 | 43 |
| 5 | DCM/H2O (10/1) | 9 equiv. | 9 equiv. | 25 °C | 4 | 65 |
| 6 | DCM/H2O (10/1) | 9 equiv. | 9 equiv. | 25 °C | 8 | 73 |
With the eleven differently protected trisaccharides 39, 40, 43, 44, and 48 - 54 in hand, they could now individually undergo selective azide reduction followed by N- and O-sulfonations. A key issue of this approach is to identify the conditions that orthogonally reduce the azido moiety as well as chemoselectively sulfate the hydroxyl and amine groups (Scheme 8). We found that utilizing trimethyl phosphine (PMe3) in THF/H2O in the absence of NaOH to avoid deacetylation of C3-acetyl of +1 GlcN unit of 40 resulted in no conversion.27 On the other hand, using Zn/AcOH conditions resulted in a complex mixture.28 Ultimately, we discovered that the smooth azido-to-amino conversion was successfully implemented using propane-1,3-dithiol in MeOH in the presence of trimethylamine at 50 °C for 10 h.29 The resulting amine 55 was immediately subjected to simultaneous N- and O-microwave sulfonation developed by the Yu group.30 Interestingly, the over-sulfated byproduct 57 was observed along with the desired product 56. No such issue was found in the previous study while methyl glycosides were utilized as substrates.30 We subsequently discovered that the amount of sulfur trioxide trimethylamine complex and reaction time play a key role in controlling the ratio of the desired polysulfated product 56 and the oversulfated product 57. The search revealed that sulfation of compound 55 using 15 equivalents of sulfur trioxide trimethylamine complex for 15 min gave almost exclusively the desired sulfated product 56 (83% yield over 2 steps) with a trace of 57. For the subsequent hydrogenolysis process to proceed smoothly, it was essential for the N-sulfate counterions to be sodium cation (Na+) as opposed to the triethylammonium (Et3NH+). Accordingly, treatment of 56 with Amberlite IR120 Na+ resin afforded the sodium sulfated product 58. Finally, 58 was subjected to hydrogenolysis over Pd(OH)2 in a mixture of MeOH and pH 7 buffer to remove the benzyl ethers followed by deacetylation31 to provide trisaccharide 1α in 88% yield over 3 steps (Scheme 8).
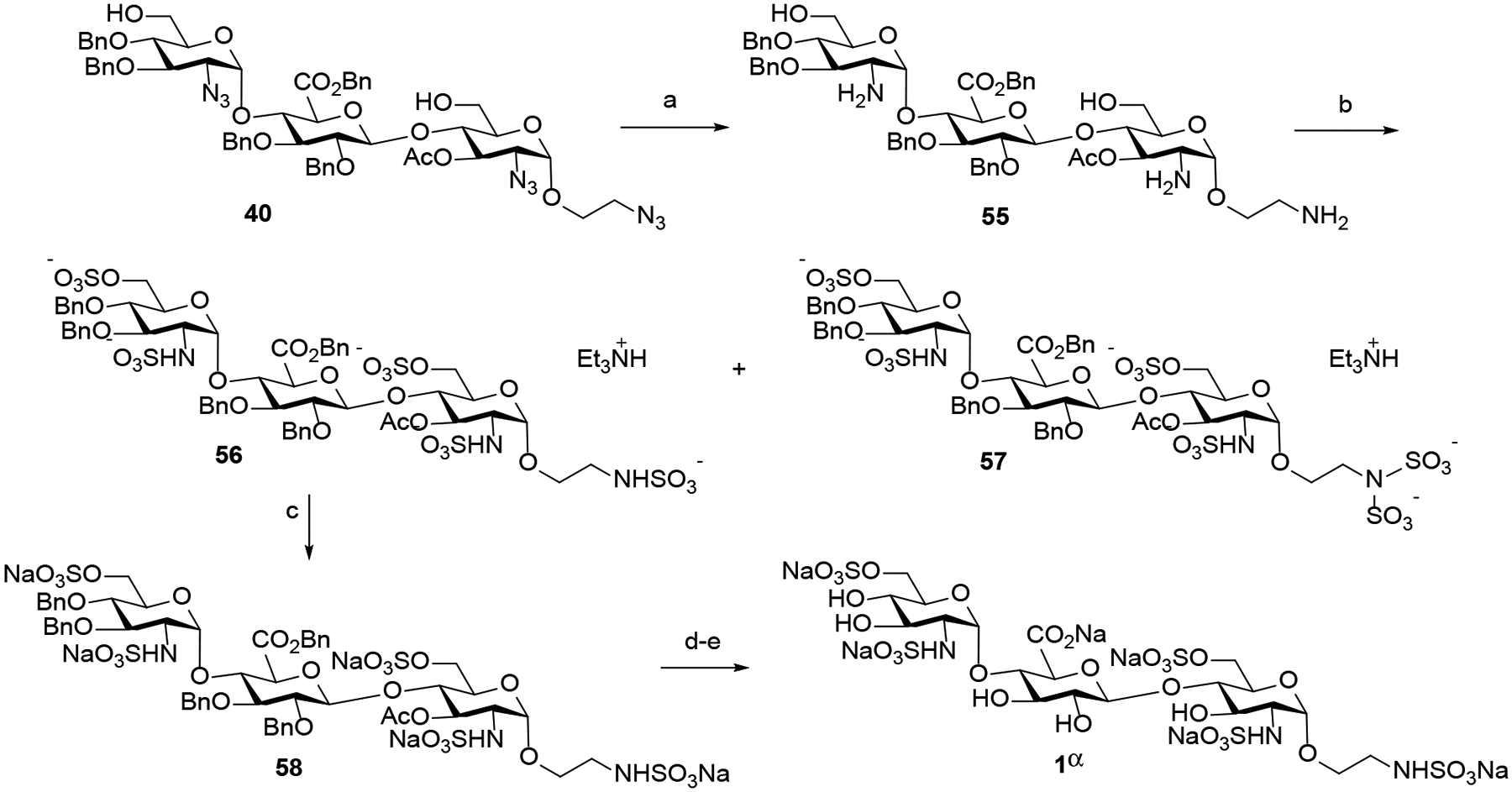
Azide reduction followed by sulfations and deprotection to form HS trisaccharide 1αa
a Reagents and conditions: (a) propane-1,3-dithiol, DIPEA, CH3OH, 50 °C, 10 h; (b) SO3.Et3N, pyridine, 100 °C, 15 min, microwave (58, 83% over 2 steps); (c) Amberlite IR120 Na+ resin, 25 °C, 24 h; (d) 100 psi H2, Pd(OH)2, CH3OH/pH 7 buffer, 25 °C, 24 h; (e) LiOH, H2O, 25 °C 24 h; then Amberlite IR120 Na+ resin, 25 °C, 24 h (1α, 88% over 3 steps).
The optimal azido-amine conversion and sulfation conditions developed in Scheme 8 were then applied to give the polysulfated trisaccharides 59 – 68 in an overall yield ranging from 73% to 85% (Scheme 9). Finally, these compounds were subjected to hydrogenolysis and deacetylation to supply the desired HS trisaccharides 1β, 2α, 2β, and 3α−9α in overall good yields. The structures of HS-liked compounds were confirmed by 1D and 2D NMR and mass spectrometry analysis (see Supporting Information). The α−1,2-cis anomeric configuration of 2-aminoglucosides was confirmed by J1,2 coupling constants. While O-sulfonation was observed by 1H NMR downfield shifts from 3.5–3.8 ppm to 4.0–4.4 ppm, N-sulfonation was observed by 13C NMR upfield shifts from 62–65 ppm to 55–58 ppm.31
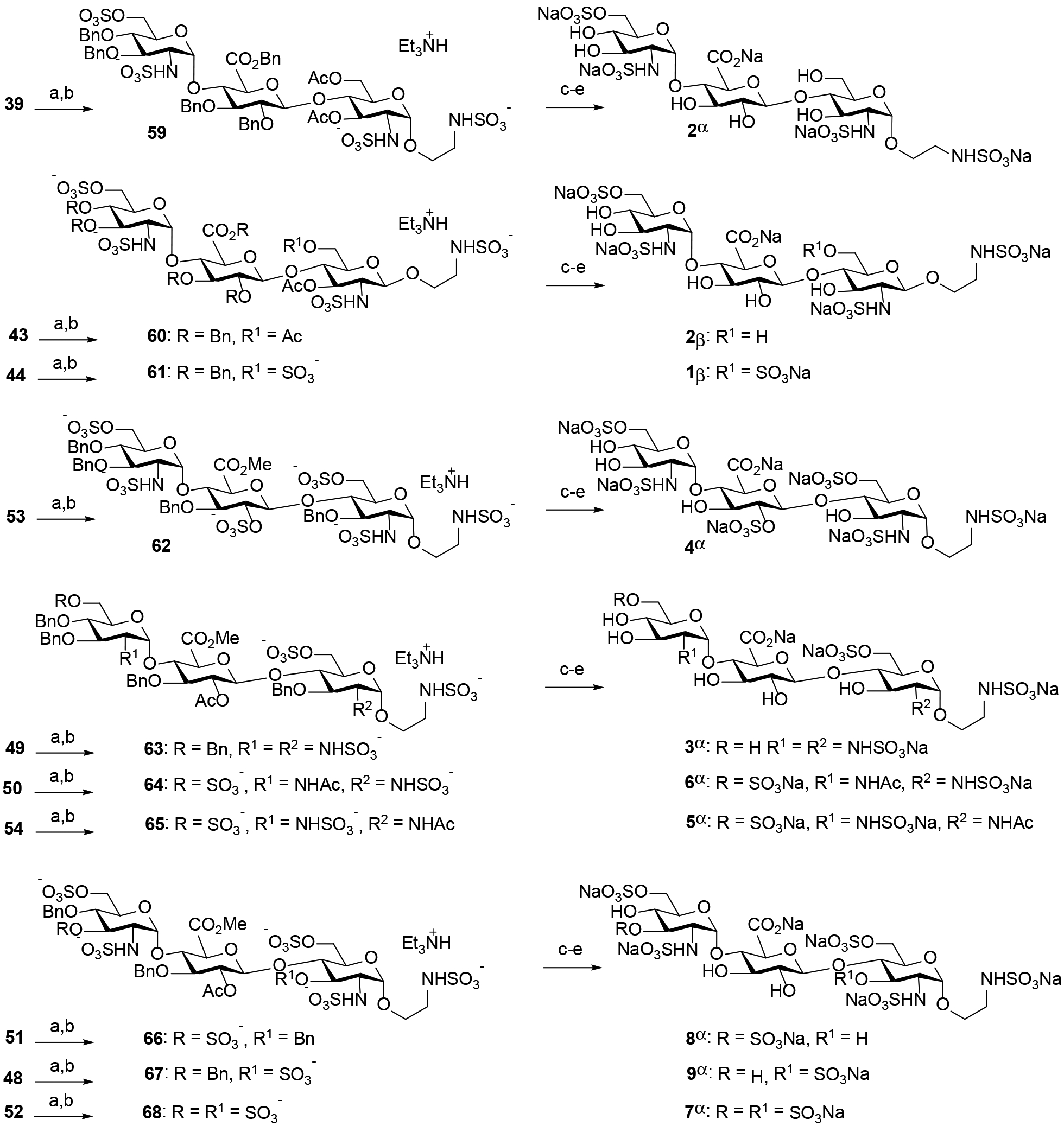
Synthesis of HS trisaccharides 1β, 2α, 2β, and 3α−9αa
a Reagents and conditions: (a) propane-1,3-dithiol, DIPEA, CH3OH, 50 °C, 10 h; (b) SO3.Et3N, pyridine, 100 °C, 15 min, microwave (59, 80%; 60, 76%; 61, 78%; 62, 81%; 63, 82%; 64, 85%; 65, 82%; 66, 84%; 67, 76%; 68, 73%; 2 steps); (c) Amberlite IR120 Na+ resin, 25 °C; (d) 100 psi H2, Pd(OH)2, CH3OH/pH 7 buffer, 25 °C, 24 h; (b) LiOH, H2O, 25 °C 24 h; then Amberlite IR120 Na+ resin, 25 °C (1β, 84%; 2α, 87%; 2β, 85%; 3α, 86%; 4α, 83%; 5α, 86%; 6α, 91%; 7α, 80%; 8α, 88%; 9α, 82%; 3 steps).
Interaction of the HS Trisaccharides with Heparanase:
With the library of trisaccharides 1α, 1β, 2α, 2β, and 3α – 9α in hand, we evaluated how their varied sulfation patterns and anomeric configuration at the reducing end modulated heparanase activity employing a time-resolved fluorescence resonance energy transfer (TR-FRET) assay against fluorescent labeled-HS.32 The assay is based on time-resolved fluorescence energy transfer (TR-FRET) between two fluorophores, a donor (europium cryptate) and an acceptor (XL665, allophycocyanin). When the donor and acceptor apart, there is no FRET signal. Once they are brought in proximity, the FRET signals are generated (for example, the emission spectrum at 665 nm). Heparan sulfate substrate labeled with both biotin and europium cryptate, then incubated with streptavidin-XL665 will give maximum emission. Cleavage of biotin-HS-cryptate substrate by heparanase results in decrease in FRET signal. If the inhibitor is present, biotin-HS-cryptate substrate remains intact resulting in high FRET signal. The FRET emission signal observed will depend on the inhibitory potency of the compound at a given concentration. As can be seen in Table 4, HS trisaccharide 1α bearing an α-anomeric linkage at the reducing end was able to inhibit heparanase with relatively high potency (entry 1, IC50 = 0.393 ± 0.010 μM). To compare, compound 1β which is composed of a β-anomeric linkage at the reducing end, displayed modest inhibitory activity (entry 2, IC50 = 42.06 ± 1.287 μM). These results could be attributed to differences in their interactions with the active site of heparanase (vide infra).11 Furthermore, these results provide the first example that the anomeric configuration at the reducing end of HS trisaccharide can modulate heparanase activity and underscore the importance of the saccharide framework for recognition by heparanase. Next, we examined whether site-defined modification to the sulfation pattern of 1α would alter its affinity for heparanase. Removal of the 6-O-sulfate group at +1 GlcN (2α, entry 3) and at −2 GlcN (3α, entry 4) decreased binding to heparanase (IC50 = 8.225 ± 0.102 μM and 3.012 ± 0.058 μM, respectively). The observation, that a) removal of 6-O-sulfation reduces potency, and b) removal of 6-O-sulfate group at +1 GlcN has a greater effect than removal of 6-O-sulfate group at −2 GlcN, suggests the importance of precise positioning of the sulfate groups on the trisaccharides for heparanase recognition. This result is also consistent with Davies’ the co-crystalized HS-heparanase structures,11 which indicate 6-O-SO3− at the +1 subsite forms a hydrogen bonding network with Gln270.15 On the other hand, compounds 5α and 6α, which have N-acetyl groups instead of N-sulfates, have 30-fold reduced potency (entries 7 and 8) compared to 1α, highlighting the importance of the −2 and +1 N-sulfates for heparanase recognition.
Table 4.
Modulation of heparanase activity by HS-like trisaccharides using a TR-FRET assay
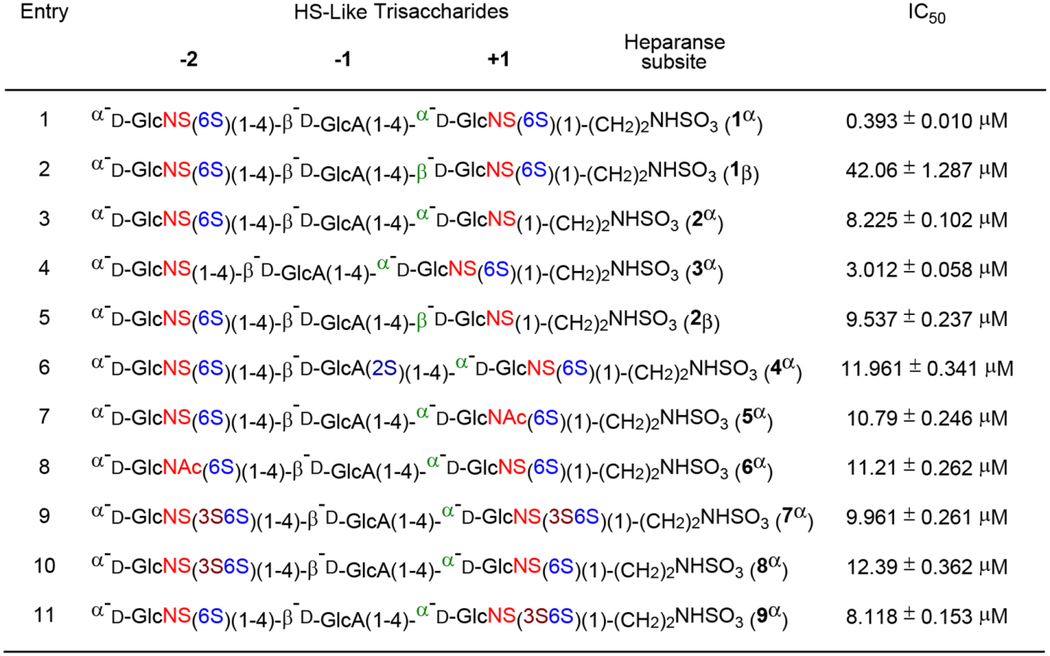 |
Previous studies have demonstrated that heparanase can recognize GlcN unit (GlcN) carrying either C6- and/or C3-O-sulfate.14–15 As can be seen in Table 4, trisaccharides bearing 3-O-sulfate at −2 GlcN (8α, entry 10) and at +1 GlcN (9α, entry 11) were significantly less active (IC50 = 12.39 ± 0.362 μM and 8.118 ± 0.153 μM, respectively) than trisaccharide 1α (entry 1). The addition of a third sulfate to the C3 position of both GlcNS6S units, forming trisaccharide 7α (entry 9, IC50 = 9.961 ± 0.261 μM), did not prove to be advantageous. Furthermore, addition of a sulfate ester at C2 of −1 glucuronic acid (4α, entry 6) unit led to a large reduction in heparanase recognition (IC50 = 11.961 ± 0.341 μM), validating that the presence of GlcA(2S) at the −1 subsite cannot be tolerated by heparanase.11 Collectively, our results illustrate that controlling the sulfation pattern and the reducing end anomeric configuration within the trisaccharides enable to modulate their heparanase-inhibitory activity.
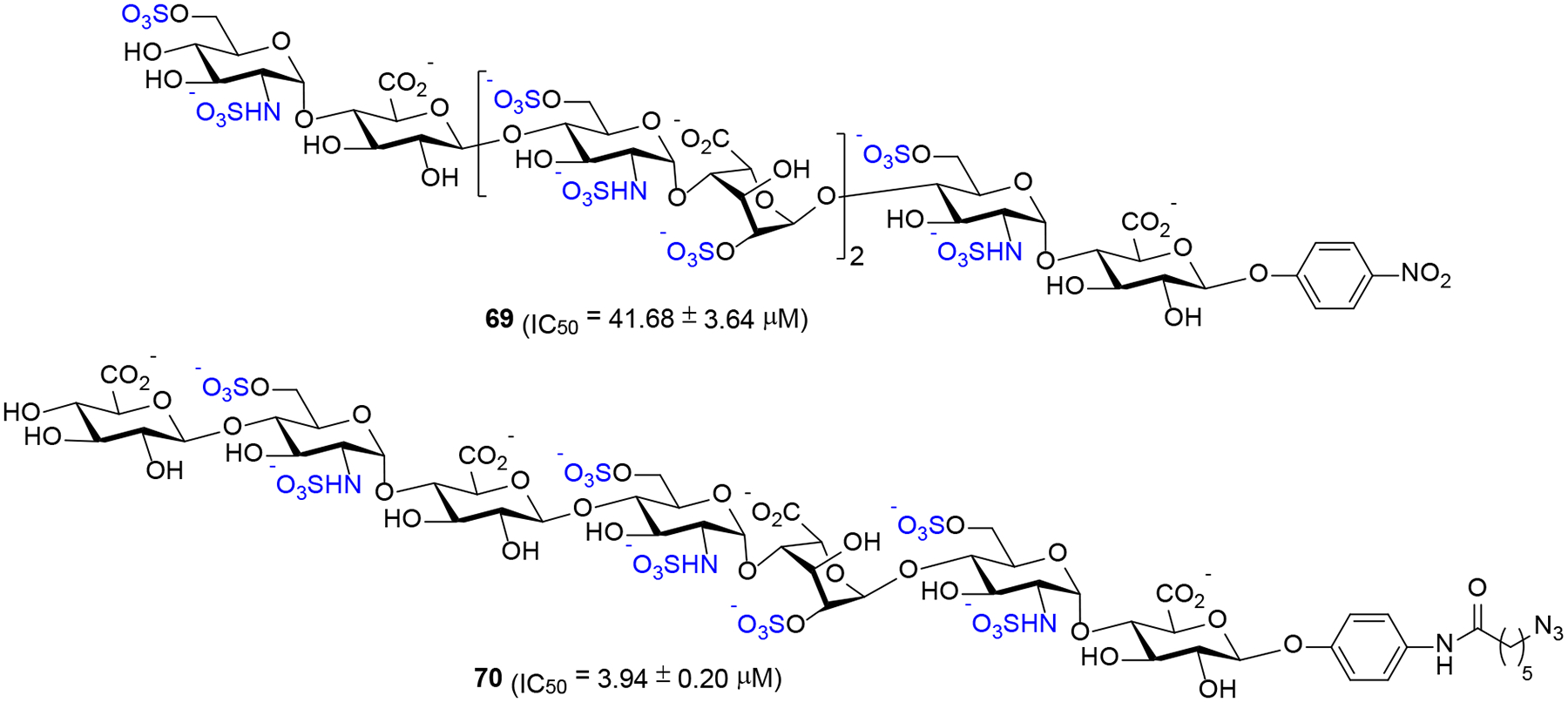
It has been previously demonstrated that HS oligosaccharides are less active than full-length HS polysaccharides.31 As such, we questioned whether relatively larger oligosaccharides exhibit higher activity than a well-defined sulfated trisaccharide 1α having an α-anomeric linkage at the reducing end linker. Surprisingly, both HS heptasaccharide 69 (IC50 = 41.68 ± 3.64 μM, Scheme 10) and octasaccharide 70 (IC50 = 3.94 ± 0.20 μM, Scheme 10) exhibited significantly decreased binding to heparanase compared to 1α.33 The low potency compound 69 is similar in activity to trisaccharide 1β (Table 4, entry 2) bearing the β-configured reducing end. Although octasaccharide 70 is more potent than 69, its potency is approximately 10-fold lower than that of trisaccharide 1α (Table 4, entry 1). The observation that trisaccharide 1α is more potent than larger oligosaccharides 69 and 70 suggests that saccharide framework, precise positioning of the sulfate groups, overall charge, and the reducing-end anomeric configuration play critical roles in determining the affinity of HS oligosaccharides for heparanase and the preferential cleavage recognized by heparanase.34
Computational Study of HS Trisaccharides 1α and 1β.
It has never been established how the anomeric configuration at the reducing end of HS oligosaccharides could have a great effect on their binding affinity for heparanase. As can be clearly demonstrated in Table 4, trisaccharide 1α with the α-configured reducing end is by far more potent than trisaccharide 1β with the β-configured reducing end. Accordingly, an in silico docking study with 1α and 1β into the apo crystal structure of heparanase (PDB code: 5E8M) was performed.11, 35–36 We initiated our study by docking the natural HS substrate, GlcNS(6S)α(1,4)GlcAβ(1,4)GlcNS(6S) trisaccharide, into heparanase to obtain a benchmark for comparison with our compounds (see Figure S6 for actual docked structures). Next, trisaccharide 1α (Figure 4A) and trisaccharide 1β (Figure 4B) were constructed in silico to be docked in the apo heparanase crystal structure. Upon examination of the docked structures for the trisaccharide-heparanase complexes, both 1α and 1β placed the GlcNS(6S)α(1,4)GlcAβ(1,4)GlcNS(6S) into subsites −2/−1/+1 (Figure 4) in a similar fashion to Davies’ the published co-crystalized HS-heparanase structures.11 The trisaccharide was oriented in the same direction as the co-crystalized structures with the non-reducing end towards HBD-1 and the reducing end closer to HBD-2. When looking at specific interactions for both trisaccharides, we found that while the N-sulfate of the aminoethyl spacer of trisaccharide 1α formed a hydrogen bond with Lys231, the spacer of trisaccharide 1β formed a hydrogen bond with Arg272. Both the +1 C6-O-sulfate of 1α and 1β formed a strong divalent hydrogen bonding network with Gln270 while both the −2 N-sulfate of 1α and 1β formed a hydrogen bond with Gly389 (Figure 4A and and4B).4B). Interestingly, while −2 C6-O-sulfate of trisaccharide 1α potentially formed a salt bridge to Lys159 (Figure 4A) in a similar fashion to Davies’ the co-crystalized HS-heparanase structures;11 −2 C6-O-sulfate of trisaccharide 1β formed a hydrogen bond with Thr97 (Figure 4B).
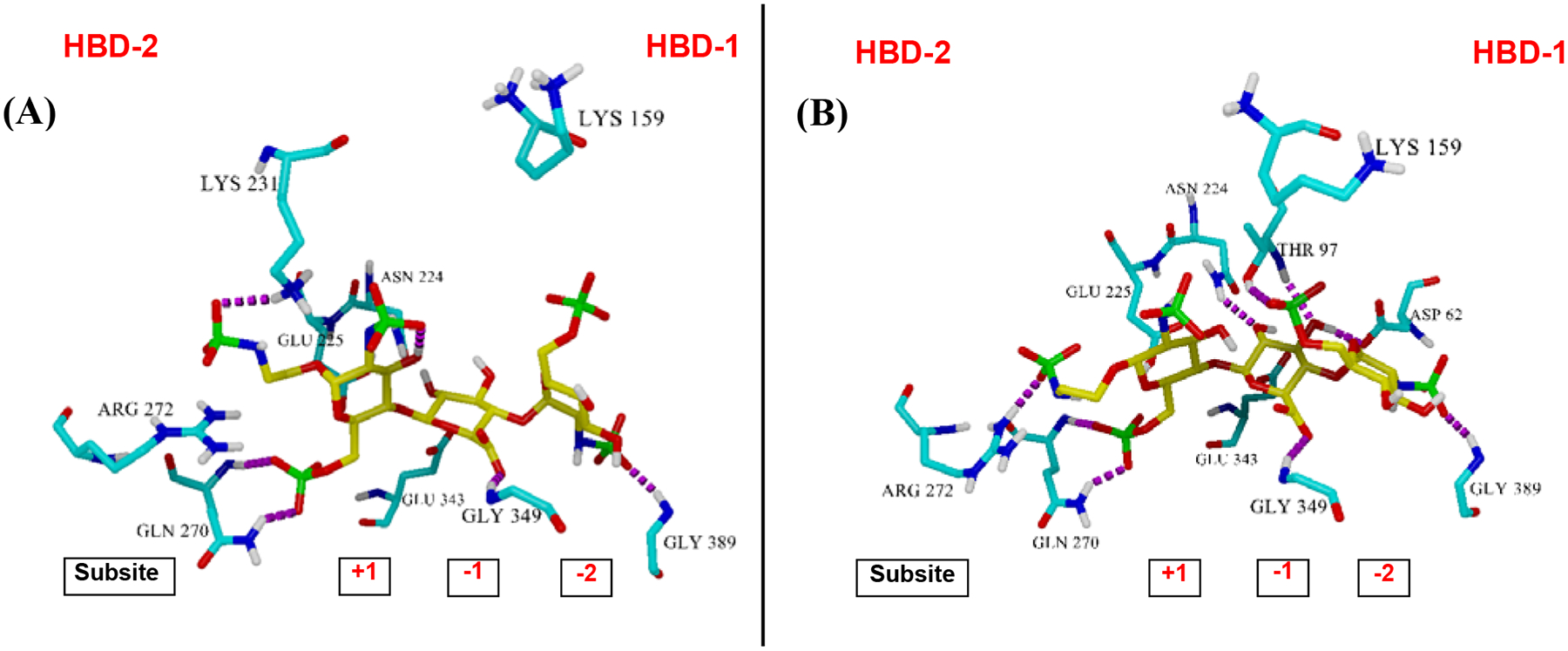
Trisaccharides docked into the apo crystal structure of heparanase (Protein Data Bank (PDB) code: 5E8M): (A) trisaccharide 1α and (B) trisaccharide 1β¸ using the Autodock Vina suite in the YASARA program. Hydrogen bonding is shown in magenta. HBD-2 is on the left and HBD-1 is on the right.
The ligand-heparanase complexes illustrated in Figure 4 were then subjected to 25000 ps molecular dynamic (MD) simulations (see Figures S7–S9) and established the complex’s stability and the distance between the scissile GlcA–GlcNS(6S) bond of trisaccharides 1α (Figure 5A) and 1β (Figure 5B) to the catalytic residues Glu225 and Glu343.11 As shown in Figure 5A, the distance of both catalytic residues to the scissile bond for 1α complex tends to be steady. However, the distance increases between Glu343 and C1 of the GlcA unit at the −1 position toward the end of the simulations (around 24000 ps). As a result, the simulation for 1α complex was further extended to 30000 ps, but no change in distance was observed (see Figure S8). The sudden increase in distance between Glu343 and the scissile bond around 24000 ps was due to a shift in hydrogen bonding interaction between Glu343 and C2/C3-hydroxyl group of the −1 GlcA unit (see superimposed snapshots in Figure 5A). During the duration of the simulations, there is a hydrogen bond interaction between Glu343 and C2-OH of the −1 GlcA unit. Near the end of the simulations, Glu343 participates in a hydrogen bond with C3-OH group of the −1 GlcA unit. Meanwhile, Glu225 maintained at approximately 5 Å away from the scissile bond as it participates in a hydrogen bond with 6-O-sulfate of +1 GlcNS6S. Collectively, these docking data illustrate that a nucleophilic residue, Glu343, moved away from the scissile bond and a proton donor, Glu225, did not participate in a hydrogen bond with C1-oxygen of the scissile bond. As a result, hydrolysis is unlikely to occur for compound 1α. On the other hand, the distance between Glu225 and the scissile GlcA–GlcNS(6S) bond of 1β fluctuated during the simulations, while Glu343 was maintained at approximately 4 Å away from the scissile bond (Figure 5B). As demonstrated in the superimposed snapshots in Figure 5B, Glu225 shifted its participation in a hydrogen bond with 6-O-sulfate of +1 GlcNS6S to C1-oxygen of the scissile bond during the fluctuation. Overall, a proton donor, Glu225, formed a hydrogen bond to C1-oyxgen of the scissile bond, while nucleophile Glu343 maintained a close distance to C1 of the scissile bond, suggesting that hydrolysis is likely to occur for compound 1β.
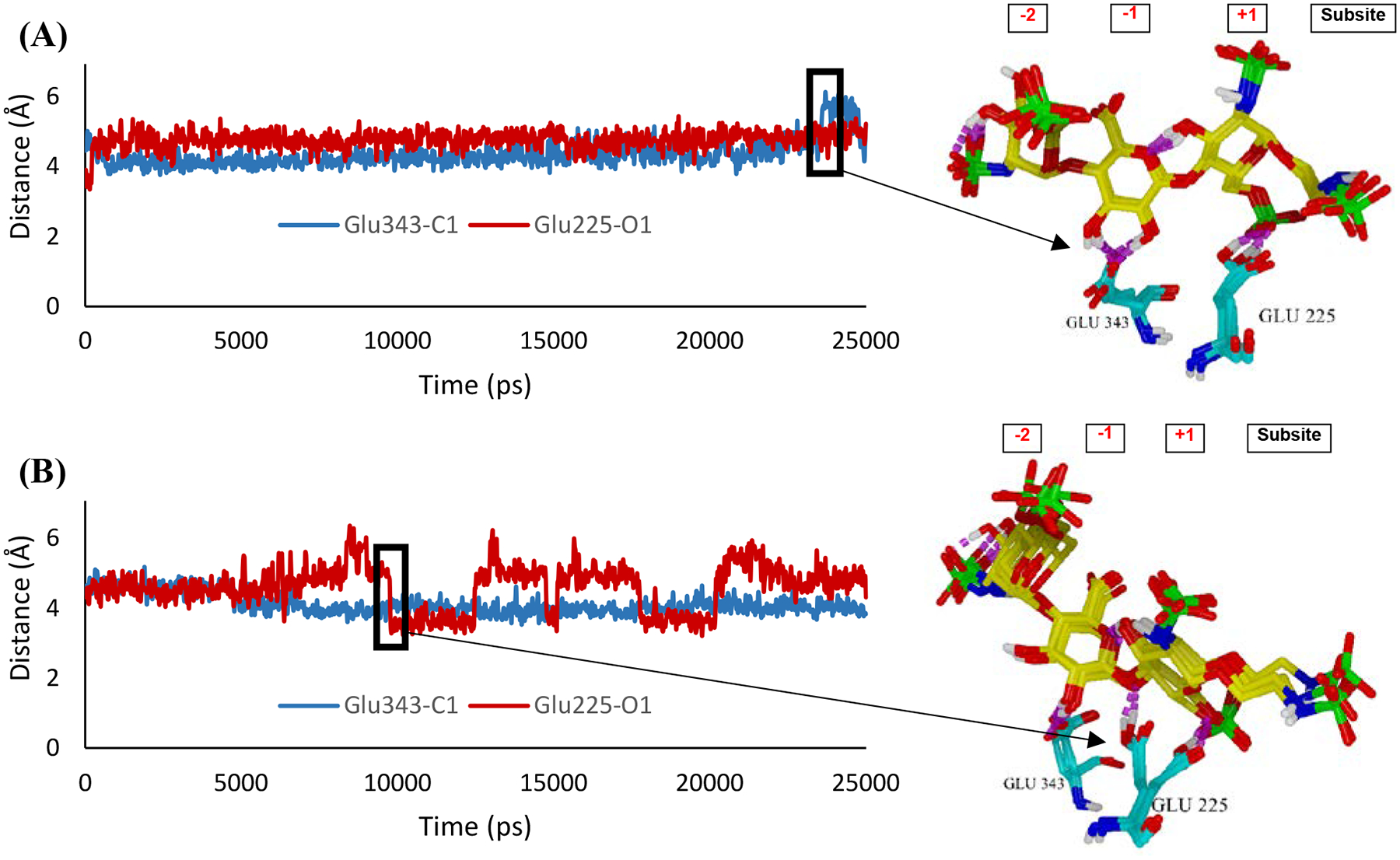
Distance of scissile bond between −1 GlcA and +1 GlcNS6S of trisaccharides 1α (A) and trisaccharide 1β (B) to the catalytic residues (Glu343 and Glu225).
Since the difference between trisaccharide 1α and 1β was the orientation of the aminoethyl linker at +1 GlcNS6S, we took a close look at the +1 region in the ligand-enzyme complex (Figure 6). Seemingly, the hydrogen bonding patterns on the aminoethyl linker are different between compound 1α and 1β. While the axial oriented linker (1α) interacts with Asn227, Ser 228, and Lys 231 (Figure 6A), the equatorial oriented linker (1β) was hydrogen-bonded to Arg272 (Figure 6B). The major difference between the two complexes is the hydrogen bonding pattern of Arg272. In 1α complex, Arg272 formed a hydrogen bond with 6-O-sulfate of the +1 subset, which allowed the catalytic residue Glu225 to interact with that 6-O-sulfate and stay away from the scissile bond (Figure 6A). On the other hand, in 1β complex, Arg272 formed a hydrogen bond with the N-sulfate of the linker, consequently shifting the position of the ligand in the binding pocket. The shifting position further leads to loose interaction of Glu225 with 6-O-sulfate of the +1 subset and allows hydrogen bonding to C1-oxygen of the scissile bond (Figure 5A).
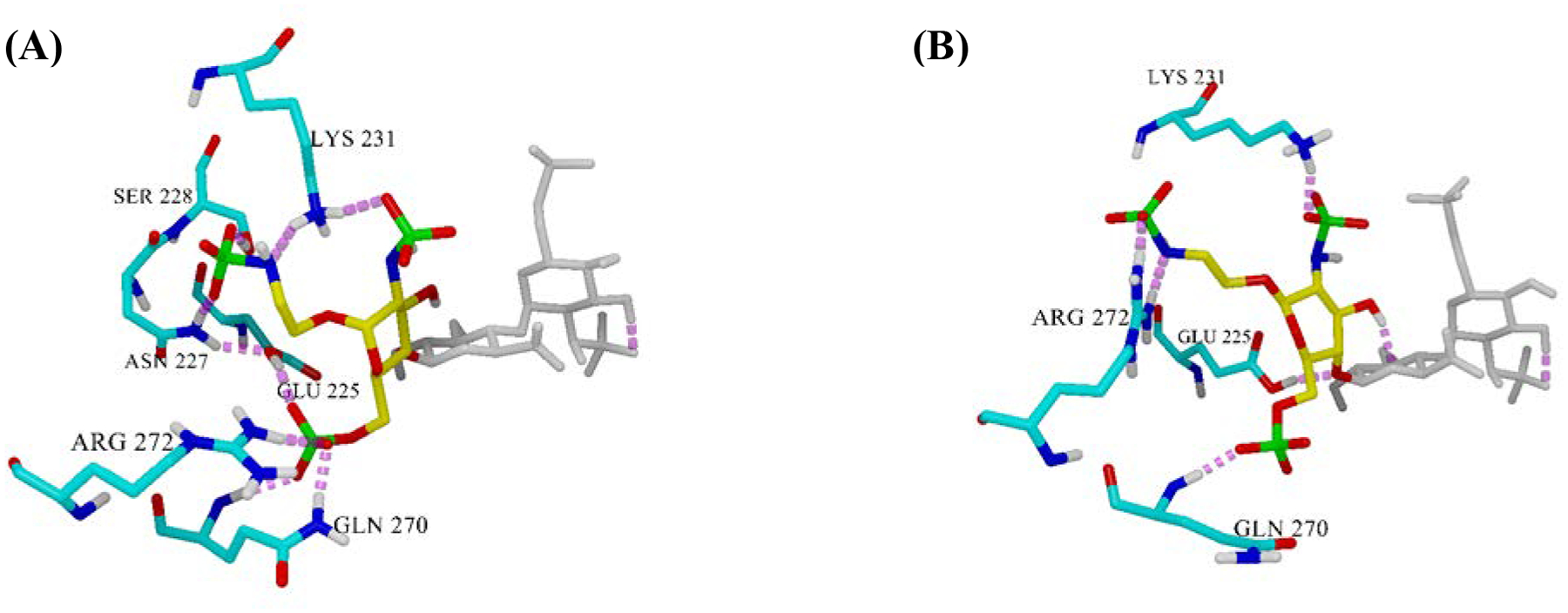
Hydrogen bonding pattern at the aminoethyl spacer region in the ligand-enzyme complex. (A) trisaccharide 1α and (B) trisaccharide 1β.
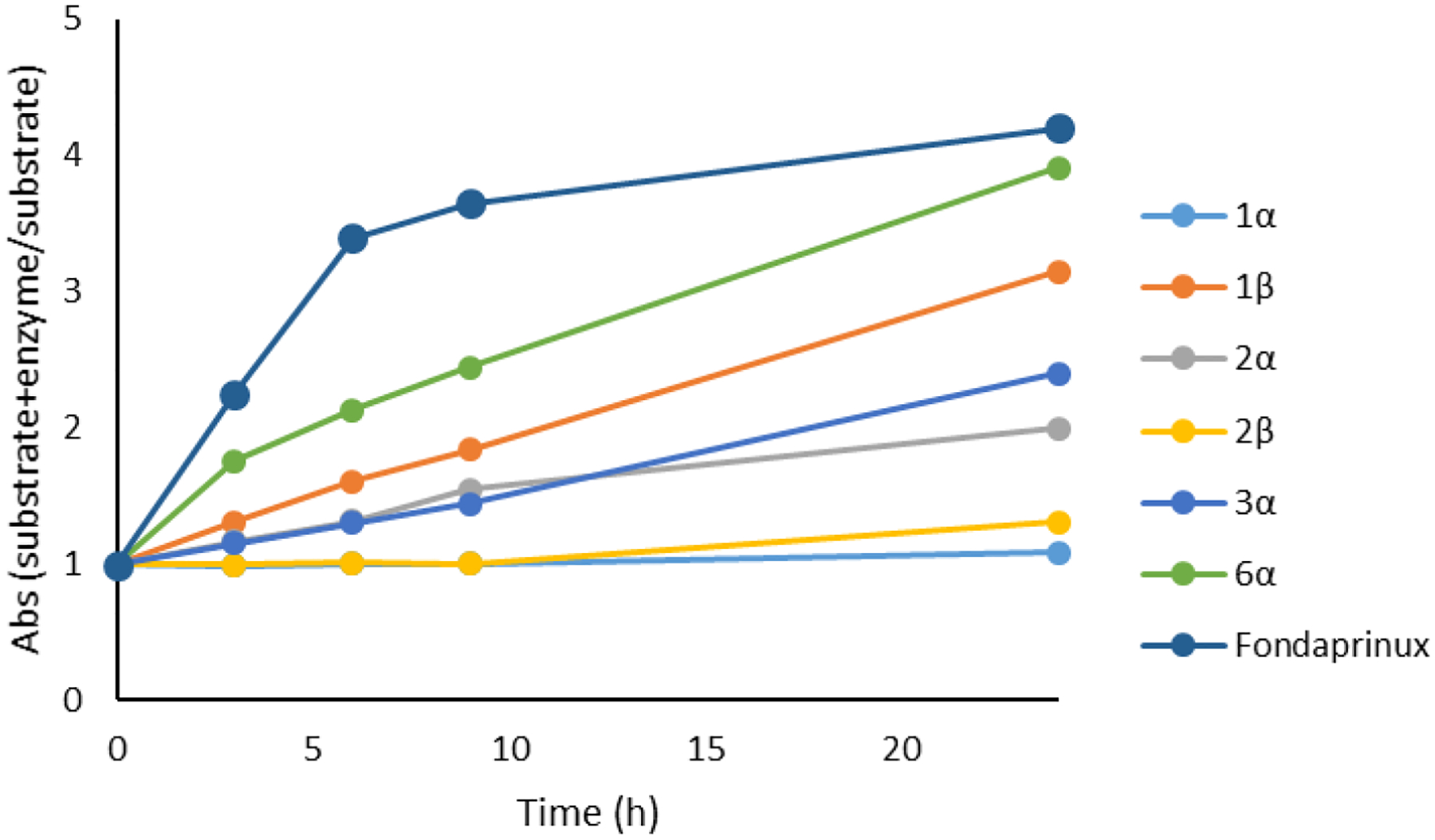
As can be seen in Figure 5B, The scissile GlcA–GlcNS(6S) bond of trisaccharide 1β is in proximity to the catalytic residues, Glu225 and Glu343 and aligned in the proper anti-orientation to these residues for cleavage by heparanase. In stark contrast, trisaccharide 1α preferred a distance further away from Glu225 and Glu343 and its GlcA–GlcNS(6S) bond is not aligned in the anti-orientation for cleavage by heparanase (Figure 5A). To confirm if cleavage of the scissile bond between −1 GlcA and +1 GlcNS6S of trisaccharides 1α and 1β is possible, a colorimetric assay was then performed (Figure 7).37–38 This assay measures the appearance of the saccharide hemiacetal product of heparanase-catalyzed cleavage utilizing a tetrazolium sodium salt (WST-1), which reacts with a newly formed anomeric hemiacetal.37 Since the ultralow molecular weight heparin, fondaparinux, is known to cleaved by heparanase,37 it was used as a control. Monitoring the degradation over 24 h showed almost no change in absorbance for the sample containing a mixture of 1α and heparanase relative to the control, suggesting that 1α is not hydrolyzed by heparanase. In stark contrast, use of 1β in this type of experiment resulted in a drastic change in absorbance over the same amount of time, suggesting that compound 1β is readily cleaved by heparanase (Figure 7). Interestingly, for trisaccharide 2β that lacks 6-O-sulfate at the +1 position, there was a small change in absorbance starting around the 11 h time point, which could be attributed to the close distance between the scissile bond and the catalytic residues (Figure 7). To compare, trisaccharide 2α undergoes hydrolysis at a faster rate than does its counterpart 2β. The results obtained with both 2α and 2β are in accordance with a docking study (see Figure S6), wherein the scissile GlcA–GlcNS(6S) bond of 2α is in closer proximity to the catalytic residues than that bond of 2β. The rate of hydrolysis of trisaccharide 3α (that lacks 6-O-sulfate at the −2 position) is similar to that of trisaccharide 2α. Among all compounds tested, sulfated trisaccharide 6α incorporating with the N-acetyl group at the +1 position had the fastest change in absorbance over the same amount of time (Figure 7), suggesting that 6α is readily cleaved by heparanase. This hydrolysis result is consistent with the docking data. We found that replacement of N-sulfate with N-acetyl at the +1 position shifts the scissile GlcA–GlcNS(6S) bond closer to the catalytic residues, Glu225 and Glu343, for cleavage by heparanase (see Figure S6). Furthermore, to determine whether any acidic autohydrolysis took place during the assay due to the acidic media, each experiment were conducted concurrently with a control of trisaccharide without heparanase. From these control experiments, acid-catalyzed hydrolysis did not occur and any change in absorbance was a result of glycosidic cleavage by heparanase. Overall, these aforementioned results suggest that heparanase preferentially cleaves a HS trisaccharide with GlcNS6S residues at the −2 and +1 position and with the β-anomeric configuration at the reducing end. Furthermore, these results suggest that changes in the sulfation pattern and the reducing end anomeric configuration disrupt the positioning of the saccharide, reducing the number of ionic salt bridges and hydrogen bonding interactions, as well as altering the distance between the scissile bond and the catalytic residues.
Cross-Bioactivity Studies:
After discovering that the GlcNS(6S)α(1,4)GlcAβ(1,4)GlcNS(6S) bearing an α-configured aminoethyl linker at the reducing end (1α) is the most effective trisaccharide to inhibit heparanase activity, we next sought to assess its potential off-target anticoagulant activity in comparison to heparin. It has been reported that heparin binds to antithrombin (AT) III to induce a conformational change that enables ATIII to inhibit the serine proteases factor Xa (FXa) and FIIa.39 As such, protease inhibition of heparin and trisaccharide 1α was assessed using chromogenic substrate assays.40 In a manner consistent with previous studies,40 heparin exhibited a significant high activity against both FXa (IC50 = 15.13 ± 0.6511 nM) and FIIa (IC50 = 5.554 ± 0.2448 nM) (Figure 8). Since trisaccharide 1α lacks an iduronic acid (IdoA) which is key in biding to ATIII,41 it is anticipated that it would bind to FXa and FIIa with low affinity. As expected, compound 1α (IC50 >2000 nM) did not inhibit either FXa or FIIa (Figure 8). To compare, a heparin pentasaccharide (fondaparinux) bearing an IdoA moiety, exhibited significantly higher activity against FXa (IC50 = 11.0 ± 0.1 nM) and limited activity against FIIa.40
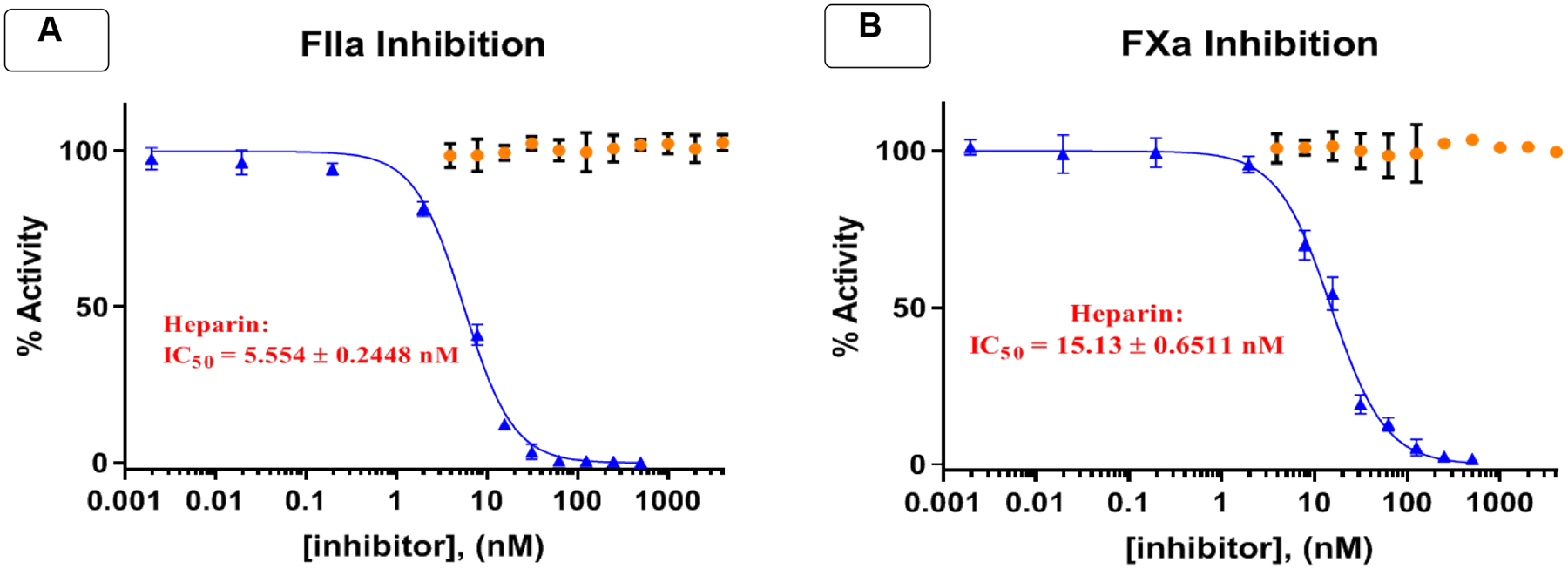
Trisaccharides 1α (orange) displayed no measurable activity against serine proteases FIIa (A) and FXa (B) in comparison to heparin (blue).
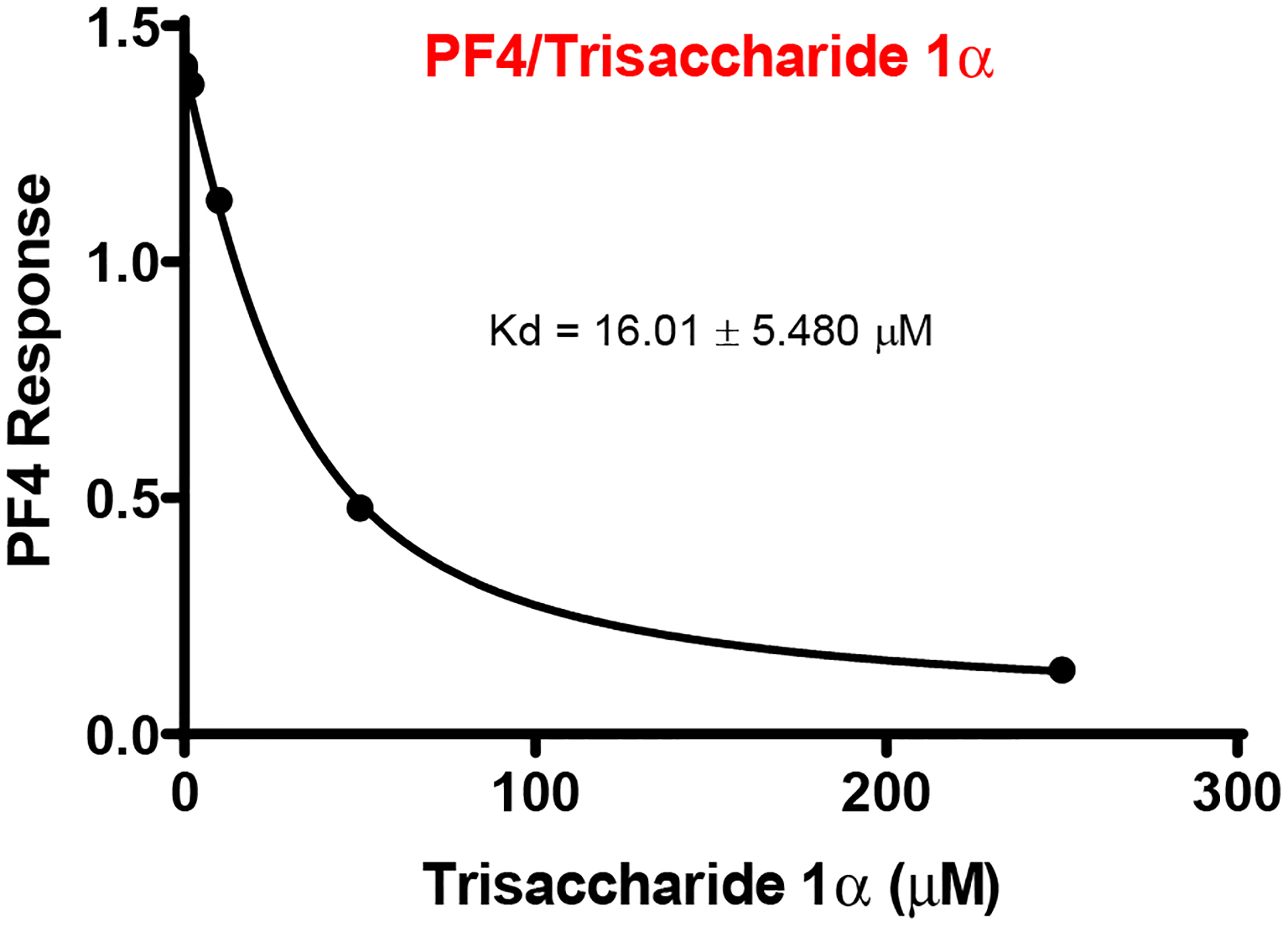
Next, we screened the ability of trisaccharide 1α to bind to the HS-binding protein, platelet factor-4 (PF4), which is responsible for causing thrombocytopenia,42–43 the main reason why clinical trials for other carbohydrate-based heparanase inhibitors were halted.44–45 To achieve this goal, a solution-based biolayer interferometry (BLI) assay was utilized to determine the apparent Kd of compound 1α to PF4 in comparison to biotinylated-heparin (18 kDa) attached to the BLI streptavidin-probe.46 Trisaccharide 1α exhibited very low affinity to PF4 (Kd = 16.01 ± 5.480 μM, Figure 9), which was significantly weaker than that of heparin (Kd = 0.3513 ± 0.0.2687 nM, see Figure S5) and that of PI-88 (Kd =16.0 ± 1.9 nM), a known oligosaccharide heparanase inhibitor.47
ECM Degradation and Pancreatic β Cells Studies.
Overall, the aforementioned data support the notion that trisaccharide 1α represents an attractive lead compound for the development of oligosaccharide small molecule inhibitors of heparanase, with a requisite sulfation pattern, size, and relative ease of synthesis. As such, we assessed the ability of sulfated trisaccharide 1α to halt ECM degradation by inhibiting heparanase enzymatic activity. Degradation of the extracellular matrix (ECM) is a critical step for tumor cell invasion and metastasis.48 Briefly, sulfate-[35S] labeled ECM,49 was incubated (5 h, 37 °C, pH 6.0) with recombinant heparanase in the absence (heparanase alone) or presence of increasing concentrations (10–50 μg/ml) of 1α.50–51 Sulfate labeled material released into the incubation medium was analyzed by gel filtration on Sepharose 6B, and HS degradation fragments were eluted at fractions 20–35. As demonstrated in Figure 10, trisaccharide 1α exhibited complete inhibition of ECM HS degradation at 50 μg/ml and nearly 50% inhibition at 10 μg/ml.
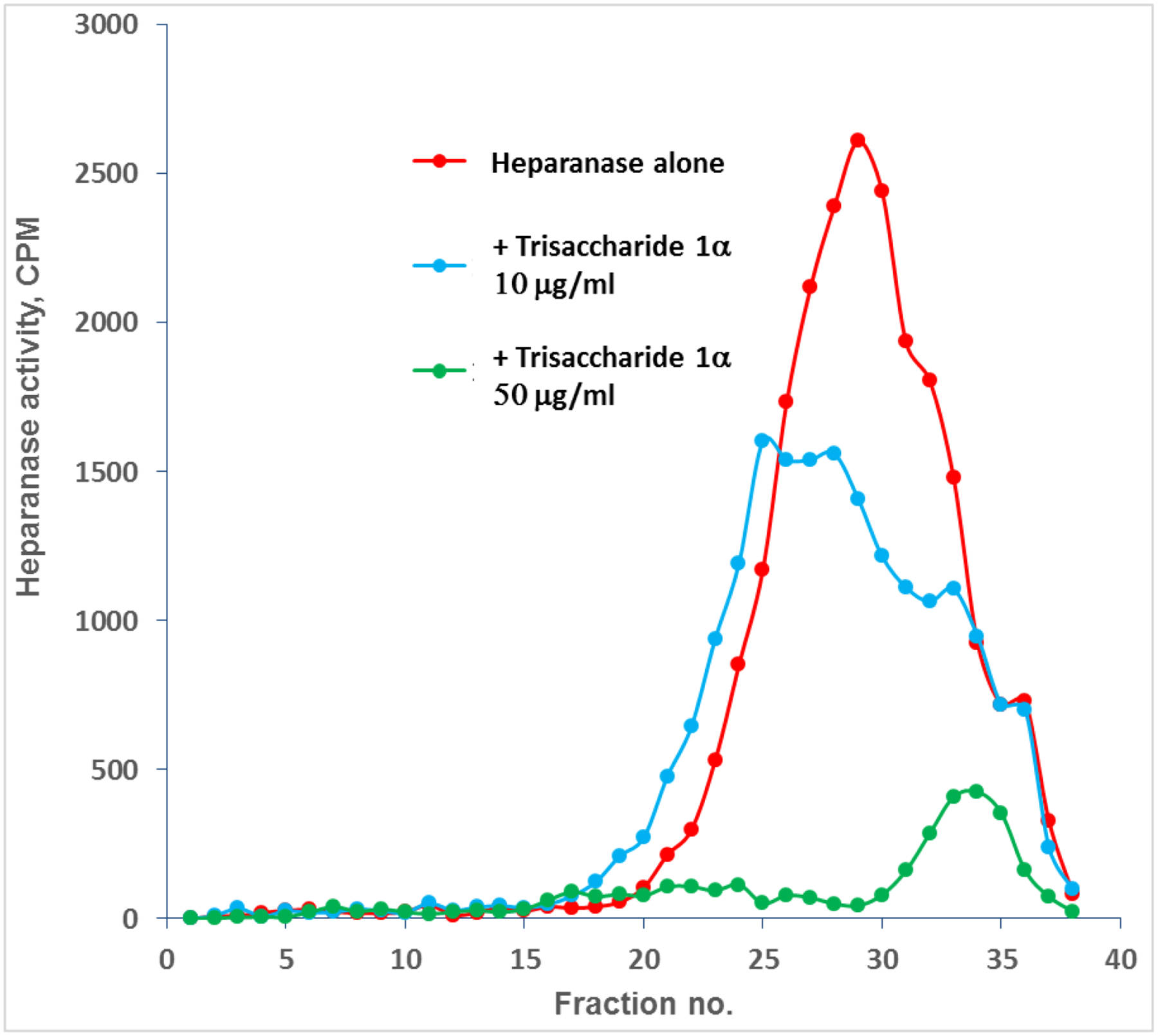
Trisaccharide 1α (10 and 50 μg/ml) inhibits heparanase mediated degradation of HS in a naturally produced ECM.
Human pancreatic β cells, like mouse pancreatic β cells, contain high levels of heparan sulfate (HS) that is lost from the β cells in Type-1 diabetes (T1D) patients. During T1D, the immune system produces heparanase that destroys HS within β cells and causes their death. To investigate the effect of trisaccharide 1α on protecting heparanase-induced damages to pancreatic β cells, mouse pancreatic β cell line Min-6 was treated with vehicle control medium, heparanase-containing medium, heparanase-containing medium plus trisaccharide 1α (15 μM), or trisaccharide 1α alone (15 μM) for 24 hours. The heparanase-containing medium was from the conditioned medium of heparanase-overexpressing CHO cells as previously described.52 Because release of high levels of mitochondrial reactive oxygen species (ROS) is a major pro-apoptotic event that triggers cell death signaling pathways,53 we investigated the levels of mitochondrial ROS release from the β cells incubated with heparanase-containing medium in the presence or absence of trisaccharide 1α (Figure 11). The levels of mitochondrial ROS released from the mouse pancreatic β cell Min-6 incubated with heparanase medium were significantly higher than those from the Min-6 cells incubated with the vehicle medium (Figure 11A–B). In contrast, the levels of mitochondrial ROS released from the Min-6 cells incubated with heparanase medium in the presence of trisaccharide 1α or the medium containing trisaccharide 1α alone were comparable to that from the Min-6 cells incubated with the vehicle medium, suggesting a role of trisaccharide 1α in preventing heparanase-triggered mitochondrial ROS release and subsequent pro-apoptotic events in β cells. Furthermore, to validate the protective role of trisaccharide 1α in repressing intracellular oxidative stress in pancreatic β cells, we examined the expression levels of the genes encoding Superoxide Dismutase 1 (SOD1) and Endoplasmic Reticulum (ER) Oxidoreductase 1α (ERO1α) in the Min-6 cells treated with vehicle, heparanase, heparanase plus trisaccharide 1α, or trisaccharide 1α alone. SOD1 is a major antioxidant enzyme in response to mitochondrial ROS release, and induction of SOD1 is an indicator of elevated mitochondrial oxidative stress.54 ERO1α is involved in oxidative protein folding process in the ER as well as ER stress-induced apoptosis.55 Elevated ERO1α indicates intracellular oxidative stress and increased apoptotic activities. As shown by quantitative real-time PCR (qPCR) analysis, expression levels of the SOD1 and ERO1α mRNAs in the Min-6 cells incubated with the heparanase medium in the presence of trisaccharide 1α or the medium containing trisaccharide 1α alone were reduced, compared to that incubated with the heparanase medium (Figure 11C), thus confirming the protective role of trisaccharide 1α in preventing oxidative stress damage in β cells. Taken together, these results indicated a discernable protective effect of the trisaccharide 1α on heparanase-caused damage to pancreatic β cells.
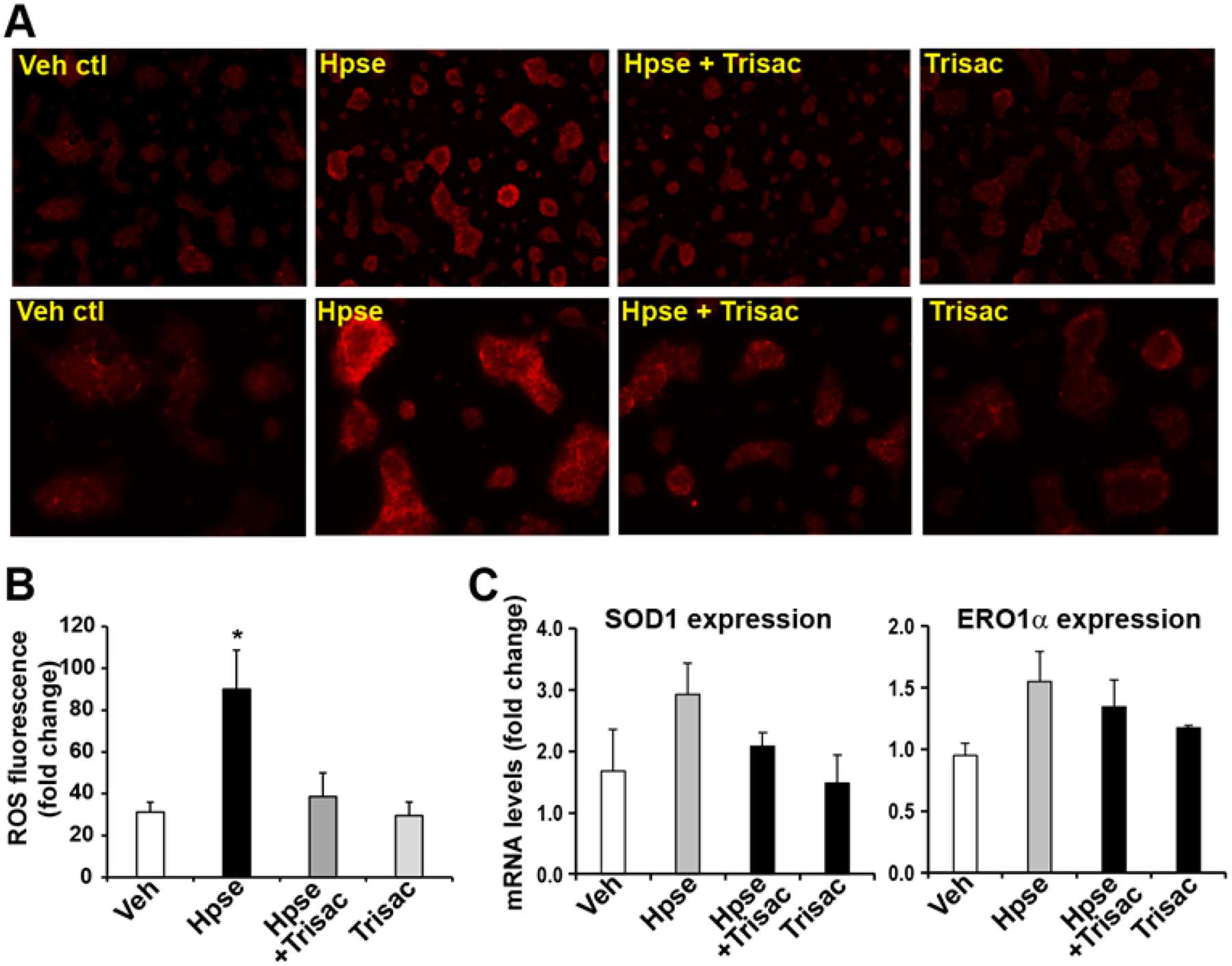
Mouse pancreatic β cell line Min-6 was incubated with medium containing vehicle (Veh ctl), heparanase-containing medium (Hpse), heparanase-containing medium plus 15 μM trisaccharide 1α (Hpse + Trisac), or 15 μM trisaccharide 1α alone (Trisac) for 24 h. The cells were stained with mitochondrial ROS probe (red) to visualize mitochondrial ROS production. (A) Representative images showing the mitochondrial ROS fluorescence activities in Min-6 cells. The upper panel shows images of 10 x magnification; and the lower panel shows images of 20 x magnification. (B) Quantification of ROS fluorescent activities in Min-6 cells. Mitochondrial ROS, staining by red fluorescence, were quantified using ImageJ software. Each bar donates mean ± SEM (n=3 biological repeats). * P ≤ 0.05. (C) Expression levels of the mRNAs encoding SOD1 and ERO1α in the Min-6 cells treated with vehicle, heparanase, heparanase plus trisaccharide 1α, or trisaccharide 1α alone, as described above, determined by qPCR analyses. Each bar donates mean ± SEM (n=3 biological replicates).
CONCLUSION
We have developed a library of eleven trisaccharides that are synthetically accessible and highly tunable in structure and sulfation pattern. By controlling the precise sulfate sequence and exploiting the anomeric configuration of the reducing end linker, the affinity of trisaccharides for heparanase and their cleavage by heparanase can be modulated. Importantly, this study illustrates the importance of the anomeric configuration of the linker at the reducing end in modulating heparanase activity. While trisaccharide bearing the β-configured linker activated heparanase and displayed hydrolysis by heparanase, their α-configured counterpart inhibited heparanase and was resistant toward hydrolysis. Furthrmore, our studies illustrate that the most potent trisaccharide, having the well-defined sulfation pattern at −2 and +1 subsites and the α-configured reducing end, exhibits minimal cross-bioactivity with serine proteases in the coagulation cascade as well as platelet factor 4. This trisaccharide also attenuates ECM degradation by inhibiting heparanase enzymatic activity and prevents heparanase-triggered mitochondrial ROS release and subsequent pro-apoptotic events in pancreatic β cells. Collectively, the results obtained in this study are significant as they combine the power of organic synthesis and biology to solve the ambiguity of how heparanase reads the sulfation pattern of HS substrates and selectively hydrolyzes HS substrates. In addition, a thorough computational study was conducted to probe the sulfated oligosaccharide-heparanase interaction mechanism. We envision that structural variations of this potent trisaccharide with α-configured linker can be synthetically tailored for the preparation of more potent heparanase inhibitors, which may be of clinical relevance for the treatment of cancer, diabetes and other disorders.
EXPERIMENTAL SECTION
General Procedures.
All reactions were performed in oven-dried flasks fitted with septa under a positive pressure of nitrogen atmosphere. Organic solutions were concentrated using a Buchi rotary evaporator below 40 °C at 25 torr. Analytical thin-layer chromatography was routinely utilized to monitor the progress of the reactions and performed using pre-coated glass plates with 230–400 mesh silica gel impregnated with a fluorescent indicator (250 nm). Visualization was then achieved using UV light, iodine, or ceric ammonium molybdate. Flash column chromatography was performed using 40–63 μm silica gel (SiliaFlash F60 from Silicycle). Dry solvents were obtained from a SG Waters solvent system utilizing activated alumina columns under an argon pressure. All other commercial reagents were used as received from Sigma Aldrich, Alfa Aesar, Acros Organics, TCI, and Combi-Blocks, unless otherwise noted. All new compounds were characterized by Nuclear Magnetic Resonance (NMR) spectroscopy and High-Resolution Mass spectrometry (HRMS). All 1H NMR spectra were recorded on either Agilent 400 or 600 MHz spectrometers. All 13C NMR spectra were recorded on either Agilent 100 or 150 MHz spectrometer. Chemical shifts are expressed in parts per million (δ scale) referenced to the residual proton in the NMR solvent (CDCl3: δ 7.26 ppm, δ 77.16 ppm). Data are presented as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, and bs = broad singlet), integration, and coupling constant in hertz (Hz). High resolution mass spectra (HRMS) were recorded using a Micromass LCT Premier XE instrument (Waters) and were determined by electrospray ionization (ESI). The purity of the final HS-liked compounds (greater than 95%) were confirmed by NMR and mass spectrometry analysis.
Materials.
The procedure for the preparation of monosaccharide building blocks (10-17), disaccharides (22, 23, and 26), 11 partially protected trisaccharides (58-68), as well as their NMR spectral data can be found in the Supporting Information.
Monosaccharide building blocks preparation.
(see pS4 in the Suppporting Information for the Synthetic Scheme).
p-Tolyl 2-azido-2-deoxy-4,6-O-(p-methoxybenzylidene)-1-thio-D-glucopyranoside (11b):
To a solution of 2-azido-2-deoxy-thioglucoside56 11a (3.7 g, 11.86 mmol) in CH3CN (100 mL) were added p-anisaldehyde dimethyl acetal (3.03 mL, 17.79 mmol) and p-TSA (225 mg, 1.186 mmol). After the reaction mixture had been stirring overnight at room temperature, it was quenched with Et3N and concentrated in vacuo. The resulting residue was subjected to silica gel column chromatography (1:6, EtOAc/hexanes) to give 11b (4.28 g, 84%) as a pale yellow syrup.
α-isomer:
Rf 0.30 (1:4, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ 7.43 (d, J = 8.7 Hz, 2H), 7.38 (d, J = 8.1 Hz, 2H), 7.13 (d, J = 7.9 Hz, 2H), 6.91 (d, J = 8.8 Hz, 2H), 5.49 (s, 1H, Ph-CH-), 5.47 (d, J = 5.6 Hz, 1H, H-1α), 4.37 (td, J = 9.9, 5.0 Hz, 1H), 4.20 (dd, J = 10.3, 5.0 Hz, 1H), 4.01 (t, J = 9.6 Hz, 1H), 3.87 – 3.84 (m, 1H), 3.80 (s, 3H, -OCH3), 3.71 (t, J = 10.3 Hz, 1H), 3.52 (t, J = 9.4 Hz, 1H), 3.00 (s, 1H, -OH), 2.33 (s, 3H, Ph-CH3). 13C NMR (150 MHz, CDCl3) δ 160.35, 138.37, 133.09, 133.07, 129.98, 129.96, 129.27, 129.13, 127.65, 113.78, 102.10, 88.08, 81.66, 70.67, 68.48, 63.87, 63.34, 55.31, 21.14. HR ESI-TOF MS (m/z): calcd for C21H23N3O5SNa [M + Na]+, 452.1251; found, 452.1260.
β-isomer:
Rf 0.40 (1:4, EtOAc–hexane); 1H NMR matches with the literature report.57
p-Tolyl 2-azido-2-deoxy-3-O-(2-naphthylmethyl)-4,6-O-(p-methoxybenzylidene)-1-thio-α-D-glucopyranoside (11c):
A solution of 11b (1.8 g, 4.19 mmol) in 45 mL of anhydrous DMF was mixted with NaH (201 mg, 8.37 mmol) at 0 °C under a N2 atmosphere. After stirring for 15 min, 2-naphtylmethyl bromide (1.39 g, 6.28 mmol) was added and the reaction was monitored by TLC. After 3 h of stirring at room temperature, the excessive NaH was quenched with saturated aq. NH4Cl solution, and the mixture was diluted with ethyl acetate. The organic layer, after being washed with saturated aq. NaCl solution, was dried with Na2SO4 and concentrated in vacuo. The residue was purified by silica gel column chromatography (1:8, EtOAc/hexanes) to give compound 11c (2.1 g, 88%) as syrup.
Rf 0.50 (1:4, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ 7.89 – 7.74 (m, 4H), 7.51 (dd, J = 8.5, 1.5 Hz, 1H), 7.48 – 7.44 (m, 2H), 7.44 – 7.41 (m, 2H), 7.41 – 7.37 (m, 2H), 7.14 (d, J = 7.9 Hz, 2H), 6.94 – 6.88 (m, 2H), 5.56 (s, 1H, Ph-CH-), 5.50 (d, J = 5.2 Hz, 1H, H-1α), 5.11 (d, J = 11.2 Hz, 1H), 4.99 (d, J = 11.2 Hz, 1H), 4.44 (td, J = 10.0, 4.9 Hz, 1H), 4.21 (dd, J = 10.4, 4.9 Hz, 1H), 4.04 – 3.95 (m, 2H), 3.83 (s, 3H, -OCH3), 3.78 – 3.72 (m, 2H), 2.34 (s, 3H, Ph-CH3). 13C NMR (150 MHz, CDCl3) δ: 160.13, 138.33, 135.22, 133.27, 133.10, 133.07, 129.97, 129.60, 129.05, 128.16, 128.00, 127.66, 127.37, 126.97, 126.09, 125.99, 125.90, 113.66, 101.53, 88.13, 82.65, 77.95, 75.16, 68.55, 63.73, 63.69, 55.31, 21.15. HR ESI-TOF MS (m/z): calcd for C32H31N3O5SNa [M + Na]+, 592.1877; found, 592.1883.
p-Tolyl 2-azido-2-deoxy-3-O-(2-naphthylmethyl)-6-O-(p-methoxybenzyl)-1-thio-α-D-glucopyranoside (11d):
To a solution of 11c (1.1 g, 1.93 mmol) in THF (30 mL) was added NaBH3CN (1.72 g, 19.3 mmol) at 0 °C. A 2N HCl/Et2O solution was then added dropwise to maintain the reaction mixture at pH 1. After the reaction mixture had been stirring at room temperature for 2 h, it was diluted with ethyl acetate, washed with aqueous NaHCO3 and brine, dried over anhydrous Na2SO4, and concentrated. The residue was subjected to flush silica gel column chromatography (1:5, EtOAc/hexanes) to give 11d (0.87 g, 79%) as syrup.
Rf 0.55 (1:3, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ 7.84 (ddd, J = 8.4, 6.8, 4.3 Hz, 4H), 7.54 (dd, J = 8.5, 1.5 Hz, 1H), 7.52 – 7.45 (m, 2H), 7.42 – 7.35 (m, 2H), 7.23 (d, J = 8.7 Hz, 2H), 7.08 (d, J = 7.9 Hz, 2H), 6.91 – 6.81 (m, 2H), 5.50 (d, J = 5.4 Hz, 1H, H-1α), 5.09 (d, J = 11.3 Hz, 1H), 5.02 (d, J = 11.3 Hz, 1H), 4.51 (d, J = 11.5 Hz, 1H), 4.43 (d, J = 11.5 Hz, 1H), 4.34 (dt, J = 9.1, 4.5 Hz, 1H), 3.90 (dd, J = 9.9, 5.4 Hz, 1H), 3.79 (s, 3H, -OCH3), 3.78 – 3.68 (m, 3H), 3.64 (dd, J = 10.4, 4.4 Hz, 1H), 2.64 (s, 1H, -OH), 2.32 (s, 3H, Ph-CH3). 13C NMR (150 MHz, CDCl3) δ 159.33, 137.95, 135.40, 133.33, 133.10, 132.71, 129.85, 129.73, 129.53, 129.38, 128.43, 128.02, 127.71, 126.96, 126.16, 126.03, 125.93, 113.82, 87.57, 81.30, 75.43, 73.28, 72.71, 70.78, 69.49, 63.58, 55.26, 21.12. HR ESI-TOF MS (m/z): calcd for C32H33N3O5SNa [M + Na]+, 594.2033; found, 594.2028.
p-Tolyl 2-azido-2-deoxy-3-O-(2-naphthylmethyl)-4-O-benzyl-6-O-(p-methoxybenzyl)-1-thio-α-D-glucopyranoside (11):
To a solution of 11d (1.0 g, 1.75 mmol) in 18 mL of anhydrous DMF was added NaH (84 mg, 3.5 mmol) at 0 °C under a N2 atmosphere. After the mixture had been stirring for15 min, benzyl bromide (311 μL, 2.62 mmol) was added. The resulting mixture was kept at room temperature for 3 h, quenched with saturated aq. NaCl solution, and diluted with ethyl acetate. The organic layer, after washing with water and brine, was dried with Na2SO4 and concentrated in vacuo. The residue was subjected to silica gel column chromatography (1:8, EtOAc/hexanes) to give 11 (1.05 g, 91%) as a yellow solid.
Rf 0.60 (1:4, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.92 – 7.72 (m, 4H), 7.55 – 7.37 (m, 5H), 7.34 – 7.28 (m, 3H), 7.28 – 7.21 (m, 2H), 7.16 (dd, J = 6.8, 2.5 Hz, 2H), 7.10 (d, J = 8.2 Hz, 2H), 6.88 – 6.78 (m, 2H), 5.58 (d, J = 5.3 Hz, 1H, H-1α), 5.09 (d, J = 10.8 Hz, 1H), 5.03 (d, J = 10.8 Hz, 1H), 4.82 (d, J = 10.9 Hz, 1H), 4.58 (d, J = 11.7 Hz, 1H), 4.52 (d, J = 10.9 Hz, 1H), 4.45 – 4.31 (m, 1H), 3.99 (dd, J = 10.3, 5.3 Hz, 1H), 3.94 – 3.86 (m, 2H), 3.83 – 3.77 (m, 2H), 3.76 (s, 3H, -OCH3), 3.63 (dd, J = 10.9, 1.8 Hz, 1H), 2.34 (s, 3H, Ph-CH3). 13C NMR (150 MHz, CDCl3) δ: 159.30, 137.93, 137.87, 135.26, 133.32, 133.07, 132.63, 129.86, 129.83, 129.66, 129.65, 128.42, 128.23, 128.01, 127.80, 127.69, 127.65, 126.85, 126.07, 125.99, 125.96, 113.77, 87.62, 81.89, 78.33, 75.74, 75.03, 73.10, 71.75, 67.83, 64.18, 55.21, 21.14. HR ESI-TOF MS (m/z): calcd for C39H39N3O5SNa [M + Na]+, 684.2503; found, 684.2507.
Phenyl 2-azido-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-1-thio-D-glucopyranoside (12):
To a solution of 2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-thioglucoside58 12a (3.0 g, 5.69 mmol) in 60 mL of anhydrous DMF was added NaH (273.36 mg, 11.39 mmol) at 0 °C under a N2 atmosphere. After the mixture had been stirring for 15 min, benzyl bromide (1.01 mL, 8.54 mmol) was added. The resulting mixture was kept at room temperature for 3 h, quenched with saturated aq. NaCl solution and diluted with ethyl acetate. The organic layer, after washing with water and brine, was dried with Na2SO4 and concentrated in vacuo. The residue was subjected to silica gel column chromatography (1:15, EtOAc/hexanes) to give 12 (3.27 g, 93%) as α/β (4/3) mixture.
Rf 0.50 (1:8, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) of α/β mixture δ: 7.92 – 7.71 (m, 5.5H), 7.68 – 7.59 (m, 1H), 7.58 – 7.42 (m, 5.8H), 7.42 – 7.11 (m, 14.6H), 7.06 (dd, J = 16.8, 6.6 Hz, 1H), 5.64 (d, J = 5.4 Hz, 1 H, H-1α), 4.97 – 4.75 (m, 6H), 4.71 (d, J = 12.2 Hz, 0.75H), 4.64 – 4.56 (m, 1.5H), 4.52 (d, J = 10.8 Hz, 1H), 4.44 (d, J = 10.1 Hz, 0.7H, H-1β), 4.42 – 4.36 (m, 1H), 3.98 (dd, J = 10.1, 5.4 Hz, 1H), 3.89 – 3.74 (m, 4H), 3.68 (dd, J = 10.9, 1.8 Hz, 1H), 3.63 (t, J = 9.4 Hz, 0.73H), 3.51 (ddd, J = 9.6, 6.0, 5.5 Hz, 1.3H), 3.38 (t, J = 9.7 Hz, 0.74H). 13C NMR (150 MHz, CDCl3) of α/β mixture δ: 137.76, 137.70, 137.64, 137.58, 135.62, 135.16, 133.60, 133.48, 133.26, 133.20, 133.02, 132.99, 132.04, 131.20, 129.06, 129.00, 128.51, 128.43, 128.39, 128.35, 128.23, 128.21, 128.17, 128.14, 128.02, 127.97, 127.88, 127.86, 127.80, 127.79, 127.71, 127.66, 127.64, 126.73, 126.32, 126.16, 126.13, 125.95, 125.90, 125.88, 125.69, 87.27, 85.97, 85.08, 81.86, 79.35, 78.24, 77.54, 75.90, 75.76, 75.08, 75.05, 73.56, 73.54, 71.79, 68.68, 68.21, 65.06, 64.11. HR ESI-TOF MS (m/z): calcd for C37H35N3O4SNa [M + Na]+, 640.2240; found, 640.2238.
2-azido-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-D-glucopyranoside (13a):
To a solution of 12 (2.18 g, 3.53 mmol) in a mixture of acetone/H2O (v/v, 50/1, 30 mL) were added NIS (1.47 g, 7.05 mmol) and AgOTf (168.6 mg, 0.705 mmol). After the reaction mixture had been stirring at room temperature for 20 min, it was quenched with Et3N, and concentrated. The residue was subjected to silica gel column chromatography (1:2, EtOAc/hexanes) to give 13a (1.52 g, 82%) as α/β (1/0.8) mixture.
Rf 0.40 (3:7, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) of α/β mixture δ: 7.80 (ddd, J = 30.8, 16.5, 12.7 Hz, 6H), 7.48 (dt, J = 15.9, 6.2 Hz, 5H), 7.42 – 7.27 (m, 7H), 7.25 – 7.11 (m, 5H), 7.04 (dd, J = 17.3, 7.2 Hz, 3H), 5.34 (t, J = 3.4 Hz, 1H, H-1α), 4.92 – 4.62 (m, 8H), 4.55 (dd, J = 7.3, 5.5 Hz, 0.78H, H-1β), 4.51 – 4.38 (m, 2H), 4.18 – 4.09 (m, 1H), 4.07 – 3.99 (m, 1H), 3.86 (d, J = 2.8 Hz, 1H), 3.73 – 3.56 (m, 4H), 3.55 – 3.31 (m, 4H). 13C NMR (150 MHz, CDCl3) of α/β mixture δ: 137.82, 137.68, 137.54, 135.00, 134.94, 133.21, 133.09, 133.08, 128.48, 128.47, 128.36, 128.34, 128.33, 128.12, 128.06, 127.91, 127.90, 127.84, 127.83, 127.78, 127.72, 126.95, 126.19, 126.18, 126.01, 126.00, 125.97, 125.96, 96.15, 92.02, 83.09, 80.12, 78.58, 77.71, 75.53, 75.52, 74.99, 74.74, 73.60, 73.57, 70.50, 68.58, 67.32, 64.00. HR ESI-TOF MS (m/z): calcd for C31H31N3O5Na [M + Na]+, 548.2156; found, 548.2166.
2-N-[(2-trifluoromethyl)benzylidene]-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-D-glucopyranoside (13c):
To a solution of 13a (1.4 g, 2.67 mmol) in CH3OH (30 mL) was added 1,3-propanedithiol (6 mL) and DIPEA (2 mL). After the reaction mixture had been stirring at 50 °C for overnight, it was concentrated in vacuo and the residue was purified by silica gel column chromatography (1:15, CH3OH/CH2Cl2) to afford 13b as colorless syrup. The resulting amine intermediate was then dissolved in pyridine/CH2Cl2 (1:10, 30 mL), and 2-(trifluoromethyl) benzaldehyde (421.82 μL, 3.2 mmol) was then added. After the reaction mixture had been stirring under reflux for overnight, it was concentrated. The resulting residue was subjected to silica gel column chromatography (1:2, EtOAc/hexanes) to give 13c (1.08 g, 62% for 2 steps) as α/β (0.28/1) mixture.
Rf 0.20 (1:2, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 8.75 (d, J = 2.0 Hz, 0.26H), 8.71 (d, J = 2.1 Hz, 1H), 8.21 (d, J = 7.6 Hz, 0.28H), 8.17 (dd, J = 5.2, 3.8 Hz, 1H), 7.86 – 7.64 (m, 6H), 7.54 – 7.41 (m, 5H), 7.23 – 7.01 (m, 10H), 5.24 (d, J = 3.4 Hz, 0.29H, H-1α), 5.04 (d, J = 7.7 Hz, 1H, H-1β), 4.84 – 4.76 (m, 1.3H), 4.75 – 4.61 (m, 3.2H), 4.57 – 4.45 (m, 2.3H), 3.91 (t, J = 9.2 Hz, 1H), 3.85 – 3.57 (m, 5H), 3.36 (dd, J = 9.4, 7.8 Hz, 1H). 13C NMR (150 MHz, CDCl3) δ: 160.69, 137.85, 135.28, 133.67, 133.20, 133.02, 131.89, 130.70, 130.38, 129.48, 128.43, 128.31, 128.29, 128.22, 127.96, 127.93, 127.89, 127.85, 127.75, 127.71, 127.69, 127.61, 127.57, 126.73, 126.08, 125.97, 125.91, 125.87, 125.62, 124.89, 123.08, 95.58 (C-1β), 93.76(C-1α), 83.23, 80.21, 78.28, 77.87, 75.38, 75.27, 75.17, 75.05, 74.95, 73.63, 73.57, 71.22, 68.98, 68.67, 60.39. HR ESI-TOF MS (m/z): calcd for C39H36F3NO5Na [M + Na]+, 678.2438; found, 678.2433.
2-N-[(2-trifluoromethyl)benzylidene]-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-D-glucopyranosyl N-phenyl-trifluoroacetimidate (13):
To a solution of 13c (950 mg, 1.45 mmol) in acetone (15 mL) was added 2,2,2-trifluoro-N-phenyl-ethanimidoyl chloride (695 μL, 4.35 mmol) and K2CO3 (400 mg, 2.9 mmol) under a N2 atmosphere After the reaction mixture had been stirring overnight at room temperature, it was filtered and concentrated in vacuum. The resulting residue was purified by silica gel column chromatography (1:10, EtOAc/hexanes + 1% triethylamine) to afford compound 13 (1.15 g, 96%) as α/β mixture.
For β anomer (majority): Rf 0.50 (1:5, EtOAc/hexanes);1H NMR (600 MHz, CDCl3) δ: 8.76 (s, 1H), 8.19 (d, J = 7.7 Hz, 1H), 7.87 – 7.77 (m, 3H), 7.73 (d, J = 7.6 Hz, 1H), 7.58 (dt, J = 29.1, 7.5 Hz, 2H), 7.53 – 7.44 (m, 3H), 7.28 – 7.00 (m, 14H), 6.75 (s, 2H), 6.03 (s, 1H), 4.87 – 4.78 (m, 2H), 4.78 – 4.65 (m, 2H), 4.59 (t, J = 11.1 Hz, 2H), 3.97 (s, 1H), 3.84 (dd, J = 21.4, 12.1 Hz, 3H), 3.67 (s, 2H). 13C NMR (150 MHz, CDCl3) δ: 161.48, 143.45, 137.85, 137.80, 135.43, 133.62, 133.24, 133.03, 131.94, 130.63, 129.65, 129.44, 128.60, 128.36, 128.27, 128.19, 127.91, 127.89, 127.87, 127.78, 127.69, 127.68, 126.67, 126.10, 125.92, 125.87, 125.76, 125.72, 124.84, 124.17, 123.02, 119.36, 83.14, 76.23, 76.10, 75.20, 75.07, 73.54, 68.14. HR ESI-TOF MS (m/z): calcd for C47H40F6N2O5Na [M + Na]+, 849.2734; found, 849.2736.
2-Chloroethyl 2-azido-2-deoxy-4,6-O-(2-naphthylmethylbenzylidene)-α-D-glucopyranoside (15c):
To a solution of 2-azido-2-deoxy-glucopyranoside59 15a (5.4 g, 26.21 mmol) in 2-chloroethanol (120 mL) was added acetyl chloride (2.79 mL, 39.32 mmol). After the reaction mixture had been stirring at 70 °C for 4 h, it was concentrated in vacuo and the residue was purified by silica gel column chromatography (1:15, CH3OH/CH2Cl2) to afford 2-chloroehyl 2-azido-2-deoxy-α-glucopyranoside 15b as colorless syrup. The intermediate 15b was dissolved in CH3CN (160 mL), and 2-naphtylaldehyde dimethyl acetal (7.94 g, 39.32 mmol) and p-TSA (497.8 mg, 2.62 mmol) were added. After the reaction mixture had been stirring overnight at 50 °C, it was quenched with Et3N and concentrated. The residue was subjected to silica gel column chromatography (1:5, EtOAc/hexanes) to give 15c (6.18 g, 58% for 2 steps) as syrup.
Rf 0.50 (1:2, EtOAc–hexane); 1H NMR (600 MHz, CDCl3) δ: 7.96 (s, 1H), 7.89 – 7.80 (m, 3H), 7.59 (dd, J = 8.5, 1.5 Hz, 1H), 7.53 – 7.45 (m, 2H), 5.68 (s, 1H, Ph-CH-), 4.95 (d, J = 3.7 Hz, 1H, H-1α), 4.31 (dd, J = 10.3, 5.0 Hz, 1H), 4.26 (t, J = 9.6 Hz, 1H), 4.03 – 3.91 (m, 2H), 3.79 (ddd, J = 27.1, 15.9, 8.0 Hz, 2H), 3.68 (t, J = 5.7 Hz, 2H), 3.55 (t, J = 9.4 Hz, 1H), 3.28 (dd, J = 10.1, 3.7 Hz, 1H), 2.79 (s, 1H, -OH). 13C NMR (150 MHz, CDCl3) δ: 134.11, 133.76, 132.85, 128.34, 128.31, 127.71, 126.58, 126.29, 125.83, 123.59, 102.17, 98.88, 81.75, 68.78, 68.74, 68.68, 62.89, 62.74, 42.36. HR ESI-TOF MS (m/z): calcd for C19H20ClN3O5Na [M + Na]+, 428.0984; found, 428.0989.
2-Azidoethyl 2-azido-2-deoxy-3-O-benzyl-4,6-O-(2-naphthylmethylbenzylidene)-α-D-glucopyranoside (15e):
To a solution of 15c (1.7 g, 4.20 mmol) in DMF (40 mL) was added NaN3 (1.36 g, 20.99 mmol). After the reaction mixture had been stirring at 80 °C for 24 h, it was diluted with CH2Cl2 and washed with water and brine, dried over anhydrous Na2SO4, and concentrated. The resulting residue 15d was dissolved in 40 mL of anhydrous DMF and NaH (201 mg, 8.39 mmol) was added at 0 °C under a N2 atmosphere. After the reaction mixture had been stirring for 15 min, benzyl bromide (747.68 μL, 6.30 mmol) was added. After the reaction mixture had been stirring at room temperature for 2 h, it was quenched with saturated aq. NaCl solution and diluted with ethyl acetate. The organic layer, after washing with water and brine, was dried with Na2SO4 and concentrated in vacuo. The residue was subjected to silica gel column chromatography (1:5, EtOAc/hexanes) to give 15e (1.81 g, 86% for 2 steps) as a yellow solid.
Rf 0.60 (1:3, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.99 (s, 1H), 7.91 – 7.84 (m, 3H), 7.61 (d, J = 8.5 Hz, 1H), 7.55 – 7.47 (m, 2H), 7.40 (d, J = 7.3 Hz, 2H), 7.31 (dt, J = 26.5, 7.3 Hz, 3H), 5.75 (s, 1H, Ph-CH-), 4.98 (d, J = 11.0 Hz, 1H, Ph-CH2-), 4.95 (d, J = 3.6 Hz, 1H, H-1α), 4.84 (d, J = 11.0 Hz, Ph-CH2-), 4.36 (dd, J = 10.3, 4.9 Hz, 1H), 4.15 (t, J = 9.5 Hz, 1H), 4.00 (td, J = 10.0, 4.9 Hz, 1H), 3.90 (ddd, J = 10.6, 7.1, 3.6 Hz, 1H), 3.80 (dt, J = 18.7, 9.8 Hz, 2H), 3.71 – 3.64 (m, 1H), 3.55 (ddd, J = 13.0, 7.1, 3.5 Hz, 1H), 3.44 (dt, J = 7.6, 4.6 Hz, 2H). 13C NMR (150 MHz, CDCl3) δ: 137.78, 134.50, 133.64, 132.90, 128.43, 128.40, 128.20, 128.16, 127.89, 127.72, 126.50, 126.25, 125.52, 123.64, 101.66, 98.80, 82.79, 75.96, 75.09, 68.92, 67.16, 63.13, 62.86, 50.60. HR ESI-TOF MS (m/z): calcd for C26H26N6O5Na [M + Na]+, 525.1857; found, 525.1864.
2-Azidoethyl 2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (15):
To a solution of 15e (1.5 g, 2.99 mmol) in THF (30 mL) was added NaBH3CN (1.72 g, 29.88 mmol) at 0 °C. A 2N HCl/Et2O solution was added dropwise to maintain the reaction mixture at pH 1. After the reaction mixture had been stirring at room temperature for 2 h, it was diluted with ethyl acetate, washed with aqueous NaHCO3 and brine, dried over anhydrous Na2SO4, and concentrated. The resulting residue was subjected to silica gel column chromatography (1:5, EtOAc/hexanes) to give 15 (1.17 g, 78%) as syrup.
Rf 0.50 (1:3, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.82 (dd, J = 9.0, 6.0 Hz, 3H), 7.76 (s, 1H), 7.50 – 7.43 (m, 3H), 7.38 (ddd, J = 13.0, 8.0, 3.9 Hz, 4H), 7.33 – 7.28 (m, 1H), 4.94 (d, J = 3.5 Hz, 1H, H-1α), 4.92 (d, J = 11.2 Hz, 1H, Ph-CH2-), 4.77 (dd, J = 14.2, 11.8 Hz, 2H, Ph-CH2-), 4.70 (d, J = 12.2 Hz, 1H, Ph-CH2-p), 3.90 – 3.81 (m, 3H), 3.79 – 3.69 (m, 3H), 3.65 (ddd, J = 10.6, 5.8, 3.5 Hz, 1H), 3.54 (ddd, J = 13.2, 7.5, 3.5 Hz, 1H), 3.41 – 3.32 (m, 2H), 2.43 (s, 1H, -OH). 13C NMR (150 MHz, CDCl3) δ: 137.99, 135.14, 133.19, 133.01, 128.65, 128.29, 128.16, 128.08, 127.85, 127.69, 126.60, 126.18, 125.98, 125.60, 98.19, 79.60, 75.09, 73.79, 71.80, 70.57, 69.49, 66.99, 62.69, 50.59. HR ESI-TOF MS (m/z): calcd for C26H28N6O5Na [M + Na]+, 527.2013; found, 527.2020.
2-Azidoethyl 2-acetamido-2-deoxy-3-O-benzyl-4,6-O-(2-naphthylmethylbenzylidene)-α-D-glucopyranoside (16c):
To a solution of 2-azidoehyl 2-acetamido-2-deoxy-α-glucopyranoside60 16a (1.04 g, 3.66 mmol) in CH3CN (30 mL) were added 2-naphtylaldehyde dimethyl acetal (1.11 g, 5.48 mmol) and p-TSA (69.5 mg, 0.366 mmol). After the reaction mixture had been stirring overnight at 50 °C, it was quenched with Et3N and concentrated. The residue was purified by silica gel column chromatography (1:10, CH3OH/CH2Cl2) to afford benzylidene 16b as white solid. The product 16b was then dissolved in DMF (30 mL), and NaN3 (1.19 g, 18.28 mmol) was added. After the reaction mixture had been stirring overnight at 80 °C, it was diluted with CH2Cl2, washed with water and brine, dried over anhydrous Na2SO4, and concentrated. The resulting intermediate was then dissolved in 30 mL anhydrous DMF, and NaH (175.7 mg, 7.32 mmol) was added at 0 °C under a N2 atmosphere. After the reaction mixture had been stirring for15 min, benzyl bromide (651.9 μL, 5.49 mmol) was added. The resulting mixture was kept at room temperature for 1 h, quenched with saturated aq. NaCl solution and diluted with ethyl acetate. The organic layer, after washing with water and brine, was dried with Na2SO4 and concentrated in vacuo. The residue was recrystallized in EtOAc/hexanes to give 16c (1.19 g, 63% for 2 steps) as a white solid.
Rf 0.50 (EtOAc); 1H NMR (600 MHz, CDCl3) δ: 7.98 (s, 1H), 7.91 – 7.77 (m, 3H), 7.60 (dd, J = 8.5, 1.4 Hz, 1H), 7.54 – 7.43 (m, 2H), 7.38 – 7.19 (m, 5H), 5.74 (s, 1H, Ph-CH-), 5.40 (d, J = 9.1 Hz, 1H, -NHAc), 4.91 (dd, J = 15.1, 8.0 Hz, 2H), 4.65 (d, J = 12.2 Hz, 1H), 4.32 (dt, J = 13.8, 4.4 Hz, 2H), 3.96 – 3.80 (m, 4H), 3.80 – 3.70 (m, 1H), 3.59 (ddd, J = 10.8, 8.2, 2.7 Hz, 1H), 3.48 (ddd, J = 13.3, 8.2, 2.8 Hz, 1H), 3.26 (ddd, J = 13.4, 5.1, 2.7 Hz, 1H), 1.90 (s, 3H, -Ac). 13C NMR (150 MHz, CDCl3) δ: 170.03, 138.29, 134.60, 133.59, 132.88, 128.41, 128.38, 128.11, 128.00, 127.75, 127.68, 126.44, 126.20, 125.46, 123.63, 101.49, 98.44, 82.69, 75.55, 74.12, 68.96, 67.37, 63.18, 52.31, 50.43, 23.26. HR ESI-TOF MS (m/z): calcd for C28H30N4O6Na [M + Na]+, 541.2058; found, 541.2055.
2-Azidoethyl 2-acetamido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (16):
To a solution of 16c (580 mg, 1.12 mmol) in THF (15 mL) was added NaBH3CN (705.4 mg, 11.2 mmol). A 2N HCl/Et2O solution was added dropwise to keep the reaction mixture at pH 1. After the mixture had been stirring at room temperature for 5 h, it was diluted with ethyl acetate, washed with aqueous NaHCO3 and brine, dried over anhydrous Na2SO4, and concentrated. The residue was subjected to silica gel column chromatography (2:1, EtOAc/hexanes) to give 16 (472 mg, 81%) as syrup.
Rf 0.50 (4:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.85 – 7.78 (m, 3H), 7.76 (s, 1H), 7.50 – 7.41 (m, 3H), 7.36 – 7.22 (m, 5H), 5.50 (d, J = 9.3 Hz, 1H, -NHAc), 4.84 (d, J = 3.6 Hz, 1H, H-1α), 4.79 – 4.64 (m, 4H), 4.27 (ddd, J = 10.5, 9.4, 3.6 Hz, 1H), 3.86 (ddd, J = 10.7, 5.3, 2.9 Hz, 1H), 3.82 – 3.69 (m, 4H), 3.64 – 3.51 (m, 2H), 3.42 (ddd, J = 13.4, 8.0, 2.9 Hz, 1H), 3.21 (ddd, J = 13.4, 5.3, 2.7 Hz, 1H), 3.03 (s, 1H, -OH), 1.86 (s, 3H, -Ac). 13C NMR (150 MHz, CDCl3) δ: 170.10, 138.35, 135.31, 133.20, 132.98, 128.58, 128.24, 128.12, 127.90, 127.87, 127.69, 126.49, 126.15, 125.93, 125.61, 97.97, 79.58, 74.01, 73.77, 71.73, 70.84, 70.04, 67.17, 51.77, 50.45, 23.29. HR ESI-TOF MS (m/z): calcd for C28H32N4O6Na [M + Na]+, 543.2214; found, 543.2205.
2-Chloroethyl 2-azido-2-deoxy-4,6-O-(p-methoxybenzylidene)-α-D-glucopyranoside (17a):
To a solution of 15b (4.0 g, 14.9 mmol) in CH3CN (100 mL) were added para-anisaldehyde dimethyl acetal (3.8 mL, 22.35 mmol) and p-TSA (283 mg, 1.49 mmol). After the reaction mixture had been stirring overnight at room temperature, it was quenched with Et3N and concentrated. The resulting residue was subjected to silica gel column chromatography (1:4, EtOAc/hexanes) to give 17a (4.43 g, 77%).
Rf 0.40 (1:2, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.40 (d, J = 8.7 Hz, 2H), 6.89 (d, J = 8.7 Hz, 2H), 5.47 (s, 1H, Ph-CH-), 4.93 (d, J = 3.7 Hz, 1H, H-1α), 4.28 – 4.15 (m, 2H), 3.97 – 3.87 (m, 2H), 3.84 – 3.75 (m, 4H), 3.72 – 3.65 (m, 3H), 3.47 (t, J = 9.4 Hz, 1H), 3.25 (dd, J = 10.1, 3.7 Hz, 1H), 2.89 (s, 1H, -OH). 13C NMR (150 MHz, CDCl3) δ: 160.31, 129.29, 127.60, 113.74, 102.02, 98.87, 81.63, 68.75, 68.62, 62.88, 62.73, 55.31, 42.35. HR ESI-TOF MS (m/z): calcd for C16H20ClN3O6Na [M + Na]+, 408.0933; found, 408.0925.
2- Azidoethyl 2-azido-2-deoxy-4,6-O-(p-methoxybenzylidene)-α-D-glucopyranoside (17b):
To a solution of 17a (4.3 g, 11.13 mmol) in DMF (100 mL) was added NaN3 (3.62 g, 55.63 mmol). After the reaction mixture had been stirring overnight at 80 °C, it was diluted with CH2Cl2, washed with water and brine, dried over anhydrous Na2SO4, and concentrated. The residue was subjected to silica gel column chromatography (1:3, EtOAc/hexanes) to give 17b (4.11 g, 94%).
Rf 0.40 (1:2, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.40 (d, J = 8.8 Hz, 2H), 6.89 (d, J = 8.8 Hz, 2H), 5.47 (s, 1H, Ph-CH-), 4.91 (d, J = 3.6 Hz, 1H, H-1α), 4.25 (dd, J = 10.3, 4.9 Hz, 1H), 4.23 – 4.16 (m, 1H), 3.87 (tdd, J = 9.7, 6.2, 3.4 Hz, 2H), 3.79 (s, 3H, -OCH3), 3.70 (t, J = 10.3 Hz, 1H), 3.64 (ddd, J = 10.5, 6.1, 3.5 Hz, 1H), 3.53 (ddd, J = 13.3, 7.2, 3.5 Hz, 1H), 3.47 (t, J = 9.3 Hz, 1H), 3.40 (ddd, J = 13.3, 6.0, 3.5 Hz, 1H), 3.27 (dd, J = 10.1, 3.7 Hz, 1H), 2.91 (s, 1H, -OH). 13C NMR (150 MHz, CDCl3) δ: 160.31, 129.30, 127.62, 113.74, 102.04, 98.77, 81.64, 68.66, 68.61, 67.22, 62.85, 62.75, 55.30, 50.60. HR ESI-TOF MS (m/z): calcd for C16H20N6O6Na [M + Na]+, 415.1337; found, 415.1338.
2- Azidoethyl 2-azido-2-deoxy-3-O-(2-naphthylmethyl)-4,6-O-(p-methoxybenzylidene)-α-D-glucopyranoside (17c):
To a solution of 17b (3.4 g, 8.65 mmol) in 80 mL of anhydrous DMF was added NaH (415.27 mg, 17.3 mmol) at 0 °C under a N2 atmosphere. After the mixture had been stirring for 15 min, NapBr (2.87 g, 12.97mmol) was added. After the resulting mixture had been stirring at room temperature for 1 h, it was quenched with saturated aq. NaCl solution and diluted with ethyl acetate. The organic layer, after washing with water and brine, was dried with Na2SO4 and concentrated in vacuo. The residue was subjected to silica gel column chromatography (1:4, EtOAc/hexanes) to give 17c (3.92 g, 85%).
Rf 0.40 (1:4, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.86 – 7.73 (m, 4H), 7.51 (dd, J = 8.5, 1.5 Hz, 1H), 7.49 – 7.36 (m, 4H), 6.95 – 6.85 (m, 2H), 5.55 (s, 1H, Ph-CH-), 5.09 (d, J = 11.3 Hz, 1H, Ph-CH2-), 4.97 (d, J = 11.3 Hz, 1H, Ph-CH2-), 4.93 (d, J = 3.6 Hz, 1H, H-1α), 4.28 (dd, J = 10.3, 4.9 Hz, 1H), 4.20 – 4.11 (m, 1H), 3.94 (td, J = 10.0, 4.9 Hz, 1H), 3.87 (ddd, J = 10.7, 7.1, 3.6 Hz, 1H), 3.82 (s, 3H, -OCH3), 3.73 (dt, J = 17.4, 9.9 Hz, 2H), 3.65 (ddd, J = 10.6, 6.1, 3.6 Hz, 1H), 3.53 (ddd, J = 13.2, 7.1, 3.6 Hz, 1H), 3.47 – 3.36 (m, 2H). 13C NMR (150 MHz, CDCl3) δ: 160.12, 135.34, 133.28, 133.07, 129.63, 128.16, 128.01, 127.64, 127.38, 127.01, 126.14, 125.95, 125.86, 113.64, 101.52, 98.77, 82.59, 76.04, 75.07, 68.78, 67.12, 63.11, 62.94, 55.30, 50.57. HR ESI-TOF MS (m/z): calcd for C27H28N6O6Na [M + Na]+, 555.1963; found, 555.1967.
2- Azidoethyl 2-azido-2-deoxy-3-O-(2-naphthylmethyl)-6-O-(p-methoxybenzyl)-α-D-glucopyranoside (17):
To a solution of 17c (3.7 g, 6.94 mmol) in THF (80 mL) was added NaBH3CN (6.56 g, 69.42 mmol) at 0 °C. A 2N HCl/Et2O solution was added dropwise to keep the mixture at pH 1. After the reaction mixture had been stirring at room temperature for 2 h, it was diluted with ethyl acetate, washed with aqueous NaHCO3 and brine, dried over anhydrous Na2SO4, and concentrated. The residue was subjected to silica gel column chromatography (1:4, EtOAc/hexanes) to give 17 (2.9 g, 78%) as syrup.
Rf 0.45 (1:2, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.94 – 7.70 (m, 4H), 7.59 – 7.36 (m, 3H), 7.31 – 7.17 (m, 2H), 6.93 – 6.78 (m, 2H), 5.06 (d, J = 11.4 Hz, 1H, Ph-CH2-), 4.97 (d, J = 11.4 Hz, 1H, Ph-CH2-), 4.94 (d, J = 3.5 Hz, 1H, H-1α), 4.53 (d, J = 11.7 Hz, 1H), 4.46 (d, J = 11.7 Hz, 1H), 3.94 – 3.84 (m, 2H), 3.84 – 3.76 (m, 4H), 3.76 – 3.60 (m, 4H), 3.54 (ddd, J = 13.2, 7.5, 3.5 Hz, 1H), 3.41 – 3.32 (m, 2H), 2.51 (s, 1H, -OH). 13C NMR (150 MHz, CDCl3) δ: 159.33, 135.45, 133.31, 133.08, 129.70, 129.38, 128.43, 128.01, 127.68, 126.99, 126.12, 126.00, 125.92, 113.84, 98.19, 79.49, 75.14, 73.32, 72.19, 70.40, 69.26, 66.99, 62.69, 55.25, 50.59. HR ESI-TOF MS (m/z): calcd for C27H30N6O6Na [M + Na]+, 557.2119; found, 557.2128.
Synthesis of Disaccharide Building Blocks
2-Azidoethyl (2-O-acetyl-3-O-benzyl-4,6-O-benzylidene-β-D-glucopyranosyl)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (18):
A mixture of thioglycoside donor 14 (1.69 g, 3.81 mmol), nucleophilic acceptor 15 (1.6 g, 3.17 mmol) and freshly activated MS 4Å in anhydrous CH2Cl2 (50 mL) was stirred at room temperature for 1 h and then cooled to 0 °C. NIS (1.71 g, 7.62 mmol) and AgOTf (97.66 mg, 0.38 mmol) were added. After 20 min of stirring, the reaction was quenched with Et3N, diluted with CH2Cl2, and filtered. The filtrate was concentrated under vacuum, and the residue was purified by silica column chromatography with EtOAc and hexanes (1:4) as the eluent to give 18 (2.39 g, 85%) as a foam.
Rf 0.40 (3:7, EtOAc–hexane); 1H NMR (600 MHz, CDCl3) δ: 7.90 – 7.75 (m, 4H), 7.53 – 7.21 (m, 16H), 7.16 (d, J = 7.1 Hz, 2H), 5.40 (s, 1H, Ph-CH-), 4.94 (dd, J = 18.0, 7.2 Hz, 2H, H-1α), 4.83 (t, J = 8.7 Hz, 1H), 4.68 (d, J = 10.6 Hz, 1H), 4.60 (d, J = 11.9 Hz, 1H), 4.52 (d, J = 12.2 Hz, 1H), 4.28 (d, J = 8.1 Hz, 1H, H-1β), 4.21 (d, J = 11.9 Hz, 1H), 4.10 (dd, J = 10.5, 4.8 Hz, 1H), 3.99 (t, J = 9.4 Hz, 1H), 3.83 (dt, J = 10.8, 6.8 Hz, 3H), 3.72 (d, J = 9.8 Hz, 1H), 3.69 – 3.60 (m, 2H), 3.52 (t, J = 9.2 Hz, 2H), 3.46 – 3.38 (m, 2H), 3.31 (t, J = 10.3 Hz, 1H), 3.08 (d, J = 4.8 Hz, 1H), 2.97 (t, J = 9.2 Hz, 1H), 1.85 (s, 3H, -OAc). 13C NMR (150 MHz, CDCl3) δ: 168.83, 138.35, 138.29, 137.24, 134.74, 133.19, 133.09, 129.00, 128.60, 128.24, 128.21, 128.17, 127.87, 127.83, 127.79, 127.69, 127.61, 127.53, 127.51, 127.36, 126.64, 126.54, 126.43, 126.03, 101.03, 100.56, 98.12, 81.32, 78.97, 77.68, 76.70, 75.12, 74.25, 73.90, 73.13, 70.77, 68.37, 67.13, 66.85, 65.96, 62.66, 50.56, 47.71, 20.66. HR ESI-TOF MS (m/z): calcd for C48H50O11N6Na [M + Na]+, 909.3430; found, 909.3435.
2-Azidoethyl (2-O-acetyl-3-O-benzyl-4,6-O-benzylidene-β-D-glucopyranosyl)-(1→4)-2-acetamido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (19):
A mixture of thioglycoside donor 14 (511 mg, 1.15 mmol), nucleophilic acceptor 16 (400 mg, 0.77 mmol) and freshly activated MS 4Å in anhydrous CH2Cl2 (10 mL) was stirred at room temperature for 1 h and then cooled to 0 °C. NIS (518 mg, 2.31 mmol) and AgOTf (29.65 mg, 0.115 mmol) were added. After 30 min of stirring, the reaction was quenched with Et3N, diluted with CH2Cl2, and filtered. The filtrate was concentrated under vacuum, and the resulting residue was purified by silica column chromatography (2:1, EtOAc/hexanes) to give 19 (666 mg, 96%) as a foam.
Rf 0.40 (4:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.90 – 7.74 (m, 4H), 7.54 – 7.35 (m, 8H), 7.36 – 7.22 (m, 8H), 7.22 – 7.14 (m, 2H), 5.42 (s, 1H), 5.28 (d, J = 8.9 Hz, 1H), 4.98 – 4.77 (m, 4H), 4.67 (d, J = 11.9 Hz, 1H), 4.54 (dd, J = 14.8, 12.2 Hz, 2H), 4.33 (dd, J = 22.8, 10.0 Hz, 2H), 4.25 – 4.13 (m, 2H), 4.04 – 3.96 (m, 1H), 3.88 – 3.78 (m, 2H), 3.69 – 3.60 (m, 2H), 3.60 – 3.51 (m, 3H), 3.48 (ddd, J = 13.3, 8.0, 3.0 Hz, 1H), 3.40 (t, J = 10.3 Hz, 1H), 3.23 (ddd, J = 13.4, 5.4, 2.8 Hz, 1H), 3.15 – 3.08 (m, 2H), 1.87 (s, 3H), 1.79 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.94, 169.05, 138.79, 138.33, 137.29, 135.09, 133.23, 133.08, 128.96, 128.48, 128.34, 128.24, 128.19, 127.89, 127.84, 127.83, 127.53, 127.52, 127.44, 127.27, 126.52, 126.37, 126.29, 126.03, 101.06, 100.71, 97.88, 81.42, 78.97, 77.21, 76.97, 74.38, 74.25, 73.81, 73.34, 71.03, 68.48, 67.41, 67.15, 65.86, 52.05, 50.41, 23.17, 20.76. HR ESI-TOF MS (m/z): calcd for C50H54O12N4Na [M + Na]+, 925.3630; found, 925.3618.
2-Azidoethyl (2-O-acetyl-3-O-benzyl-β-D-glucopyranosyl)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (20):
Disaccharide 18 (2.0 g, 2.26 mmol) was dissolved in a mixture of acetic acid/H2O (4:1, 30 mL). After the reaction mixture had been stirring at 80 °C for 5 h, it was concentrated. The residue was subjected to silica gel column chromatography (1:1, EtOAc/hexanes) to give 20 (1.30 g, 72%) as a syrup.
Rf 0.40 (1.5:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.94 – 7.76 (m, 4H), 7.55 – 7.43 (m, 3H), 7.39 – 7.22 (m, 8H), 7.17 (d, J = 7.1 Hz, 2H), 5.02 – 4.88 (m, 3H), 4.81 – 4.67 (m, 2H), 4.53 (d, J = 12.2 Hz, 1H), 4.28 – 4.16 (m, 3H), 3.93 (t, J = 9.5 Hz, 1H), 3.83 (ddd, J = 10.9, 10.2, 6.2 Hz, 3H), 3.74 (d, J = 10.0 Hz, 1H), 3.71 – 3.60 (m, 2H), 3.58 – 3.48 (m, 2H), 3.48 – 3.31 (m, 3H), 3.20 (dd, J = 12.0, 6.0 Hz, 1H), 3.09 – 3.00 (m, 1H), 2.77 (t, J = 9.2 Hz, 1H), 2.09 (s, 1H), 1.81 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.93, 138.35, 138.03, 134.71, 133.24, 133.13, 128.65, 128.51, 128.37, 127.93, 127.86, 127.83, 127.75, 127.67, 127.32, 127.17, 126.74, 126.58, 126.43, 100.04, 98.15, 87.32, 82.96, 77.69, 76.59, 75.02, 74.95, 74.43, 73.97, 73.24, 70.74, 70.67, 67.14, 66.88, 62.83, 62.16, 50.56, 20.69. HR ESI-TOF MS (m/z): calcd for C41H46O11N6Na [M + Na]+, 821.3117; found, 821.3111.
2-Azidoethyl (2-O-acetyl-3-O-benzyl-β-D-glucopyranosyl)-(1→4)-2-acetamido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (21):
Compound 21 (888 mg, 82%) was prepared from 19 (1.2 g, 1.33 mmol) using the same procedure for the synthesis of 20.
Rf 0.60 (1:10, CH3OH–CH2Cl2); 1H NMR (600 MHz, CDCl3) δ: 7.93 – 7.75 (m, 4H), 7.54 – 7.43 (m, 3H), 7.35 – 7.14 (m, 10H), 5.30 (d, J = 9.1 Hz, 1H), 4.93 (d, J = 12.3 Hz, 1H), 4.87 (d, J = 3.7 Hz, 1H), 4.84 – 4.77 (m, 2H), 4.54 (dd, J = 19.7, 12.1 Hz, 2H), 4.42 (d, J = 11.7 Hz, 1H), 4.30 (t, J = 9.8 Hz, 2H), 4.22 (ddd, J = 10.6, 9.2, 3.7 Hz, 1H), 4.00 – 3.93 (m, 1H), 3.84 (ddd, J = 10.8, 5.4, 2.9 Hz, 1H), 3.77 (dd, J = 10.6, 3.2 Hz, 1H), 3.71 – 3.61 (m, 3H), 3.60 – 3.50 (m, 2H), 3.50 – 3.38 (m, 3H), 3.23 (ddd, J = 13.4, 5.4, 2.8 Hz, 1H), 3.16 – 3.09 (m, 1H), 2.95 (t, J = 9.2 Hz, 1H), 2.61 (s, 1H), 1.81 (s, 3H), 1.79 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.18, 169.19, 138.49, 138.24, 135.01, 133.25, 133.10, 128.54, 128.47, 128.45, 127.92, 127.83, 127.75, 127.73, 127.71, 127.38, 126.53, 126.45, 126.26, 100.10, 97.86, 87.32, 82.89, 77.18, 76.72, 75.35, 74.55, 74.50, 73.86, 73.42, 71.08, 70.75, 67.39, 67.22, 62.13, 52.04, 50.39, 23.17, 20.77. HR ESI-TOF MS (m/z): calcd for C43H50O12N4Na [M + Na]+, 837.3317; found, 837.3312.
2-Azidoethyl (methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (22):
To a solution of 20 (1.52 g, 1.90 mmol) in CH2Cl2/H2O (2:1, 30 mL) were added BAIB (1.53 g, 4.76 mmol) and TEMPO (58.5 mg, 0.38 mmol) at 0 °C. After the reaction mixture had been stirring at room temperature for 2 h, it was diluted with CH2Cl2, washed with aq. Na2S2O3, dried over anhydrous Na2SO4, and concentrated. The resulting residue was then dissolved in DMF (30 mL), KHCO3 (760 mg, 7.6 mmol) and MeI (712 μL, 11.4 mmol) were added. After the reaction mixture had been stirring at room temperature for 8 h, it was diluted with EtOAc, washed with water and brine, dried over anhydrous Na2SO4, and concentrated. The residue was subjected to silica gel column chromatography (1:2, EtOAc/hexanes) to give 22 (1.23 g, 78%) as a syrup.
Rf 0.70 (1:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.94 – 7.76 (m, 4H), 7.53 – 7.42 (m, 3H), 7.39 – 7.13 (m, 10H), 5.06 (d, J = 10.9 Hz, 1H), 4.96 (d, J = 12.2 Hz, 1H), 4.92 (d, J = 3.6 Hz, 1H), 4.82 (dd, J = 9.6, 8.2 Hz, 1H), 4.62 (d, J = 10.9 Hz, 1H), 4.53 (dd, J = 12.0, 9.0 Hz, 2H), 4.33 (d, J = 8.1 Hz, 1H), 4.22 (d, J = 11.7 Hz, 1H), 4.03 (dd, J = 9.9, 8.9 Hz, 1H), 3.91 – 3.61 (m, 6H), 3.56 – 3.47 (m, 4H), 3.44 – 3.32 (m, 2H), 2.92 (d, J = 2.8 Hz, 1H), 2.84 (t, J = 9.2 Hz, 1H), 1.83 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.33, 168.80, 138.49, 138.26, 134.78, 133.23, 133.12, 128.69, 128.33, 128.12, 127.92, 127.89, 127.58, 127.56, 127.34, 127.31, 126.59, 126.52, 126.41, 100.31, 98.24, 81.42, 77.68, 75.10, 74.45, 73.98, 73.93, 72.42, 71.98, 70.67, 67.12, 67.05, 62.60, 52.55, 50.57, 20.65. HR ESI-TOF MS (m/z): calcd for C42H46O12N6Na [M + Na]+, 849.3066; found, 849.3075.
2-Azidoethyl (methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-acetamido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (23):
Compound 23 (286 mg, 80%) was prepared from 21 (345 mg, 0.42 mmol) using the same procedure for the synthesis of 22.
Rf 0.50 (EtOAc); 1H NMR (600 MHz, CDCl3) δ: 7.96 – 7.68 (m, 4H), 7.59 – 7.39 (m, 3H), 7.39 – 7.07 (m, 10H), 5.21 (d, J = 8.7 Hz, 1H), 4.97 – 4.78 (m, 4H), 4.63 – 4.47 (m, 3H), 4.35 (d, J = 8.1 Hz, 1H), 4.29 (d, J = 11.8 Hz, 1H), 4.20 (ddd, J = 10.7, 8.8, 3.7 Hz, 1H), 4.04 – 3.98 (m, 1H), 3.85 – 3.76 (m, 3H), 3.63 (dt, J = 5.0, 2.1 Hz, 2H), 3.60 – 3.50 (m, 5H), 3.45 (ddd, J = 13.4, 7.9, 2.9 Hz, 1H), 3.21 (ddd, J = 13.4, 5.4, 2.8 Hz, 1H), 3.00 (s, 1H), 2.94 (t, J = 9.2 Hz, 1H), 1.84 (s, 3H), 1.74 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.04, 169.52, 169.02, 139.04, 138.27, 135.10, 133.23, 133.07, 128.54, 128.33, 128.32, 127.97, 127.87, 127.59, 127.40, 127.28, 126.48, 126.40, 126.29, 100.53, 97.79, 81.48, 77.31, 77.25, 74.49, 74.08, 73.86, 73.81, 72.48, 72.10, 70.89, 67.43, 67.18, 52.54, 51.99, 50.37, 23.13, 20.76. HR ESI-TOF MS (m/z): calcd for C44H50N4O13Na [M + Na]+, 865.3267; found, 865.3279.
2-Azidoethyl (2-O-acetyl-3-O-benzyl-4,6-O-benzylidene-β-D-glucopyranosyl)-(1→4)-2-azido-2-deoxy-3-O-(2-naphthylmethyl)-6-O-(p-methoxybenzyl)-α-D-glucopyranoside (24):
Compound 24 (1.57 g, 87%) was prepared from 14 (1.307 g, 2.94 mmol) and 17 (1.05 g, 1.96 mmol) using the same procedure for the synthesis of 18.
Rf 0.30 (1:2, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.92 – 7.72 (m, 4H), 7.59 – 7.15 (m, 15H), 6.96 – 6.79 (m, 2H), 5.31 (s, 1H), 5.12 (d, J = 10.8 Hz, 1H), 4.96 (dd, J = 9.2, 8.2 Hz, 1H), 4.92 (d, J = 3.6 Hz, 1H), 4.87 (t, J = 11.5 Hz, 2H), 4.72 (d, J = 11.9 Hz, 1H), 4.60 (d, J = 12.0 Hz, 1H), 4.32 – 4.24 (m, 2H), 4.05 (dd, J = 10.5, 4.9 Hz, 1H), 4.03 – 3.97 (m, 1H), 3.95 – 3.88 (m, 1H), 3.86 – 3.78 (m, 2H), 3.76 – 3.71 (m, 1H), 3.69 – 3.63 (m, 4H), 3.61 – 3.49 (m, 3H), 3.46 – 3.33 (m, 3H), 3.20 (t, J = 10.3 Hz, 1H), 3.04 (td, J = 9.8, 5.0 Hz, 1H), 1.98 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.94, 159.53, 138.38, 137.18, 135.91, 133.25, 132.96, 130.10, 129.52, 128.95, 128.29, 128.17, 127.90, 127.86, 127.70, 127.58, 127.53, 126.58, 126.11, 125.98, 125.95, 125.72, 114.01, 101.01, 100.49, 98.12, 81.44, 78.83, 77.83, 76.50, 75.31, 74.20, 73.23, 73.18, 70.83, 68.37, 67.13, 66.67, 65.92, 62.68, 55.18, 50.56, 20.82. HR ESI-TOF MS (m/z): calcd for C49H52N6O12Na [M + Na]+, 939.3535; found, 939.3533.
2-Azidoethyl (2-O-acetyl-3-O-benzyl-β-D-glucopyranosyl)-(1→4)-2-azido-2-deoxy-3-O-(2-naphthylmethyl)-6-O-(p-methoxybenzyl)-α-D-glucopyranoside (25):
Compound 25 (302 mg, 75%) was prepared from 24 (445 mg, 0.49 mmol) using the same procedure for the synthesis of 20.
Rf 0.25 (1.5:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.80 (dd, J = 9.2, 3.8 Hz, 4H), 7.49 (dd, J = 8.4, 1.6 Hz, 1H), 7.47 – 7.39 (m, 2H), 7.37 – 7.31 (m, 2H), 7.31 – 7.24 (m, 5H), 6.95 – 6.86 (m, 2H), 5.12 (d, J = 11.1 Hz, 1H), 4.95 – 4.83 (m, 3H), 4.72 (d, J = 11.9 Hz, 1H), 4.63 (s, 2H), 4.32 (d, J = 11.9 Hz, 1H), 4.25 (d, J = 8.1 Hz, 1H), 3.93 (dt, J = 18.8, 9.0 Hz, 2H), 3.82 (ddd, J = 10.9, 7.5, 3.6 Hz, 1H), 3.79 – 3.71 (m, 4H), 3.65 (ddd, J = 10.6, 5.8, 3.6 Hz, 1H), 3.58 (dd, J = 10.6, 1.8 Hz, 1H), 3.53 – 3.36 (m, 5H), 3.18 (dt, J = 11.4, 7.6 Hz, 2H), 3.06 (ddd, J = 9.4, 5.9, 3.3 Hz, 1H), 2.42 (s, 1H), 1.95 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.10, 159.48, 138.19, 135.76, 133.24, 132.91, 130.23, 129.48, 128.57, 128.10, 127.89, 127.86, 127.69, 127.50, 126.08, 126.05, 125.80, 125.48, 114.07, 100.07, 98.14, 82.97, 77.66, 76.59, 75.10, 75.05, 74.57, 73.31, 73.26, 70.83, 70.81, 67.13, 66.66, 62.81, 62.15, 55.23, 50.55, 20.88. HR ESI-TOF MS (m/z): calcd for C42H48N6O12Na [M + Na]+, 851.3222; found, 851.3226.
2-Azidoethyl (methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-(2-naphthylmethyl)-6-O-(p-methoxybenzyl)-α-D-glucopyranoside (26):
Compound 26 (708 mg, 83%) was prepared from 25 (823 mg, 0.99 mmol) using the same procedure for the synthesis of 22.
Rf 0.50 (1:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.86 – 7.75 (m, 4H), 7.52 (dd, J = 8.4, 1.6 Hz, 1H), 7.44 (tt, J = 6.8, 5.2 Hz, 2H), 7.38 – 7.26 (m, 7H), 6.96 – 6.90 (m, 2H), 5.24 (d, J = 11.1 Hz, 1H), 4.97 – 4.89 (m, 2H), 4.85 – 4.72 (m, 3H), 4.64 (d, J = 11.8 Hz, 1H), 4.32 (dd, J = 11.3, 10.2 Hz, 2H), 4.08 – 4.02 (m, 1H), 3.92 (dt, J = 19.3, 9.7 Hz, 2H), 3.86 – 3.78 (m, 5H), 3.74 (dd, J = 11.9, 2.0 Hz, 1H), 3.67 (ddd, J = 10.6, 5.7, 3.6 Hz, 1H), 3.62 – 3.49 (m, 3H), 3.44 – 3.39 (m, 2H), 3.34 (s, 3H), 3.23 – 3.17 (m, 1H), 2.99 (s, 1H), 1.96 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.50, 168.91, 159.53, 138.34, 136.01, 133.29, 132.87, 130.19, 129.53, 128.41, 128.00, 127.74, 127.68, 127.54, 127.49, 126.57, 126.22, 125.76, 125.58, 114.10, 100.34, 98.22, 81.39, 77.66, 76.91, 75.18, 74.51, 73.93, 73.28, 72.39, 72.18, 70.69, 67.13, 66.68, 62.61, 55.23, 52.51, 50.56, 20.80. HR ESI-TOF MS (m/z): calcd for C43H48N6O13Na [M + Na]+, 879.3172; found, 879.3168.
Synthesis of Protected Trisaccharides
2-Azidoethyl [2-azido-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranosyl]-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-acetamido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (27):
A mixture of thioglycoside donor 12 (278.5 mg, 0.45 mmol), disaccharide acceptor 23 (190 mg, 0.22 mmol), DMF (104.7 μL, 1.35 mmol) and freshly activated MS 4Å in anhydrous CH2Cl2 (8 mL) was stirred at room temperature for 1 h and. NIS (203.1 mg, 0.90 mmol) and AgOTf (116 mg, 0.45 mmol) were added. After 12 hours of stirring at 35 °C, the reaction was quenched with Et3N, diluted with CH2Cl2, and filtered. The filtrate was concentrated under vacuum, and the residue was purified by silica column chromatography (1:2, acetone/hexanes) to give 27 (155 mg, 51%) as a foam.
Rf 0.45 (4:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.94 (d, J = 8.4 Hz, 1H), 7.89 – 7.86 (m, 2H), 7.81 (s, 1H), 7.76 – 7.72 (m, 2H), 7.55 – 7.25 (m, 23H), 7.16 (dd, J = 11.4, 4.4 Hz, 4H), 7.01 (d, J = 6.9 Hz, 1H), 5.37 (d, J = 3.7 Hz, 1H), 5.07 (d, J = 8.6 Hz, 1H), 4.98 – 4.83 (m, 7H), 4.75 (dd, J = 11.7, 5.1 Hz, 2H), 4.54 (dd, J = 14.5, 9.8 Hz, 3H), 4.47 (dd, J = 11.8, 4.4 Hz, 2H), 4.36 (d, J = 8.1 Hz, 1H), 4.12 – 4.04 (m, 2H), 3.98 – 3.95 (m, 1H), 3.87 – 3.73 (m, 7H), 3.61 – 3.43 (m, 9H), 3.36 – 3.32 (m, 1H), 3.24 – 3.17 (m, 2H), 1.77 (s, 3H), 1.70 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.96, 168.84, 168.08, 138.94, 138.00, 137.78, 137.63, 135.13, 134.93, 133.25, 133.14, 133.12, 132.97, 128.66, 128.45, 128.44, 128.37, 128.36, 128.33, 128.24, 128.19, 128.06, 127.99, 127.88, 127.85, 127.64, 127.61, 127.57, 127.48, 127.29, 127.08, 126.81, 126.61, 126.55, 126.29, 126.10, 125.96, 125.88, 100.41, 97.80, 97.67, 82.34, 79.89, 77.83, 77.65, 75.38, 74.92, 74.70, 74.59, 74.50, 74.25, 73.94, 73.70, 73.20, 71.28, 70.77, 69.48, 67.45, 66.94, 63.34, 52.47, 52.07, 50.37, 23.12, 20.67. HR ESI-TOF MS (m/z): calcd for C75H79N7O17Na [M + Na]+, 1372.5425; found, 1372.5436.
2-Azidoethyl (2-azido-2-deoxy-3,4,6-tri-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (28):
A mixture of thioglycoside donor 10 (263.5 mg, 0.45 mmol), disaccharide acceptor 22 (187 mg, 0.226 mmol), DMF (105 μL, 1.36 mmol) and freshly activated MS 4Å in anhydrous CH2Cl2 (8 mL) was stirred at room temperature for 1 h. NIS (203.8 mg, 0.90 mmol) and AgOTf (116 mg, 0.45 mmol) were added. After 5 hours of stirring at 35 °C, the reaction was quenched with Et3N, diluted with CH2Cl2, and filtered. The filtrate was concentrated under vacuum, and the residue was purified by silica column chromatography (1:3, EtOAc/hexanes) as the eluent to give 28 (183 mg, 63%) as a foam.
Rf 0.40 (1:2, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.94 (d, J = 8.4 Hz, 1H), 7.88 (t, J = 8.7 Hz, 2H), 7.82 (s, 1H), 7.56 – 7.47 (m, 3H), 7.39 – 7.21 (m, 21H), 7.18 – 7.12 (m, 4H), 5.34 (d, J = 3.7 Hz, 1H), 5.04 (d, J = 10.9 Hz, 1H), 4.98 (d, J = 12.2 Hz, 1H), 4.93 – 4.86 (m, 4H), 4.78 (d, J = 11.0 Hz, 1H), 4.59 – 4.52 (m, 4H), 4.48 (d, J = 11.0 Hz, 1H), 4.43 (d, J = 12.1 Hz, 1H), 4.37 (d, J = 8.0 Hz, 1H), 4.06 (dd, J = 19.0, 9.8 Hz, 2H), 3.98 (dd, J = 9.8, 9.0 Hz, 1H), 3.87 – 3.79 (m, 4H), 3.76 – 3.70 (m, 4H), 3.63 (dddd, J = 25.1, 12.8, 8.3, 2.0 Hz, 3H), 3.52 (ddd, J = 13.2, 7.5, 3.6 Hz, 1H), 3.48 – 3.44 (m, 4H), 3.43 – 3.39 (m, 1H), 3.34 (ddd, J = 10.1, 6.2, 3.7 Hz, 2H), 3.13 (t, J = 9.1 Hz, 1H), 1.76 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.62, 167.81, 138.24, 138.15, 137.80, 137.77, 137.63, 134.57, 133.24, 133.13, 128.79, 128.46, 128.35, 128.34, 128.32, 128.19, 128.05, 128.02, 127.90, 127.89, 127.77, 127.71, 127.65, 127.58, 127.47, 127.38, 127.06, 126.66, 126.61, 126.36, 100.21, 98.25, 97.73, 82.19, 79.94, 77.87, 77.58, 75.38, 75.20, 74.77, 74.53, 74.40, 74.07, 73.58, 73.07, 71.34, 70.62, 67.58, 67.13, 66.90, 63.41, 62.60, 52.42, 50.57, 20.56. HR ESI-TOF MS (m/z): calcd for C69H73N9O16Na [M + Na]+, 1306.5067; found, 1306.5063.
2-Azidoethyl [2-azido-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranosyl]-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (29):
Compound 29 (241 mg, 65%) was prepared from 12 (343.6 mg, 0.56 mmol) and 22 (230 mg, 0.28 mmol) using the same procedure for the synthesis of 28.
Rf 0.50 (1:1.5, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.94 (d, J = 8.4 Hz, 1H), 7.88 (dd, J = 8.6, 7.6 Hz, 2H), 7.82 (s, 1H), 7.79 – 7.73 (m, 2H), 7.56 – 7.41 (m, 5H), 7.38 – 7.08 (m, 22H), 7.07 – 7.02 (m, 1H), 5.37 (d, J = 3.7 Hz, 1H), 5.04 (d, J = 10.9 Hz, 1H), 4.98 (d, J = 12.2 Hz, 1H), 4.91 (ddd, J = 22.5, 14.9, 9.6 Hz, 4H), 4.79 – 4.73 (m, 2H), 4.60 – 4.53 (m, 3H), 4.52 – 4.46 (m, 2H), 4.37 (d, J = 8.0 Hz, 1H), 4.10 – 4.02 (m, 2H), 3.98 (dd, J = 9.8, 8.9 Hz, 1H), 3.87 – 3.79 (m, 4H), 3.74 (ddd, J = 20.2, 15.9, 8.9 Hz, 4H), 3.69 – 3.58 (m, 3H), 3.55 – 3.29 (m, 8H), 3.14 (t, J = 9.1 Hz, 1H), 1.77 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.63, 167.82, 138.23, 138.02, 137.78, 137.64, 135.18, 134.57, 133.25, 133.16, 133.14, 133.00, 128.79, 128.70, 128.54, 128.46, 128.39, 128.36, 128.30, 128.26, 128.22, 128.18, 128.06, 128.02, 127.91, 127.89, 127.86, 127.78, 127.66, 127.58, 127.37, 127.35, 127.06, 126.81, 126.66, 126.62, 126.36, 126.13, 125.97, 125.91, 100.22, 98.25, 97.76, 82.20, 79.97, 77.87, 77.58, 75.39, 75.22, 74.75, 74.53, 74.41, 74.07, 73.73, 73.08, 71.34, 70.62, 69.48, 68.18, 67.55, 67.13, 66.90, 63.43, 62.61, 52.43, 50.57, 20.57. HR ESI-TOF MS (m/z): calcd for C73H75N9O16Na [M + Na]+, 1356.5224; found, 1356.5227.
2-Azidoethyl [2-azido-2-deoxy-3-O-(2-naphthylmethyl)-4-O-benzyl-6-O-(p-methoxybenzyl)-α-D-glucopyranosyl]-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (30):
Compound 30 (111 mg, 65%) was prepared from 11 (175 mg, 0.26 mmol) and 22 (109 mg, 0.13 mmol) using the same procedure for the synthesis of 28.
Rf 0.50 (1:1.5, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.95 – 7.76 (m, 8H), 7.56 – 7.38 (m, 8H), 7.35 – 7.12 (m, 15H), 6.81 (d, J = 8.6 Hz, 2H), 5.37 (d, J = 3.7 Hz, 1H), 5.07 (dd, J = 18.5, 7.6 Hz, 3H), 5.01 – 4.97 (m, 1H), 4.96 – 4.91 (m, 2H), 4.78 (d, J = 11.1 Hz, 1H), 4.62 – 4.55 (m, 3H), 4.49 (dt, J = 10.5, 9.9 Hz, 2H), 4.38 (ddd, J = 14.6, 11.4, 8.6 Hz, 2H), 4.13 – 3.99 (m, 3H), 3.92 (d, J = 10.1 Hz, 1H), 3.88 – 3.65 (m, 13H), 3.58 – 3.47 (m, 4H), 3.40 (dddd, J = 19.5, 13.8, 7.8, 3.6 Hz, 4H), 3.14 (t, J = 9.1 Hz, 1H), 1.78 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.64, 167.85, 159.28, 100.21, 98.25, 97.73, 82.19, 82.07, 79.89, 78.94, 77.88, 77.62, 77.59, 75.41, 75.38, 75.16, 74.80, 74.72, 74.54, 74.48, 74.45, 74.40, 74.07, 73.96, 73.18, 73.09, 72.93, 71.34, 70.70, 70.62, 67.14, 63.44, 62.61, 62.59, 55.17, 52.44, 50.58, 20.57. HR ESI-TOF MS (m/z): calcd for C74H77N9O17Na [M + Na]+, 1386.5330; found, 1386.5324.
2-Azidoethyl [2-azido-2-deoxy-3-O-(2-naphthylmethyl)-4-O-benzyl-6-O-(p-methoxybenzyl)-α-D-glucopyranosyl]-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-(2-naphthylmethyl)-6-O-(p-methoxybenzyl)-α-D-glucopyranoside (31):
Compound 31 (247 mg, 57%) was prepared from 11 (412 mg, 0.62 mmol) and 26 (266 mg, 0.31 mmol) using the same procedure for the synthesis of 28.
Rf 0.60 (1:1.5, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.92 – 7.72 (m, 8H), 7.57 (d, J = 8.4 Hz, 1H), 7.51 – 7.43 (m, 4H), 7.38 – 7.20 (m, 13H), 7.17 – 7.11 (m, 2H), 6.99 (d, J = 8.6 Hz, 2H), 6.80 (d, J = 8.6 Hz, 2H), 5.47 (d, J = 3.7 Hz, 1H), 5.28 – 5.23 (m, 1H), 5.08 (dd, J = 9.2, 8.3 Hz, 1H), 5.03 (d, J = 2.0 Hz, 2H), 4.92 (d, J = 3.5 Hz, 1H), 4.85 (d, J = 10.8 Hz, 1H), 4.81 – 4.73 (m, 3H), 4.58 (dd, J = 11.2, 6.6 Hz, 2H), 4.50 (dd, J = 9.6, 6.1 Hz, 2H), 4.38 (t, J = 11.7 Hz, 2H), 4.20 (t, J = 9.2 Hz, 1H), 4.02 (d, J = 9.7 Hz, 1H), 3.95 – 3.89 (m, 2H), 3.86 – 3.65 (m, 14H), 3.55 (ddd, J = 29.9, 19.6, 11.9 Hz, 5H), 3.44 – 3.34 (m, 5H), 1.93 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.81, 167.96, 159.59, 159.27, 138.19, 137.66, 135.80, 135.33, 133.38, 133.27, 133.02, 132.99, 130.20, 130.09, 129.75, 129.74, 129.67, 128.47, 128.42, 128.30, 128.16, 127.94, 127.88, 127.85, 127.73, 127.71, 127.65, 127.60, 127.54, 127.50, 127.38, 127.27, 127.07, 126.98, 126.75, 126.56, 126.54, 126.04, 125.93, 125.92, 125.64, 125.55, 114.15, 113.76, 100.29, 98.21, 97.74, 82.48, 79.96, 77.84, 77.58, 75.44, 75.40, 75.36, 75.04, 74.98, 74.70, 74.63, 74.59, 74.56, 74.46, 73.43, 73.27, 73.20, 73.12, 71.33, 70.74, 70.68, 67.12, 67.07, 66.78, 63.41, 62.67, 62.60, 55.24, 55.15, 52.39, 50.57, 20.78. HR ESI-TOF MS (m/z): calcd for C75H79N9O18Na [M + Na]+, 1416.5435; found, 1416.5442.
2-Azidoethyl [2-azido-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranosyl]-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-(2-naphthylmethyl)-6-O-(p-methoxybenzyl)-α-D-glucopyranoside (32):
Compound 32 (188 mg, 60%) was prepared from 12 (283 mg, 0.46 mmol) and 26 (196 mg, 0.23 mmol) using the same procedure for the synthesis of 28.
Rf 0.50 (1:1.5, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.89 – 7.72 (m, 8H), 7.54 (dd, J = 8.4, 1.6 Hz, 1H), 7.47 – 7.39 (m, 5H), 7.38 – 7.25 (m, 13H), 7.19 (ddd, J = 15.8, 7.8, 2.0 Hz, 3H), 7.05 – 6.96 (m, 3H), 5.47 (d, J = 3.7 Hz, 1H), 5.24 (d, J = 11.1 Hz, 1H), 5.08 (dd, J = 9.3, 8.1 Hz, 1H), 4.94 – 4.71 (m, 8H), 4.59 (dd, J = 11.6, 6.5 Hz, 2H), 4.53 – 4.47 (m, 2H), 4.39 (d, J = 11.8 Hz, 1H), 4.22 – 4.16 (m, 1H), 4.05 – 3.99 (m, 1H), 3.94 – 3.70 (m, 11H), 3.69 – 3.46 (m, 7H), 3.44 – 3.32 (m, 5H), 1.93 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.81, 167.93, 159.59, 138.00, 137.79, 137.66, 135.77, 135.17, 133.36, 133.16, 132.99, 132.95, 130.07, 129.46, 128.47, 128.43, 128.24, 128.22, 128.13, 128.01, 127.86, 127.73, 127.66, 127.56, 127.52, 127.35, 127.27, 127.02, 126.78, 126.50, 126.12, 125.94, 125.91, 125.61, 125.52, 114.14, 100.30, 98.21, 97.76, 82.49, 79.99, 77.82, 77.56, 75.42, 75.37, 75.08, 74.73, 74.60, 74.56, 73.74, 73.42, 73.11, 71.34, 70.68, 67.58, 67.12, 66.80, 63.39, 62.66, 55.25, 52.39, 50.56, 20.78. HR ESI-TOF MS (m/z): calcd for C74H77N9O17Na [M + Na]+, 1386.5330; found, 1386.5337.
2-Azidoethyl {2-N-[(2-trifluoromethyl)benzylidene]-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranosyl}-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (33):
A mixture of donor 13 (250.5 mg, 0.30 mmol), acceptor 22 (167 mg, 0.20 mmol), DMF (92.8 μL, 1.2 mmol) and freshly activated MS 4Å in anhydrous CH2Cl2 (8 mL) was stirred at room temperature for 1 h and. TfOH (26.6 μL, 0.30 mmol) was added. After 8 hours of stirring at rt, the reaction was quenched with Et3N, diluted with CH2Cl2, and filtered. The filtrate was concentrated under vacuum, and the resulting residue was purified by silica column chromatography (1:2, EtOAc/hexanes) to give 33 (168 mg, 57%) as a foam.
Rf 0.30 (1:2, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 8.67 (d, J = 2.1 Hz, 1H), 8.20 (d, J = 7.8 Hz, 1H), 7.92 (d, J = 8.4 Hz, 1H), 7.87 – 7.70 (m, 7H), 7.65 (d, J = 7.9 Hz, 1H), 7.54 (dd, J = 8.4, 1.6 Hz, 1H), 7.46 (dddd, J = 19.2, 13.7, 7.8, 4.4 Hz, 5H), 7.36 (dd, J = 11.2, 4.2 Hz, 2H), 7.32 – 7.27 (m, 2H), 7.26 – 7.02 (m, 16H), 6.91 (dd, J = 6.5, 3.0 Hz, 2H), 5.23 (d, J = 3.4 Hz, 1H), 5.05 (d, J = 10.8 Hz, 1H), 4.97 – 4.89 (m, 3H), 4.87 – 4.77 (m, 3H), 4.70 – 4.44 (m, 8H), 4.21 – 4.15 (m, 2H), 4.07 – 3.98 (m, 2H), 3.92 – 3.61 (m, 10H), 3.56 (dd, J = 10.0, 3.3 Hz, 1H), 3.53 – 3.46 (m, 4H), 3.44 – 3.37 (m, 1H), 3.34 (dd, J = 10.3, 3.6 Hz, 1H), 3.20 (t, J = 9.2 Hz, 1H), 1.70 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 168.78, 167.84, 138.43, 138.23, 138.20, 137.99, 135.45, 134.81, 133.23, 133.20, 133.06, 133.01, 131.85, 130.42, 128.58, 128.39, 128.34, 128.22, 128.20, 128.17, 127.97, 127.90, 127.88, 127.83, 127.69, 127.56, 127.46, 127.42, 127.41, 127.10, 126.77, 126.54, 126.42, 126.28, 126.26, 126.12, 126.04, 125.88, 100.49, 99.86, 98.24, 82.27, 80.24, 77.57, 77.48, 77.09, 76.90, 75.75, 75.38, 75.02, 74.96, 74.71, 73.98, 73.72, 72.70, 72.01, 70.72, 67.93, 67.10, 52.44, 50.57, 20.54. HR ESI-TOF MS (m/z): calcd for C81H80F3N7O16Na [M + Na]+, 1486.5506; found, 1486.5513.
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-6-O-acetyl-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3,6-di-O-acetyl-D-glucopyranoside (37 and 38):
To a solution of 3418 (600 mg, 0.524 mmol) in anhydrous CH2Cl2 (10 mL) was added trichloroacetonitrile (524 μL, 5.24 mmol) and DBU (5 drops) at 0 °C under N2 protection. After 2 h of stirring at rt, the mixture was concentrated in vacuum, and the residue was subjected to silica gel column with EtOAc and hexanes (1:4, EtOAc/hexanes + 1% Et3N) to afford compound 35 (608 mg, 90%) as a syrup. To a stirred mixture of resulting trichloroacetimidate 35 (608 mg, 0.47 mmol), acceptor 36 (82 mg, 0.94 mmol), and freshly activated MS 4Å in anhydrous CH2Cl2 (6 mL) was added TMSOTf (8.53 μL, 0.047 mmol) under N2 protection at 0 °C. After the reaction mixture had been stirring for 15 min, it was neutralized with Et3N, filtered, and concentrated. The residue was subjected to silica gel column chromatography (1:4, EtOAc/hexanes) to afford 37 (292 mg, 51%) and 38 (172 mg, 30%) as foam.
For 37: Rf 0.35 (1:3, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.48 – 7.05 (m, 25H), 5.55 (d, J = 3.7 Hz, 1H), 5.49 (dd, J = 10.6, 9.3 Hz, 1H), 5.22 (d, J = 12.4 Hz, 1H), 5.06 (d, J = 12.4 Hz, 1H), 4.99 (d, J = 10.7 Hz, 1H), 4.94 (d, J = 3.6 Hz, 1H), 4.85 – 4.62 (m, 6H), 4.54 (d, J = 10.8 Hz, 1H), 4.35 (ddd, J = 14.1, 13.2, 1.9 Hz, 3H), 4.20 (ddd, J = 16.9, 12.2, 4.2 Hz, 2H), 4.14 – 4.04 (m, 1H), 3.92 (ddd, J = 9.7, 5.2, 3.0 Hz, 2H), 3.87 – 3.81 (m, 1H), 3.78 (dd, J = 10.2, 8.9 Hz, 1H), 3.74 – 3.63 (m, 3H), 3.60 – 3.53 (m, 2H), 3.46 (ddd, J = 13.2, 10.8, 7.0 Hz, 3H), 3.29 – 3.19 (m, 2H), 2.05 (s, 3H), 2.04 (s, 3H), 2.00 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.67, 170.20, 170.13, 167.91, 137.93, 137.60, 137.56, 137.40, 134.46, 128.79, 128.62, 128.49, 128.47, 128.39, 128.36, 128.21, 128.08, 128.04, 128.01, 127.94, 127.89, 127.79, 127.71, 127.57, 127.29, 103.11, 98.00, 97.27, 83.92, 81.92, 80.13, 77.52, 75.81, 75.43, 75.27, 75.22, 75.12, 74.37, 74.26, 69.84, 69.33, 69.21, 67.77, 67.25, 63.16, 62.24, 61.53, 60.76, 50.54, 20.87, 20.85, 20.69. HR ESI-TOF MS (m/z): calcd for C61H67N9O18Na [M + Na]+, 1236.4496; found, 1236.4493.
For 38: Rf 0.45 (1:3, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.46 – 7.10 (m, 25H), 5.54 (d, J = 3.7 Hz, 1H), 5.22 (d, J = 12.3 Hz, 1H), 5.06 (d, J = 12.4 Hz, 1H), 4.99 – 4.93 (m, 2H), 4.85 – 4.66 (m, 6H), 4.53 (d, J = 10.8 Hz, 1H), 4.42 (dd, J = 12.1, 1.8 Hz, 1H), 4.38 – 4.29 (m, 3H), 4.21 (dd, J = 12.1, 3.8 Hz, 1H), 4.12 – 4.03 (m, 2H), 3.96 (dd, J = 5.7, 3.8 Hz, 1H), 3.89 (d, J = 9.7 Hz, 1H), 3.81 – 3.64 (m, 4H), 3.56 (s, 1H), 3.50 – 3.36 (m, 6H), 3.24 (dd, J = 10.4, 3.7 Hz, 1H), 2.04 (s, 3H), 2.03 (s, 3H), 2.00 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.66, 170.16, 167.84, 137.90, 137.57, 137.54, 137.38, 134.43, 128.69, 128.62, 128.56, 128.49, 128.47, 128.40, 128.35, 128.08, 128.05, 127.96, 127.93, 127.77, 127.72, 127.63, 127.58, 127.38, 127.28, 102.97, 101.90, 97.27, 83.92, 82.02, 80.10, 77.52, 75.43, 75.37, 75.24, 75.12, 74.30, 74.20, 73.07, 71.63, 69.85, 68.89, 67.80, 63.83, 63.14, 62.24, 61.38, 50.68, 20.84, 20.67. HR ESI-TOF MS (m/z): calcd for C61H67N9O18Na [M + Na]+, 1236.4496; found, 1236.4491.
Selective Hydrolysis of Protected Trisaccharides
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3,6-di-O-acetyl-α-D-glucopyranoside (39)
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-acetyl-α-D-glucopyranoside (40):
To a solution of 37 (230 mg, 0.19 mmol) in CH3OH/CH2Cl2 (v/v, 1/1, 9.5 mL) was added CH3ONa (2.05 mg, 0.038 mmol). After the reaction mixture had been stirring at room temperature for 5 h, it was quenched with Amberlyst 15 hydrogen resin, and concentrated. The resulting residue was subjected to silica gel column chromatography (1:2, EtOAc/hexanes) to give 39 (93 mg, 42%) and 40 (71 mg, 33%).
For 39: Rf 0.50 (1:1.5, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.45 – 7.03 (m, 25H), 5.54 (d, J = 3.5 Hz, 1H), 5.49 (t, J = 10.0 Hz, 1H), 5.20 (d, J = 12.5 Hz, 1H), 5.06 – 4.91 (m, 3H), 4.74 (ddd, J = 33.1, 20.8, 9.7 Hz, 6H), 4.51 (dd, J = 16.0, 9.3 Hz, 2H), 4.31 (d, J = 12.0 Hz, 1H), 4.20 (dd, J = 12.1, 3.7 Hz, 1H), 4.08 (t, J = 9.2 Hz, 1H), 3.96 (d, J = 9.7 Hz, 1H), 3.89 – 3.63 (m, 8H), 3.56 (d, J = 6.8 Hz, 2H), 3.49 – 3.40 (m, 3H), 3.22 (dt, J = 10.0, 3.8 Hz, 2H), 2.03 (s, 3H), 1.99 (s, 3H). 13C NMR (150 pMHz, CDCl3) δ: 170.67, 170.31, 168.09, 138.00, 137.71, 137.58, 137.41, 134.52, 128.58, 128.52, 128.47, 128.45, 128.41, 128.35, 128.28, 128.06, 128.01, 127.93, 127.80, 127.76, 127.55, 127.28, 102.97, 98.29, 97.27, 83.98, 81.95, 80.01, 75.38, 75.22, 75.13, 75.08, 74.80, 74.47, 74.08, 71.32, 69.79, 69.37, 67.64, 67.18, 63.12, 62.26, 60.93, 60.08, 50.56, 47.91, 20.83, 20.71. HR ESI-TOF MS (m/z): calcd for C59H65N9O17Na [M + Na]+, 1194.4391; found, 1194.4382.
For 40: Rf 0.40 (1:1.5, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.41 – 7.08 (m, 25H), 5.52 – 5.43 (m, 2H), 5.21 (d, J = 12.4 Hz, 1H), 5.07 (d, J = 12.4 Hz, 1H), 4.96 (dd, J = 20.0, 6.9 Hz, 2H), 4.80 (dd, J = 17.7, 7.0 Hz, 3H), 4.74 – 4.67 (m, 3H), 4.63 (d, J = 11.0 Hz, 1H), 4.49 (d, J = 7.9 Hz, 1H), 4.09 (t, J = 9.2 Hz, 1H), 3.97 (d, J = 9.7 Hz, 1H), 3.89 – 3.61 (m, 8H), 3.60 – 3.48 (m, 2H), 3.47 – 3.40 (m, 3H), 3.20 (ddd, J = 20.6, 10.6, 3.4 Hz, 2H), 1.97 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.26, 168.35, 138.01, 137.73, 137.68, 134.41, 128.62, 128.44, 128.41, 128.38, 128.32, 127.92, 127.86, 127.78, 127.76, 127.60, 127.34, 103.02, 98.29, 97.35, 83.91, 81.90, 79.80, 77.68, 75.32, 75.22, 75.07, 74.90, 74.76, 74.27, 72.03, 71.32, 69.39, 67.74, 67.18, 63.20, 61.18, 60.93, 60.07, 50.56, 20.72. HR ESI-TOF MS (m/z): calcd for C57H63N9O16Na [M + Na]+, 1152.4285; found, 1152.4290.
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3,6-di-O-acetyl-β-D-glucopyranoside (43)
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3O-acetyl-β-D-glucopyranoside (44):
Compounds 43 (73 mg, 41%) and 44 (60 mg, 35%) were prepared from 38 (185 mg, 0.15 mmol) using the same procedure for the synthesis of 39 and 40.
For 43: Rf 0.45 (3:7, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.45 – 7.07 (m, 25H), 5.53 (d, J = 3.7 Hz, 1H), 5.21 (d, J = 12.4 Hz, 1H), 5.04 (d, J = 12.4 Hz, 1H), 5.01 – 4.92 (m, 2H), 4.83 – 4.62 (m, 6H), 4.51 (dd, J = 20.3, 9.3 Hz, 2H), 4.39 (d, J = 8.0 Hz, 1H), 4.31 (dd, J = 12.0, 1.8 Hz, 1H), 4.20 (dd, J = 12.1, 3.8 Hz, 1H), 4.10 – 4.03 (m, 1H), 4.03 – 3.93 (m, 2H), 3.87 – 3.66 (m, 6H), 3.56 (d, J = 8.6 Hz, 1H), 3.51 – 3.34 (m, 5H), 3.30 – 3.17 (m, 2H), 2.04 (s, 3H), 1.99 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.66, 170.34, 168.05, 137.99, 137.68, 137.58, 137.41, 134.51, 128.59, 128.55, 128.47, 128.45, 128.41, 128.34, 128.32, 128.06, 128.02, 127.94, 127.75, 127.69, 127.55, 127.28, 102.81, 102.03, 97.28, 83.99, 82.03, 79.99, 77.55, 75.24, 75.19, 75.14, 75.09, 74.43, 74.22, 74.03, 71.70, 69.80, 68.88, 67.67, 63.94, 63.11, 62.27, 60.09, 50.66, 20.83, 20.69. HR ESI-TOF MS (m/z): calcd for C59H65N9O17Na [M + Na]+, 1194.4391; found, 1194.4386.
For 44: Rf 0.35 (3:7, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.48 – 7.04 (m, 25H), 5.48 (d, J = 3.6 Hz, 1H), 5.22 (d, J = 12.4 Hz, 1H), 5.08 (d, J = 12.4 Hz, 1H), 5.02 – 4.92 (m, 2H), 4.85 – 4.57 (m, 7H), 4.50 (d, J = 7.9 Hz, 1H), 4.39 (d, J = 8.0 Hz, 1H), 4.08 (t, J = 9.2 Hz, 1H), 4.04 – 3.93 (m, 2H), 3.78 (ddt, J = 21.2, 17.9, 9.3 Hz, 7H), 3.67 (s, 1H), 3.52 (t, J = 9.4 Hz, 1H), 3.41 (ddt, J = 34.0, 32.0, 16.1 Hz, 5H), 3.26 (d, J = 9.8 Hz, 1H), 3.19 (dd, J = 10.4, 3.7 Hz, 1H), 1.97 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.30, 168.32, 138.00, 137.74, 137.70, 134.40, 128.64, 128.44, 128.42, 128.38, 128.36, 127.93, 127.87, 127.76, 127.69, 127.60, 127.33, 102.85, 102.02, 97.37, 83.93, 81.98, 79.78, 77.69, 75.33, 75.25, 75.20, 75.07, 74.72, 74.32, 74.21, 72.03, 71.73, 68.88, 67.78, 63.94, 63.19, 61.18, 60.08, 50.66, 20.71. HR ESI-TOF MS (m/z): calcd for C57H63N9O16Na [M + Na]+, 1152.4285; found, 1152.4295.
2-Azidoethyl [2-azido-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranosyl]-(1→4)-(methyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (45):
To a solution of 29 (150 mg, 0.11 mmol) in CH3OH/CH2Cl2 (v/v, 2/1, 6 mL) was added CH3ONa (24.3 mg, 0.45 mmol). After the reaction mixture had been stirring at room temperature for 24 h, it was quenched with Amberlyst 15 hydrogen resin, and concentrated. The resulting residue was subjected to silica gel column chromatography (1:3, EtOAc/hexanes) to give 45 (123 mg, 85%).
Rf 0.70 (1:1.5, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.96 – 7.59 (m, 9H), 7.54 – 6.97 (m, 25H), 5.52 (d, J = 3.7 Hz, 1H), 5.01 (d, J = 10.8 Hz, 1H), 4.93 (d, J = 3.6 Hz, 1H), 4.88 – 4.83 (m, 2H), 4.77 (dd, J = 11.6, 6.1 Hz, 2H), 4.67 (dd, J = 22.3, 11.1 Hz, 3H), 4.57 (d, J = 12.3 Hz, 1H), 4.50 – 4.46 (m, 2H), 4.08 – 4.01 (m, 3H), 3.92 (dd, J = 10.1, 8.9 Hz, 1H), 3.88 – 3.79 (m, 3H), 3.77 – 3.69 (m, 4H), 3.67 – 3.59 (m, 2H), 3.56 – 3.43 (m, 6H), 3.41 – 3.29 (m, 4H). 13C NMR (150 MHz, CDCl3) δ: 168.21, 138.32, 138.07, 138.04, 137.80, 135.19, 134.61, 133.21, 133.17, 133.09, 133.00, 128.51, 128.43, 128.38, 128.32, 128.27, 128.24, 128.21, 128.03, 128.00, 127.96, 127.87, 127.84, 127.66, 127.59, 127.55, 127.52, 127.34, 127.28, 126.82, 126.35, 126.16, 126.12, 125.98, 125.90, 102.73, 98.09, 97.77, 83.79, 79.99, 78.37, 77.84, 77.35, 75.36, 75.16, 75.05, 74.92, 74.79, 74.75, 74.61, 73.93, 73.71, 71.19, 70.51, 69.48, 68.04, 67.60, 67.00, 63.39, 62.91, 52.38, 50.59. HR ESI-TOF MS (m/z): calcd for C71H73N9O15Na [M + Na]+, 1314.5118; found, 1314.5113.
2-Azidoethyl [2-acetamido-2-deoxy-3,4-di-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranosyl]-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-6-O-(2-naphthylmethyl)-α-D-glucopyranoside (47):
To a solution of 33 (270 mg, 0.185 mmol) in acetone (15 mL) were added 6N HCl (300 μL). After the reaction mixture had been stirring at room temperature for 20 min, it was diluted with acetone and concentrated. The resulting residue was subjected to silica gel column chromatography (1:20, CH3OH–CH2Cl2) to give 46. The amine intermediate 46 was then dissolved in CH2Cl2 (5 mL), Ac2O (87.15 μL, 0.92 mmol) and Et3N (128 μL, 0.92 mmol) were added. After the reaction mixture had been stirring at room temperature for 5 h, it was quenched with CH3OH and concentrated. The residue was subjected to silica gel column chromatography (1:2, EtOAc/hexanes) to give 47 (159 mg, 64% for 2 steps) as a syrup.
Rf 0.40 (1:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.98 – 7.73 (m, 8H), 7.59 – 7.02 (m, 26H), 5.67 (d, J = 9.7 Hz, 1H), 5.01 (dd, J = 17.9, 11.5 Hz, 2H), 4.94 – 4.79 (m, 5H), 4.75 (d, J = 12.3 Hz, 1H), 4.67 – 4.59 (m, 3H), 4.51 (dd, J = 24.9, 11.5 Hz, 2H), 4.26 (dd, J = 9.9, 3.3 Hz, 1H), 4.22 (d, J = 8.0 Hz, 1H), 4.04 – 3.97 (m, 2H), 3.94 – 3.79 (m, 7H), 3.75 – 3.62 (m, 5H), 3.56 – 3.51 (m, 2H), 3.45 – 3.38 (m, 4H), 3.37 – 3.33 (m, 1H), 1.83 (s, 3H), 1.18 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.08, 168.57, 167.01, 138.38, 138.36, 138.03, 136.70, 135.47, 134.89, 133.32, 133.21, 133.16, 132.99, 128.73, 128.62, 128.40, 128.31, 128.29, 128.21, 128.07, 128.03, 127.93, 127.88, 127.68, 127.64, 127.61, 127.56, 127.49, 127.41, 127.25, 127.09, 126.85, 126.59, 126.55, 126.14, 125.88, 125.86, 100.30, 100.27, 98.24, 81.01, 80.72, 78.44, 77.52, 75.41, 75.36, 74.99, 74.85, 74.65, 73.96, 73.60, 72.89, 72.62, 70.61, 68.08, 67.21, 66.86, 62.51, 52.69, 52.64, 50.60, 22.23, 20.58. HR ESI-TOF MS (m/z): calcd for C75H79N7O17Na [M + Na]+, 1372.5425; found, 1372.5234.
General Procedure for Simultaneous Nap & PMB Removal
To a solution of trisaccharide substrate (30~50 mg) in CH2Cl2/H2O (10:1, 1.1 mL) were added DDQ (3 equiv. per Nap or PMB) and β-pinene (3 equiv. per Nap or PMB). After the reaction mixtured was completed based on TLC monitor, it was diluted with CH2Cl2, washed with aq. NaHCO3, brine, dried over anhydrous Na2SO4, and concentrated. The resulting residue was subjected to silica gel column chromatography to give the corresponding product.
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-α-D-glucopyranoside (48):
foam, 73% yield (35 mg).
Rf 0.40 (2:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.43 – 7.14 (m, 15H), 5.39 (d, J = 3.7 Hz, 1H), 5.10 – 5.04 (m, 1H), 4.90 – 4.77 (m, 5H), 4.72 – 4.61 (m, 3H), 4.29 – 4.23 (m, 1H), 4.13 (d, J = 8.9 Hz, 1H), 4.11 – 4.05 (m, 1H), 3.93 (s, 1H), 3.87 (dd, J = 10.2, 8.9 Hz, 1H), 3.83 – 3.72 (m, 7H), 3.71 – 3.60 (m, 4H), 3.58 – 3.53 (m, 1H), 3.53 – 3.44 (m, 2H), 3.44 – 3.38 (m, 1H), 3.28 (dd, J = 10.4, 3.8 Hz, 1H), 3.16 (dd, J = 10.4, 3.6 Hz, 1H), 2.01 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.20, 168.19, 137.69, 137.62, 137.28, 128.52, 128.50, 128.46, 128.00, 127.97, 127.93, 127.80, 127.53, 101.14, 98.31, 97.55, 81.82, 80.48, 79.68, 77.60, 75.41, 75.01, 74.41, 74.23, 74.21, 72.71, 72.23, 70.22, 69.39, 67.19, 63.21, 62.17, 61.11, 60.52, 53.21, 50.58, 20.74. HR ESI-TOF MS (m/z): calcd for C44H53N9O16Na [M + Na]+, 986.3502; found, 986.3511.
2-Azidoethyl (2-azido-2-deoxy-3,4,6-tri-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-α-D-glucopyranoside (49):
foam, 82% yield (40 mg).
Rf 0.45 (1:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.42 – 7.36 (m, 2H), 7.35 – 7.18 (m, 21H), 7.13 (dd, J = 7.5, 1.8 Hz, 2H), 5.42 (d, J = 3.7 Hz, 1H), 5.12 (dd, J = 8.7, 8.0 Hz, 1H), 5.08 (d, J = 10.9 Hz, 1H), 4.90 – 4.82 (m, 3H), 4.78 – 4.73 (m, 2H), 4.67 (dd, J = 21.1, 10.9 Hz, 2H), 4.59 (d, J = 12.1 Hz, 1H), 4.51 (d, J = 11.0 Hz, 1H), 4.43 (d, J = 12.1 Hz, 1H), 4.24 (t, J = 8.7 Hz, 1H), 3.97 (d, J = 9.0 Hz, 1H), 3.92 – 3.87 (m, 2H), 3.86 – 3.77 (m, 4H), 3.76 – 3.70 (m, 2H), 3.63 (dddd, J = 25.3, 12.8, 7.9, 2.1 Hz, 3H), 3.54 – 3.47 (m, 4H), 3.44 – 3.39 (m, 1H), 3.32 (dd, J = 10.3, 3.7 Hz, 1H), 3.28 – 3.24 (m, 1H), 2.00 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.15, 168.05, 138.25, 138.06, 137.77, 137.53, 128.44, 128.42, 128.35, 128.34, 128.24, 128.10, 127.99, 127.91, 127.85, 127.77, 127.71, 127.67, 127.52, 127.46, 127.42, 100.80, 98.16, 97.79, 82.21, 79.85, 77.81, 77.54, 77.12, 75.34, 75.20, 75.09, 74.80, 74.51, 73.58, 73.26, 71.37, 71.35, 69.49, 67.61, 67.07, 63.26, 62.87, 60.39, 52.47, 50.57, 20.78. HR ESI-TOF MS (m/z): calcd for C58H65N9O16Na [M + Na]+, 1166.4441; found, 1166.4435.
2-Azidoethyl (2-acetamido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-α-D-glucopyranoside (50):
foam, 75% yield (28 mg).
Rf 0.55 (1:1, acetone/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.52 – 7.05 (m, 20H), 5.73 (d, J = 9.5 Hz, 1H), 5.13 (t, J = 8.3 Hz, 1H), 5.07 (d, J = 10.8 Hz, 1H), 4.91 (d, J = 3.6 Hz, 1H), 4.87 (d, J = 3.6 Hz, 1H), 4.84 – 4.76 (m, 3H), 4.67 – 4.56 (m, 4H), 4.52 (d, J = 11.0 Hz, 1H), 4.21 (td, J = 10.0, 3.6 Hz, 1H), 4.08 (t, J = 8.4 Hz, 1H), 3.96 (d, J = 8.4 Hz, 1H), 3.92 – 3.86 (m, 2H), 3.83 – 3.71 (m, 4H), 3.64 (ddd, J = 15.6, 14.2, 9.4 Hz, 5H), 3.57 – 3.46 (m, 5H), 3.40 (ddd, J = 13.3, 5.7, 3.5 Hz, 1H), 3.28 (dd, J = 9.6, 3.9 Hz, 1H), 1.96 (s, 3H), 1.44 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.16, 169.04, 167.88, 138.21, 138.16, 137.78, 136.80, 128.74, 128.51, 128.44, 128.35, 128.22, 128.00, 127.98, 127.96, 127.92, 127.76, 127.73, 127.48, 100.50, 98.87, 98.16, 80.88, 80.02, 77.79, 77.46, 75.82, 75.21, 75.02, 74.93, 74.71, 73.35, 73.19, 71.31, 67.12, 62.79, 61.65, 60.27, 52.81, 52.59, 50.57, 22.61, 20.73. HR ESI-TOF MS (m/z): calcd for C53H63N7O17Na [M + Na]+, 1092.4173; found, 1092.4176.
2-Azidoethyl (2-azido-2-deoxy-4-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-α-D-glucopyranoside (51):
foam, 71% yield (25 mg).
Rf 0.40 (2:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.41 – 7.21 (m, 15H), 5.36 (d, J = 3.8 Hz, 1H), 5.10 (d, J = 8.6 Hz, 1H), 5.06 (d, J = 10.8 Hz, 1H), 4.87 (d, J = 3.6 Hz, 1H), 4.82 (d, J = 11.3 Hz, 1H), 4.76 – 4.62 (m, 5H), 4.21 (t, J = 8.7 Hz, 1H), 3.97 – 3.89 (m, 4H), 3.82 – 3.76 (m, 5H), 3.68 – 3.62 (m, 3H), 3.54 – 3.47 (m, 4H), 3.42 – 3.36 (m, 3H), 3.28 – 3.25 (m, 1H), 3.12 (dd, J = 10.4, 3.8 Hz, 1H), 1.98 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.18, 168.24, 138.17, 137.85, 137.49, 128.71, 128.66, 128.46, 128.44, 128.24, 128.21, 128.18, 128.10, 128.06, 127.97, 127.82, 127.77, 127.76, 127.55, 127.50, 127.47, 127.36, 100.71, 98.13, 97.57, 82.25, 81.24, 77.72, 77.64, 77.53, 75.22, 75.07, 74.89, 74.84, 74.66, 74.58, 74.53, 74.09, 73.28, 72.58, 72.13, 71.67, 71.58, 71.36, 71.29, 67.08, 62.88, 62.82, 61.17, 60.40, 60.33, 52.65, 50.56, 20.79. HR ESI-TOF MS (m/z): calcd for C44H53N9O16Na [M + Na]+, 986.3502; found, 986.3501.
2-Azidoethyl (2-azido-2-deoxy-4-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-α-D-glucopyranoside (52):
foam, 76% yield (37 mg).
Rf 0.25 (2:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.43 – 7.11 (m, 10H), 5.38 (d, J = 3.8 Hz, 1H), 5.07 (t, J = 7.8 Hz, 1H), 4.87 (d, J = 3.6 Hz, 1H), 4.75 – 4.72 (m, 2H), 4.68 (d, J = 7.4 Hz, 1H), 4.63 (d, J = 11.2 Hz, 1H), 4.26 (t, J = 8.6 Hz, 1H), 4.10 (d, J = 8.8 Hz, 1H), 4.07 – 4.04 (m, 1H), 3.97 (t, J = 9.5 Hz, 1H), 3.84 (s, 1H), 3.82 – 3.70 (m, 7H), 3.68 – 3.60 (m, 4H), 3.51 – 3.46 (m, 2H), 3.40 – 3.35 (m, 2H), 3.18 (ddd, J = 13.9, 9.0, 3.7 Hz, 2H), 1.94 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 169.34, 168.56, 137.88, 137.31, 128.67, 128.50, 128.42, 128.16, 128.02, 127.92, 127.90, 127.82, 127.55, 100.75, 98.25, 97.57, 82.10, 77.55, 74.95, 74.71, 74.67, 74.08, 73.99, 72.69, 72.02, 71.86, 71.65, 70.38, 69.35, 67.18, 62.68, 62.15, 60.93, 60.37, 53.20, 50.56, 20.68. HR ESI-TOF MS (m/z): calcd for C37H47N9O16Na [M + Na]+, 896.3033; found, 896.3026.
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-azido-2-deoxy-3-O-benzyl-α-D-glucopyranoside (53):
foam, 71% yield (31 mg).
Rf 0.50 (2:1, EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.51 – 7.12 (m, 20H), 5.48 (d, J = 3.8 Hz, 1H), 4.98 – 4.89 (m, 3H), 4.88 – 4.75 (m, 5H), 4.69 (d, J = 7.5 Hz, 1H), 4.64 (d, J = 11.2 Hz, 1H), 4.08 – 3.94 (m, 4H), 3.90 (d, J = 9.5 Hz, 1H), 3.87 – 3.71 (m, 5H), 3.72 – 3.58 (m, 7H), 3.56 – 3.48 (m, 2H), 3.38 (ddd, J = 13.0, 5.5, 3.3 Hz, 2H), 3.35 – 3.32 (m, 1H), 3.26 (dd, J = 10.3, 3.8 Hz, 1H). 13C NMR (150 MHz, CDCl3) δ: 168.62, 138.11, 137.78, 137.62, 128.49, 128.48, 128.45, 128.39, 128.05, 127.94, 127.93, 127.92, 127.80, 127.78, 127.75, 127.71, 102.83, 98.09, 97.55, 83.67, 79.77, 78.51, 77.61, 75.58, 75.41, 75.17, 74.98, 74.95, 74.79, 74.70, 74.39, 71.98, 71.44, 66.98, 63.36, 63.25, 61.18, 60.70, 52.77, 50.58. HR ESI-TOF MS (m/z): calcd for C49H57N9O15Na [M + Na]+, 1034.3866; found, 1034.3872.
2-Azidoethyl (2-azido-2-deoxy-3,4-di-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-acetamido-2-deoxy-3-O-benzyl-α-D-glucopyranoside (54):
foam, 65% yield (28 mg).
Rf 0.65 (2:1, acetone/hexanes); 1H NMR (600 MHz, CDCl3) δ: 7.37 – 7.12 (m, 20H), 5.38 (d, J = 3.8 Hz, 1H), 5.16 – 5.13 (m, 1H), 4.90 – 4.79 (m, 6H), 4.74 – 4.62 (m, 3H), 4.51 (d, J = 12.4 Hz, 1H), 4.21 – 4.17 (m, 1H), 4.06 (ddd, J = 10.7, 8.7, 3.7 Hz, 1H), 3.99 (d, J = 9.3 Hz, 1H), 3.90 – 3.74 (m, 6H), 3.63 – 3.50 (m, 8H), 3.47 – 3.39 (m, 2H), 3.27 – 3.19 (m, 2H), 2.00 (s, 3H), 1.71 (s, 3H). 13C NMR (150 MHz, CDCl3) δ: 170.12, 169.26, 168.29, 138.78, 137.76, 137.64, 137.57, 128.48, 128.47, 128.45, 128.43, 128.25, 128.03, 127.97, 127.91, 127.83, 127.79, 127.73, 127.58, 127.40, 101.02, 97.72, 97.69, 82.43, 79.73, 77.85, 77.64, 75.42, 75.26, 74.92, 74.75, 74.65, 74.43, 73.30, 72.07, 71.42, 67.46, 63.32, 61.16, 60.46, 52.68, 52.30, 50.36, 23.10, 20.86. HR ESI-TOF MS (m/z): calcd for C53H63N7O17Na [M + Na]+, 1092.4173; found, 1092.4166.
General Procedure for Azide Reduction Followed by Simultaneous N,O-Sulfonations
To a solution of trisaccharide substrate (10~20 mg) in CH3OH (1 mL) were added 1,3-propanedithiol (150 μL) and DIPEA (50 μL). After the reaction mixture had been stirring at 50 °C for 10 h, it was concentrated in vacuo. The resulting residue was kept under high vacuum for overnight to afford the amine intermediate as white solid, which was then dissolved in pyridine (2 mL), SO3· Et3N (3 equiv. per OH or NH2) and Et3N (0.2 mL) were added. After the reaction mixture was stirred under microwave for 15 min at 100 °C, it was quenched with CH3OH and concentrated. The resuting residue was subjected to RP-18 column chromatography (v/v, 1/0 →1/5, H2O/CH3OH) to give thecorresponding product.
Note: higher concentration or SO3· Et3N equiv. would produce more over-sulfonated side-product, which is difficult to separate from the desired product.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3-O-acetyl-6-O-sulfo-α-D-glucopyranoside (56):
pale yellow foam, 85% yield (65.3 mg).
1H NMR (600 MHz, CD3OD) δ: 7.48 – 7.09 (m, 25H), 5.60 (d, J = 3.3 Hz, 1H, H-1α), 5.34 (d, J = 12.7 Hz, 1H, Bn), 5.16 (d, J = 3.6 Hz, 1H, H-1’α), 5.12 – 4.97 (m, 3H, Bn, H-3 -OAc), 4.92 (d, J = 11.2 Hz, 1H, Bn), 4.88 – 4.82 (m, 4H, Bn, H-1β), 4.75 (q, J = 10.4 Hz, 2H, Bn), 4.63 (d, J = 11.5 Hz, 1H, Bn), 4.50 (d, J = 11.2 Hz, 1H, Bn), 4.36 (ddd, J = 16.4, 10.8, 2.3 Hz, 2H, H-6a, H-6b), 4.20 (dd, J = 10.6, 1.9 Hz, 1H, H-6b’), 4.13 (d, J = 9.6 Hz, 1H), 4.08 – 3.97 (m, 2H, H-6a’), 3.85 (ddd, J = 19.1, 14.0, 9.1 Hz, 3H), 3.71 (dd, J = 9.9, 2.1 Hz, 2H), 3.58 (dd, J = 16.0, 6.2 Hz, 2H), 3.54 – 3.49 (m, 1H), 3.42 – 3.31 (m, 3H), 3.23 (dt, J = 6.4, 5.0 Hz, 2H), 1.87 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 171.86, 168.81, 139.15, 138.62, 138.47, 138.37, 135.36, 128.34, 128.10, 127.95, 127.89, 127.83, 127.81, 127.77, 127.74, 127.67, 127.67, 127.11, 126.94, 126.86, 102.23, 98.14, 97.69, 82.53, 81.96, 80.09, 77.13, 76.23, 74.85, 74.68, 74.65, 74.59, 74.08, 70.74, 70.29, 69.04, 67.23, 66.97, 65.53, 64.82, 58.65, 56.29, 43.14, 20.13. Waters ZQ ESI-TOF MS (m/z): calcd for C57H66N3O31S5 [M – 3H]3-, 482.74; found, 482.98.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3,6-di-O-acetyl-α-D-glucopyranoside (59):
pale yellow foam, 80% yield (31.5 mg).
1H NMR (600 MHz, CD3OD) δ: 7.46 – 7.01 (m, 25H), 5.60 (d, J = 3.4 Hz, 1H), 5.24 (d, J = 12.6 Hz, 1H), 5.15 (d, J = 3.6 Hz, 1H), 5.06 (d, J = 10.1 Hz, 1H), 4.98 (dd, J = 20.4, 11.1 Hz, 2H), 4.86 (d, J = 10.4 Hz, 3H), 4.80 (d, J = 11.5 Hz, 1H), 4.76 (d, J = 11.0 Hz, 1H), 4.61 (d, J = 11.6 Hz, 1H), 4.52 (d, J = 11.0 Hz, 1H), 4.43 (d, J = 11.0 Hz, 1H), 4.38 (dd, J = 11.0, 2.3 Hz, 1H), 4.32 (dd, J = 11.9, 2.0 Hz, 1H), 4.16 (d, J = 9.6 Hz, 1H), 4.13 – 3.99 (m, 3H), 3.91 – 3.80 (m, 3H), 3.69 (dd, J = 7.0, 5.1 Hz, 2H), 3.61 – 3.49 (m, 2H), 3.42 – 3.33 (m, 3H), 3.25 – 3.22 (m, 1H), 1.99 (s, 3H), 1.89 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 171.94, 171.25, 168.75, 139.03, 138.40, 138.32, 135.16, 128.27, 128.24, 128.10, 127.95, 127.92, 127.91, 127.89, 127.86, 127.84, 127.72, 127.63, 127.62, 127.40, 127.22, 127.04, 126.86, 126.82, 104.52, 102.21, 97.81, 97.72, 82.50, 82.20, 82.03, 81.70, 80.42, 77.33, 76.97, 76.36, 75.94, 75.30, 74.90, 74.84, 74.82, 74.62, 74.36, 74.33, 74.22, 73.98, 70.01, 69.90, 69.32, 69.05, 68.99, 67.22, 67.18, 67.03, 65.60, 64.83, 64.14, 62.48, 59.82, 58.60, 56.86, 56.25, 54.79, 43.13, 40.77, 20.10, 19.43. Waters ZQ ESI-TOF MS (m/z): calcd for C59H68N3O29S4 [M – 3H]3-, 470.09; found, 470.37.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3,6-di-O-acetyl- β -D-glucopyranoside (60):
pale yellow foam, 76% yield (19.2 mg).
1H NMR (600 MHz, CD3OD) δ: 7.41 – 7.12 (m, 25H), 5.63 (s, 1H), 5.20 (d, J = 12.1 Hz, 1H), 5.09 – 4.95 (m, 4H), 4.84 (d, J = 19.5 Hz, 4H), 4.75 (d, J = 11.2 Hz, 1H), 4.60 (d, J = 11.6 Hz, 1H), 4.52 (d, J = 11.2 Hz, 1H), 4.46 – 4.30 (m, 4H), 4.21 – 4.04 (m, 5H), 3.98 (s, 1H), 3.89 (dd, J = 19.4, 9.5 Hz, 2H), 3.75 (s, 1H), 3.66 – 3.62 (m, 1H), 3.49 (dd, J = 18.3, 9.1 Hz, 2H), 3.40 (d, J = 9.3 Hz, 2H), 3.16 (dd, J = 46.0, 7.2 Hz, 2H), 1.99 (s, 3H), 1.94 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 172.87, 172.00, 169.22, 141.25, 139.03, 138.66, 138.33, 135.00, 128.69, 128.17, 128.09, 128.04, 127.94, 127.89, 127.86, 127.84, 127.81, 127.72, 127.68, 127.65, 127.62, 127.47, 127.41, 127.24, 127.02, 126.85, 126.81, 126.54, 125.89, 124.98, 103.44, 102.34, 102.04, 84.80, 80.50, 77.95, 77.06, 75.50, 74.88, 74.83, 74.48, 73.06, 72.95, 72.45, 72.13, 68.97, 68.71, 68.35, 67.22, 64.48, 61.83, 58.72, 58.52, 58.46, 58.29, 58.18, 57.20, 43.00, 20.11, 19.07. Waters ZQ ESI-TOF MS (m/z): calcd for C59H68N3O29S4 [M – 3H]3-, 470.09; found, 470.28.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(benzyl-2,3-di-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3-O-acetyl-6-O-sulfo-β-D-glucopyranoside (61):
pale yellow foam, 78% yield (36.7 mg).
1H NMR (600 MHz, CD3OD) δ: 7.48 – 6.98 (m, 25H), 5.62 (s, 1H), 5.30 – 5.23 (m, 1H), 5.12 – 4.97 (m, 3H), 4.93 (d, J = 11.1 Hz, 1H), 4.82 – 4.70 (m, 5H), 4.59 (d, J = 11.2 Hz, 1H), 4.46 – 4.35 (m, 3H), 4.13 (ddd, J = 40.6, 29.6, 9.8 Hz, 5H), 3.97 (s, 1H), 3.87 (dd, J = 16.4, 8.5 Hz, 2H), 3.71 (ddd, J = 26.8, 15.8, 7.7 Hz, 3H), 3.64 – 3.59 (m, 1H), 3.53 – 3.34 (m, 5H), 3.22 – 3.09 (m, 2H), 1.90 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 172.37, 169.05, 139.01, 138.66, 138.35, 138.33, 135.15, 128.19, 128.08, 128.03, 127.99, 127.92, 127.90, 127.89, 127.86, 127.81, 127.72, 127.69, 127.63, 127.47, 127.45, 127.04, 126.98, 126.85, 126.80, 102.34, 102.08, 98.11, 82.45, 81.71, 80.47, 76.96, 75.81, 74.91, 74.61, 74.54, 73.94, 73.71, 72.94, 70.26, 68.69, 67.29, 65.53, 64.75, 58.39, 58.21, 43.00, 20.10. Waters ZQ ESI-TOF MS (m/z): calcd for C57H66N3O31S5 [M – 3H]3-, 482.74; found, 482.98.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-sulfo-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3-O-benzyl-6-O-sulfo-α-D-glucopyranoside (62):
pale yellow foam, 81% yield (18.5 mg).
1H NMR (600 MHz, CD3OD) δ: 7.67 (d, J = 7.4 Hz, 2H), 7.45 (d, J = 7.4 Hz, 2H), 7.39 (d, J = 7.2 Hz, 2H), 7.33 – 7.12 (m, 16H), 5.51 (d, J = 3.2 Hz, 1H), 5.20 (d, J = 3.5 Hz, 1H), 5.11 (dd, J = 23.1, 9.0 Hz, 2H), 5.02 (d, J = 11.1 Hz, 1H), 4.93 (dd, J = 17.3, 10.5 Hz, 2H), 4.79 – 4.66 (m, 3H), 4.63 (d, J = 11.1 Hz, 1H), 4.43 (t, J = 8.1 Hz, 1H), 4.28 (dd, J = 10.6, 2.7 Hz, 1H), 4.16 – 4.01 (m, 4H), 3.94 (dd, J = 19.6, 10.5 Hz, 2H), 3.88 – 3.82 (m, 1H), 3.76 – 3.65 (m, 2H), 3.65 – 3.56 (m, 4H), 3.52 (dd, J = 8.0, 4.8 Hz, 2H), 3.41 – 3.31 (m, 2H), 3.23 (t, J = 5.5 Hz, 2H). 13C NMR (150 MHz, CD3OD) δ: 168.96, 139.39, 139.22, 138.71, 138.21, 129.57, 128.15, 127.88, 127.79, 127.71, 127.70, 127.65, 127.10, 126.99, 126.83, 126.69, 100.92, 98.27, 97.50, 88.15, 80.14, 77.67, 77.13, 74.77, 74.46, 73.82, 70.16, 69.03, 67.11, 65.87, 65.56, 58.67, 58.19, 57.50, 56.19, 51.83, 43.14. Waters ZQ ESI-TOF MS (m/z): calcd for C49H60N3O33S6 [M – 3H]3-, 470.05; found, 470.15.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4,6-tri-O-benzyl-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3-O-benzyl-6-O-sulfo-α-D-glucopyranoside (63):
pale yellow foam, 82% yield (22.6 mg).
1H NMR (600 MHz, CD3OD) δ: 7.50 – 7.06 (m, 25H), 5.51 (d, J = 3.3 Hz, 1H), 5.19 (d, J = 3.1 Hz, 1H), 5.02 – 4.92 (m, 3H), 4.86 (d, J = 10.8 Hz, 1H), 4.69 (ddd, J = 34.9, 21.7, 11.3 Hz, 4H), 4.48 (dt, J = 20.3, 12.0 Hz, 3H), 4.30 (dd, J = 10.9, 3.3 Hz, 1H), 4.21 – 4.13 (m, 2H), 4.07 (d, J = 9.1 Hz, 1H), 3.99 (t, J = 8.8 Hz, 1H), 3.87 – 3.84 (m, 1H), 3.76 (d, J = 9.9 Hz, 1H), 3.71 – 3.52 (m, 7H), 3.45 (s, 3H), 3.39 (ddd, J = 24.4, 10.3, 3.3 Hz, 2H), 3.21 (dd, J = 11.6, 6.6 Hz, 2H), 1.97 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 170.16, 168.52, 139.23, 138.92, 138.63, 138.19, 138.14, 128.01, 127.98, 127.97, 127.96, 127.88, 127.77, 127.70, 127.68, 127.66, 127.31, 127.26, 127.11, 127.01, 126.94, 126.67, 100.21, 98.52, 97.51, 81.42, 81.00, 79.79, 79.69, 79.50, 77.55, 77.53, 77.18, 77.07, 76.81, 76.55, 74.99, 74.83, 74.54, 74.45, 74.35, 74.32, 74.18, 73.65, 73.11, 72.97, 72.90, 71.46, 71.33, 69.91, 69.22, 68.59, 67.97, 67.36, 67.09, 66.01, 65.35, 59.85, 58.60, 57.81, 57.48, 55.19, 51.52, 42.07, 19.73. Waters ZQ ESI-TOF MS (m/z): calcd for C58H68N3O28S4 [M – 3H]3-, 461.14; found, 460.82.
2-N-sulfo-ethyl (2-acetamido-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3-O-benzyl-6-O-sulfo-α-D-glucopyranoside (64):
pale yellow foam, 85% yield (25.1 mg).
1H NMR (600 MHz, CD3OD) δ: 7.47 (d, J = 7.4 Hz, 2H), 7.37 – 7.15 (m, 18H), 5.33 (d, J = 3.8 Hz, 1H), 5.20 (d, J = 3.3 Hz, 1H), 5.10 (d, J = 8.2 Hz, 1H), 4.99 – 4.93 (m, 1H), 4.90 (d, J = 10.6 Hz, 1H), 4.82 – 4.78 (m, 3H), 4.76 – 4.64 (m, 3H), 4.57 (d, J = 10.8 Hz, 1H), 4.42 (dd, J = 11.0, 2.3 Hz, 1H), 4.33 (dd, J = 10.7, 2.7 Hz, 1H), 4.23 – 4.12 (m, 4H), 4.13 – 4.07 (m, 1H), 4.04 – 3.98 (m, 1H), 3.95 – 3.84 (m, 2H), 3.79 (d, J = 10.1 Hz, 1H), 3.71 – 3.49 (m, 8H), 3.37 (dd, J = 10.4, 3.3 Hz, 1H), 3.22 (dd, J = 9.7, 4.6 Hz, 2H), 2.05 (s, 3H), 1.89 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 171.97, 170.15, 168.98, 139.08, 138.63, 138.37, 137.56, 128.17, 128.13, 127.89, 127.85, 127.76, 127.73, 127.51, 127.48, 127.20, 127.12, 127.10, 126.83, 99.84, 97.61, 97.13, 82.32, 80.20, 77.52, 77.45, 76.51, 74.76, 74.46, 74.33, 74.10, 73.92, 73.84, 73.13, 70.19, 69.25, 67.63, 67.16, 65.32, 65.18, 57.41, 52.71, 51.85, 43.24, 21.52, 19.72. Waters ZQ ESI-TOF MS (m/z): calcd for C53H64N3O29S4 [M – 3H]3-, 445.11; found, 444.79.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-acetamido-2-deoxy-3-O-benzyl-6-O-sulfo-α-D-glucopyranoside (65):
pale yellow foam, 82% yield (16.3 mg).
1H NMR (600 MHz, CD3OD) δ: 7.50 – 7.06 (m, 20H), 5.47 (d, J = 3.3 Hz, 1H), 5.01 – 4.93 (m, 3H), 4.72 – 4.61 (m, 3H), 4.47 – 4.42 (m, 1H), 4.32 – 4.27 (m, 1H), 4.10 (dddd, J = 39.2, 35.4, 19.6, 11.9 Hz, 6H), 3.86 – 3.80 (m, 2H), 3.77 (d, J = 9.3 Hz, 1H), 3.74 – 3.67 (m, 2H), 3.58 (d, J = 8.4 Hz, 3H), 3.53 – 3.46 (m, 2H), 3.39 (dd, J = 10.6, 7.2 Hz, 1H), 3.20 – 3.16 (m, 2H), 1.96 (s, 3H), 1.89 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 171.88, 170.15, 168.56, 139.12, 138.94, 138.62, 138.21, 128.09, 127.95, 127.94, 127.86, 127.83, 127.74, 127.72, 127.71, 127.65, 127.46, 127.18, 127.05, 126.95, 126.86, 100.40, 98.74, 97.67, 80.90, 79.42, 78.42, 77.66, 77.33, 77.21, 74.97, 74.62, 74.48, 74.39, 73.21, 71.86, 70.38, 69.51, 68.14, 67.06, 65.44, 65.11, 58.55, 52.49, 51.94, 43.20, 21.36, 19.69. Waters ZQ ESI-TOF MS (m/z): calcd for C53H64N3O29S4 [M – 3H]3-, 445.11; found, 444.79.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-4-O-benzyl-3,6-di-O-sulfo-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3-O-benzyl-6-O-sulfo-α-D-glucopyranoside (66):
pale yellow foam, 84% yield (20.3 mg).
1H NMR (600 MHz, CD3OD) δ: 7.54 (d, J = 7.1 Hz, 2H), 7.42 (t, J = 7.1 Hz, 3H), 7.32 –f 7.13 (m, 10H), 5.56 (d, J = 3.0 Hz, 1H), 5.37 (d, J = 11.8 Hz, 1H), 5.18 (d, J = 3.5 Hz, 1H), 5.14 (d, J = 9.5 Hz, 1H), 5.01 (d, J = 8.1 Hz, 1H), 4.87 (dd, J = 9.4, 8.1 Hz, 2H), 4.73 – 4.66 (m, 2H), 4.62 (dd, J = 13.1, 10.8 Hz, 2H), 4.35 (dd, J = 10.5, 2.1 Hz, 1H), 4.30 (dd, J = 10.9, 3.1 Hz, 1H), 4.14 (ddd, J = 13.2, 11.1, 2.4 Hz, 3H), 4.04 (d, J = 9.5 Hz, 1H), 3.89 – 3.81 (m, 3H), 3.78 – 3.72 (m, 2H), 3.71 – 3.66 (m, 1H), 3.64 – 3.53 (m, 5H), 3.36 (ddd, J = 25.1, 10.4, 2.9 Hz, 2H), 3.21 (dd, J = 8.4, 3.8 Hz, 2H), 2.01 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 170.43, 168.64, 139.29, 138.97, 138.74, 128.92, 128.05, 128.00, 127.69, 127.60, 127.51, 126.96, 126.76, 126.66, 100.23, 98.82, 97.43, 81.42, 78.64, 77.73, 76.84, 76.26, 74.95, 74.90, 74.60, 73.76, 72.73, 70.59, 69.19, 67.02, 65.39, 65.30, 57.49, 57.35, 51.94, 43.16, 19.79. Waters ZQ ESI-TOF MS (m/z): calcd for C44H55N3O34S5 [M – 4H]4-, 340.57; found, 340.45.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,4-di-O-benzyl-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3,6-di-O-sulfo-α-D-glucopyranoside (67):
pale yellow foam, 76 % yield (15.2 mg).
1H NMR (600 MHz, CD3OD) δ: 7.62 (d, J = 7.1 Hz, 2H), 7.40 (d, J = 7.2 Hz, 2H), 7.32 – 7.13 (m, 11H), 5.57 (s, 1H), 5.15 – 5.01 (m, 3H), 4.72 (ddd, J = 32.0, 20.1, 9.7 Hz, 4H), 4.60 (d, J = 8.9 Hz, 1H), 4.48 (t, J = 7.1 Hz, 1H), 4.28 (d, J = 8.6 Hz, 1H), 4.25 – 4.21 (m, 1H), 4.15 (dd, J = 8.9, 4.1 Hz, 3H), 4.02 (s, 1H), 3.94 (d, J = 9.6 Hz, 1H), 3.86 – 3.80 (m, 1H), 3.79 – 3.67 (m, 5H), 3.56 (ddd, J = 45.6, 15.3, 7.5 Hz, 4H), 3.42 (dd, J = 10.2, 3.3 Hz, 1H), 3.26 – 3.19 (m, 3H), 1.98 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 170.18, 168.62, 138.61, 138.24, 129.22, 127.98, 127.94, 127.82, 127.76, 127.73, 127.69, 127.65, 127.10, 127.02, 126.83, 100.08, 97.60, 97.58, 79.98, 77.21, 74.74, 74.47, 70.98, 70.17, 69.62, 68.51, 67.09, 65.79, 60.12, 58.50, 57.96, 53.64, 52.20, 43.14, 19.71.Waters ZQ ESI-TOF MS (m/z): calcd for C44H55N3O34S5 [M – 4H]4-, 340.57; found, 340.41.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-4-di-O-benzyl-3,6-di-O-sulfo-α-D-glucopyranosyl)-(1→4)-(methyl-2-O-acetyl-3-O-benzyl-β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-3,6-di-O-sulfo-α-D-glucopyranoside (68):
pale yellow foam, 73% yield (12.5 mg).
1H NMR (600 MHz, CD3OD) δ: 7.48 (d, J = 7.2 Hz, 2H), 7.39 – 7.14 (m, 8H), 5.51 (d, J = 3.3 Hz, 1H), 5.15 – 5.10 (m, 1H), 5.06 (dd, J = 12.2, 6.8 Hz, 2H), 4.97 (d, J = 8.0 Hz, 1H), 4.78 (d, J = 10.9 Hz, 1H), 4.74 – 4.60 (m, 4H), 4.32 – 4.16 (m, 6H), 4.08 – 4.02 (m, 1H), 3.91 (d, J = 5.1 Hz, 2H), 3.88 – 3.82 (m, 1H), 3.77 (s, J = 9.2 Hz, 3H), 3.73 – 3.68 (m, 1H), 3.67 – 3.61 (m, 2H), 3.48 (dd, J = 9.9, 3.7 Hz, 1H), 3.21 (d, J = 4.8 Hz, 2H), 2.04 (s, 3H). 13C NMR (150 MHz, CD3OD) δ: 170.18, 169.32, 138.21, 137.65, 128.68, 128.02, 127.66, 127.48, 127.31, 127.19, 99.92, 95.19, 81.43, 77.08, 74.87, 74.60, 74.55, 74.19, 73.85, 71.81, 71.67, 70.81, 70.59, 70.16, 69.37, 67.92, 67.13, 65.98, 65.56, 65.36, 64.90, 61.53, 54.24, 53.92, 53.86, 52.72, 52.44, 42.96, 39.97, 19.68. Waters ZQ ESI-TOF MS (m/z): calcd for C37H49N3O37S7 [M – 4H]4-, 338.05; found, 337.88.
General Procedure for Global Deprotection
Step 1: To a solution of trisaccharide substrates, 58 – 68, (15~25 mg) in CH3OH (5 mL) were added Amberlite IR120 Na+ resin. After being stirred at room temperature for 24 h, the mixture was filtered and concentrated to afford the trisaccharide sodium salt quantitatively; Step 2: The resulting residue was then dissolved in a mixture of CH3OH/pH 7 Buffer (1:1, 3 mL), Pd(OH)2/C (20%, 3 times the weight of starting material) was added. After the reaction mixture had been stirring under 100 psi H2 at room temperature for 24 h, it was filtered and concentrated. The resulting residue was subjected to Sephadex LH-20 column chromatography (H2O) to give the corresponding product; Step 3: To a solution of the intermediate generated from step 2 in H2O (9 mL) was added 1 M LiOH (1 mL) at 0 °C. The reaction mixture was gradually warmed to room temperature and stirred for 24 h. The mixture was lypholized and loaded to to Sephadex LH-20 column and eluted with H2O. The fractions containing product were again charged with Na+ exchange resin for 24 h followed by filtration and lypholizing to deliver the final product.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D glucopyranosyl-uronate)-(1→4)-2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranoside (1α).
Semi white solid, 88% yield (28.5 mg).
1H NMR (600 MHz, D2O) δ: 5.48 (d, J = 3.7 Hz, 1H, H-1α), 5.01 (d, J = 3.4 Hz, 1H, H-1α’), 4.44 (d, J = 7.9 Hz, 1H, H-1β), 4.24 – 4.11 (m, 3H, H6a, H6b, H6a’), 3.99 (d, J = 10.6 Hz, 1H, H6b’), 3.88 (d, J = 9.2 Hz, 1H), 3.75 – 3.52 (m, 7H), 3.50 – 3.34 (m, 3H), 3.28 – 2.98 (m, 5H). 13C NMR (150 MHz, D2O) δ: 174.86, 102.00, 97.30, 97.00, 78.12, 76.30, 76.21, 76.00, 72.81, 70.98, 69.64, 69.55, 68.86, 68.22, 66.79, 66.18, 57.81, 57.21, 42.87. HR ESI-ORBI MS (m/z): calcd for C20H35N3O30S5 [M − 6Na + 4H]2-, 478.4960; found, 478.4961; calcd for C20H34N3O30S5Na [M − 5Na + 3H]2-, 489.4870; found, 489.4870; calcd for C20H33N3O30S5Na2 [M − 4Na + 2H]2-, 500.4779; found, 500.4781; C20H32N3O30S5Na3 [M − 3Na + 1H]2-, 511.4689; found, 511.4690; calcd for C20H31N3O30S5Na4 [M − 2Na]2-, 522.4599; found, 522.4599.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-uronate)-(1→4)-2-N-sulfo-2-deoxy-6-O-sulfo-β-D-glucopyranoside (1β).
Semi white soldi, 84% yield (15.9 mg).
1H NMR (600 MHz, D2O) δ: 5.48 (d, J = 3.6 Hz, 1H), 4.45 (dd, J = 8.2, 4.1 Hz, 2H), 4.25 – 4.14 (m, 3H), 4.00 (dd, J = 11.1, 1.9 Hz, 1H), 3.88 (dd, J = 10.1, 5.6 Hz, 1H), 3.74 – 3.53 (m, 8H), 3.47 – 3.40 (m, 2H), 3.21 (dd, J = 9.3, 8.1 Hz, 1H), 3.10 (dd, J = 10.0, 3.8 Hz, 1H), 3.06 – 2.98 (m, 2H), 2.92 (dd, J = 10.1, 8.4 Hz, 1H). 13C NMR (150 MHz, D2O) δ: 174.88, 101.85, 101.75, 97.28, 77.85, 76.30, 76.17, 76.03, 72.85, 72.50, 72.28, 70.97, 69.65, 68.88, 68.77, 66.21, 59.55, 57.80, 42.84. HR ESI-ORBI MS (m/z): calcd for C20H36N3O30S5 [M − 6Na + 5H]−, 957.9993; found, 957.9975; C20H35N3O30S5Na [M − 5Na + 4H]−, 979.9812; found, 979.9795; calcd for C20H34N3O30S5Na2 [M − 4Na + 3H]−, 1001.9632; found, 1001.9612; calcd for C20H33N3O30S5Na3 [M − 3Na + 2H]−, 1023.9451; found, 1023.9427; calcd for C20H32N3O30S5Na4 [M − 2Na + 1H]−, 1045.9270; found, 1045.9248; calcd for C20H31N3O30S5Na5 [M − 2Na]−, 1067.9090; found, 1067.9071.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-uronate)-(1→4)-2-N-sulfo-2-deoxy-α-D-glucopyranoside (2α).
Semi white solid, 87% yield (12.6 mg).
1H NMR (600 MHz, D2O) δ: 5.48 (d, J = 3.7 Hz, 1H), 5.03 (d, J = 3.6 Hz, 1H), 4.47 (d, J = 7.9 Hz, 1H), 4.22 (qd, J = 11.2, 3.0 Hz, 2H), 3.94 – 3.88 (m, 1H), 3.79 – 3.54 (m, 9H), 3.52 – 3.44 (m, 2H), 3.35 – 3.30 (m, 1H), 3.24 (dd, J = 9.3, 8.0 Hz, 1H), 3.17 (dd, J = 10.0, 3.7 Hz, 1H), 3.09 (dt, J = 8.6, 4.3 Hz, 3H). 13C NMR (150 MHz, D2O) δ: 174.93, 101.99, 97.30, 97.04, 78.12, 76.46, 76.30, 75.97, 72.89, 71.50, 71.15, 69.59, 68.24, 66.82, 66.27, 60.04, 58.00, 57.25, 42.91. HR ESI-ORBI MS (m/z): calcd for C20H36N3O27S4 [M − 5Na +4H]−, 878.0424; found, 878.0409; calcd for C20H35N3O27S4Na [M − 4Na +3H]−, 900.0244; found, 900.0229; calcd for C20H34N3O27S4Na2 [M − 3Na + 2H]−, 922.0063; found, 922.0048; calcd for C20H33N3O27S4Na3 [M − 2Na + H]−, 943.9883; found, 943.9864; calcd for C20H32N3O27S4Na4 [M − Na]−, 965.9702; found, 965.9681.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-uronate)-(1→4)-2-N-sulfo-2-deoxy-β-D-glucopyranoside (2β).
Semi white solid, 85% yield (7.7 mg).
1H NMR (600 MHz, D2O) δ: 5.46 (d, J = 3.7 Hz, 1H), 4.45 (dd, J = 8.1, 2.3 Hz, 2H), 4.25 – 4.21 (m, 1H), 4.16 (dd, J = 11.1, 4.4 Hz, 1H), 3.90 – 3.85 (m, 1H), 3.74 – 3.51 (m, 10H), 3.47 – 3.42 (m, 1H), 3.31 (t, J = 9.7 Hz, 1H), 3.21 (dd, J = 9.3, 8.1 Hz, 1H), 3.10 – 2.97 (m, 3H), 2.95 – 2.89 (m, 1H). 13C NMR (150 MHz, D2O) of δ: 174.95, 101.80, 101.73, 97.27, 77.81, 76.43, 76.26, 75.97, 72.89, 72.48, 72.28, 71.48, 71.10, 69.54, 68.77, 66.24, 60.00, 59.57, 57.97, 42.84. HR ESI-ORBI MS (m/z): calcd for C20H35N3O27S4Na [M − 4Na + 3H]−, 900.0244; found, 900.0229; calcd for C20H34N3O27S4Na2 [M − 3Na + 2H]−, 922.0063; found, 922.0049; calcd for C20H33N3O27S4Na3 [M − 2Na + 1H]−, 943.9883; found, 943.9867; calcd for C20H32N3O27S4Na4 [M − 2Na]−, 965.9702; found, 965.9684.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranoside (3α).
Semi white solid, 86% yield (9.6 mg).
1H NMR (600 MHz, D2O) δ: 5.49 (d, J = 3.6 Hz, 1H), 5.04 (d, J = 3.5 Hz, 1H), 4.47 (d, J = 7.9 Hz, 1H), 4.28 – 4.17 (m, 2H), 3.91 (d, J = 9.1 Hz, 1H), 3.79 – 3.54 (m, 9H), 3.52 – 3.44 (m, 2H), 3.33 (t, J = 9.7 Hz, 1H), 3.27 – 3.22 (m, 1H), 3.17 (dd, J = 9.9, 3.5 Hz, 1H), 3.09 (dt, J = 9.1, 4.6 Hz, 3H). 13C NMR (150 MHz, D2O) δ: 173.56, 100.61, 95.93, 95.67, 76.74, 75.09, 74.94, 74.60, 71.52, 70.14, 69.77, 68.21, 66.87, 65.46, 64.89, 58.68, 56.62, 55.88, 41.54. HR ESI-ORBI MS (m/z): calcd for C20H36N3O27S4 [M − 5Na + 4H]−, 878.0425; found, 878.0405; calcd for C20H35N3O27S4Na [M − 4Na + 3H]−, 900.0244; found, 900.0224; calcd for C20H34N3O27S4Na2 [M − 3Na + 2H]−, 922.0063; found, 922.0044; calcd for C20H33N3O27S4Na3 [M − 2Na + 1H]−, 943.9883; found, 943.9862; calcd for C20H32N3O27S4Na4 [M − Na]−, 965.9702; found, 965.9678.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(2-O-sulfo-β-D gluco-pyranosyluronate)-(1→4)-2-N-sulfo-2-deoxy-6-O-sulfo- α-D-glucopyranoside (4α).
Semi white solid, 83% yield (7.1 mg).
1H NMR (600 MHz, D2O) δ: 5.51 (d, J = 3.6 Hz, 1H), 5.03 (d, J = 3.6 Hz, 1H), 4.62 (d, J = 7.6 Hz, 1H), 4.47 – 4.41 (m, 1H), 4.22 (dd, J = 11.1, 2.1 Hz, 1H), 4.10 (d, J = 9.6 Hz, 1H), 4.06 – 3.96 (m, 2H), 3.91 – 3.81 (m, 2H), 3.80 – 3.68 (m, 4H), 3.66 – 3.58 (m, 2H), 3.54 – 3.38 (m, 3H), 3.16 (dd, J = 10.0, 3.6 Hz, 1H), 3.13 – 3.01 (m, 3H). 13C NMR (150 MHz, D2O) δ: 174.74, 99.94, 97.68, 97.08, 79.76, 77.69, 76.30, 74.94, 71.15, 69.80, 69.33, 68.91, 68.16, 66.81, 66.23, 65.78, 57.82, 57.24, 42.81. HR ESI-ORBI MS (m/z): calcd for C20H34N3O33S6Na [M − 6Na + 4H]2-, 529.4654; found, 529.4646; calcd for C20H33N3O33S6Na2 [M − 5Na + 3H]2-, 540.4563; found, 540.4555; calcd for C20H32N3O33S6Na3 [M − 4Na + 2H]2-, 551.4473; found, 551.4464; calcd for C20H31N3O33S6Na4 [M − 3Na + 1H]2-, 562.4383; found, 562.4373; calcd for C20H30N3O33S6Na5 [M − 2Na]2-, 573.4293; found, 573.4282.
2-N-sulfo-ethyl(2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-uronate)-(1→4)-2-acetamido-2-deoxy-6-O-sulfo-α-D-glucopyranoside (5α).
Semi white solid, 86% yield (5.8 mg).
1H NMR (600 MHz, D2O) δ: 5.51 (d, J = 3.5 Hz, 1H), 4.74 (d, J = 3.3 Hz, 1H), 4.46 (d, J = 7.9 Hz, 1H), 4.21 (d, J = 9.5 Hz, 3H), 4.02 (d, J = 10.5 Hz, 1H), 3.95 (d, J = 9.8 Hz, 1H), 3.86 – 3.67 (m, 7H), 3.58 (d, J = 9.4 Hz, 1H), 3.48 – 3.43 (m, 3H), 3.28 – 3.22 (m, 1H), 3.13 (dd, J = 9.9, 3.7 Hz, 1H), 3.06 (t, J = 5.1 Hz, 2H), 1.90 (s, 3H). 13C NMR (150 MHz, D2O) δ: 174.74, 174.32, 102.10, 97.29, 96.83, 78.41, 76.27, 76.13, 76.01, 72.84, 71.01, 69.66, 69.38, 68.90, 68.49, 66.70, 66.26, 66.22, 57.82, 52.98, 42.90, 21.84. HR ESI-ORBI MS (m/z): calcd for C22H38N3O28S4 [M − 5Na + 4H]−, 920.0530; found, 920.0513; calcd for C22H37N3O28S4Na [M − 4Na + 3H]−, 942.0350; found, 942.0331; calcd for C22H36N3O28S4Na2 [M − 3Na + 2H]−, 964.0169; found, 964.0151; calcd for C22H35N3O28S4Na3 [M − 2Na + 1H]−, 985.9988; found, 985.9967; calcd for C22H34N3O28S4Na4 [M − Na]−, 1007.9808; found, 1007.9783.
2-N-sulfo-ethyl(2-acetamido-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-uronate)-(1→4)-2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranoside (6α).
Semi white solid, 91% yield (11.3 mg).
1H NMR (600 MHz, D2O) δ: 5.30 (d, J = 3.7 Hz, 1H), 5.04 (d, J = 3.6 Hz, 1H), 4.45 (d, J = 7.9 Hz, 1H), 4.30 – 4.14 (m, 3H), 4.04 (d, J = 9.7 Hz, 1H), 3.91 (d, J = 8.9 Hz, 1H), 3.83 – 3.53 (m, 9H), 3.48 (ddd, J = 32.5, 14.9, 7.8 Hz, 2H), 3.21 (t, J = 8.6 Hz, 1H), 3.17 (dd, J = 9.8, 3.6 Hz, 1H), 3.08 (dd, J = 9.9, 4.4 Hz, 2H), 1.92 (s, 3H). 13C NMR (150 MHz, D2O) δ: 173.55, 173.00, 100.62, 95.71, 95.46, 76.69, 75.06, 75.00, 74.39, 72.11, 69.13, 68.58, 68.20, 67.61, 66.88, 65.48, 64.86, 64.82, 55.89, 52.07, 41.55, 20.49. HR ESI-ORBI MS (m/z): calcd for C22H38N3O28S4 [M − 5Na +4H]−, 920.0530; found, 920.0514; calcd for C22H37N3O28S4Na [M − 4Na +3H]−, 942.0350; found, 942.0332; calcd for C22H36N3O28S4Na2 [M − 3Na +2H]−, 964.0169; found, 964.0151; calcd for C22H35N3O28S4Na3 [M − 2Na +1H]−, 985.9988; found, 985.9969; calcd for C22H34N3O28S4Na4 [M − Na]−, 1007.9808; found, 1007.9785.
2-N-sulfo-ethyl(2-N-sulfo-2-deoxy-3,6-di-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-uronate)-(1→4)-2-N-sulfo-2-deoxy-3,6-di-O-sulfo-α-D-glucopyranoside (7α).
Semi white solid, 80% yield (5.3 mg).
1H NMR (600 MHz, D2O) δ: 5.34 (d, J = 2.9 Hz, 1H), 4.81 (d, J = 3.1 Hz, 1H), 4.48 (d, J = 7.7 Hz, 1H), 4.35 (t, J = 9.5 Hz, 1H), 4.30 – 4.18 (m, 4H), 4.04 – 3.96 (m, 2H), 3.83 – 3.74 (m, 3H), 3.70 – 3.62 (m, 4H), 3.56 – 3.50 (m, 3H), 3.29 – 3.25 (m, 1H), 3.09 (s, 2H). 13C NMR (150 MHz, D2O) δ: 175.08, 100.96, 98.76, 98.24, 82.34, 79.99, 77.15, 76.97, 73.03, 72.62, 69.93, 69.46, 67.30, 66.73, 66.22, 66.09, 54.56, 54.23, 42.88. HR ESI-ORBI MS (m/z): calcd for C20H33N3O30S5Na2 [M – 2SO3 - 4Na + 2H]2-, 500.4779; found, 500.4772; calcd for C20H32N3O30S5Na3 [M – 2SO3 - 3Na + 1H]2-, 511.4689; found, 511.4682; calcd for C20H35N3O33S6Na2 [M – 1SO3 - 7Na + 5H]2-, 518.4744; found, 518.4760; calcd for C20H31N3O30S5Na4 [M – 2SO3 - 2Na]2-, 522.4599; found, 522.4591; calcd for C20H34N3O33S6Na [M – 1SO3 - 6Na + 4H]2-, 529.4654; found, 529.4669.
2-N-sulfo-ethyl (2-N-sulfo-2-deoxy-3,6-di-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-uronate)-(1→4)-2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranoside (8α).
Semi white solid, 88% yield (7.8 mg).
1H NMR (600 MHz, D2O) δ: 5.47 (d, J = 3.5 Hz, 1H), 5.04 (d, J = 3.6 Hz, 1H), 4.47 (d, J = 7.9 Hz, 1H), 4.23 (dd, J = 13.3, 7.1 Hz, 4H), 4.04 (d, J = 9.4 Hz, 1H), 3.95 – 3.89 (m, 1H), 3.84 (d, J = 10.2 Hz, 1H), 3.79 – 3.71 (m, 3H), 3.67 – 3.56 (m, 4H), 3.52 – 3.46 (m, 1H), 3.29 – 3.24 (m, 2H), 3.18 (dd, J = 10.1, 3.6 Hz, 1H), 3.09 (dd, J = 8.8, 4.7 Hz, 2H). 13C NMR (150 MHz, D2O) δ: 174.73, 102.11, 98.23, 97.01, 78.40, 77.95, 76.30, 75.82, 72.69, 69.84, 69.61, 68.26, 67.78, 66.81, 66.34, 66.08, 57.23, 56.64, 42.90. HR ESI-ORBI MS (m/z): calcd for C20H34N3O33S6Na [M − 6Na + 4H]2-, 529.4654; found, 529.4647; calcd for C20H33N3O33S6Na2 [M − 5Na + 3H]2-, 540.4563; found, 540.4556; calcd for C20H32N3O33S6Na3 [M − 4Na + 2H]2-, 551.4473; found, 551.4465; calcd for C20H31N3O33S6Na4 [M − 3Na + 1H]2-, 562.4383; found, 562.4374; calcd for C20H30N3O33S6Na5 [M − 2Na]2-, 573.4293; found, 573.4282.
2-N-sulfo-ethyl(2-N-sulfo-2-deoxy-6-O-sulfo-α-D-glucopyranosyl)-(1→4)-(β-D-glucopyranosyl-ronate)-(1→4)-2-N-sulfo-2-deoxy-3,6-di-O-sulfo-α-D-glucopyranoside (9α).
Semi white solid, 82% yield (6.4 mg).
1H NMR (600 MHz, D2O) δ: 5.51 (d, J = 3.6 Hz, 1H), 5.03 (d, J = 3.5 Hz, 1H), 4.45 (d, J = 9.5 Hz, 1H), 4.22 (d, J = 10.6 Hz, 1H), 4.09 (d, J = 9.7 Hz, 1H), 4.06 – 3.98 (m, 2H), 3.89 – 3.82 (m, 2H), 3.72 (dt, J = 16.6, 9.5 Hz, 4H), 3.63 (t, J = 8.4 Hz, 2H), 3.53 – 3.42 (m, 3H), 3.18 – 3.05 (m, 4H). 13C NMR (150 MHz, D2O) δ: 174.74, 99.93, 97.67, 97.06, 79.75, 77.68, 76.29, 74.93, 71.14, 69.79, 69.32, 68.90, 68.15, 66.80, 65.76, 57.81, 57.23, 42.80. HR ESI-ORBI MS (m/z): calcd for C20H34N3O33S6Na [M − 6Na + 4H]2-, 529.4654; found, 529.4646; calcd for C20H33N3O33S6Na2 [M − 5Na +4H]2-, 540.4563; found, 540.4555; calcd for C20H32N3O33S6Na3 [M − 4Na + 2H]2-, 551.4473; found, 551.4464; calcd for C20H31N3O33S6Na4 [M − 3Na + 1H]2-, 562.4383; found, 562.4374; calcd for C20H30N3O33S6Na5 [M − 2Na]2-, 573.4293; found, 573.4282.
IC50 Determination Using the TR-FRET Heparanase Inhibition Assay.
42 μL of HS trisaccharide solution in Milli-Q water (0.0038–500 μM) or just Milli-Q water (as a control), and 42 μL of heparanase (5.3 nM, R&D Systems) solution in pH 7.5 triz buffer (consisting of 20 mM TrisHCl, 0.15 M NaCl and 0.1% CHAPS) or just buffer as blank were added into microtubes and pre-incubated at 37 °C for 10 min bringing the [heparanase] to 0.5 nM. Next, 84 μL of biotin-heparan sulfate-Eu cryptate (Cisbio, Cat #: 61BHSKAA) (58.6 ng in pH 5.5 0.2 M NaOAc buffer) was added to the microtubes, and the resulting mixture was incubated for 60 min at 37 °C. The reaction mixture was stopped by adding 168 μL of Streptavidin-XLent! (Cisbio, Cat #: 611SAXLA) (1.0 μg/ml) solution in pH 7.5 dilution buffer made of 0.1 M NaH2PO4, 0.8 M KF, 0.1% BSA. After the mixture had been stirring at room temperature for 15 min, 100 μL (per well) of the reaction mixture was transferred to a 96 well microplate (Corning #3693 96 well, white polystyrene, half-area) in triplicates and HTRF emissions at 616 nm and 665 nm were measured by exciting at 340 nm using a SpectraMax iD5 Microplate Reader (Molecular Devices).
Computational Docking.
In the docking study, the enzyme structure was inherited from previously modified apo heparanase structure (PDB code: 5E8M).11, 33 The ligand saccharide backbone was obtained from Glycam GAGs builders (www.glycam.com), then the sulfation patterns and aliphatic portion was modified on GaussView 5.0.61 The modified ligand pdb files were then subjected to energy minimization in YASARA, and saved in .yob format. Each ligand was docked separately into heparanase within the simulation cell (80 × 61 × 76 Å) using Autodock VINA36 default parameters. The trisaccharide ligand (1α−9α, 1β, and 2β) and heparanase were subjected to 100 docking runs, where AMBER14 force field62 was applied to the protein, and GLYCAM0663 and GAFF/Am1BCC force filed was applied to each ligand. During docking study, the ligands and receptor binding pocket residues were kept flexible. The most populated clusters of the docking runs was subjected to MD simulation.
Trisaccharides Hydrolysis Assay.
Micro centrifuge tubes were pretreated for 2h at 37 °C by using a solution of phosphate buffered saline comprising of 0.05% Tween 20 (PBST) and 4% bovine serum albumin. Tubes were then washed three times with PBST, dried, stored at 4 °C, and used for assay within a week. A 100 μM solution of trisaccharide 1α, 1β or 2β with heparanase enzyme (3 nM) in pH 5.0 sodium acetate buffer (40 mM) were incubated at 37 °C in a microtube. The enzymatic reaction was stopped at different time points by adding equal volumes of reaction mixture and freshly made 1.69 mM WST‐1 (Toronto Research Chemicals) in 0.1 M NaOH into a new microtube and developed at 60 °C for 60 min. A 200 μl (per well) solution of developed reaction solution was transferred to a clear 96‐well microplate in triplicates and absorbance was measured at 584 nm with a SpectraMax iD5 Microplate Reader (Molecular Devices). For the controls, exactly the same procedure was followed for a 100 μM solution of trisaccharide 1α, 1β or 2β without heparanase enzyme to rationalize non‐enzymatic autohydrolysis.
Two-Stage Chromogenic Assay to Evaluate Anticoagulant Activity.
BIOPHEN Heparin AntiXa (2 stages) USP/EP (Cat #: A221005‐USP) kits from Aniara and BIOPHEN Heparin Anti‐IIa (2 stages) USP/EP (Cat #: A220005‐USP) were utilized to assess FXa and FIIa activity, respectively.
Factor Xa activity.
All the reagents for this assay were reconstituted and prepared according to the manufacturer’s instructions and incubated at 37 °C for 15 min. Different concentrations of heparin (0.002‐500 nM; 40 μL) or trisaccharide 1α (3.9‐4000 nM; 40 μL) and ATIII (0.04 IU; 40 μL) were added to a deep‐well block (Nunc 96 DeepWell 1.0 mL/well, clear), mixed, and incubated at 37 °C for 2 min. To the reaction mixture, FXa (0.32 μg; 40 μL) was added by multichannel pipette and was incubated at 37 °C for another 2 min (stage 1), then FXa specific chromogenic substrate (0.048 mmol; 40 μL) was added. The reaction was stopped by adding citric acid (240 μL; 20 g/L) exactly after 2 min. A 100 μL solution was then transferred to a clear 96‐well microplate in triplicate, and absorbance at 405 nm was measured with a SpectraMax iD5 Microplate Reader (Molecular Devices). The sample blank was measured by mixing the reagents in reverse order from that of the test, i.e. citric acid, FXa substrate, FXa, ATIII, and sample. The sample blank value was deducted from the absorbance measured for the corresponding assay.
Factor IIa activity.
All the reagents for this assay were reconstituted and prepared according to the manufacturer’s instructions and incubated at 37 °C for 15 min. Different concentrations of heparin (0.002‐500 nM; 40 μL) or trisaccharide 1α (3.9‐4000 nM; 40 μL) and ATIII (0.01 IU; 40 μL) were added to a deep‐well block (Nunc 96 DeepWell 1.0 mL/well, clear), mixed, and incubated at 37 °C for 2 min. To the reaction mixture, FIIa (1.2 nkat; 40 μL) was added by multichannel pipette and was incubated at 37 °C for another 2 min (stage 1), then FIIa specific chromogenic substrate (0.048 mmol; 40 μL) was added. The reaction was stopped by adding citric acid (240 μL; 20 g/L) exactly after 2min. 100 μL was transferred to a 96‐well microplate in triplicate, and absorbance at 405 nm was measured with SpectraMax iD5 Microplate Reader (Molecular Devices). The sample blank was measured by mixing the reagents in reverse order from that of the test, i.e. citric acid, FXa substrate, FXa, ATIII, and sample. The sample blank value was deducted from the absorbance measured for the corresponding assay.
Kd Determination Using Platelet Factor 4 Binding Test by Biolayer Interferometry (BLI) Assay.34
Assays were performed on an Octet Red Instrument (fortéBIO) at 25 °C. Immobilization and binding analysis were carried out at 1000 rpm using HBS-EP buffer [10 mM HEPES, pH 7.4, 150 mM NaCl, 3.0 mM EDTA, and 0.005% (v/v) surfactant tween20]. A solution affinity assay, used to determine affinities of ligands by SPR analysis was adopted to BLI.46 In this method, PF 4 (100 nM; 100 μL) was mixed with various concentrations of trisaccharide 1α (0.08‐250 μM; 100 μL) or heparin (0.4‐400 nM; 100 μL). Free protein in this equilibrium mixture is tested for binding against immobilized heparin (all proteins are carrier-free and purchased from R&D Systems). Heparin-biotin (Creative PEGworks, 18 kDa, 1 biotin per HP polymer), 5 μg /mL was immobilized on to streptavidin biosensors (fortéBio) for 5 min. Binding experiments were carried under conditions of mass transport. Binding was fitted to equation 1 of ref 10 using Graphpad Prism. BLI response was used in place of F and ligand (heparin /trisaccharide 1α) concentration was used in place of [metal].
Inhibition of ECM Degradation Assay.
Briefly, sulfate [35S] labeled ECM coating the surface of 35 mm tissue culture dishes,49 is incubated (5 h, 37°C, pH 6.0, 1 ml final volume) with recombinant human heparanase (0.5 ng/ml) in the absence and presence of trisaccharide 1α. The reaction mixture contains: 50 mM NaCl, 1 mM DTT, 1 mM CaCl2, and 10 mM buffer Phosphate-Citrate, pH 6.0. To evaluate the occurrence of proteoglycan degradation, the incubation medium is collected and applied for gel filtration on Sepharose 6B columns (0.9 × 30 cm). Fractions (0.2 ml) are eluted with PBS and counted for radioactivity. The excluded volume (Vo) is marked by blue dextran and the total included volume (Vt) by phenol red. Degradation fragments of HS side chains, characterized as previously described,50 are eluted from Sepharose 6B at 0.5 < Kav < 0.8 (fractions 20–35). Results are best represented by the actual gel filtration pattern.64–66 Each experiment was performed at least three times and the variation of elution positions (Kav values) did not exceed ±15%.
Inhibition of Heparanase in Pancreatic β Cells:
Culture of mouse pancreatic β cell line MIN6 and detection of cellular mitochondrial ROS using mitochondrial ROS trackers.
Mouse pancreatic β cell line (insulinoma) MIN6 was cultured as we previously described.67 Specifically, MIN6 cells were cultured with high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), 50μM β-mercaptoethanol, and antibotics. Before experiments, MIN6 cells were seeded in 4-well slide-chambers for immunofluorescence analyses. When the cells reached approximately 50% confluency, they were cultured in the conditioned medium from heparanase-producing CHO cells or control CHO cells. Heparanase-producing CHO cells were provided by Dr. Israel Vlodavsky (Technion, Israel). Meanwhile, the cells were treated with vehicle (PBS) or the trisaccharide 1α inhibitor for 24 h. The cells were then stained with mitochondrial ROS tracker probe using the concentration of 100 nM according to the manufacturer’s instruction (Molecular Probes, Invitrogen). Mitochondrial ROS in the MIN6 cells were visualized by fluorescence microscopy, and the extents and intensities of fluorescent activities, which reflect intracellular mitochondrial ROS, were quantified using ImageJ software.
Quantitative real-time PCR (qPCR):
Total RNA from Min-6 cells was extracted using Trizol by following manufacture’s instruction and complementary DNA was synthesized from 500 ng of total RNA to with High Capacity cDNA Reverse Transcription Kit (purchased from Applied Biosystems). The abundance of mRNA was measured by q PCR analysis using the SYBR Green PCR Master mix (also from Applied Biosystems). The sequences of the primers for SOD1: 5’-GCGGTGAACCAGTTGTGTTGTC-3’ and 5’-CAGTCACATTGCCCAGGTCTCC-3’; and for ERO1α: 5’-ATGGACTGTGTTGGCTGCTT-3’ and 5’-GCTTTCCGGCATATTTGCGA-3’.
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health (R01 GM098285) and Wayne State University. The authors acknowledge the Wayne State Proteomic Core and Chemistry Lumigen Instrument Center for assistance. The authors would also like to thank Professor Jian Liu at the University of North Carolina at Chapel Hill for providing HS oligosaccharides 69 and 70.
ABBREVIATION USED
| DCM | dichloromethane |
| DMF | N,N-dimethylformamide |
| DBU | diazabicyclo[5.4.0]undec-7-ene |
| Cl3CCN | trichloroacetonitrile |
| Et3N | trimethylamine |
| DIPEA | diisopropylethylamine |
| TFA | trifluoroacetic acid |
| DDQ | 2,3-dichloro-5,6-dicyano-p-benzoquinone |
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.
Molecular docking, synthetic schemes, biological assays, NMR spectra and mass spectra (PDF).
Molecular formula strings (CSV).
Access Codes
PBD code for human heparanase is 5E8M.
The authors declare no competing financial interests.
Supporting Information Availability: Include Molecular Formula Strings
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1021/acs.jmedchem.0c00156
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc7376576?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1021/acs.jmedchem.0c00156
Article citations
Trauma promotes heparan sulfate modifications and cleavage that disrupt homeostatic gene expression in microvascular endothelial cells.
Front Cell Dev Biol, 12:1390794, 24 Jul 2024
Cited by: 0 articles | PMID: 39114570 | PMCID: PMC11303185
Chemical Synthesis of Δ-4,5 Unsaturated Heparan Sulfate Oligosaccharides for Biomarker Discovery.
Org Lett, 26(12):2462-2466, 18 Mar 2024
Cited by: 0 articles | PMID: 38498917 | PMCID: PMC10985652
Heparan Sulfate-Mimicking Glycopolymers Bind SARS-CoV-2 Spike Protein in a Length- and Sulfation Pattern-Dependent Manner.
ACS Med Chem Lett, 14(10):1411-1418, 29 Sep 2023
Cited by: 1 article | PMID: 37849547 | PMCID: PMC10577887
Role of heparanase in pulmonary hypertension.
Front Pharmacol, 14:1202676, 11 Aug 2023
Cited by: 0 articles | PMID: 37637421 | PMCID: PMC10450954
Review Free full text in Europe PMC
Rational Design and Expedient Synthesis of Heparan Sulfate Mimetics from Natural Aminoglycosides for Structure and Activity Relationship Studies.
Angew Chem Int Ed Engl, 62(32):e202304325, 29 Jun 2023
Cited by: 0 articles | PMID: 37285191 | PMCID: PMC10527013
Go to all (7) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(3 citations)
PDBe - 5E8MView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Multi-faceted substrate specificity of heparanase.
Matrix Biol, 32(5):223-227, 13 Mar 2013
Cited by: 48 articles | PMID: 23499529
Review
Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol splitting.
J Biol Chem, 280(13):12103-12113, 12 Jan 2005
Cited by: 130 articles | PMID: 15647251
Unraveling the specificity of heparanase utilizing synthetic substrates.
J Biol Chem, 285(19):14504-14513, 24 Feb 2010
Cited by: 46 articles | PMID: 20181948 | PMCID: PMC2863188
Molecular Aspects of Heparanase Interaction with Heparan Sulfate, Heparin and Glycol Split Heparin.
Adv Exp Med Biol, 1221:169-188, 01 Jan 2020
Cited by: 1 article | PMID: 32274710
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (1)
Grant ID: R01 GM098285
National Institute of General Medical Sciences (1)
Grant ID: R01 GM098285