Abstract
Free full text

Systematic analysis of lysine acetylome and succinylome reveals the correlation between modification of H2A.X complexes and DNA damage response in breast cancer
Abstract
Abnormal protein acetylation and succinylation in lysine residues can cause the initiation and development of numerous different types of tumors. However, to the best of our knowledge, there is currently a lack of systematic investigation in breast cancer. Using proteomic techniques, the present study systematically investigated the two modifications of all proteins in invasive ductal carcinoma tissues to identify potential targets. The results revealed significantly higher modification levels for the majority of proteins in breast cancer tissue when compared with para-carcinomous normal tissue. The bioinformatic analysis demonstrated that either highly acetylated or succinylated proteins were significantly enriched in histone H2A.X (H2A.X) complexes and nucleophosmin (NPM1) may be the key member among them. The results of further analyses revealed that H2A.X complexes were associated with DNA damage response (DDR), and the proteomic results for protein quantification provided further evidence for the abnormal DDR condition in breast cancer tissues. Later, the western blotting results validated the high acetylation and succinylation levels of the majority of proteins, including the modification of NPM1 and its correlation with cell viability. Finally, the upregulation of H2A.X in breast cancer tissues further demonstrated the association between H2A.X complex modification and DDR in breast cancer. Overall, the present study systematically investigated the protein acetylation and succinylation in breast cancer and provided evidence to support H2A.X complexes as potential targets. These results broaden the horizon for breast cancer investigation and link it with epigenetics.
Introduction
Recently, breast cancer has become the primary cause of mortality among women worldwide (1,2). Although gene mutations have been regarded as the main cause of breast cancer and a number of these mutations have been identified (3–5), numerous setbacks have occurred during the investigation into cancer and effective therapy remains to be determined. With the development of epigenetics, abnormal modification of genes and proteins can also affect a number of biological processes in breast cancer cells without any gene mutations and have been regarded as another important pathogenesis of breast cancer (6–9).
Protein acetylation and succinylation can cause significant changes in the alteration, structure and function of proteins, and are closely associated with the formation and development of a number of different types of tumors (10–12). At present, acetylation has been one of the most focused-on modifications, and many studies have demonstrated that enzymes regulating acetylation and deacetylation are involved in apoptosis, proliferation, aging and certain other biological processes of breast cancer (13–15), indicating the close association between protein acetylation and breast cancer. However, these enzymes can simultaneously modulate the acetylation or deacetylation of many proteins (16,17), thus these studies cannot determine the key player among them. Therefore, it is essential to determine and fully understand the level of protein acetylation in breast cancer and to identify the key targets among them. Lysine succinylation has been discovered in recent years (18) and has been reported to be involved in the initiation and development of a number of different types of tumors, such as lung and gastric cancer (19,20). In addition to the aforementioned types of cancer, Liu et al revealed that succinylation may play a significant role in regulating pentose phosphate and endoplasmic reticulum protein processing pathway (21). However, this result remains to be confirmed. Recently, an increasing number of studies have demonstrated that numerous acetylated lysine sites can also be succinylated (22,23). Meanwhile, various enzymes that regulate acetylation can also regulate succinylation (19,24–26), indicating the similarity in function between protein acetylation and succinylation. Thus, it is essential to investigate protein acetylation and succinylation collaboratively.
The present study used proteomic techniques to detect the overall protein acetylation and succinylation levels in breast cancer tissues and adjacent normal tissues (hereinafter referred to as normal tissues). The results revealed that the acetylation and succinylation levels of the majority of proteins in breast cancer tissue were significantly higher than those in normal tissue. Further bioinformatic analyses indicated that the modified proteins were significantly enriched in three H2A.X complexes, and that nucleophosmin (NPM1) may be a key player in these complexes. The function of the H2A.X complexes in the DNA damage response (DDR) revealed via biological process analyses, and the abnormal DDR statues determined by protein quantification linked the modification of H2A.X complexes and DDR in breast cancer. Finally, the western blotting results demonstrated high acetylation and succinylation levels in the majority of proteins, including the modification of NPM1 and its correlation between cell viability. In addition, the high expression level of H2A.X in breast cancer tissues further suggested a certain association between the modification of H2A.X complexes and DDR in breast cancer. In conclusion, the results of the present study highlighted the importance of protein acetylation and succinylation in the initiation and development of breast cancer, and revealed the potential target: H2A.X complexes and its member NPM1. These results provide a foundation for the investigation of protein acetylation and succinylation, and provide new research targets for breast cancer.
Materials and methods
Sample acquisition
Ten pairs of tumor tissue and normal tissue samples were obtained from patients at the Second Affiliated Hospital of Qiqihar Medical University (Qiqihar, Heilongjiang), who were diagnosed with invasive ductal breast carcinoma from January 2018 to December of 2018 but who received no previous chemotherapy or radiotherapy. The age range of the patients was from 38 to 64 years old and their average age was 51.9 years old. The tissue samples were immediately stored in liquid nitrogen. All methods were performed in accordance with relevant guidelines. All experimental protocols were approved by the Qiqihar Medical Ethics Committee, approval no. [2018]27. Patients provided informed consent for the use of their samples.
Proteomic detection
Protein extraction and digestion
Tissue samples were ground quickly and dissolved in lysis buffer [8 M urea, 1% Triton X-100, 65 mM DTT, 1% Protease Inhibitor Cocktail, 3 µM TSA (Trichostatin A), 50 mM NAM (nicotinamide), 50 mM Tris-HCl] for sonication three times using a high intensity ultrasonic processor (Scientz). After centrifugation for 10 min at 12,000 × g and 4°C, the proteins in the supernatant were precipitated by 15% trichloroacetic acid (TCA) for 2 h at −20°C and centrifuged for 10 min at 4°C. The precipitate was then washed with cold acetone and dissolved in 8 M urea, 100 mM triethylamine borane (TEAB), pH 8.0. Prior to digestion, the protein solution was reduced in 10 mM DTT for 1 h at 37°C and then alkylated with 20 mM iodoacetamide (IAA) for 45 min in the dark at room temperature. After adding 100 mM TEAB to reduce the urea concentration to <2 M, trypsin at a ratio of 1:50 trypsin-to-protein (w/w) was added into the solution for the first digestion overnight at 37°C, and a second digestion at a ratio of 1:100 trypsin-to-protein (w/w) for 4 h at 37°C followed.
Tandem mass tag (TMT) labeling
Tryptic peptide samples were desalted using a Strata X C18 SPE column (Phenomenex) and reconstituted in 0.5 M TEAB. The TMT labeling was performed according to the manufacturer's protocol for the 6-plex TMT kit. For the acetylation and succinylation assessment, 126- and 127-TMT labeling reagents were used for the cancer and normal tissue labeling, respectively. For the quantitative study, three cancer tissues were labeled using 126-, 127-, 128-TMT labeling reagents, and three normal tissues were labeled using 129-, 130-, 131-TMT labeling reagents. After labeling, equal amounts of sample from each group were mixed. The peptide mixtures were then desalted and dried by vacuum centrifugation.
Affinity enrichment for acetylated and succinylated proteins
Affinity enrichment was applied for collecting the acetylated and succinylated peptides. First, tryptic peptides were dissolved in NP-40 lysis buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl and 0.5% NP-40, pH 8.0) and incubated with pre-washed corresponding antibody beads (PTM Biolabs) at 4°C overnight with gentle shaking. The beads were then washed four times with NP-40 lysis buffer and twice with ddH2O. The eluting buffer was selected as 0.1% TFA and the eluted fractions were combined and vacuum-dried. After desalting using C18 ZipTips (EMD Millipore), the samples were ready to undergo high performance liquid chromatography (HPLC) separation.
Separation of peptides
Peptides for the acetylation and succinylation analysis were separated using an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific, Inc.). Samples were first dissolved in 0.1% formic acid (FA) and loaded onto a reverse-phase pre-column (Acclaim PepMap 100; Thermo Fisher Scientific, Inc.), and then separated in solvent B by loading onto a reverse-phase analytical column (Acclaim PepMap RSLC; Thermo Fisher Scientific, Inc.). The solvent B gradient was set as follows: 6 to 22% over 24 min; 22 to 36% for 8 min; 80% for 4 min; and finally 80% for 4 min; all performed at a constant flow rate of 280 nl/min.
Peptides for quantification were separated via high pH reverse-phase HPLC with an Agilent 300Extend C18 column (Agilent). Briefly, the peptides were first separated in a gradient from 2 to 60% acetonitrile in 10 mM ammonium bicarbonate, pH 10.0 over 80 min. Meanwhile, the solution was collected every 1 min. A total of 80 fractions were obtained and then combined into 18 fractions and dried by vacuum centrifuging.
MS/MS analysis
MS/MS analysis for separated peptides was performed by Q Exactive™ Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Inc.) equipped with an NSI (Nanospray Ionization) source. The parameters were set as follows: Resolution for intact and ion fragment peptides was 70,000 and 17,500, respectively; normalized collision energy was set at 30%; electro spray voltage was set to 2.0 kV; threshold of ion count in MS survey scan was set to 2E4 under a 30 sec dynamic exclusion; automatic gain control was 5E4; the m/z scan range was from 350–1,800; and the first fixed mass was 100.
Database search
For the acetylation and succinylation experiments, the resulting MS/MS data was processed using MaxQuant (27) integrated with the Andromeda search engine (version 1.4.1.2) (28). Tandem mass spectra were searched for against the SwissProt_Human database (29) (20,203 sequences) concatenated with reverse decoy database. Trypsin/P was specified as a cleavage enzyme allowing up to 4 missing cleavages, 5 modifications per peptide and 5 charges. The mass error was set to 20 ppm for the first search, 5 ppm for the main search and 0.02 Da for the fragment ions. Carbamidomethylation on Cys was specified as fixed modification and oxidation on Met, acetylation and succinylation on Lys, and the protein N-terminal was specified as variable modifications. The false discovery rate (FDR) threshold for the protein, peptide and modification sites were specified at 1%. Minimum peptide length was set at 7. All the other parameters in MaxQuant were set to default values. The site localization probability was set as >0.75.
The Mascot search engine (version 2.3.0) was applied for the quantification analysis. The resulting MS/MS data were processed using the Mascot search engine (version 2.3.0). Tandem mass spectra were searched against the Uniprot_Human database (29) (8,274 sequences). Trypsin/P was specified as a cleavage enzyme allowing up to 2 missing cleavages. The mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys, TMT-6plex (N-term) and TMT-6plex (K) were specified as fixed modification, and oxidation on Met was specified as variable modifications. FDR was adjusted to <1% and peptide ion score was set as >20.
QC validation of MS data
The MS data validation are presented in Fig. S1. First, the mass error of all the identified peptides were checked. The distribution of mass error was near zero and the majority were <0.02 Da, which indicated that the mass accuracy of the MS data fit the requirement (Fig. S1A-C). Secondly, the lengths of the majority of the peptides were distributed between 8 and 20, which was consistent with the lengths of tryptic peptides (Fig. S1D-F), indicating that the sample preparation had met the standard.
Bioinformatic analysis
Complex enrichment analysis
Manually the curated CORUM protein complexes database (30) for humans was used for the protein complexes analysis. Enrichment complexes were identified using a hypergeometric test for each category of proteins. A two-tailed Fisher's exact test was employed to test the enrichment of the proteins containing SwissProt entries against all SwissPort human proteins. Correction for multiple hypothesis testing was performed using standard FDR control methods and complexes. Corrected P-value <0.05 was considered to indicate a statistically significant difference.
Analysis of H2A.X complexes
The interaction, Venn and biological process analyses of all members in H2A.X complexes were performed using FunRich software (version 3.1.3) (31). Briefly, all members in the H2A.X complexes were identified in the CORUM database (http://mips.helmholtz-muenchen.de/corum/) and the corresponding protein ID was loaded onto the Fun_Rich software. The Venn and biological process analyses were then performed in the background of the UniProt database. Biological process entries with a corrected P-value <0.05 were considered to indicate a statistically significant difference.
The sequence conservatism analysis of NPM1 was performed using MEGA software (version 7.0.26) (32). Briefly, the protein sequences of NPM1 from different species were identified in the NCBI database and saved in the Fasta file. The file was then loaded onto MEGA software and the sequences were aligned with ClustalW to analyze their conservatism.
Analysis of proteins associated with DDR
The present study used Functional Annotation Tool of DAVID Bioinformatics Resources 6.7 (33) to analyze the enriched Gene Ontology (GO) proteins that were upregulated (fold change≥1.5) or downregulated (fold change ≤1/1.5) in breast cancer tissues. The GO with a corrected P-value<0.05 was considered to be significantly different. The GO terms associated with DDR were listed and a heat map of the proteins in these terms was created using RGui software (version 3.4.2) (34).
Validation of proteomic results
Cell culture
MCF-7 and BT-549 cells were purchased from the Cell Bank of the Chinese Academy of Science. MCF-7 cells were cultured in MEM (HyClone) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare), 1 mM sodium pyruvate (Sigma-Aldrich; Merck KGaA) and 1.74 µM bovine insulin (Sigma-Aldrich; Merck KGaA). BT-549 cells were cultured in RPMI-1640 (HyClone; GE Healthcare) supplement with 10% fetal bovine serum (FBS), 14 mM glucose (Sigma-Aldrich; Merck KGaA), 1 mM sodium pyruvate and 0.023 IU/ml insulin (Sigma-Aldrich; Merck KGaA). Both cells were cultured in a humidified atmosphere of 5% CO2 at 37°C.
Antibodies
Human polyclonal antibodies against H2A.X and NPM1 were purchased from Abcam (cat. nos. ab10530 and ab20669, respectively), rabbit polyclonal antibody against pan-acetylation proteins of all species, and rabbit monoclonal antibodies against human GAPDH were purchased from Cell Signaling Technology (cat. nos. 9441 and 8884, respectively). Rabbit polyclonal antibodies against pan-succinylation proteins of all species were obtained from PTM Biolab (cat. nos. PTM-401), and the secondary antibodies against mouse IgG and rabbit IgG were purchased from Cell Signaling Technology (cat. nos. 7076 and 7074, respectively).
Plasmid constructs and transfection
Gene expression sequence of NPM1 was amplified using the designed primers (Table I) via Ex Taq HS enzyme (Takara). The PCR conditions were set as 94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min and the above step was repeated for 30 cycles finally followed by 72°C for 10 min. After that, the sequence was cloned into the PC-DNA3.0 and PCI2 vector respectively via T4 DNA ligase (TransGen Biotech) according to the manufacturer's instructions. The NPM1-K27R variant was constructed using PCI2-NPM1 as the template with Quick Mutation Site-Directed Mutagenesis Kit (Beyotime Institute of Biotechnology) according to the manufacturer's instructions; the primers used are presented in Table I. All above plasmids were transfected via Liposomal Transfection Reagent (HAN Biotechnology) according to the manufacturer's instructions.
Table I.
Sequence of the primers for NPM1 and its variant NPM1-K27R.
| Gene | Primer sequences |
|---|---|
| NPM1 | Forward: CCAAGCTTTGCCGCCACCCGATGGAAGATTC |
| Reverse: GGGGTACCTTAAAGAGACTTCCTCCACTGCC | |
| NPM1-K27R | Forward: GGTTGTGAACTAAAGGCCGACAGAGATTATCACTTTAAGGTGG |
| Reverse: CCACCTTAAAGTGATAATCTCTGTCGGCCTTTAGTTCACAACC |
NPM1, nucleophosmin.
Protein extraction and western blotting
Cells and tissues were collected and dissolved in RIPA lysis buffer (Beyotime Biotechnology) and the proteins were extracted according to the manufacturer's protocol. Proteins were separated via 12% SDS-PAGE and transferred to nitrocellulose filters (EMD Millipore). The membranes were cut into small pieces according to the protein ladder (Thermo Scientific, Inc.) and the parts with the target proteins were reserved. After that, the membranes were blocked with 5% non-fat milk in TBST buffer (20 mM Tris-HCl, pH 7.6, 150 mM NaCl and 0.05% Tween-20) for 1 h at room temperature and then incubated overnight at 4°C with the following primary antibodies: H2A.X (dilution 1:500), NPM1 (dilution 1:500), pan-acetylation (dilution 1:1,000), pan-succinylation (dilution 1:1,000), GAPDH (dilution 1:1,000). The membranes were then washed with TBST buffer and incubated with horseradish peroxidase-conjugated secondary antibody (dilution 1:2,500) for 30 min at room temperature. After extensive washing, the proteins were detected using an Electrochemiluminescence-Plus kit (CoWin Biotechnology) and imaged using a Gel-imaging system (Bio-Rad) with Image Lab Software (version 5.2; Bio-Rad Laboratories).
Cell viability detection
BT-549 cells were seeded into a 96-well at a density of 0.8×104 cells/well. After being cultured for 24 h, the cells were transfected into control vector PCI2, NPM1 and its variant NPM1-K27R with Liposomal Transfection Reagent (HAN Biotechnology) respectively and the culture was continued for 36 h. After that, 20 µl/well MTT (5 mg/ml; Sigma-Aldrich, Merck KGaA) was added into the cells and incubated for 4 h. Finally, the supernatant was removed and 150 µl/well DMSO was added (Sigma-Aldrich, Merck KGaA) and the absorbance was detected at a wavelength of 570 nm using a microscope reader (Olympus Corp.).
Statistical analysis
Enrichment analysis was performed using a Fisher' exact test. Average modification level analysis between breast cancer and normal tissue was performed by Wilcoxon Signed Rank Test. Cell viability analysis was performed by Student's t-test. The significance level was set at P<0.05 and P<0.01. Error bars denote the standard deviation.
Results
High protein acetylation and succinylation levels in breast cancer tissue
Using TMT labeling and affinity enrichment followed by high-resolution LC-MS/MS analysis, quantitative lysine acetylation and succinylation analyses were performed in the present study. Altogether, 364 lysine acetylation sites from 229 proteins and 392 succinylation sites from 226 proteins were identified, among which 267 acetylation sites in 167 proteins and 257 succinylation sites in 130 proteins were quantified; both the acetylation and succinylation levels showed a statistically significant difference (Fig. 1A and Fig. S2). Among the quantified proteins, when setting 1.5 as the fold threshold change in modification levels (cancer vs. normal tissue), 170 acetylation sites in 113 proteins and 134 succinylation sites in 94 proteins with higher acetylation and succinylation levels (Fig. 1A), respectively, reached 68 and 72% of all identified proteins (fold change≥1.5, Fig. 1B and C). However, only 33 acetylation sites in 13 proteins and 20 succinylation sites in 15 proteins exhibited lower acetylation and succinylation levels (Fig. 1A), accounting for 8 and 12% of all identified proteins (fold change ≤1/1.5; Fig. 1B and C). All the quantified protein sites and their modification levels in breast cancer and normal tissues are presented in Fig. 1D and E. Among the proteins whose acetylation or succinylation levels were altered more than 1.5 fold, 41 of them were increased in both of these two modifications (Fig. 1F), 16 of the 41 proteins changed in the same lysine sites (Table SI); 7 of the proteins were decreased in both of these two modifications (Fig. 1G), 4 of the 7 proteins changed in the same lysine sites (Table SII). These results suggest that protein acetylation and succinylation may work together and play an important role in the initiation and development of breast cancer.
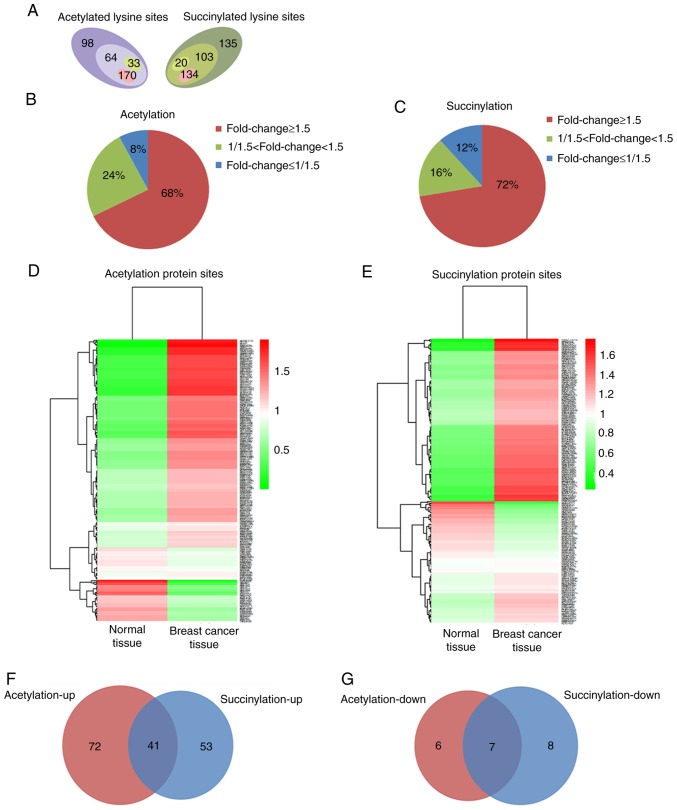
Acetylation and succinylation data of proteins in breast cancer and adjacent normal tissues. (A) Numbers of protein sites whose acetylation and succinylation were identified (sum of all numbers respectively) and quantified (acetylation: 64+170+33; succinylation: 103+20+134) in cancer and adjacent normal tissues, as well as protein sites whose modification levels were upregulated (acetylation: 170; succinylation: 134) and downregulated (acetylation: 33; succinylation: 20) in breast cancer tissue. (B and C) Statistics of the changes in modification levels of quantified proteins when setting 1.5 as the significant fold change (cancer vs. normal). (D and E) All quantified protein sites and their modification levels in breast cancer and adjacent normal tissues. (F) Venn diagram of proteins with significantly higher acetylation and succinylation levels in breast cancer tissue. (G) Venn diagram of proteins with significant lower acetylation and succinylation level in breast cancer tissue.
Highly acetylated or succinylated proteins are enriched in H2A.X complexes
The present study performed bioinformatic analyses in order to determine the potential common functions of the acetylation and succinylation proteins in the breast cancer tissue. The protein complex enrichment analysis revealed that both high acetylation and succinylation proteins in breast cancer tissue were enriched in numerous complexes, such as H2A.X, HCF-1, Nop56p-associated pre-rRNA complexes and others (Fig. 2A and B). Due to the small protein number, the low modification proteins in breast cancer tissue were only observed to be enriched in 5 complexes, including SERPINA1-CTSG, SERPINA1-ELA2, LRP-1-α-2-M-annexin VI, DJ-1-SNCA and PPD complexes (Fig. 2C and D). It is worth noting that either highly acetylated or succinylated proteins in breast cancer were most commonly enriched in three H2A.X complexes (Fig. 2A and B), and proteins with lower modification levels were both more commonly enriched in the SERPINA1-CTSG and SERPINA1-ELA2 complexes (Fig. 2C and D), indicating that the protein acetylation and succinylation may exert similar functions in these complexes. In order to investigate the function of highly modified proteins, H2A.X complexes were selected for further analysis in the present study.
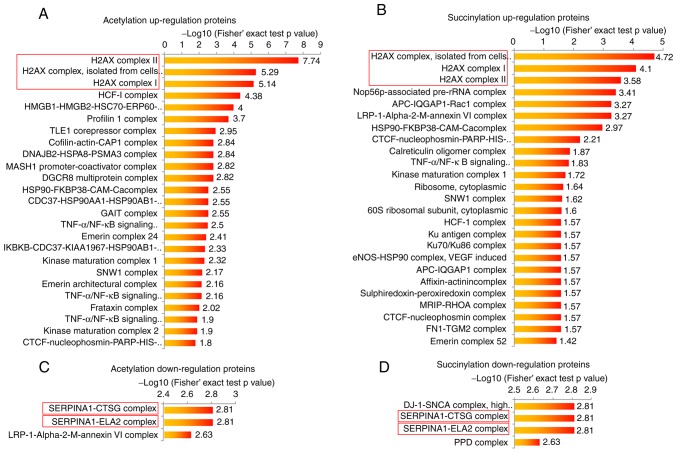
Protein complex enrichment analysis of acetylation and succinylation proteins. (A and B) Complex enrichment analysis of proteins with significant higher (A) acetylation and (B) succinylation level in breast cancer tissue. (C and D) Complex enrichment analysis of proteins with significant lower (C) acetylation and (D) succinylation levels in breast cancer tissue.
Acetylation and succinylation of H2A.X complexes may affect DDR in breast cancer
The present study further analyzed the function of three H2A.X complexes. It has been reported that the three H2A.X complexes are formed in different ionizing radiation conditions, which can cause DDR (35). Meanwhile, the further analysis demonstrated that all members in the H2A.X complexes were enriched in certain biology processes, such as nucleosome assembly, DNA repair, cellular senescence and cellular response to γ radiation (Fig. 3A), all of which are closely associated with DDR (36–38). This result indicated that acetylation and succinylation of proteins in H2A.X complexes may affect DDR in breast cancer. For a comprehensive understanding of the DDR condition in breast cancer, the present study quantified all protein expression levels in breast cancer tissues and compared them with those in normal tissues. Proteins whose expression level changed significantly (fold change≥1.5 or fold change≤1/1.5) in breast cancer tissues are presented in Tables SIII and SIV. The GO analysis of these proteins revealed those biological process entries that were associated with DDR (P<0.05; Fig. 3B and C), corresponding proteins and their expression levels in breast cancer and normal tissues, and these are presented in Fig. 3D and E. Among them, the proteins involved in DNA repair were significantly upregulated, and those proteins that were associated with the response to certain stimuli that can induce DNA damage were significantly downregulated. These results indicated an abnormal form of breast cancer that may be associated with the abnormal modification of H2A.X complexes.
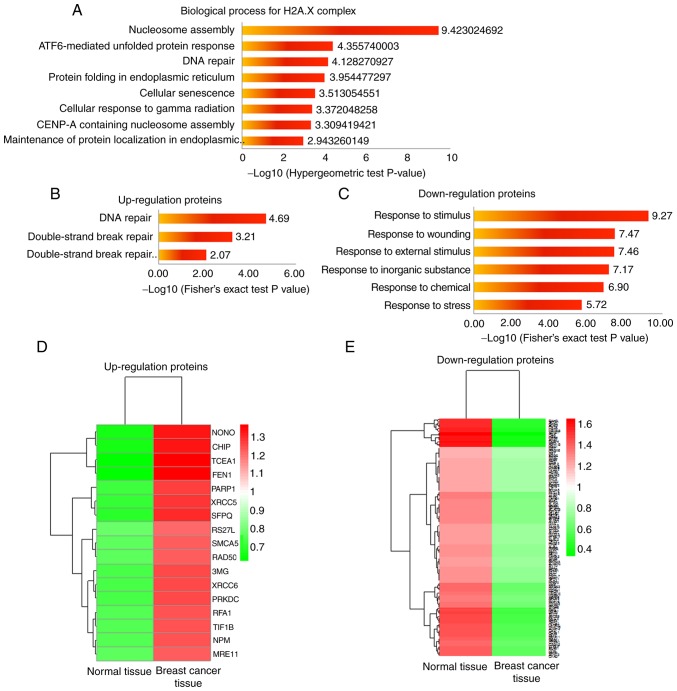
Acetylation and succinylation of H2A.X-complexes may affect DDR in breast cancer. (A) Biological process analysis of H2A.X-complexes; the first eight entries with a corrected P-value<0.05 are presented. (B) Biological process entries associated with DDR (P<0.05) of proteins with significantly higher expression levels in breast cancer tissues (fold change≥1.5 compared with adjacent normal tissues). (C) Biological process entries associated with DDR (P<0.05) of proteins with significantly lower expression levels in breast cancer tissues (fold change≤1/1.5 compared with adjacent normal tissues). (D and E) Relative expression levels of proteins corresponding to B and C in breast cancer tissues and adjacent normal tissues. H2A.X, histone H2A.X; DDR, damage response.
NPM1 may play an important role in H2A.X complexes
The present study then performed further analyses of H2A.X complexes in order to determine the key factors among them. The CORUM database revealed the 19 proteins in the H2A.X complexes, the majority of which can interact with H2A.X (encoded by H2A.FX) directly, and a few of them interact with H2A.X indirectly by combining with other proteins in these complexes (Fig. 4A, among the 19 proteins, Histone H4 can be encoded by three kinds of genes: HIST1H4C, HIST1H4A and HIS4H4, so there are 21 gene members in the figure). Histone H4 (encoded by HIST1H4A), Histone H2Btype1B (HIST1H2BB) and Histone H2Btype3B (HIST3H2BB) were highly acetylated, and CALM was highly succinylated. It is worth noting that NPM1, HSPA5 and CALR were upregulated in both their acetylation and succinylation (Fig. 4A). In addition, the Venn analysis revealed that NPM1, CALR and HSPA5 were also the common members of the three H2A.X complexes (Fig. 4B). The results indicated that the modification of these three proteins may play an important role in H2A.X complexes. The acetylation and succinylation at the lysine sites of proteins in the H2A.X complexes are presented in Fig. 4C and D; notably, the 27th lysine site of NPM1 was highly acetylated and succinylated simultaneously and it is a highly conserved amino acid site in a number of different species (Fig. 4E). All the aforementioned results indicated that NPM1 may play a more important role in H2A.X complexes.
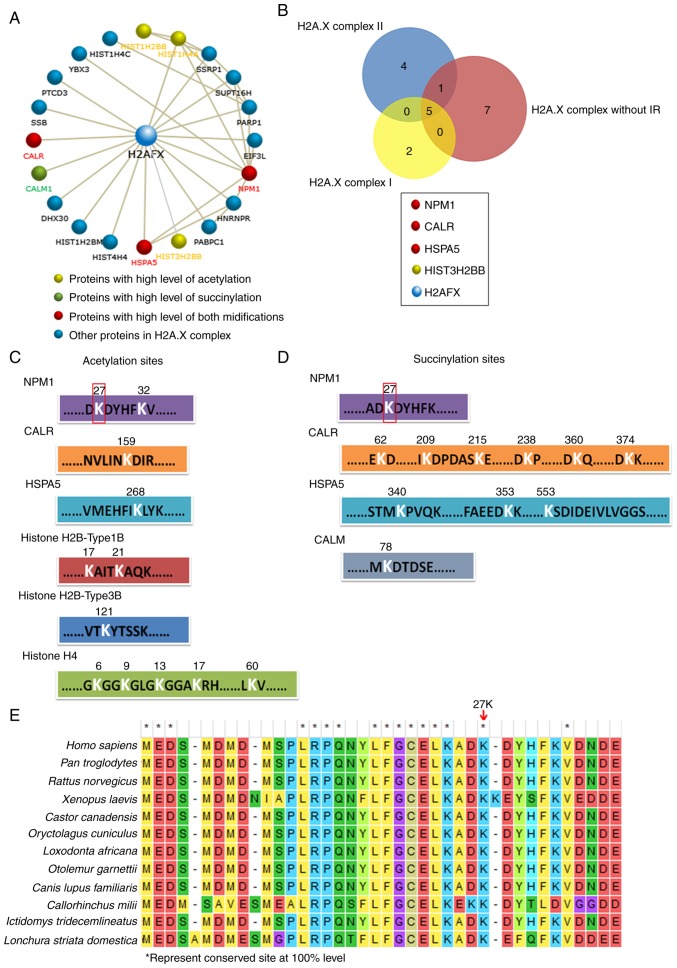
NPM1 may play an important role in H2A.X-complexes. (A) All members and their interactions in H2A.X-complexes. The acetylation and succinylation proteins are highlighted with different colors. (B) Venn analysis of three H2A.X-complexes, the common members and their modification information are provided below. (C) Acetylation and (D) succinylation lysine sites of proteins in H2A.X-complexes. (E) Sequence conservatism analysis of 27th lysine sites of NPM1 in different species. *represent conserved site at 100% level. NPM1, nucleophosmin; H2A.X, histone H2A.X; CALR, calreticulin; HSPA5, endoplasmic reticulum chaperone BiP; HIST3H2BB, Histone H2B type 3-B; CALM, calmodulin-1.
Verification of proteomic results
Finally, the present study validated the proteomics results via western blotting. The results revealed that the acetylation and succinylation levels of the majority of proteins in breast cancer tissues were significantly higher than that of normal tissues (Fig. 5A and B). The acetylation and succinylation of 40 kDa protein weights indicated the obvious modification of NPM1 in breast cancer tissues (Fig. 5C). Meanwhile, the acetylation and succinylation levels of the 40 kDa proteins increased following the overexpression of NPM1 in MCF-7 and BT-549 cells (Fig. 5D and E). Moreover, when the 27th lysine was mutated into arginine, which cannot be acetylated or succinylated, the modification of the protein weight in 40 kDa was restored to the normal level (Fig. 5E). This provides further evidence to support the acetylation and succinylation of NPM1 in breast cancer cells. In addition, the overexpression of NPM1 increased the viability of BT-549 cells, while that of its variant NPM1-K27R lost this function (Fig. 5F). This indicated that the acetylation and succinylation can affect some function related to cell proliferation. Finally, it was revealed that H2A.X, the core member of the H2A.X complexes and the marker of DDR (39,40), was significantly upregulated in breast cancer tissues (Fig. 5G). This further proved that breast cancer is in an abnormal DDR condition, which may be associated with the modification of H2A.X complexes.
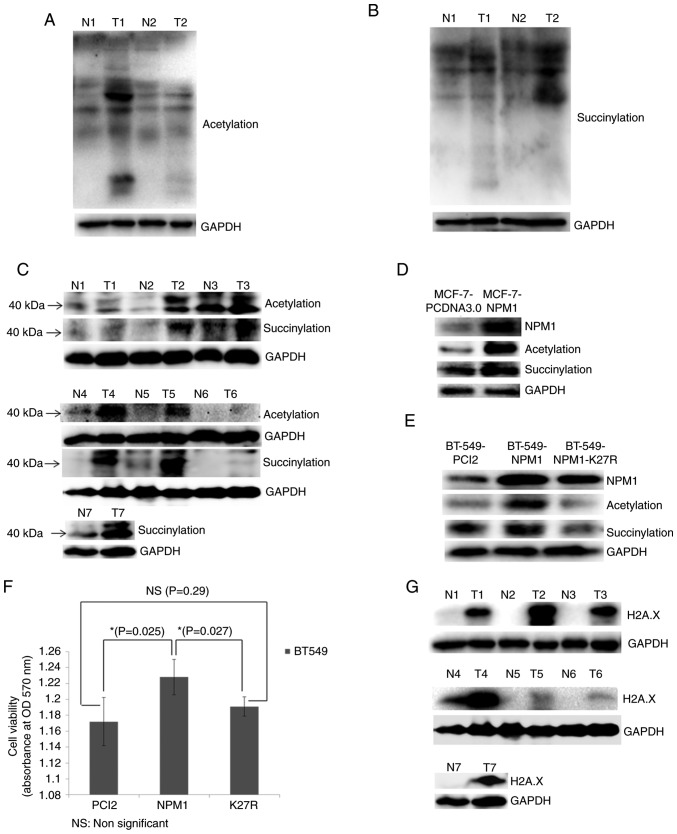
Validation of proteomic results. (A) Acetylation and (B) succinylation of whole proteins in breast cancer tissues (T) and adjacent normal tissues (N) examined via western blotting. GAPDH was used as a loading control. (C) Acetylation and succinylation of proteins ~40 kDa (used for indicating NPM1) in breast cancer tissues (T) and adjacent normal tissues (N) examined via western blotting. GAPDH was used as a loading control. (D) MCF-7 cells overexpressing NPM1 (MCF-7-NPM1) and PCDNA3.0 (MCF-7-PCDNA3.0, used as control) were collected for the protein extraction. Expression level of NPM1 and its acetylation and succinylation were detected via western blotting. GAPDH was used as a loading control. (E) BT549 cells overexpressing NPM1 (BT549-NPM1), PCI2 (BT549-PCI2, used as control) and the variants of NPM1 (NPM1-K27R, lysine mutated into arginine) were collected for protein extraction. Expression level of NPM1 and its acetylation and succinylation were detected via western blotting. GAPDH was used as a loading control. (F) Viability of BT549 cells overexpressing NPM1 (NPM1), PCI2 (PCI2, used as control) and the variants of NPM1 (K27R, lysine mutated into arginine) was detected via MTT methods. *P<0.05. (G) Expression of H2A.X in breast cancer tissues (T) and adjacent normal tissues (N) were detected via western blotting. GAPDH was used as a loading control. The H2A.X and the succinylation bands from N4 to T6 shows non-adjacent bands from the same gel, therefore they used the same GAPDH result. NPM1, nucleophosmin; H2A.X, histone H2A.X.
Discussion
Lysine acetylation and succinylation can change the function of proteins as well as their target genes and proteins without any gene mutation, thus, taking on another role with a genetic code that can play an important role in the initiation and development of a number of different types of tumors (9,41–43). However, the information regarding the majority of proteins, particularly for non-histone proteins, remains unknown. In addition, extensive overlap of protein acetylation and succinylation (22,23), as well as their common regulators, such as SIRT5 (19,25), SIRT7 (26) and KAT2A (24) have demonstrated similarity in their function. Thus, it is necessary to investigate protein acetylation and succinylation both simultaneously and systematically. Through proteomic investigation, the present study demonstrated a significantly higher acetylation and succinylation level in the majority of proteins in breast cancer, and observed a high repetition rate and a similar alteration tendency of the two kinds of modified proteins. These results proved that it is feasible to identify potential targets for breast cancer among the modified proteins. At present, some of the common functions of protein acetylation and succinylation have been reported, such as abnormal spermatogenesis, metabolism and DNA damage process. However, there is an urgent need for precise targets for cancer investigation. Our systematic analysis results can provide useful clues for the double-modified target proteins in breast cancer research. Moreover, compared with only one modification, double-modification can also narrow the scope of the regulators, which can help us to find or design potential anticancer drugs.
The enrichment analysis of the complexes provide evidence to support the common functions of protein acetylation and succinylation. The results revealed that both acetylation and succinylation proteins were most significantly enriched in three H2A.X complexes and they are involved in a number of processes associated with DNA damage response (DDR). This builds a connection between protein modification and DDR. It has previously been reported that H2A.X complexes are the dynamic foci formed at DNA break sites under different DNA damage conditions (35). When DNA is damaged, H2A.X is activated and then many other proteins are recruited to form protein foci to mediate DNA damage repair (40). What's more, this process can be regulated by protein modification (37,44,45). Thus, the over-acetylation and succinylation of proteins in H2A.X complexes may affect DNA damage repair by regulating the formation of protein foci in breast cancer. The results of the present study demonstrated that those proteins associated with DNA damage repair were upregulated, but those sensitive to DNA damage were downregulated, suggesting a resistance characteristic to DNA damage of breast cancer. This may be caused by the over-modification of H2A.X complexes and may help to explain the resistance to chemotherapy and radiotherapy for breast cancer.
The present study revealed that nucleophosmin (NPM1) may be a key player of the complexes, for the following reasons: i) NPM1 is one of the core members that were both acetylated and succinylated in the three H2A.X complexes; ii) NPM1 can also interact with numerous proteins in the three complexes, forming a second regulation center in these complexes; iii) NPM1 was the only protein whose acetylation and succinylation occurred on the same lysine site, which is highly conserved in a number of other different species; iv) upregulation of NPM1 expression and its modification in breast cancer further demonstrated the importance of NPM1 in H2A.X complexes. It has previously been reported that NPM1 is a type of histone chaperonin that can transfer histones to the chromosome and then stabilize the structure of the chromosome. However, the stable structure of the chromosome can form a barrier for the recognition of DNA damage and this can be regulated by the modification of NPM1. Thus, the acetylation and succinylation of NPM1 may affect DDR through the regulation of chromosome structure. In addition, NPM1 is a type of protein that undergoes a variety of post-translational modifications, such as phosphorylation, acetylation, sumoylation and so on, all of these can direct its various functions (46,47). Some of the modifications, such as its phosphorylation and acetylation, have been reported to play an important role in a number of different types of tumor (46,48–50). To the best of our knowledge, there have not yet been any reports regarding the succinylation of NPM1, and thus, the way in which its acetylation and succinylation work in breast cancer remains unknown. Thus, the results of the present study may provide a new potential target for breast cancer investigation and therapy.
Acknowledgements
We thank all the patients for their participation in the study and the support funding.
Glossary
Abbreviations
| H2A.X | histone H2A.X |
| NPM1 | nucleophosmin |
| DDR | DNA damage response |
| TMT | tandem mass tag |
| TEAB | triethylamine borane |
| TFA | trifluoroacetic acid |
| FA | formic acid |
| HPLC | high performance liquid chromatography |
| FDR | false discovery rate |
Funding
The present study was supported by the National Natural Science Foundation of China (nos. 81802834 and 81972491), the Education Department of Heilongjiang Province (no. 2017-KYYWF-0696), the Scientific Research Startup Foundation for Postdoctoral of Heilongjiang Province (no. LBH-Q17178), Qiqihar Science and Technology Bureau (no. SFGG-201912) and the Doctoral Research Startup Fund of Qiqihar Medical University (QY2016B-04).
Availability of data and materials
Data and materials are available upon request to the corresponding author.
Authors' contributions
LY conceived and designed the experiments. HB conducted the participant recruitment and the tissue collection. LZ conducted the data analysis. The experiments were performed by XG, WZ and LL. XG and HB wrote the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All experimental protocols were approved by Qiqihar Medical Ethics Committee [NO. (2018)27]. The patients provided informed consent for the use of tissue samples.
References
Articles from Oncology Reports are provided here courtesy of Spandidos Publications
Full text links
Read article at publisher's site: https://doi.org/10.3892/or.2020.7554
Read article for free, from open access legal sources, via Unpaywall:
https://www.spandidos-publications.com/10.3892/or.2020.7554/download
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.3892/or.2020.7554
Article citations
Acetylation of PGK1 at lysine 323 promotes glycolysis, cell proliferation, and metastasis in luminal A breast cancer cells.
BMC Cancer, 24(1):1054, 27 Aug 2024
Cited by: 0 articles | PMID: 39192221 | PMCID: PMC11348675
Systematic proteomics analysis revealed different expression of laminin interaction proteins in breast cancer: lower in luminal subtype and higher in claudin-low subtype.
Transl Cancer Res, 13(5):2108-2121, 29 May 2024
Cited by: 0 articles | PMID: 38881926 | PMCID: PMC11170511
HIST3H2A promotes the progression of prostate cancer through inhibiting cell necroptosis.
BMC Cancer, 24(1):544, 29 Apr 2024
Cited by: 1 article | PMID: 38684944 | PMCID: PMC11059659
Roles of Histone H2A Variants in Cancer Development, Prognosis, and Treatment.
Int J Mol Sci, 25(6):3144, 09 Mar 2024
Cited by: 2 articles | PMID: 38542118 | PMCID: PMC10969971
Review Free full text in Europe PMC
Lysine Acetylome of Breast Cancer-Derived Small Extracellular Vesicles Reveals Specific Acetylation Patterns for Metabolic Enzymes.
Biomedicines, 11(4):1076, 02 Apr 2023
Cited by: 4 articles | PMID: 37189694 | PMCID: PMC10135746
Go to all (11) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Regulation of DNA methyltransferase 1 transcription in BRCA1-mutated breast cancer: a novel crosstalk between E2F1 motif hypermethylation and loss of histone H3 lysine 9 acetylation.
Mol Cancer, 13:26, 06 Feb 2014
Cited by: 22 articles | PMID: 24502362 | PMCID: PMC3936805
Changes in the Acetylome and Succinylome of Bacillus subtilis in Response to Carbon Source.
PLoS One, 10(6):e0131169, 22 Jun 2015
Cited by: 72 articles | PMID: 26098117 | PMCID: PMC4476798
Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells.
BMC Cancer, 13:37, 29 Jan 2013
Cited by: 45 articles | PMID: 23356739 | PMCID: PMC3583671
Histone H2A ubiquitination in transcriptional regulation and DNA damage repair.
Int J Biochem Cell Biol, 41(1):12-15, 26 Sep 2008
Cited by: 41 articles | PMID: 18929679
Review




