Abstract
Free full text

Genomic diversity generated by a transposable element burst in a rice recombinant inbred population
Significance
Transposable elements (TEs) represent the largest component of the genomes of higher eukaryotes. Among this component are some TEs that have attained very high copy numbers, with hundreds, even thousands, of elements. By documenting the spread of mPing elements throughout the genomes of a rice population, we demonstrate that such bursts of amplification generate functionally relevant genomic variations upon which selection can act. Specifically, continued mPing amplification increases the number of tightly linked elements that, in turn, increases the frequency of structural variations that appear to be derived from aberrant transposition events. The significance of this finding is that it provides a TE-mediated mechanism that may generate much of the structural variation represented by pan-genomes in plants and other organisms.
Abstract
Genomes of all characterized higher eukaryotes harbor examples of transposable element (TE) bursts—the rapid amplification of TE copies throughout a genome. Despite their prevalence, understanding how bursts diversify genomes requires the characterization of actively transposing TEs before insertion sites and structural rearrangements have been obscured by selection acting over evolutionary time. In this study, rice recombinant inbred lines (RILs), generated by crossing a bursting accession and the reference Nipponbare accession, were exploited to characterize the spread of the very active Ping/mPing family through a small population and the resulting impact on genome diversity. Comparative sequence analysis of 272 individuals led to the identification of over 14,000 new insertions of the mPing miniature inverted-repeat transposable element (MITE), with no evidence for silencing of the transposase-encoding Ping element. In addition to new insertions, Ping-encoded transposase was found to preferentially catalyze the excision of mPing loci tightly linked to a second mPing insertion. Similarly, structural variations, including deletion of rice exons or regulatory regions, were enriched for those with break points at one or both ends of linked mPing elements. Taken together, these results indicate that structural variations are generated during a TE burst as transposase catalyzes both the high copy numbers needed to distribute linked elements throughout the genome and the DNA cuts at the TE ends known to dramatically increase the frequency of recombination.
Transposable elements (TEs) represent the largest component of the genomes of higher eukaryotes, comprising almost 50% of the human genome and over 80% of many plant genomes (1–4). Making up the TE component are hundreds, sometimes thousands, of TE families, containing autonomous elements (encoding the enzymes that catalyze transposition) and the more numerous nonautonomous members (5). A subset of TE families attain very high copy numbers, with hundreds, thousands, even tens of thousands of copies (5). Phylogenetic analysis of high copy number family members often reveals a star-phylogeny, indicative of a burst of amplification that is repressed (either by mutation or host-mediated silencing), and then the sequences of all copies drift into oblivion (5).
The prevalence of evidence for ancient bursts in plant and animal genomes implies that TE families have evolved mechanisms to insert copies throughout the genome while avoiding host silencing (6). To unravel these mechanisms requires the identification of TE families that are in the midst of a burst—where the biochemical features responsible for successful amplification are still active and their insertion sites have not been obscured by negative selection acting over evolutionary time.
To determine the features of a successful burst, we characterized the Ping/mPing TE family, found to be amplifying in four accessions (EG4, HEG4, A119, and A123) of Oryza sativa (rice) (7–9). The TE family is comprised of the autonomous Ping, a member of the PIF/Harbinger superfamily of class II elements, and the miniature inverted-repeat transposable element (MITE) mPing (10). MITEs are a subset of nonautonomous DNA elements characterized by their short length (<600 base pairs [bp]) and ability to amplify from one or a few elements to hundreds or thousands of copies (11, 12). MITEs are the most abundant TEs associated with the genes of higher plants where they populate noncoding regions (introns, 5′ and 3′ flanking sequences) and are a major contributor to allelic diversity (13, 14). While most characterized rice accessions have 0 to 1 Ping and 1 to 50 mPings (15), the four accessions have 7 to 10 Pings, ~230 to 500 mPings, and up to ~40 new mPing insertions per plant per generation (7–9).
Prior studies of the four inbred accessions where the Ping/mPing family has been active over decades revealed key features of successful bursts (9). First, although mPing has a preference for genic insertion sites, it can dramatically increase in copy number without having a major impact on the phenotype because of its preference for AT-rich target sites while rice exons are GC-rich (8, 16). Thus, mPing rarely inserts into rice exons (8). Second, because mPing is a deletion derivative of Ping but does not share any coding sequences, host recognition of mPing does not silence Ping expression (9). Despite robust host epigenetic regulation, the bursts have been ongoing for decades and are likely to continue until Ping transposes into a region that elicits host silencing or the TE load destabilizes the genome (9).
The focus of this study is another dimension of the mPing burst, that being impacts of its spread through a population. While TE bursts are rare phenomena, invasions of naive populations by TE bursts in real time are exceedingly rare, with the most prominent example being the worldwide invasion of Drosophila melanogaster and, more recently, Drosophila simulans by P elements (17, 18). All available evidence indicates that bursts of both P elements and the Ping/mPing family occurred during the past century (9, 17, 18); however, significant increases in mPing copy number are likely restricted to a few related accessions as no bursts were detected in over 3,000 sequenced rice accessions (15, 19). This likely reflects the fact that, unlike D. melanogaster, which is a wild species and an outcrosser, rice is a domesticated crop that propagates by self- or sib-pollination.
In this study, a recombinant inbred line (RIL) population, previously constructed to assess the phenotypic consequences of mPing insertions (20), was repurposed to model the spread of the Ping/mPing burst. The parents of the population were the reference Nipponbare (NB) (where the Ping/mPing family rarely transposes) and HEG4, one of the four bursting accessions. F1s from this initial cross were used to establish 272 inbred lines following 10 generations of self- or sib-pollination (Fig. 1). As such, the RIL population serves as a proxy for the spread of a TE burst in a largely selfing species. Of note was that burst activity was maintained throughout the population as there was no evidence of silencing. In addition, comparative analysis of RIL genome sequences provided a high-resolution picture of the extent of diversity generated by a TE burst, including mPing insertions, excisions, and structural rearrangements.
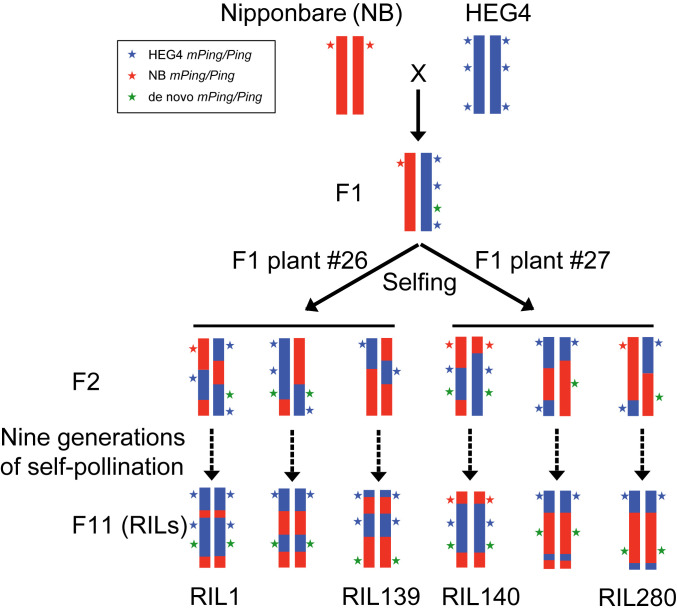
Schematic diagram of RIL construction. The RIL population was constructed by crossing NB (maternal) with HEG4 (paternal). Two F1 plants (no. 26 and no. 27) were used to breed F2s via self-pollination. F2 progeny were self-crossed for nine generations to develop the RILs. HEG4 contains 7 Pings and 422 mPings whereas NB contains 1 Ping and 51 mPings. After 10 generations of self-pollination, Ping and mPing elements from HEG4 (blue stars) and NB (red stars) segregated in the RILs; new mPing/Ping transpositions (green stars) are not in the RIL parents.
Results
Sequence and Analysis of Recombination Map in the RIL Population.
A collection of 272 RILs derived from a cross between accessions HEG4 and NB (Materials and Methods) were sequenced using Illumina paired-end reads. A total of ~1 Tb (terrabase) of sequences were generated with an average depth of ~11-fold coverage per RIL (SI Appendix, Fig. S1A and Supplementary Data 1). Sequence reads from each RIL were aligned to the NB reference genome (MSUv7) (21), and the genotype was scored for the 105,900 high-quality single-nucleotide polymorphisms (SNPs) that distinguish the parents (9). This approach determined 93.5% of parental SNP genotypes in every RIL, ranging from 60.9 to 99.7% (SI Appendix, Fig. S1B and Supplementary Data 1). Recombination bins were inferred using a hidden Markov model (HMM) approach (22). The resulting recombination map contained 2,572 bins and an average bin length of 142.76 kilobases (kb) (SI Appendix, Fig. S1C and Table S1), which is comparable to other sequenced RIL populations in rice (23, 24) and enabled efficient quantitative trait locus (QTL) dissection of genetic loci controlling mPing transposition.
De Novo mPing and Ping Insertion Sites.
The genomic locations of mPing elements in each RIL were determined with RelocaTE2 (25). A total of 87,450 mPing insertions were identified across the 272 RILs and simplified to 16,914 unique mPing loci in the population (Table 1 and SI Appendix, Supplementary Data 2). Of these 16,914 loci, 3% were parental while the remaining 16,448, or 97% of mPing loci in the population were nonparental and defined herein as de novo insertions. Among the de novo insertions, 88% (14,534 of 16,448) are unique to a single individual, indicating that most new insertions occurred during or after the F2 generations that produced the recombinant inbreds (Table 1). The rest of the de novo insertions (1,914 of 16,448) are shared among 2 to 145 RILs, representing either early insertions that occurred in F1 or late insertions that share target sites in the genome (SI Appendix, Fig. S2). Of those found in a single individual, 72% (10,527 of 14,534) are homozygous and likely transposed in early generations of RIL self-pollination while the remaining 28% (4,007 of 14,534) are heterozygous (Table 1) and represent either more recent mPing insertions or mPing alleles that are lethal or less fit when homozygous. To distinguish these possibilities, SNPs flanking heterozygous mPing loci were analyzed to determine genotypes of genomic regions underlying mPing insertions as recent insertions are more likely to be in homozygous (inbred) regions. Of the 4,007 heterozygous insertions, we were able to unambiguously genotype 2,964, of which 96% (2,864 of 2,964) were homozygous (SI Appendix, Table S2). These data suggest that the vast majority of heterozygous mPing insertions are late events. Finally, like previously reported de novo mPing insertions (7, 8), 45% of all insertions (both homozygous and heterozygous) identified in this study are within 5 kb upstream of protein-coding genes (SI Appendix, Fig. S3).
Table 1.
Classification of parental and de novo mPing, Ping, and Pong loci in RILs
| Classification | No. of mPing loci | No. of Ping loci | No. of Pong loci |
| Parental | 466 | 8 | 6 |
 Shared Shared | 7 | 0 | 5 |
 Unique HEG4 Unique HEG4 | 415 | 7 | 0 |
 Unique NB Unique NB | 44 | 1 | 1 |
| De novo | 16,448 | 17 | 0 |
 Shared Shared | 1,914 | 0 | 0 |
 Unique homozygous Unique homozygous | 10,527 | 17 | 0 |
 Unique heterozygous Unique heterozygous | 4,007 | 0 | 0 |
Analysis of Ping elements identified only 17 nonparental (de novo) Pings, and over half of these (8 of 17) are within 5 kb of protein-coding genes (Table 1 and SI Appendix, Table S3). Among the population, RIL270 had two de novo Ping insertions (SI Appendix, Supplementary Data 2) while each of the 15 RILs had a single new Ping insertion. De novo Ping insertions were only found in RILs with two or more parental Ping loci (SI Appendix, Supplementary Data 2).
Correlation between Ping Copy Number and Number of New mPing Insertions.
Among the 272 RILs, significant variation (0 to 138) was found in the number of unique mPing insertions (from here on restricted to de novo unique homozygous mPing insertions) (SI Appendix, Supplementary Data 2), suggesting that multiple genetic loci control mPing transposition activity. To identify these loci, QTL mapping was performed using the number of unique mPing insertions as the trait (SI Appendix, Fig. S4). Three major loci were identified with the logarithm of the odds (LOD) scores greater than 3.31 and accounting for 49% of the phenotypic variation (SI Appendix, Fig. S5 and Table S4). Two of these loci contain multiple Ping elements (PingA and PingB on chromosome 1 and PingE, PingF, and PingG on chromosome 9), confirming prior data that all bursting accessions had multiple Pings (7–10 Pings) (9). A third QTL located on chromosome 4 (accounting for 11.82% of phenotypic variance) does not contain a Ping element (SI Appendix, Fig. S5 and Table S4), suggesting that additional factors may contribute to the rate of transposition. These potential host factors are beyond the scope of this study and will not be further discussed.
To date, quantification of the impact of Ping loci on mPing transposition has been restricted to accessions with either the single Ping locus in NB (PingH) or the collective impact of the 7 to 10 Ping loci in the bursting accessions (HEG4, EG4, A123, and A119) (9). In NB, mPing rarely transposes, and Ping transcript levels are very low (9). In contrast, there is significant amplification of mPing and transcription of Ping in the accessions with 7 to 10 Pings, including the RIL parent, HEG4 (9). This population allows for higher resolution analyses of Ping copy number and transposition and transcription activity because the eight parental Pings are segregating and, theoretically, should contain most combinations of Ping loci (SI Appendix, Fig. S6). In fact, several RILs were found to contain from 0 to 8 Ping loci, and a positive correlation was found to exist between Ping copy number and the number of new mPing insertions (Fig. 2A) (two-tailed Pearson’s correlation test, r = 0.71, P = 1e−42). For example, there are approximately 5 unique mPing insertions in RILs with one Ping whereas RIL with seven Ping elements have, on average, 65 unique mPing insertions. In addition, transcript levels of both ORF1 (Fig. 2B, blue) and TPase (Fig. 2B, red) increased linearly with Ping copy number (Fig. 2B; two-tailed Pearson’s correlation test, ORF1: r = 0.9, P = 6e−14; TPase: r = 0.85, P = 4e−11), suggesting that a simple dosage relationship exists between Ping expression and transposition activity.

Accumulation of unique mPing insertions is dependent on Ping dosage. (A) Correlation between unique mPing insertions and Ping copy number in the RILs. The 272 RILs were grouped by Ping copy number ranging from 0 to 7, and the scatterplot shows the number of unique mPing insertions in each group. The number of RILs in each group is in parentheses (n=). Green lines are the group median. The significance of correlation was tested by a two-tailed Pearson’s correlation test and is indicated by P value. (B) qRT-PCR analysis (three replicates) of Ping ORF1 and TPase transcription levels. RILs with Ping copy numbers of 1 to 7 were randomly selected. Relative transcription levels of ORF1 (blue) and TPases (red) were normalized with rice actin. Colored lines are the best-fit line of the linear regression model. The significance of correlation was tested by a two-tailed Pearson’s correlation test and is indicated by P values for both ORF1 (blue) and TPase (red). Sampled RILs with 1 to 7 Ping copies used in B were as follows: 1 (RIL12, -100, -166, -179); 2 (RIL16, -19, -36, -123); 3 (RIL5, -18, -22, -37, -111); 4 (RIL8, -13, -87, -119, -177, -219); 5 (RIL10, -15, -23, -58, -73, -92, -94); 6 (RIL30, -44, -54, -118, -134); and 7 (RIL11, -21, -34, -69, -198).
Ping Activity at Eight Different Loci.
The RIL population provided a unique opportunity to explore the activity of various combinations of the eight segregating Ping loci present in the two parents. This was of particular interest because, although all Ping elements are identical, one Ping locus has been implicated in initiating the bursts (9, 15). Specifically, PingA_Stow on chromosome 1 (called PingA in this study) was shown previously to be the only Ping shared by the four bursting accessions (9) and found preferentially in the genomes of rice accessions with higher than background mPing copies (15). Taken together, these correlative data suggest that the PingA locus differs from the other Ping loci, perhaps by catalyzing more transposition.
The activity of individual Ping loci was quantified by first identifying RIL with one Ping. Among the RIL population are 23 lines with a single Ping that, collectively, harbor all parental Pings except PingF. PingA is present as a single Ping locus in RIL60. Because mPing rarely transposes in accessions with one Ping (like NB), two high-resolution independent methodologies, transposon display and deep sequencing, were employed to assess the number of new mPing insertions.
To identify new mPing insertions in individual progeny, transposon display (TD) was performed using DNA isolated from eight plants derived by self-pollination from each analyzed single-Ping RIL (Fig. 3A and SI Appendix, Fig. S7 and Table S5). The mPing transposition activity was estimated by counting new amplicons present in one individual but absent from its siblings. New mPing insertions were detected in all single Ping RILs analyzed except RIL179 (PingB) (Fig. 3A); however, the progeny of RIL60 (with PingA), had significantly more (4 to 8 vs. 0 to 3 in the other single-Ping RIL) (Fig. 3A) (one-way ANOVA with Tukey’s honestly significant difference [HSD] test, P < 9.48e−8).
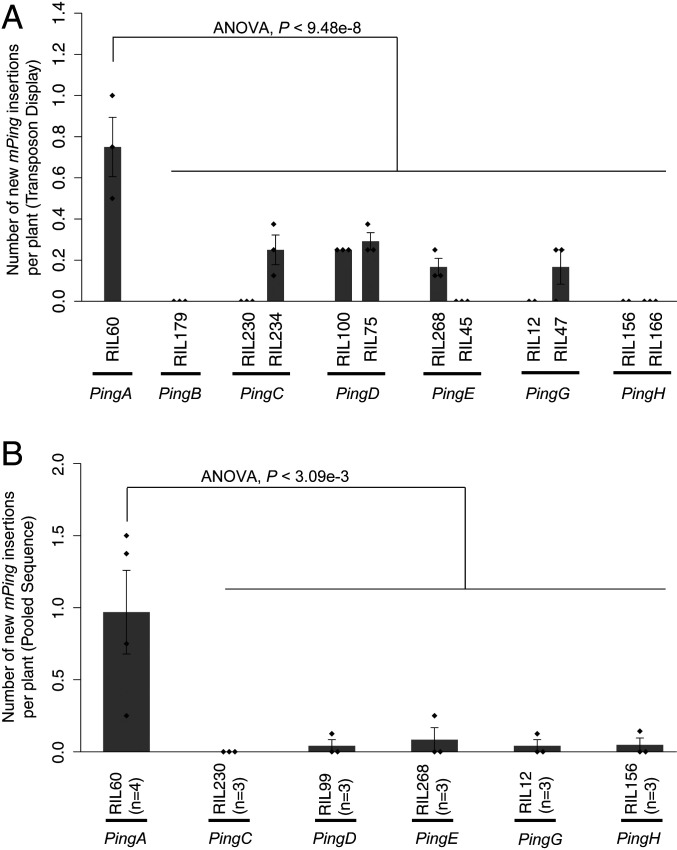
New mPing insertions in single-Ping RILs. (A) New mPing insertions estimated by transposon display. New insertions were estimated by counting the number of new bands displayed by single plants. Eight sibling plants from a single-seed descent were used for each selected single-Ping RIL. Error bars show SE of 2 to 3 independent biological replicates. (B) New mPing insertions estimated by a pooled-sequencing approach with multiple progeny from a single-Ping RIL. RIL179 (PingB) was excluded due to contamination. Error bars show SE of 3 to 4 independent biological replicates represented by black diamonds. Differences between PingA and other Ping loci were tested by a one-way ANOVA followed by Tukey’s HSD post hoc test.
In the second approach, new insertions were directly counted following deep sequencing of DNA samples isolated from eight-pooled progeny of each RIL. Detection of new mPing insertions for these RILs confirmed that all Pings tested are active, thus providing a simple explanation for the Ping dosage series reported above. Similarly, the finding that PingA promotes more transposition than the other Pings (Fig. 3B and SI Appendix, Table S6) (one-way ANOVA with Tukey’s HSD test, P < 3.09e−3) is consistent with the hypothesis that it initiated the bursts, but the underlying mechanism is beyond the scope of this study (Discussion).
Loss of Parental mPing Insertion Loci in the RIL Population.
Ping and mPing elements, like other DNA transposons, can both insert into new loci and excise from existing sites. By tracking the fate of the 466 parental loci in the RIL population (44 from NB, 415 from HEG4, and 7 shared) (Materials and Methods), we sought to determine whether some mPing loci were lost at a higher frequency than others. Loss of TEs over time has been attributed to many factors, the two most prominent being negative selection and structural instability (26, 27). To discriminate between these possibilities, we developed methodologies to identify all parental mPing excision events among the RIL population, determine excision frequencies at each locus, and analyze the genome context of loci that excised frequently. To this end, we characterized empty (excised) mPing sites in RILs within haplotype blocks classified by parent of origin and expected to contain parental mPing loci (Fig. 4A). On average, 127 RILs (ranging from 71 to 159) were analyzed for each parental mPing.

Identification of mPing loci with high excision frequencies. (A) Workflow for identification of mPing excisions (see details in Materials and Methods). Red and blue colors represent NB and HEG4 genotypes in parental accessions. mPing insertions (orange boxes) are exclusively in HEG4 genotypes. Sequencing reads aligned to pseudoreference sequences with or without the mPing insertion are indicated by light blue bars. (B) Example illustrating the method used to detect independent excision events at mPing locus Chr1:6816415 to 6816417. Red letters in NB indicate the 3-bp TSDs upon mPing insertion. Footprints (deletions and insertions) are indicated by dots or arrows. CIGAR-like strings are used to record the footprints. Excision footprints are characterized as deletions (D) or insertions (A) with the length (e.g., D-X bp) and, in parenthesis, the number of RILs with this footprint. (C) Distribution of parental mPing loci and number of independent excision events. The P value is based on a two-tailed binomial test. The dashed line is the cutoff for significant high frequency excision. The number of different mPing loci with that many excisions is at the top of each bar. (D) mPing loci with more excision events are in close proximity to another mPing. The box shows a zoom of the 0.04- to 100-kb range.
In total, 742 excisions were detected from 177 parental mPing loci (17 NB, 160 HEG4), and no excisions were detected from the remaining 289 loci (34 NB, 255 HEG4). To validate the accuracy of the computer-assisted excision data, 30 loci were randomly selected and assayed by PCR with DNA isolated from the RIL where the excision was detected. Using this approach, 26 of 30 events were confirmed (SI Appendix, Fig. S8). Next, to assess the excision frequency at each of the 177 parental loci, it was necessary to establish the independence of the excision events. For example, an excision event occurring early in the generation of the RIL population might be propagated in several RILs. Two approaches were used to eliminate dependent events: 1) a standard method where all excisions were analyzed after removal of early events, and 2) a conservative method where excision footprints were exploited to identify independent events. The latter is likely to underestimate independent events because perfect excision, which is common for mPing, will only be counted once per locus per RIL. Because both approaches produced very similar results, only the conservative method is described below. However, the results of both are found in Materials and Methods.
For the conservative method, 367 of the 742 excisions from parental mPing loci have excision footprints (SI Appendix, Supplementary Data 3). This reduced to 322 independent events following removal of excisions from the same locus with the same footprint (Fig. 4B and SI Appendix, Supplementary Data 3). Of the 177 loci, 114 had only a single excision, 51 had fewer than five (Fig. 4C), and 12 loci experienced five or more independent excision events (Fig. 4C and SI Appendix, Table S7), which is significantly more than expected by chance (two-tailed binomial test, P = 7.1e−4). Significantly, a majority of these loci (7 of 12) have another mPing insertion within 10 kb. Of these seven loci, five have a nearby mPing in HEG4, including two pairs of high frequency excision loci located close to each other (2.8 kb between Chr1:36267659 and Chr1:36270511; 2.6 kb between Chr5:15210313 and Chr5:15213006) and one (Chr1:6806761) located 9.6 kb from another parental mPing (Chr1:6816415) with four independent excisions (Fig. 4D and SI Appendix, Table S7). Two of the seven loci have a nearby mPing (1.9 kb and 2.3 kb away) that is not a parental locus but rather present in 45% of the RILs (SI Appendix, Table S7). For the remaining 5 of 12 high excision mPing loci, the nearest mPing is from 22.3 to 335.8 kb (SI Appendix, Table S7). Taken together, these findings indicate that the loss of mPing among the RILs can be attributable to structural instability, not negative selection. In support of this claim is the finding that expression analysis of the protein-coding genes associated with the 12 high frequency excision loci identified only a single differentially expressed gene between the two parental accessions (LOC_Os01g07300) (SI Appendix, Table S8).
mPing-Mediated Sequence Rearrangements.
Another measure of the impact of high copy mPings on genome stability is the number of structural rearrangements in the RIL population and their proximity to mPing insertions. A prior study, limited to a single comparison between two accessions with high mPing copy numbers (HEG4 and EG4), identified a 120-kb inversion with multiple mPing at the break point (9).
The RIL population, with 97% having 200 or more mPing copies (SI Appendix, Supplementary Data 2), provided an opportunity to investigate the impact, if any, of an active burst on the generation of structural rearrangements. A limitation of this analysis is that the use of short sequence reads precludes the identification of inversions mediated by mPing or other TEs; only deletions and small insertions (duplications) could be detected with confidence. Positions of rearrangements, irrespective of their proximity to mPing elements, were determined by aligning and comparing RIL sequences with the NB reference genome using a read depth strategy implemented in CNVnator (28). Rearrangements only in the RILs, were identified by first analyzing HEG4 using the same method to detect and eliminate from consideration rearrangements in the parental accession. In total, 16 rearrangements (15 deletions and 1 deletion plus duplication), ranging from ~700 bp to 56 kb, were resolved (Table 2 and SI Appendix, Table S9). Fourteen of the 16 were only in a single RIL while 2 are shared in several RILs and found to derive from a single F1 plant (no. 27) (Materials and Methods) that segregated in F2 generations (SI Appendix, Table S10). Of note, 12 of the 16 rearrangements have mPings in close proximity to the break points, with 9 starting precisely at mPing sequences (Fig. 5, SI Appendix, Figs. S9 and S10, and Table 2). Eleven mPing loci were identified at the break points of these 9 rearrangements. Of these 11 loci, 5 were also characterized as high frequency excision loci (SI Appendix, Table S11). The 9 rearrangements with mPing sequences at the break points were all validated by PCR (SI Appendix, Fig. S11 and Table 2).
Table 2.
Features of SVs in RILs
| SV | RIL | Ping no. | SV type | Length, bp | Break point (BP) feature | Gene annotation | Parental genotype |
| SV1 | RIL22 | 3 | Deletion | 1,732 | mPing at both BP | —* | HEG4 |
| SV2 | RIL11 | 7 | Deletion | 8,148 | mPing at both BP | WRKY66† | HEG4 |
| SV3 | RIL26 | 5 | Deletion | 1,138 | mPing at left BP | —* | HEG4 |
| SV4 | RIL31 | 5 | Deletion plus duplication | 701 | mPing at left BP | —* | HEG4 |
| SV5 | RIL222 | 5 | Deletion | 2,694 | mPing at both BP | Expressed protein‡ | HEG4 |
| SV6 | RIL242 | 4 | Deletion | 2,694 | mPing at both BP | Expressed protein‡ | HEG4 |
| SV7 | RIL155 | 7 | Deletion | 2,313 | mPing at both BP | —* | HEG4 |
| SV8 | RIL22 | 3 | Deletion | 9,242 | mPing at left BP | Transmembrane receptor‡ | HEG4 |
| SV9 | RIL198 | 7 | Deletion | 56,012 | mPing at right BP | Transcription factor†; expressed protein† | NB |
| SV10 | RIL274 | 3 | Deletion | 5,758 | Homology | Cinnamoyl CoA reductase‡ | HEG4 |
| SV11 | RIL158 | 3 | Deletion | 3,153 | mPing at 12 bp upstream right BP | Transmembrane protein 16K‡ | HEG4 |
| SV12 | RIL263 | 4 | Deletion | 1,024 | mPing at 113 bp downstream left BP | —* | HEG4 |
| SV13 | RIL61 | 5 | Deletion | 10,818 | No | Aldehyde oxidase‡ | HEG4 |
| SV14 | RIL68 | 4 | Deletion | 2,751 | Microhomology; mPing at 843 bp upstream right BP | —* | HEG4 |
| SV15 | RIL78 | 4 | Deletion | 1,461 | Microhomology | —* | NB |
| SV16 | RIL78 | 4 | Deletion | 4,150 | Filler DNA | —* | HEG4 |

Structural variations mediated by mPing elements. (A) SV2. (B) SV1. (C) SV6. (D) SV4. (E) SV7 and SV8. (F) SV9. mPing-associated SVs in select RILs were aligned with either NB or HEG4 based on the chromosome of origin. Lines indicate DNA fragments flanking mPing insertions or the break point of SV. Letters (a–f) on lines label the DNA fragment end. Orange boxes indicate mPing insertions. Blue boxes indicate exons of protein-coding genes. TSDs of mPing insertions are highlighted in red. Filler DNA at break points is italicized. Red arrows indicate the break points where SV occurred. Allele frequency (AF) is indicated if mPing is present in RILs but is absent in parental accessions.
Close examination of these nine rearrangements implicate aberrant transposition as the underlying mechanism. For example, transposase acting at the distal termini of mPings upstream and downstream of rice gene LOC_Os02g47060 (Fig. 5A and SI Appendix, Table S12) likely generated a megatransposon that includes both elements and 8.1 kb of intervening DNA. Similarly, termini of linked mPings appear to be involved in the deletion of potential regulatory sequences upstream of LOC_Os01g51290 (Fig. 5B and SI Appendix, Table S12) and intragenic sequences of LOC_Os05g26140 (Fig. 5C and SI Appendix, Table S12) leaving at the break point, a single mPing in the former and a rearranged mPing in the latter. Deletion of sequences downstream of LOC_Os03g12260 (Fig. 5D and SI Appendix, Table S12) also correlates with the presence of tightly linked mPings although only one mPing terminus appears to be directly involved in the rearrangement. Tightly linked mPings are also involved in distinct intergenic rearrangements in RIL155 and RIL22, with the termini of both mPings implicated in the former, but only one of the two in the latter (Fig. 5E). The largest rearrangement with mPing at a break point (a 56-kb deletion including two rice genes) (Fig. 5F and SI Appendix, Table S12) involves only a single, unlinked mPing that is derived from the NB parent. Its participation in a rearrangement may be due to high transposase concentration—as RIL198 has seven Pings (Table 2). In total, rearrangements were associated with deletion of four protein-coding genes and partial deletion of six protein-coding genes (SI Appendix, Table S12).
Discussion
In this study, RILs generated by crossing a bursting accession and the reference NB were exploited to characterize the spread of TEs through a small population and the resulting impact on genome diversity. To quantify family (transposase) activity, the number of new mPing insertions was used as a trait that varied across the RILs. In this way, the number of new mPing insertions per RIL served as a high-resolution proxy for Ping activity and led to a deeper understanding of the features of this TE family and its interaction with the host. To understand the impact on gene and genome diversity, all Ping and mPing insertions were localized, and a determination was made of their contribution to structural variation (SV). Conclusions from these dual analyses are summarized below.
Assessing Ping Activity.
Ping dosage.
Prior studies of Ping activity were restricted to accessions, like NB, with a single Ping element, and the bursting accessions, HEG4, EG4, A119, and A123, with 7 to 10 Pings (9). Transposition of mPing was extremely rare in the former and robust in the latter, with up to ~40 new insertions per plant per generation (7, 8). In this study, segregation of the eight parental Ping loci generated lines with 0 to 8 Pings, often with many RILs for each Ping dosage (SI Appendix, Supplementary Data 2). Variation in the number of new mPing insertions per RIL (0 to 138) led to the identification of two QTLs that each contain multiple linked Ping elements: on chromosomes 1 (PingA and PingB) and 9 (PingE, PingF, and PingG) (SI Appendix, Fig. S5). That these QTLs contain the only linked Pings raises the question of whether their contribution to mPing transposition is due to the effect of Ping dosage or something more subtle. We think the former for the following reasons. First, RILs with PingC, PingD, and PingH contain comparable numbers of new mPing insertions as RILs with Ping loci identified by QTL mapping (SI Appendix, Fig. S6 B–G). This suggests that single Ping loci also contribute to the dosage effect on transposition activity. Second, two other bursting accessions (A119 and A123) with high Ping copy numbers (7 and 10 respectively) only share the PingA locus (9). That is, their bursts are robust even though they do not have the two QTLs reported in this study. Thus, we conclude that variation in the number of new mPing insertions across the RILs reflects increasing Ping copy number (Fig. 2A) and Ping transcripts (Fig. 2B). These results indicate that incremental increases in Ping-encoded products promote incremental increases in Ping activity. Just how high Ping dosage can be increased before activity plateaus is the goal of a future study to investigate new RIL populations where both parents are in the midst of a burst and many more Pings and mPings are segregating.
Individual Ping activity.
Segregation of the seven (out of eight) Ping loci (containing identical Ping elements) into single Ping-containing RILs facilitated a comparative analysis of their ability to catalyze mPing transposition. To overcome the limitation of low germinal activity associated with single Ping-containing lines, we exploited the higher frequency of somatic transposition coupled with deep sequencing. Consistent with prior studies where PingA was found to be the only locus shared by all bursting and high copy accessions (9), we found that PingA catalyzes more transposition than the other Ping loci (Fig. 3). Although these data could be interpreted to mean that the burst was initiated by higher PingA activity, future studies are needed to determine the underlying mechanism.
Ping is not silenced.
The Ping/mPing family was shown previously to successfully amplify by eluding epigenetic silencing for decades of sib- or self-pollination of bursting inbred lines (9). In this study, we extend that finding by showing that this TE family remains active after outcrossing (of a bursting accession to NB) and subsequent rounds of sib- or self-pollination of the RIL population. First, demonstration that there are no new insertions of Pong, which is active but silenced (9), in any RIL indicates that epigenetic regulation is maintained throughout the population (Table 1 and SI Appendix, Supplementary Data 2). Despite robust epigenetic regulation, all RIL with Ping elements, especially lines with many Pings, have new mPing insertions, indicating that Ping has not been silenced in any of these lines (SI Appendix, Supplementary Data 2).
Data from this study provide important clues to why Ping activity has persisted in the population after outcrossing and why, as the burst continues, it is increasingly unlikely to be silenced. A prior study (9) demonstrated that, although mPing is recognized by host epigenetic machinery, Ping is not silenced because mPing does not harbor any of Ping’s coding sequences. Thus, only Ping terminal sequences, that are shared with mPing, are methylated (9). These results suggested that, for Ping to be silenced, it has to transpose into a genomic region that generates small interfering RNA (siRNA). Here, we show that Ping rarely transposes, making Ping silencing unlikely. Only 17 new Ping insertions were detected among the RIL population—compared to over 14,000 new mPing insertions (SI Appendix, Supplementary Data 2). A close examination of the RILs with transposed Pings identified from 21 to 96 new mPing insertions, including 4 to 60 heterozygous insertions that are likely to be new and provide evidence of recent activity (SI Appendix, Table S3). In addition, of the 17 new Ping insertion sites, 14 are into euchromatic loci and 3 into heterochromatin (SI Appendix, Table S3). Among this latter class (which is more likely to silence Ping), new mPing insertions vary from 63 (21 are heterozygous) to 67 (15 are heterozygous) to 111 (15 are heterozygous), again inconsistent with the loss of Ping activity (SI Appendix, Supplementary Data 2).
Taken together, these data suggest that, as the burst continues, the ratio of mPing to Ping copies will increase—which is likely to further decrease the frequency of Ping transposition and its subsequent silencing. In addition, the finding that the single SNP adjacent to its terminal inverted repeats (TIRs) makes mPing a better substrate for transposase than Ping (15) will further reduce the chances of Ping silencing as the burst continues.
Genome Instability as the Burst Continues.
If the burst is not likely to end with Ping silencing, how will it end? Data from this study strongly suggest that increases in mPing copy number destabilize the genome by elevating the chances of potentially deleterious SV. TEs have long been known to generate SVs. The first TE discovered by McClintock, Dissociation (Ds), was initially detected as a site of chromosome breakage (29). These so-called “breaking” Ds elements are a structurally distinct minority of characterized elements, often composed of tightly linked Ds copies (30, 31). Double-Ds elements form frequently because of the preference of the Ac transposase to catalyze local transposition (32, 33). Similar local hops and chromosome breakage are associated with TEs from all three kingdoms, including the Drosophila P and the bacterial Tn10 elements (34, 35).
In contrast, Ping does not promote local transposition of mPing as new insertions from a single integrated donor site in transgenic Arabidopsis thaliana, yeast, and soybean were not linked to that site (16, 36, 37). In operational terms, this means that mPing bursts scatter copies randomly throughout the genome, with only a few near existing insertion sites. However, as the burst continues, the probability that new insertions will be linked to existing mPing elements increases. Whether linked mPing elements are more likely to generate SVs was investigated in the RIL population in two ways: 1) by quantifying the relative frequencies of parental mPing excisions, and 2) by identifying SVs in the RIL population and determining whether any had mPing at one or both break points. For the analysis of excisions, the RILs were scored for the frequency of excision among the 466 parental mPing loci. The fact that 7 of the 12 loci with five or more excisions had another mPing within 10 kb suggests that tightly linked elements are unusually active with regard to transposition (SI Appendix, Table S7).
Whether higher transposition activity also contributes to the formation of a subset of SVs was assessed by identifying deletions and small duplications among the RILs. As mentioned in Results, a limitation of this analysis is that the use of short sequence reads precludes the identification of inversions and many larger SVs. Overall, 16 SVs were detected, and 9 were shown to have mPing at one (4 of 9) or both (5 of 9) break points (Table 2). Furthermore, the positions of the break points—at mPing termini—implicate Ping transposase activity in SV formation. This situation is reminiscent of the occurrence of Ds breakage only if Ac is in the genome (29), and that aberrant transposition is the likely mechanism (38, 39).
Conclusions and Implications.
While most genomic changes resulting from TE bursts are likely neutral or deleterious, the results of this study suggest that there may be an evolutionary benefit to the host when a TE family bursts, scattering copies throughout the genome. Specifically, continued mPing amplification increases the number of tightly linked elements, which, in turn, increases the number of functionally relevant SVs upon which selection can act. Most of the SVs reported in this study are deletions that are unlikely to be of evolutionary benefit. However, this limitation probably reflects the use of short read sequences that have been reported to miss most of the SVs larger than 2 kb, including duplications and inversions (40). In fact, a prior study that compared the assembled genomes of two virtually identical rice accessions undergoing independent mPing bursts, HEG4 and EG4, detected an inversion of ~120 kb with mPings at the break points (9).
The significance of this finding is that it provides a TE-mediated mechanism that may generate much of the SVs represented by pan-genomes in plants and other organisms (41, 42). Specifically, it is generally accepted that a species, or even a population, cannot be adequately represented by a single reference genome (43). Rather, members of a species share core genes, but variation exists in the form of sequence rearrangements that alter so-called dispensable (or accessory) gene content. Associations of dispensable genomic regions with more TEs suggest that TEs are involved in generating observed presence, absence, and copy number variations (44). However, until this study, the only evidence was correlative; questions remained concerning when and how TEs generate the raw material for pan-genomes. Here, we demonstrate that “when” is during an active TE burst, and “how” is through the scattering of tightly linked elements throughout the gene space, thereby seeding the genome with regions susceptible to transposase mediated SVs. One could imagine that, in a sufficiently large population undergoing similar bursts, SVs that involve components of the core genome will be eliminated by negative selection while SVs that alter dispensable genes could be retained in the population.
Materials and Methods
RIL Plants, DNA Extractions, and Sequencing.
RILs were developed through a single-seed descent approach from the F2 generation of a cross between NB (maternal parent) and an mPing active accession HEG4 (paternal parent) (see details in Fig. 1). RIL seeds were germinated at 37 °C and moved to soil 5 d after germination. Seedlings of 3-wk-old plants were harvested to extract genomic DNA using the cetyl trimethylammonium bromide (CTAB) method (45). Purified genomic DNA was fragmented to 300 bp using a Covaris S220 Ultrasonicator (Covaris), and paired-end libraries were constructed using an Illumina TruSeq DNA sample prep kit (Illumina) or KAPA LTP library preparation kit (Kapa Biosystems). Libraries were multiplexed and sequenced on HiSeq 2500 or NextSeq 500 (Illumina).
RNA Extraction and qRT-PCR.
RILs were grown as described above. Seedlings of 3-wk-old plants were used to extract total RNAs using the RNeasy Plant Mini Kit (Qiagen). Total RNAs were treated with amplification-grade RNase-free DNase I (Qiagen) to remove contaminating DNAs. The treated DNA-free total RNAs were reverse transcribed into complementary DNAs (cDNAs) using SuperScript III first-strand supermix (Invitrogen). The resulting cDNAs were used as templates to perform qRT-PCR with iQ SYBR Green Supermix (Bio-Rad) on a CFX96 system (Bio-Rad). Transcript levels of ORF1 and TPase were normalized to rice actin. Primers for qRT-PCR are in SI Appendix, Supplementary Data 4.
Construction of Recombination Bin and Linkage Map.
A total of 105,900 SNPs between parental accessions HEG4 and NB were identified and filtered using the Genome Analysis Toolkit (GATK) (46) as described previously (9). High-quality SNPs were used to genotype the RILs and a hidden Markov model (HMM) algorithm was applied to construct recombination bins (22). Briefly, paired-end reads of each RIL were mapped to the NB reference genome (MSUv7) by Burrows–Wheeler Aligner (BWA) aligner v0.6.2 (21, 47). Read alignments were sorted, and PCR duplicates were removed using MarkDuplicate as implemented in PicardTools (broadinstitute.github.io/picard/). Potential SNP sites were identified by the GATK toolkit following the best practice workflow of GATK (46). Only SNP sites matching parental SNPs were considered as informative sites. A genotyping error rate of 1.4% was estimated by counting the number of incorrectly genotyped SNPs from independently sequenced parental samples, NB and HEG4. Of these 105,900 SNPs, 1,311 were misgenotyped as HEG4 or heterozygous SNPs in the NB sample (864 HEG4 and 447 heterozygous, respectively) whereas 1,656 were misgenotyped as NB or heterozygous SNPs in the HEG4 sample (333 NB and 1,323 heterozygous, respectively). The resulting RIL SNP genotypes were imported into the R environment to construct a recombination bin map using the HMM approach as described in the Maximum Parsimony of Recombination (MPR) package (22). The expected proportion of genotypes for each locus was 49.75:0.50:49.75, with an error rate of 1.4%. Recombinant bins were used as markers, and the missing data were imputed using an argmax method in the R/qtl package (48). A linkage map was built using the haldane function in the R/qtl package (48).
Identification of mPing and Ping Copies in the RILs.
mPing copies, including elements present in the NB reference genome and elements present in the RILs or HEG4, were identified by RelocaTE2 (25). Briefly, RelocaTE2 identified paired-end read pairs that match the ends of mPing (junction reads) or internal sequences of mPing (supporting reads) using BLAT (49). After trimming mPing sequences, the remaining sequences from the original read pairs were searched against MSUv7 by BWA v0.6.2 to identify potential mPing insertions either present in the reference genome or in the RILs. Raw results from RelocaTE2 were filtered by removing mPing insertions with junction reads from only one end or mPing insertions without 3 bp target site duplications (TSDs), which is a hallmark of mPing transposition. Heterozygous mPing insertions were characterized by analyzing BAM files of raw read pairs aligned to MSUv7 (50). These mPing insertions with reads aligned to both sides of the flanking sequences were characterized as heterozygous mPing insertions. Ping copy number in RILs was characterized as described for mPing. Read pairs from each mPing/Ping insertion site were analyzed to distinguish mPing and Ping sequence differences. All candidate Ping insertions were confirmed by PCR or Southern blot.
Determination of mPing Insertion Sites in Single-Ping RILs.
Two experimental approaches (transposon display and pooled DNA sequencing) were performed on multiple individuals to determine de novo mPing insertions. Because mPing transpositions are rare in single-Ping RILs, multiple individuals were pooled to maximize detection of new insertions catalyzed by each Ping.
Transposon display.
Single-Ping RILs used in the experiment were as follows: PingA: RIL60; PingB: RIL179; PingC: RIL230, RIL234, RIL43; PingD: RIL100, RIL25, RIL75; PingE: RIL268, RIL45; PingG: RIL12, RIL47; and PingH: RIL156, RIL166, RIL259. Genomic DNA was extracted using the CTAB method (45). Transposon display of mPing was performed on eight progeny of each RIL as described (51). Briefly, 50 ng of genomic DNA was digested with MseI and ligated with adapters overnight at 37 °C. Preselective amplifications were performed with primers complementary to the adapters (MseI + 0) and internal mPing sequences (mPing P1). Selective amplifications were performed with one selective base of the adapter primer (MseI + T/A/G/C) and mPing P2 primers. PCR reactions were loaded on 6% denaturing acrylamide-bisacrylamide gels to visualize mPing transpositions. New bands that are present exclusively in one individual are counted as de novo mPing insertions. Transposition frequency of mPing was calculated using the number of new mPing insertions per individual from all of the selective primer combinations. The primers for transposon display are in SI Appendix, Supplementary Data 4.
Pooled DNA sequencing.
Single-Ping RILs used in the experiment were as follows: PingA: RIL60; PingC: RIL230; PingD: RIL99; PingE: RIL268; PingG: RIL12; and PingH: RIL156. Genomic DNA from 7 to 9 plants per RIL were extracted and fragmented as described above, and paired-end libraries were constructed with the KAPA LTP library preparation kit. Libraries were multiplexed and sequenced on a HiSeq 2500 or NextSeq 500. Three or four biological replicates were performed for each experiment. RelocaTE2 was employed to identify mPing insertion sites in each sequence replicate. Only heterozygous mPing insertions that are exclusively present in one replicate were counted as de novo insertions.
Analysis of mPing Excision Events.
Parental mPing insertions segregating in the RILs were identified. First, 51 mPing insertions exclusively in NB or shared between HEG4 and NB were identified as parental mPing loci. These loci have allele frequencies of ~50% (35 to 55%) for 44 NB-specific mPing loci or ~90% (89 to 95%) for 7 shared mPing loci. Second, 415 nonreference mPing insertions were identified from four independent sequenced HEG4 libraries (Sequence Read Archive [SRA] accession numbers: SRR1619147, SRR833485, SRR833529, and SRR1619373). Determination of the allele frequency of the 415 mPing loci in the RILs revealed that they were present in more than 10% of the 272 RILs (ranging from 16 to 74%), suggesting these mPing loci were segregating in the population. Together, these 466 mPing loci were used as parental mPing loci in the analysis.
The recombination bin map and SNPs that distinguish the parental genotypes were used to phase the haplotypes of mPing loci in each RIL. Empty mPing loci within an NB or HEG4 haplotype were considered as potential excision sites and further confirmed by analyzing the alignments of reads covering the mPing insertion sites. Excisions were characterized only if there were two or more reads covering the 20 bp flanking an mPing insertion. Footprints of excision events were characterized by small insertion/deletions (indels) in the Compact Idiosyncratic Gapped Alignment Report (CIGAR) alignments. Two approaches were applied to identify independent excision events: 1) a standard method that counts each excision as an independent event assuming that all RILs are independent; 2) a conservative method that counts independent excisions from each locus according to the configuration of footprints in each RIL. Excisions that have the same footprint were counted as dependent events. A binomial test was used to test for the significance of high excision frequency mPing insertions based on a frequency of 0.0051 excision events per locus per plant. Two methods identified similar sets of mPing loci with high frequency excision events (SI Appendix, Table S13). Excisions of high excision frequency mPing loci identified by both the standard method and conservative method were manually inspected and confirmed by a PCR approach.
Structural Variation in the RILs and Proximity to mPing Elements.
A bioinformatics approach was employed to detect potential sequence rearrangements in the RILs. Only deletions could be identified with confidence because of the limitations associated with the resolution of short reads (75 bp and 100 bp in this study) and small insertion size libraries (~200 bp in this study). CNVnator was used to analyze the read depth of bam alignment files in 272 RILs (28). A series of filters were used to obtain high-confident deletions from the raw results of CNVnator: 1) considering only those regions with normalized read depth less than 0.05 as candidate deletions; 2) excluding centromeres, telomere chromosome ends, and low coverage regions in NB and HEG4 genomes; and 3) any candidate regions should have at least one read supporting the junction of at least one end of deletions. Candidate SVs were further analyzed by a PCR approach or sequencing with additional primers (SI Appendix, Supplementary Data 4).
Differential Gene Expression Analysis.
RNAseq reads from NB and HEG4 were obtained from SRA BioProject PRJNA264731. Each sample has three biological replicates with 51 nucleotide (nt) single-end reads. Reads were aligned to MSUv7 with tophat v2.1.1 (52) using default parameters. Read counts of protein-coding genes were estimated by the htseq-count tool from HTSeq v0.7.2 (53). Differential gene expression analysis was performed with edgeR v2.99.8 (54) using the generalized linear model (GLM). Genes with a false discovery rate (FDR) ≤ 0.05 were reported as differentially expressed genes.
Statistical Analysis.
Pearson’s correlation test, Mann–Whitney U test, one-way ANOVA, and Tukey’s HSD test were performed in R with “cor.test,” “wilcox.test,” “aov,” and “TukeyHSD” functions. Samples sizes, statistical tests, and P values are described in figures or figure legends.
Acknowledgments
We thank Dr. Anna M. McClung (US Department of Agriculture) for maintaining the RILs; and Dr. Venkateswari J. Chetty for preparation of plant materials and sequencing libraries. This work was supported by NSF Grant IOS-1027542 (to S.R.W. and J.E.S.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/10.1073/pnas.2015736117/-/DCSupplemental.
Data Availability.
Scripts used in this study are available on GitHub (https://github.com/stajichlab/Dynamic_rice_publications) or Zenodo (https://zenodo.org/record/3662095#.X2Jk6y2ZNTI). The raw sequences have been deposited in the National Center for Biotechnology Information SRA (accession no. SRP072364) or BioProject (accession no. PRJNA316308). All study data are included in the article and SI Appendix.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.2015736117
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/117/42/26288.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.2015736117
Article citations
A decade of dinoflagellate genomics illuminating an enigmatic eukaryote cell.
BMC Genomics, 25(1):932, 04 Oct 2024
Cited by: 0 articles | PMID: 39367346 | PMCID: PMC11453091
Review Free full text in Europe PMC
Forces driving transposable element load variation during Arabidopsis range expansion.
Plant Cell, 36(4):840-862, 01 Mar 2024
Cited by: 5 articles | PMID: 38036296
Recent reactivation of a pathogenicity-associated transposable element is associated with major chromosomal rearrangements in a fungal wheat pathogen.
Nucleic Acids Res, 52(3):1226-1242, 01 Feb 2024
Cited by: 2 articles | PMID: 38142443 | PMCID: PMC10853768
Phased secondary small interfering RNAs in Camellia sinensis var. assamica.
NAR Genom Bioinform, 5(4):lqad103, 24 Nov 2023
Cited by: 0 articles | PMID: 38025046 | PMCID: PMC10673657
Transposable Elements as a Source of Novel Repetitive DNA in the Eukaryote Genome.
Cells, 11(21):3373, 26 Oct 2022
Cited by: 14 articles | PMID: 36359770 | PMCID: PMC9659126
Review Free full text in Europe PMC
Go to all (9) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (1 citation) BioProject - PRJNA264731
Nucleotide Sequences (4)
- (1 citation) ENA - SRR1619373
- (1 citation) ENA - SRR833529
- (1 citation) ENA - SRR833485
- (1 citation) ENA - SRR1619147
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Tracking the genome-wide outcomes of a transposable element burst over decades of amplification.
Proc Natl Acad Sci U S A, 114(49):E10550-E10559, 20 Nov 2017
Cited by: 21 articles | PMID: 29158416 | PMCID: PMC5724284
Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana.
Proc Natl Acad Sci U S A, 104(26):10962-10967, 19 Jun 2007
Cited by: 71 articles | PMID: 17578919 | PMCID: PMC1904124
Tracking the origin of two genetic components associated with transposable element bursts in domesticated rice.
Nat Commun, 10(1):641, 07 Feb 2019
Cited by: 20 articles | PMID: 30733435 | PMCID: PMC6367367
mPing: The bursting transposon.
Breed Sci, 64(2):109-114, 01 Jun 2014
Cited by: 6 articles | PMID: 25053919 | PMCID: PMC4065317
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIH HHS (1)
Grant ID: S10 OD016290
National Science Foundation (1)
Grant ID: IOS-1027542





