Abstract
Free full text

The role of sequestosome 1/p62 protein in amyotrophic lateral sclerosis and frontotemporal dementia pathogenesis
Abstract
Amyotrophic lateral sclerosis and frontotemporal lobar degeneration are multifaceted diseases with genotypic, pathological and clinical overlap. One such overlap is the presence of SQSTM1/p62 mutations. While traditionally mutations manifesting in the ubiquitin-associated domain of p62 were associated with Paget’s disease of bone, mutations affecting all functional domains of p62 have now been identified in amyotrophic lateral sclerosis and frontotemporal lobar degeneration patients. p62 is a multifunctional protein that facilitates protein degradation through autophagy and the ubiquitin-proteasome system, and also regulates cell survival via the Nrf2 antioxidant response pathway, the nuclear factor-kappa B signaling pathway and apoptosis. Dysfunction in these signaling and protein degradation pathways have been observed in amyotrophic lateral sclerosis and frontotemporal lobar degeneration, and mutations that affect the role of p62 in these pathways may contribute to disease pathogenesis. In this review we discuss the role of p62 in these pathways, the effects of p62 mutations and the effect of mutations in the p62 modulator TANK-binding kinase 1, in relation to amyotrophic lateral sclerosis-frontotemporal lobar degeneration pathogenesis.
Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
Amyotrophic lateral sclerosis (ALS), the most common form of motor neuron disease, is a severe neurodegenerative disorder marked by progressive muscle weakness and paralysis caused by selective loss of motor neurons in the brain and spinal cord. The average survival time post-diagnosis is 2–3 years, with death most commonly caused by respiratory failure (Zarei et al., 2015). ALS can be classified as either sporadic, with no previously reported family history of the disease, or as familial. Frontotemporal lobar degeneration (FTLD), the third most common cause of dementia (Bang et al., 2015), is an umbrella term that encompasses a range of diseases marked by progressive loss of neurons in either one or both the frontal and temporal lobes (Mann et al., 1993; Rosen et al., 2002). FTLD patients generally present with behavioral and personality changes, and similarly to ALS, can be classified as either familial or sporadic (Seelaar et al., 2011). Originally considered unrelated diseases the two diseases are now thought to exist on a disease continuum due to pathological, genetic and clinical overlap (Neumann et al., 2006; Vance et al., 2006; Hasegawa et al., 2008; Lillo et al., 2012; Majounie et al., 2012).
While the main symptom of ALS is progressive muscle paralysis, up to 50% of patients will also develop cognitive impairment, and up to 15% of ALS patients meet the FTLD diagnostic criteria (Lomen-Hoerth et al., 2003; Ringholz et al., 2005). Conversely, up to 40% of FTLD patients exhibit ALS symptoms, while 15% meet the diagnostic criteria for ALS (Lomen-Hoerth et al., 2002). Additionally there is overlap in protein pathology. Pathological protein inclusions containing TAR DNA-binding protein 43 (TDP-43) or tau have been identified in both ALS and FTLD patients (Lippa et al., 2000; Morris et al., 2001; Yang et al., 2003; Neumann et al., 2006; Behrouzi et al., 2016). On a genetic level, while mutations in the superoxide dismutase 1 (SOD1) gene appear to be unique to ALS and mutations in MAPT (coding for tau) are identified only in FTLD patients (Bennion Callister and Pickering-Brown, 2014), commonly associated genes have been identified. For example, the hexanucleotide repeat expansion in chromosome 9 open reading frame 72 (C9ORF72) is the most common genetic cause of both ALS and FTLD, having been identified in 30–40% of familial ALS and up to 25% of FTLD cases (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Repeat expansions have also been identified in sporadic ALS and FTLD (Renton et al., 2011; Majounie et al., 2012).
Other genes implicated in both diseases include those coding for the autophagy receptors (ARs) sequestosome 1/p62 protein (SQSTM1/p62), optinuerin (OPTN), valosin-containing protein (VCP) and ubiquilin 2 (UBQLN2) (Synofzik et al., 2012; Le Ber et al., 2013; Kwok et al., 2014; Deng et al., 2017; Blauwendraat et al., 2018; Pottier et al., 2018), the function of these ARs is to sequester and remove old or damaged proteins and organelles via the autophagy-lysosome system. In addition to variants in ARs, mutations in tank-bingling kinase (TBK1), a kinase that modifies OPTN and SQSTM1/p62 function, have also been identified in ALS and FTLD (Freischmidt et al., 2015; Le Ber et al., 2015; Cui et al., 2018; McCombe et al., 2018). We searched for the terms p62 and SQSTM1 in combination with neurodegeneration, amyotrophic lateral sclerosis, frontotemporal degeneration, ALS and FTD. Searches were carried out on September 1st, 2019.
Sequestosome 1/p62 Protein in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
SQSTM1/p62, henceforth referred to as p62, is a scaffold protein that has roles in various signaling pathways and protein degradation. Loss of p62 enhances the rate of neurodegeneration in a number of disease models; p62 knockout mice exhibited increased accumulation of hyperphosphorylated tau and subsequent neurodegeneration (Babu et al., 2008), while absence of p62 in SOD1H46R mice lead to increased motor neuron degeneration (Hadano et al., 2016), enhanced α-synuclein pathology in a Lewy body disease mouse model (Tanji et al., 2015), and exacerbated motor phenotypes and neuropathological outcomes in a Spinal and bulbar muscular atrophy mouse model (Doi et al., 2013).
Mutations in the SQSTM1 gene are commonly identified in Paget’s disease of bone (PDB) (Laurin et al., 2002; Falchetti et al., 2004; Hocking et al., 2004), but have more recently been identified in both ALS and FTLD patients (Fecto et al., 2011; Rubino et al., 2012; Hirano et al., 2013; Le Ber et al., 2013; Chen et al., 2014; Bartolome et al., 2017; Blauwendraat et al., 2018). Unlike mutations associated with PDB, which cluster within the C-terminal ubiquitin-associated (UBA) domain, ALS and FTLD-associated mutations affect various domains throughout the entire protein (Additional Table 1). Further evidence of p62 involvement in ALS and FTLD is the observation of p62-positive inclusions in patient samples (Arai et al., 2003; Mann et al., 2013). While the role of ALS and FTLD-associated p62 variants and their contribution to disease onset and progression remains unclear, recent research demonstrates that pathogenic p62 mutant proteins alter signaling pathways involved in cell survival and differentiation.
Additional Table 1
SQSTM1/p62 variants identified in ALS or FTLD patients
| Mutation | Exon | Change (bp) | p62 Domain | ALS | FTLD | PDB | Controls |
|---|---|---|---|---|---|---|---|
| A16V | 1 | c.47C>T | – | Yesh | Yesh | – | – |
| A33V | 1 | c.98C>T | PB1 | Yesb | Yesbk | – | Yesh |
| A53T | 1 | c.157G>A | PB1 | Yesc | – | – | – |
| D80E | 1 | c.240C>G | PB1 | – | Yesh | – | – |
| E81K | 2 | c.241G>A | PB1 | Yesi | – | – | – |
| M87V | 2 | c.259A>G | PB1 | Yese | – | – | – |
| V90M | 2 | c.268G>A | PB1 | Yesdh | – | – | – |
| I99L | 2 | c.295A>C | PB1 | Yesf | – | – | – |
| K102E | 3 | c.304A>G | C-terminal to PB1 | Yesc | – | – | – |
| R107W | 3 | c.319C>T | – | Yesh | – | – | – |
| R110C | 3 | c.328C>T | – | – | Yesk | – | – |
| D129N | 3 | c.358G>A | ZZ | Yesh | – | – | – |
| V153I | 3 | c.457G>A | ZZ | Yesbd | – | – | Yesh |
| E155K | 3 | c.463G>A | ZZ | Yesg | – | – | |
| R212C | 4 | c.634C>T | – | Yesh | Yesh | – | – |
| G219V | 4 | c.656G>T | – | – | Yesh | – | – |
| S226P | 5 | c.676T>C | TBS | – | Yesh | – | – |
| P228L | 5 | c.683C>T | TBS | Yesb | – | – | Yesagk |
| P232T | 5 | c.694C>A | TBS | – | Yesh | – | – |
| V234V | 5 | c.702G>A | TBS | Yesb | – | – | – |
| K238E | 5 | c.712A>G | TBS | Yesa | Yesk | – | Yeshk |
| K238del | 5 | c.714–716delGAA | TBS | Yesb | – | – | Yesh |
| N239K | 5 | c.717C>A | TBS | Yesi | – | – | – |
| D258N | 6 | c.772G>A | – | Yesh | – | – | – |
| V259L | 6 | c.775G>C | – | – | Yesa | – | – |
| H261H | 6 | c.783C>T | Close to PEST1 | Yesb | – | – | Yesb |
| E274D | 6 | c.822G>C | PEST 1 | Yesagh | Yesahk | – | Yesahk |
| E280del | 6 | c.838-840delGAG | PEST1 | – | Yesh | – | – |
| P296P | 6 | c.888G>T | Close to PEST1 | Yesg | – | – | Yesg |
| G297S | 6 | c.889G>A | Close to PEST1 | Yesi | – | – | – |
| R312H | 6 | c.1029A>G | – | – | Yesk | – | – |
| S318P | 6 | c.952T>C | – | Yesb | – | – | |
| S318S | 6 | c.954C>T | – | Yesg | – | – | Yesg |
| E319K | 6 | c.955G>A | – | – | Yesa | – | Yesh |
| R321C | 6 | c.961C>T | – | Yesb | – | – | Yesh |
| R321H | 6 | c.962G>A | – | – | Yesh | – | – |
| D329G | 7 | c.986A>G | LIR | – | Yesh | – | – |
| D337E | 7 | c.1011C>G | LIR | Yesf | – | – | – |
| L341V | 7 | c.1021C>G | LIR | Yesf | – | – | – |
| K344E | 7 | c.1032A>G | Close to LIR/KIR | – | Yesa | – | – |
| V346V | 7 | c.1038G>A | KIR | Yesg | – | – | Yesg |
| P348L | 7 | c.1044C>T | KIR | Yesa | Yesh | – | – |
| S370P | 7 | c.1108T>C | PEST 2 | Yesb | – | – | Yesh |
| E372D | 7 | c.1116G>C | PEST2 | Yesi | – | – | – |
| A381V | 7 | c.1142C>T | Close to UBA | – | Yesk | Yes | – |
| P387L | 7 | c.1160C>T | Close to UBA | – | Yeshk | – | – |
| P388S | 7 | c.1162C>T | Close to UBA | Yesi | – | – | – |
| A390X | 7 | c.1165+1G>A | Intronic | Yese | – | Yes | – |
| P392A | 7 | c.1174C>G | UBA | – | Yesj | – | – |
| P392L | 8 | c.1175C>T | UBA | Yesbcghi | Yeshk | Yesk | Yeshk |
| G411S | 8 | c.1231G>A | UBA | Yesb | – | Yes | – |
| G425R | 8 | c.1273G>A | UBA | Yesb | – | Yes | Yesh |
| T430P | 8 | c.1288AA>C | UBA | – | Yesh | – | – |
| P438L | 8 | c.1313C>T | C-terminal region | Yesa | – | – | Yeseh |
| P439L | 8 | c.1316C>T | C-terminal region | Yesc | Yesh | – | Yesh |
ALS: Amyotrophic lateral sclerosis; FTLD: frontotemporal lobar degeneration; KIR: Keap1-interacting region; LIR: LC3-interacting region; PB1: Phox and Bem1p; PEST: proline, glutamate, serine and threonine rich sequence; TBS: TRAF6 binding sequence; UBA: ubiquitin-associated; ZZ: zinc finger. a: Rubino et al., 2012; b: Fecto et al., 2011; c: Hirano et al., 2013; d: Shimizu et al., 2013; e: Teyssou et al., 2013; f: Chen et al., 2014; g: Kwok et al., 2014; h: van der Zee et al., 2014; i: Yang et al., 2015; j: Blauwendraat et al., 2018; k: Le Ber et al., 2013.
p62 structure and function
The functions of p62 are executed through protein-protein interactions, facilitated by various domains (Figure 1). The role of p62 as an autophagy receptor is mediated through several domains; p62 first binds to ubiquitinated substrates via its UBA domain and subsequently binds to the autophagosome membrane-protein LC3 via its LC3-interacting region (LIR) (Pankiv et al., 2007). The PB1 domain of p62 mediates self-oligomerisation, which can play an important role in the degradation of specific substrates by either macroautophagy (Jain et al., 2010; Itakura and Mizushima, 2011) or the ubiquitin-proteasome system (UPS) (Seibenhener et al., 2004).
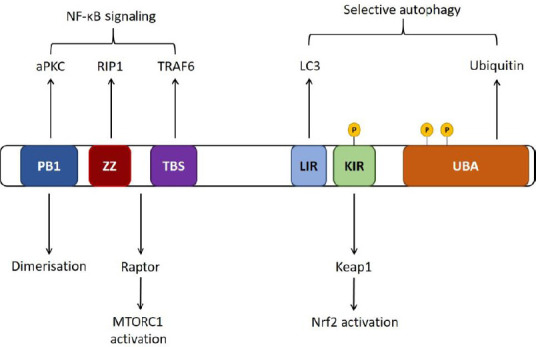
Schematic of p62 domains.
p62 self-oligomerizes via its PB1 domain. The LC3-interacting region (LIR) is required for p62 binding to the autophagosome membrane protein LC3, and p62 binds to ubiquitinated substrates via the ubiquitin associated (UBA) domain. p62 regulates Nrf2 signaling through interaction with inhibitory protein Keap1 via the Keap1 interacting region (KIR) domain, and regulates nuclear factor-kappa B (NF-κB) signaling via TRAF6 binding through the TRAF6 binding sequence (TBS) and interaction with RIP via the ZZ domain (Ichimura and Komatsu, 2018).
p62 also regulates several signaling pathways important for neuronal health, namely those that activate the transcription factors Nrf2 and nuclear factor kappa B (NF-κB). The Nrf2 pathway is the main response to oxidative stress in neurons and knockdown of Nrf2 leaves cells susceptible to neurotoxic insult (Ishii et al., 2000; Mimoto et al., 2012). Under normal conditions Kelch-like receptor protein 1 (Keap1) binds to and inhibits the action of the transcription factor, Nrf2. Upon cellular exposure to stress conditions, p62 expression is increased and p62 binds to Keap1 via the Keap1-interacting region (KIR) (Ishii et al., 2000; Copple et al., 2010; Jain et al., 2010; Lau et al., 2010). This promotes translocation of Nrf2 to the nucleus and subsequently transcription of antioxidant and protective genes such as Heme Oxygenase 1 and NAD(P)H quinone oxidase 1 (Itoh et al., 1997; Dinkova-Kostova et al., 2002; Kobayashi et al., 2004, 2006). The other main signaling pathway that p62 regulates is NF-κB, which is pathologically upregulated in ALS (Frakes et al., 2014). The role of p62 in this pathway has mainly been characterized with respect to mutations that cause PDB, a chronic and progressive skeletal disorder that involves increased osteoclastic bone resorption and deposition, leading to focal lesions of increased bone turnover. PDB-associated mutations primarily affect the UBA domain of p62, as either missense or truncating mutations that result in the removal of most or the entire domain. Very few PDB-associated mutations have been identified outside of the UBA domain, however those that have are located within approximately 50 residues of the UBA domain (Rea et al., 2014). PDB research shows that overexpressed mutant p62 proteins do not inhibit NF-κB, in contrast to over-expression of wild type p62 (Rea et al., 2006, 2009). The effect of increased NF-κB signaling due to SQSTM1/p62 mutations in an ALS and FTLD context could increase the production of pro-inflammatory cytokines with adverse effects on neuronal health (Shih et al., 2015).
It is important to note that p62 has been shown to be involved in positive regulation of NF-κB. Thus, p62 has a dual role in NF-κB regulation that may be dependent on p62 concentration or may be temporally regulated. The overall effect of these mutations on PDB pathogenesis is still under investigation, however there is strong evidence to suggest they contribute significantly to the disease, as patients with SQSTM1 mutations experience greater disease severity compared to patients without (Visconti et al., 2010; Rea et al., 2014). The main effect of SQSTM1/p62 mutations appears to be on autophagy (Hocking et al., 2004; Daroszewska et al., 2011) and NF-κB signaling (Najat et al., 2009; Rea et al., 2009), both of which play a role in the pathogenesis of ALS and FTLD.
p62 and proteostasis
The cell utilizes two systems to degrade misfolded or damaged proteins; the UPS and macroautophagy, or simply autophagy. The UPS degrades short-lived proteins, while autophagy degrades long-lived or aggregated proteins, bacteria, and organelles, both systems have the ability to degrade misfolded proteins, and both systems recognize ubiquitin as a marker for degradation. A major hallmark of ALS and FTLD is the accumulation of misfolded, ubiquitinated proteins in damaged neurons (Leigh et al., 1991; Bucciantini et al., 2002; Strong et al., 2005; Mackenzie and Neumann, 2016), indicating a role for malfunctioning protein degradation systems in pathogenesis. The accumulation of protein aggregates are linked with cell death (Giordana et al., 2010; Ticozzi et al., 2010; Brettschneider et al., 2014). p62 is a cargo protein for both systems (Seibenhener et al., 2004).
p62 and aggregate/inclusion body formation
p62 has been identified as a major component of aggregates from a number of neurodegenerative diseases; from neurofibrillary tangles in Alzheimer’s disease (Kuusisto et al., 2001a, 2002), and ubiquitin-positive inclusions in Parkinson’s disease (Kuusisto et al., 2003), FTLD (Kuusisto et al., 2002), and dementia with Lewy bodies (Kuusisto et al., 2001a), to SOD1 aggregates in ALS (Gal et al., 2007), and even Mallory bodies in hepatocytes (Zatloukal et al., 2002). While the presence of toxic protein aggregates inevitably leads to further damage to cellular organelles and subsequent cell death, it is possible that protein aggregate formation is an attempt at cellular protection. p62 was found to selectively form aggregates with mutant SOD1, but not wild-type SOD1, and these aggregates were not found to be directly toxic to the cell (Gal et al., 2007). Likewise, p62 was observed to form a shell around mutant huntingtin, and interference with p62 function lead to increased cell death (Bjorkoy et al., 2005). Depletion of p62 in spinal muscular bulbar atrophy mouse models exacerbated motor phenotypes via accumulation of toxic mutant androgen receptor protein, whereas increased p62 expression induced cytoprotective inclusion formation and subsequent improved phenotype (Doi et al., 2013). In SOD1H46R mice, p62 suppression lead to an increase in insoluble mutant SOD1 and ubiquitinated proteins (Hadano et al., 2016).
It is not clear whether p62 initiates aggregate formation, however it does appear to bind to toxic misfolded proteins in an attempt to isolate them from the cell, a process that is enhanced by substrate ubiquitination (Zatloukal et al., 2002; Gal et al., 2007). Thus, p62 may play a protective role in preventing further cell damage by toxic misfolded proteins, as cells that retain this p62 function exhibit increased survivability over cells that do not (Nakaso et al., 2004; Bjorkoy et al., 2005; Gal et al., 2007; Komatsu et al., 2007). On one hand p62 may bind to aggregates in order to prevent further accumulation of misfolded proteins to the aggregate (Zatloukal et al., 2002), or more likely, binds toxic proteins and holds them in a less active state in order to prevent them from causing further damage to the cell in their unbound form until clearance is possible.
p62, the ubiquitin proteasome system and neurodegeneration
The UPS facilitates the degradation of K48- and K63-linked polyubiquitinated substrates that are shuttled to the proteasome by cargo proteins (Seibenhener et al., 2004). UPS dysfunction is a hallmark of a number of neurodegenerative diseases (Keller et al., 2000; Seo et al., 2004; Bukhatwa et al., 2010), including ALS and FTLD (Bendotti et al., 2012; Kabashi et al., 2012; Myeku et al., 2016). Accumulation of UPS-specific substrates and proteins involved in UPS function into cellular inclusions provides evidence of UPS dysfunction (Farrawell et al., 2018). Further, aggregation of ALS-associated proteins leads to an accumulation of UPS-specific substrates, indicating that protein aggregation directly impedes UPS function (Farrawell et al., 2018) and deletion of a proteasome subunit, Rpt3, but not the critical autophagy protein Atg7, was sufficient to cause an ALS-like phenotype in mice (Tashiro et al., 2012). Thus, proteasome function may be critical to ALS and FTLD pathogenesis.
p62 is a cargo protein that shuttles the Alzheimer’s and FTLD-associated protein tau to the proteasome for degradation (Babu et al., 2005). Through the PB1 domain p62 directly interacts with the 26S proteasome (Seibenhener et al., 2004; Cohen-Kaplan et al., 2016), whereas the UBA domain of p62 recognizes ubiquitinated substrates. Thus it is possible that a mutation affecting either domain may impede p62-mediated degradation of substrates via the proteasome. Mutations to the UBA domain of p62, some of which have been identified in ALS and FTLD patients, reduce or abolish p62 ubiquitin-binding (Cavey et al., 2005; Garner et al., 2011). Thus, shuttling of ubiquitinated substrates to the UPS or autophagy is likely to be impeded. Intriguingly, either a loss of p62 (Seibenhener et al., 2004) or an increase in p62 (Korolchuk et al., 2009) can abrogate UPS function, and both p62 suppression and overexpression can cause accelerated disease onset and shortened lifespan in ALS-mouse models (Hadano et al., 2016; Mitsui et al., 2018). Together, these studies indicate a fine balance between p62 levels and both UPS function and disease pathogenesis. Upon proteasomal inhibition, p62 expression and autophagic activity is upregulated as a compensatory mechanism (Kuusisto et al., 2001b; Myeku and Figueiredo-Pereira, 2011; Choe et al., 2014; Fraiberg et al., 2017; Sha et al., 2018). ALS and FTLD-associated SQSTM1 mutations that affect the cargo function of p62, or perhaps p62 protein levels, may contribute to disease pathogenesis by upsetting this balance.
p62, autophagy and neurodegeneration
Basal autophagy is vital for maintaining neuronal health, and suppression of basal autophagy is sufficient to cause neurodegeneration in mice (Hara et al., 2006; Komatsu et al., 2006). Deficiencies in autophagy have been implicated in several neurodegenerative diseases; including FTLD (Gotzl et al., 2016), ALS (Xie et al., 2015), Alzheimer’s disease (Nixon et al., 2005), Huntingtin’s disease (Shibata et al., 2006; Fu et al., 2017), spinocerebellar ataxia type 7 (Alves et al., 2014) and across several tauopathies including corticobasal degeneration, and progressive supranuclear palsy (Lin et al., 2003; Piras et al., 2016). Inhibition of autophagy leads to accumulation of a number of proteins involved in neurodegenerative diseases including hyperphosphorylated tau (Babu et al., 2008; Tang et al., 2013), α-synuclein (Minakaki et al., 2018), and TDP-43 (Brady et al., 2011), and accelerates disease onset in SOD1G93A mice (Rudnick et al., 2017). In contrast, pharmacological enhancement of autophagy in several mouse models has proven effective at enhancing clearance of mutant SOD1 (Perera et al., 2018), as well as TDP-43 and FUS-positive inclusions (Brady et al., 2011; Wang et al., 2012; Cheng et al., 2015).
The formation of polyubiquitinated-inclusion bodies and their subsequent removal by autophagy are dependent on p62 (Pankiv et al., 2007), this includes the clearance of mutant SOD1 via autophagy, which is facilitated by p62 binding to mutant SOD1 (Gal et al., 2009). p62 itself is degraded by autophagy, but not the proteasome (Myeku and Figueiredo-Pereira 2011), and as such autophagy inhibition leads to an accumulation of p62 (Bjorkoy et al., 2009). This is problematic, as increased p62 can delay the delivery of ubiquitinated substrates to the proteasome, eventually compromising the UPS (Korolchuk et al., 2009). Unlike the ability of autophagy to compensate for a UPS failure, the UPS is unable to compensate for a failure in autophagy. The effect of p62-overexpression on protein degradation was demonstrated in p62-overexpressing SOD1H46R mice. These mice had accelerated disease onset and reduced turnover of SOD1H46R by either the UPS or autophagy (Mitsui et al., 2018). However, p62-ablation in the same mouse model lead to a worsened disease phenotype and shortened lifespan (Hadano et al., 2016). This highlights that alterations in p62 levels may affect protein degradation, overall autophagic flux, and ALS and FTLD disease progression.
Due to the significant role of p62 in autophagy, SQSTM1 mutations that may affect p62 function are of interest in ALS and FTLD. A number of mutations affecting the PB1 domain of p62 have been identified in both ALS and FTLD patients (Le Ber et al., 2013; Chen et al., 2014; Yang et al., 2015), and deletion of the PB1 domain prevents p62 binding to mutant SOD1 (Gal et al., 2009). p62 utilizes its PB1 domain to facilitate self-oligomerization during autophagy (Itakura and Mizushima, 2011). Due to the proximity of functional binding sites, p62 is not always able to bind multiple substrates at once. One such example is Keap1, a Nrf2 inhibitory protein that is degraded by autophagy in a p62-dependent manner. Due to the proximity of the Keap1-interacting region and the LC3-interacting region, one p62 monomer bound to LC3 will bind to another p62 monomer that is bound to Keap1, and in this way facilitates Keap1 degradation via autophagy (Jain et al., 2010). Mutations affecting the PB1 domain of p62 may affect functions other than dimerization; an FTLD-associated p62 variant, p.R110C, located just outside of the PB1 domain, is unable to promote Nrf2 signaling in line with p62 wild-type, and is associated with increased NF-κB signaling, while having no observed effect on dimerization (Foster et al., 2019), thus indicating further roles for the PB1 domain in p62 function. p62 can bind to LC3 on the isolation membrane through the LIR as well as ubiquitinated cargo through the UBA domain (Wurzer et al., 2015). Mutations affecting the LIR of p62 have been identified in ALS patients, p.D337E, and p.L341V (Chen et al., 2014). In vivo analyses demonstrated that a reduction in p62 LC3-binding ability leads to p62 accumulation in ubiquitinated-inclusions, yet the mutant p62 proteins were unable to be degraded by autophagy (Ichimura et al., 2008). Further, removal of the UBA domain or the LIR domain of p62 reduces the ability of p62 to clear TDP-43 aggregates (Brady et al., 2011). Thus, mutations to the LIR or UBA domains of p62, or the PB1 domain, which is also essential for autophagy, may contribute to ALS and FTLD pathology through a build-up of aggregated TDP-43.
However, upregulation of autophagy does not provide a simple solution to reducing disease progression. Inhibition of autophagy in SOD1G93A mice lead to increased survival time via reduced glial inflammation (Rudnick et al., 2017), while upregulated autophagy increased symptom progression and motor neuron degeneration via increased mitochondrial depletion and increased apoptosis in the same mouse model (Zhang et al., 2011; Perera et al., 2018). Overexpression of p62 in SOD1H46R mice lead to accelerated disease onset (Mitsui et al., 2018), indicating a fine balance between over- and underactive autophagy and p62 levels.
Additionally, p62 plays a role in the interplay between autophagy and the UPS. Of note, proteotoxic stress caused by proteasome inhibition can activate autophagy through p62 phosphorylation, however increased p62 levels can lead to delayed delivery of ubiquitinated substrates to the proteasome, causing UPS impairment despite an intact proteasome (Liu et al., 2016). This indicates that p62 variants that lead to abrogated p62 function may simultaneously affect both autophagy and the UPS by upsetting the interplay between the two. For a more in depth review of the interplay between the UPS and autophagy (Liu et al., 2016).
p62 and mitophagy
Fragmentation and changes to mitochondrial morphology have been extensively documented in ALS (Sasaki and Iwata, 2007; Jin et al., 2013), and are linked with aberrant oxidative metabolism and greater production of reactive oxygen species (ROS) (Mattiazzi et al., 2002; Jin et al., 2013). Clearance of mitochondria occurs by a form of autophagy known as mitophagy. Defects in mitophagy are observed in ALS, and lysosomal defects were observed along with accumulation of damaged mitochondria in motor neurons of SOD1G93A mice (Xie et al., 2015). The role of p62 in the removal of dysfunctional mitochondria is controversial. While some studies indicated that p62 is essential for the final clearance of mitochondria (Geisler et al., 2010; Lee et al., 2010), other studies suggest that p62 is responsible for the aggregation and autophagosomal engulfment of mitochondria, but is not essential for their ultimate removal via mitophagy (Narendra et al., 2010; Okatsu et al., 2010; Matsumoto et al., 2015). It is possible that, like its role in inclusion body formation, p62-mediated aggregation of mitochondria into tight clusters is a cytoprotective attempt at preventing further mitochondrial access to substrates and thereby limiting the spread of mitochondria-derived ROS throughout the cell (Narendra et al., 2010). Mutations that cause a loss of p62 function could prevent mitochondrial aggregation and reduce turnover of dysfunctional mitochondria, exposing the cell to further assault by ROS. Mutations in modifiers of p62, such as TBK1, may also decrease p62 function as ablation of TBK1-mediated phosphorylation of Serine residue 403 (Ser-403) of p62 results in reduced autophagosomal engulfment of mitochondria (Matsumoto et al., 2015). Thus, it is possible that TBK1-mutations lead to ALS pathogenesis partly via reduced p62 phosphorylation. Of note, the p.R110C and p.G427R mutations of p62 were reported to lead to decreased phosphorylation at Ser-403 and Ser-349, which was associated with dysregulated cell signaling (Deng et al., 2019; Foster et al., 2019).
The Effect of p62 Mutations on the Oxidative Stress Response
Nuclear erythroid 2-related factor 2 signaling
Nuclear-erythroid factor-2 (Nrf2) is a transcription factor that is normally sequestered in the cytosol, however under conditions of oxidative stress it is translocated to the nucleus where it promotes the expression of genes involved in the stress response, including glutathione-s-transferase and nuclear respiratory factor-1 (Nrf-1), Heme oxygenase 1, and NAD(P)H quinone oxidase 1 (NQO1) (Itoh et al., 1997; Ishii et al., 2000). The Nrf2 response is the primary mechanism utilised by neurons to protect against oxidative stress (Lau et al., 2010). SOD1G93A mice display impaired induction of Nrf2-induced protective genes (Mimoto et al., 2012; Guo et al., 2013), while overexpression of Nrf2 in the same mouse model delays symptom onset and extends survival time (Vargas et al., 2008). Further, Nrf2-deficient cells are more prone to cell death upon to exposure to reactive oxygen species (Ishii et al., 2000). It is therefore likely that the Keap1-Nrf2 system is a crucial pathway in ALS-FTLD pathogenesis. As an early stress response gene p62 promotes Nrf2 signaling. Under basal conditions Nrf2 is bound to the inhibitory protein Keap1, which shuttles Nrf2 to the proteasome for degradation (Ishii et al., 2000; Cullinan et al., 2004; Kobayashi et al., 2004; Furukawa and Xiong, 2005). p62 has a KIR that mediates p62 binding to Keap1 (Jain et al., 2010; Lau et al., 2010). The interaction between p62 and Keap1 is essential for p62-dependent Nrf2 signaling, and ALS and FTLD-associated mutant proteins that exhibit reduced p62-Keap1 binding also have a reduced ability to activate Nrf2 (Copple et al., 2010; Jain et al., 2010; Lau et al., 2010; Foster et al., 2019). This includes mutations affecting the KIR region of p62, such as p.P348L and p.G351A (Goode et al., 2016), and p.R110C, a mutation affecting the PB1 domain (outside of the KIR) (Foster et al., 2019). In the absence of a PB1 domain, or in the presence of point mutations within the PB1 domain (p.R21A and p.D69A), activation of the NQO1 anti-oxidant response element luciferase reporter was greatly diminished, indicating that a fully functioning PB1 domain is required for p62-dependent upregulation of Nrf2 (Jain et al., 2010). Pharmacological induction of Nrf2 was sufficient to restore NADH and FAD pools and cellular respiration in p62-deficient SH-SY5Y cells and FTLD patient fibroblasts expressing p62 variants p.A381V and p.K238del (Bartolome et al., 2017). Thus, activation of Nrf2 may be an important avenue of research for ALS and FTLD therapeutics.
Selective autophagy and p62-dependent induction of Nrf2 may also be linked to p62 phosphorylation at Serine residue 349 (Ser-349), which is phosphorylated in an mTORC1-dependent manner, and increases p62 binding affinity for Keap1 and subsequently increases Nrf2 signaling (Ichimura et al., 2013). Inhibition of mTORC kinase by rapamycin (and subsequently induction of autophagy) decreased p62 Ser-349 phosphorylation and resulted in decreased Heme oxygenase-1 expression and Nrf2 activity (Ichimura et al., 2013). Further, autophagy inhibition increases p62-dependent Nrf2 activity (Jain et al., 2010). Thus, it is clear that autophagy and the Nrf2 response are linked via p62 activity.
Mutations in genes that code for post-translational modifiers of p62, such as TBK1, may also affect p62-mediated Nrf2 signaling. The phosphorylation of Ser-349 of p62 enhances the binding affinity of p62 for Keap1 (Ichimura et al., 2013), and thus promotes Nrf2 signaling (Figure 2). A protein with the FTLD-associated variant p.R110C substitution had reduced phosphorylation of serine residues 403 and 349 (351 in mice), and while it did not demonstrate altered autophagic flux, did exhibit reduced ability to stimulate Nrf2 (Foster et al., 2019). Mutations in TBK1, which is responsible for the phosphorylation of p62 at Ser-403 (Matsumoto et al., 2011, 2015; Pilli et al., 2012), have been identified in ALS and FTLD patients (Freischmidt et al., 2015; van der Zee et al., 2017). Phosphorylation of Ser-403 (Matsumoto et al., 2011, 2015; Pilli et al., 2012) is a preceding step for Ser-349 phosphorylation (Ichimura et al., 2013), therefore mutations in kinases such as TBK1 may play a role in altered cell signaling in ALS/FTLD via changes to p62 phosphorylation.
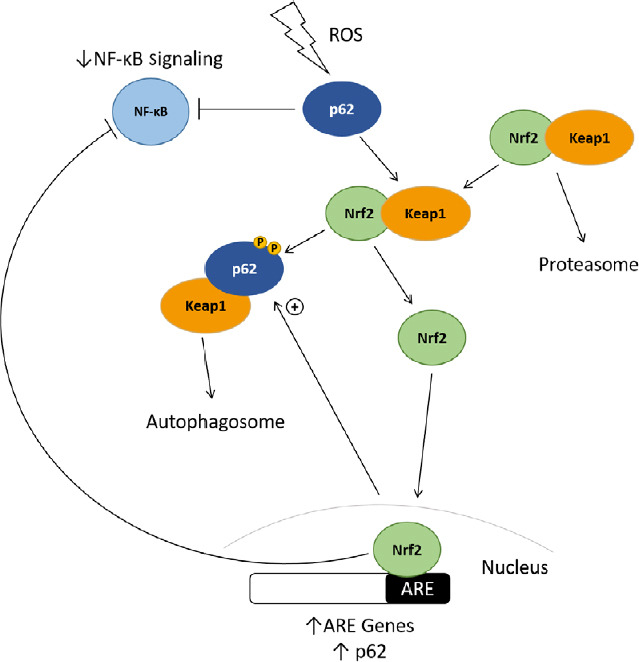
The effect of p62 on Nrf2 and NF-κB signaling.
Upon exposure to oxidative stress, p62 is phosphorylated at Serine residues 403 and 349, and binds to the Nrf2-inhibitory protein Keap1, shuttling it to the autophagosome for degradation. Nrf2 can then enter the nucleus and promote transcription of ARE-genes, including p62, which can also downregulate NF-κB signaling. Keap1: Kelch-like receptor protein 1; NF- κB: nuclear factor-kappa B; Nrf2: nuclear-erythroid factor-2; TRAF6: tumor necrosis factor receptor associated factor 6; ZZ: zinc finger.
Nuclear factor-kappa B signaling
NF-κB is a transcription factor responsible for promoting expression of both pro-survival and pro-inflammatory genes. Under basal conditions, NF-κB is bound to the inhibitory proteins IκBα, β, and ε, which prevent translocation of NF-κB into the nucleus and thereby block transcriptional effects. Under stimulation, the IκB proteins are phosphorylated and degraded via the proteasome, allowing NF-κB to move into the nucleus and promote transcription. Constitutive NF-κB signaling is required for neuronal survival and induction of NF-κB activity occurs in response to oxidative stress (Mattson et al., 2000; Bhakar et al., 2002).
The potential role of NF-κB signaling in ALS is complex. NF-κB is upregulated in the spinal cords of ALS patients and SOD1G93A mice (Swarup et al., 2011; Frakes et al., 2014). However, while inhibition of NF-κB in ALS astrocytes did not prevent motor neuron death, selective inhibition of NF-κB in microglia prevented microglial-mediated death in vitro, impaired pro-inflammatory microglial activation and increased survival time in ALS mice (Frakes et al., 2014). Constitutive expression of NF-κB in microglia is sufficient to induce gliosis and motor neuron death (Frakes et al., 2014), while increased NF-κB activity promoted survival of cells exposed to oxidative stress and NF-κB inhibition resulted in greater toxicity (Heck et al., 1999). Astrocytic NF-κB activation upregulated microglial proliferation in SOD1G93A mice, and this response delayed muscle denervation and prolonged the presymptomatic phase, while inhibition of the early microglial response resulted in acute detrimental effects (Ouali Alami et al., 2018). Interestingly, astrocytic NF-κB activation accelerated disease progression in the symptomatic phase of SOD1G93A mice (Ouali Alami, Schurr et al., 2018), however inhibition of NF-κB in astrocytes in the same mouse model did not show any change in disease onset or progression (Crosio et al., 2011). ALS-associated proteins FUS, TDP-43 (Swarup et al., 2011; Zhao et al., 2015), and UBQLN2 (Picher-Martel et al., 2015) all promote NF-κB signaling, and either astrocytic- (Kia et al., 2018) or microglial-induced motor neuron death (Zhao et al., 2015).
p62 has a dual role in the regulation of NF-κB signaling and dysregulation of NF-κB has been associated with PDB associated UBA domain p62-variants, which have a decreased ability to inhibit NF-κB compared with p62 wild type (Rea et al., 2014). While over-expressed wild type p62 inhibits NF-κB, p62 has also been reported to play an essential role in positively regulating NF-κB signaling in relation to osteoclastogenesis, bone homeostasis, and in cancer (Duran et al., 2004, 2008; Guo et al., 2011). Genetic ablation of p62 was shown to decrease IκB kinase activation and subsequently inhibited NF-κB translocation both in vitro and in vivo in mice (Duran, Serrano et al., 2004), and p62 downregulation abrogated tumour necrosis factor receptor 6 (TRAF6) and IL-1 mediated NF-κB signaling (Sanz et al., 2000), further demonstrating a role for p62 in NF-κB activation outside of neurodegeneration. Ubiquitin-binding affinity affects p62-regulated NF-κB signaling as p62 dimerization via the UBA domain and ubiquitin-binding are mutually exclusive (Isogai et al., 2011). Non-disease associated mutations within the UBA domain that prevent dimer formation increase p62 binding to polyubiquitin and subsequently decrease NF-κB signaling. Consistent with this finding, p62 UBA domain variants have a reduced ability to bind ubiquitin and impaired negative regulation of NF-κB signaling (Long et al., 2010). However, the PDB-associated non-UBA domain mutant p.P364S exhibits increased NF-κB signaling similar to p62 UBA-domain variants, despite retaining full affinity for ubiquitin-binding (Rea et al., 2009), indicating that the role of p62 in regulating NF-κB signaling is not solely due to its ability to bind ubiquitin.
p62 interacts with TRAF6, an adaptor protein and E3 ubiquitin-ligase that regulates NF-κB, via its TRAF6-binding sequence (TBS), and mutations within this domain of p62 have been identified in ALS and FTLD (Rubino et al., 2012; Le Ber et al., 2013). TRAF6 polyubiquitination is associated with increased NF-κB and is promoted by p62, as evident in p62 knockout mice, which exhibit no polyubiquitinated TRAF6 in brain tissue and also exhibit neurofibrillary tangles and neurodegeneration (Wooten et al., 2005). However, p62 also facilitates TRAF6 de-ubiquitination by forming a scaffolding complex linking the de-ubiquitinating enzyme CYLD with TRAF6 (Jin et al., 2008; Wooten et al., 2008). Expression of a p62 UBA-deletion construct abolished TRAF6 polyubiquitination, as did a p62 PB1-deletion construct. Nerve growth factor treatment of PC12 cells induces TRAF6 polyubiquitination and formation of the p62-TRAF6-IκB kinase-PKC iota complex, which leads to NF-κB activation, and inhibition of p62-TRAF6 interaction blocks both TRAF6 polyubiquitination and complex formation (Wooten et al., 2005). Deletion of the TRAF6 binding region of p62 was sufficient to block TRAF6-mediated NF-κB induction. TRAF6 exhibits a low basal level of polyubiquitination, however co-expression with p62 enhances TRAF6 polyubiquitination (Wooten et al., 2005). Therefore, TBS domain variants in ALS-FTLD may alter NF-κB signaling and thereby contribute to ALS-FTLD pathogenicity.
Concluding Remarks
In this review, we have summarized the current understanding of the role of p62 in ALS and FTLD pathogenesis, and how mutations in various domains of the protein may contribute to disease onset (Figure 3).
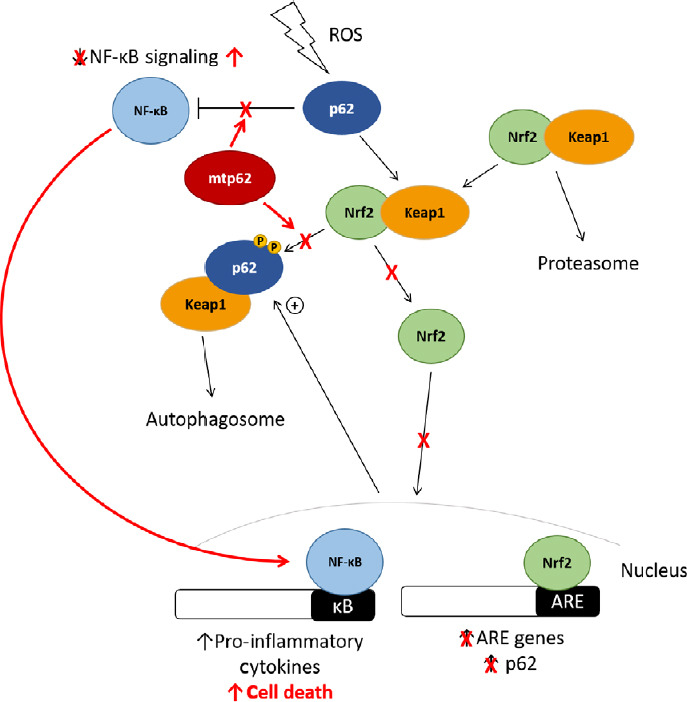
The effect of mutant p62 on Nrf2 and NF-κB signaling in ALS and FTLD pathogenesis.
Several ALS and FTLD-associated variants exhibit reduced Keap1-binding, preventing Nrf2 from entering the nucleus and promoting protective genes, predisposing the cell death upon exposure to ROS. Upregulation of Nrf2 signaling downregulates NF-κB signaling, indicating that p62 variants that fail to promote Nrf2 signaling may also lead to a concomitant increase in NF-κB signaling. Additionally, several p62 variants are unable to regulate NF-κB signaling, which could potentially lead to increased transcription of pro-inflammatory and pro-apoptotic factors, predisposing the cell to death. ALS: Amyotrophic lateral sclerosis; FTLD: frontotemporal lobar degeneration; FronNrf2: nuclear-erythroid factor-2; NF-κB: nuclear factor kappa-B; ROS: reactive oxygen species.
While SQSTM1 mutations identified in PDB cases primarily affect the UBA domain, the variants identified in ALS or FTLD patients span the entirety of the protein. As p62 is a multifunctional protein that exerts its effects through numerous functional domains, mutations affecting these domains can have an effect on both autophagy and cell signaling pathways such as the Nrf2 and NF-κB pathways, and are likely to also affect mitophagy and aggregate/inclusion body formation. ALS and FTLD-associated p62 variants may therefore contribute to ALS and FTLD pathogenesis via reduced degradation of toxic proteins as well as via a reduced ability to mount a suitable stress response, leaving cells more susceptible to neurotoxic insult. The role of p62 and modifiers such as TBK1 in a functioning autophagy lysosome system are well established, however further research into how mutations that affect mitochondrial turnover and altered NF-κB signaling in regards to neuronal health and survival is required. As failure of autophagy can disturb the UPS and lead to increased dysfunctional mitochondria and increased ROS production, further investigations into modulation of autophagy and mitophagy to identify potential therapeutic approaches for ALS-FTLD are needed.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Financial support: This work was supported by the NHMRC-ARC Dementia Research Development Fellowship Grant (AP1102977) and an Australian Government Research Training Program (RTS) Scholarship.
Copyright license agreement: The Copyright License Agreement has been signed by both authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Funding: This work was supported by the NHMRC-ARC Dementia Research Development Fellowship Grant (AP1102977) and an Australian Government Research Training Program (RTS) Scholarship.
C-Editors: Zhao M, Li JY; T-Editor: Jia Y
References
Articles from Neural Regeneration Research are provided here courtesy of Wolters Kluwer -- Medknow Publications
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4103/1673-5374.284977
Article citations
Aberrant protein aggregation in amyotrophic lateral sclerosis.
J Neurol, 271(8):4826-4851, 13 Jun 2024
Cited by: 0 articles | PMID: 38869826
Review
PLK2-mediated phosphorylation of SQSTM1 S349 promotes aggregation of polyubiquitinated proteins upon proteasomal dysfunction.
Autophagy, 20(10):2221-2237, 19 Jun 2024
Cited by: 0 articles | PMID: 39316746
Circ-Bptf Ameliorates Learning and Memory Impairments via the miR-138-5p/p62 Axis in APP/PS1 Mice.
Mol Neurobiol, 61(11):8575-8589, 25 Mar 2024
Cited by: 0 articles | PMID: 38528305
Therapeutic value of homeoprotein signaling pathways.
Front Neurosci, 18:1359523, 14 Mar 2024
Cited by: 0 articles | PMID: 38550565 | PMCID: PMC10973124
Review Free full text in Europe PMC
Mitophagy in human health, ageing and disease.
Nat Metab, 5(12):2047-2061, 30 Nov 2023
Cited by: 35 articles | PMID: 38036770
Review
Go to all (15) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The converging roles of sequestosome-1/p62 in the molecular pathways of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
Neurobiol Dis, 166:105653, 07 Feb 2022
Cited by: 7 articles | PMID: 35143965
Review
PTK2/FAK regulates UPS impairment via SQSTM1/p62 phosphorylation in TARDBP/TDP-43 proteinopathies.
Autophagy, 16(8):1396-1412, 05 Nov 2019
Cited by: 28 articles | PMID: 31690171 | PMCID: PMC7469499
Functional and structural consequences of TBK1 missense variants in frontotemporal lobar degeneration and amyotrophic lateral sclerosis.
Neurobiol Dis, 174:105859, 13 Sep 2022
Cited by: 9 articles | PMID: 36113750
Review
SQSTM1 mutations--bridging Paget disease of bone and ALS/FTLD.
Exp Cell Res, 325(1):27-37, 30 Jan 2014
Cited by: 87 articles | PMID: 24486447
Review




