Abstract
Background
Recent findings in neuroimaging and epigenetics offer important insights into brain structures and biological pathways of altered gene expression associated with posttraumatic stress disorder (PTSD). However, it is unknown to what extent epigenetic mechanisms are associated with PTSD and its neurobiology in youth.Methods
In this study, we combined a methylome-wide association study and structural neuroimaging measures in a Dutch cohort of youths with PTSD (8-18 years of age). We aimed to replicate findings in a similar independent U.S. cohort.Results
We found significant methylome-wide associations for pediatric PTSD (false discovery rate p < .05) compared with non-PTSD control groups (traumatized and nontraumatized youths). Methylation differences on nine genes were replicated, including genes related to glucocorticoid functioning. In both cohorts, methylation on OLFM3 gene was further associated with anterior hippocampal volume.Conclusions
These findings point to molecular pathways involved in inflammation, stress response, and neuroplasticity as potential contributors to neural abnormalities and provide potentially unique biomarkers and treatment targets for pediatric PTSD.Free full text

Differential DNA Methylation is associated with Hippocampal Abnormalities in Pediatric Posttraumatic Stress Disorder
Abstract
Background:
Recent findings in neuroimaging and epigenetics offer important insights into brain structures and biological pathways of altered gene expression associated with posttraumatic stress disorder (PTSD). However, it is unknown to what extent epigenetic mechanisms are associated with PTSD and its neurobiology in youth.
Methods:
In this study we combined a methylome-wide association study and structural neuroimaging measures in a Dutch cohort of youth with PTSD (ages 8-18 years). We aimed to replicate findings in a similar independent American cohort.
Results:
We found significant methylome-wide associations for pediatric PTSD (FDR p <0.05) compared to non-PTSD control groups (traumatized and non-traumatized youth). Methylation differences on 9 genes were replicated, including genes related to glucocorticoid functioning. In both cohorts, methylation on OLFM3 gene was further associated with anterior hippocampal volume.
Conclusions:
These findings point to molecular pathways involved in inflammation, stress response, and neuroplasticity as potential contributors to neural abnormalities and provide potentially unique biomarkers and treatment targets for pediatric PTSD.
Introduction
Childhood trauma is common and imposes a substantial psychological burden on youth. Approximately two-thirds of youth are exposed to psychological trauma before they reach adulthood, and up to 16% of all youth develop post-traumatic stress disorder (PTSD) by the age of 18 (1, 2). Youth suffering with PTSD often show lower academic achievement than their non-affected counterparts; this decrease in achievement is coupled with increased rates of depression, suicide attempts, and substance abuse into adulthood (3, 4). These statistics highlight the need for additional research into the neurodevelopmental underpinnings of pediatric PTSD, with the goals of improving detection, prognosis, and treatment.
The developmental origins of health and disease hypothesis posits that early life exposures, including exposure to childhood trauma, will have prolonged effects on child health, including neurodevelopmental outcomes. Epigenetic alterations provoked by exposure to childhood trauma are likely to play a central role in the molecular mechanisms underlying the emergence of PTSD in youth. Specifically, trauma-related alterations in DNA methylation (DNAm) at cytosine-guanine junctions (CpGs) may influence the developmental programming of neural circuitry underlying stress responses, including the hippocampus, amygdala, and medial prefrontal cortex (mPFC) (5).
Important strides have been made in identifying epigenetic substrates of adult PTSD using peripheral DNAm markers (6-8). Other studies have also reported associations between DNAm and brain phenotypes in predominantly healthy cohorts (9-12). However, despite these promising results, it remains unclear whether similar DNAm markers are present in a pediatric PTSD, which is of critical importance given ongoing physiological and neurodevelopment in youth. It is also unclear whether these DNAm markers are related to neural abnormalities previously identified in pediatric PTSD, which is important to identify potential molecular/cellular pathways associated with altered neurodevelopment contributing to pediatric PTSD (13). In addition, previous studies examining childhood trauma are mostly based on a priori searches of candidate genes(14). However, as is common with a priori searches, this approach increases the risk of confirmation biases, thereby increasing the risk of false negatives for useful epigenetic biomarkers of developmental psychopathology after adversity.
To address these knowledge gaps, we performed a Methylome-wide association study (MWAS) using a discovery cohort of youth with PTSD, compared with two control groups. The first group (Dutch sample) consisted of trauma-exposed youth without PTSD (a resilient group), and a second group of healthy non traumatized youth. We then attempted to replicate these DNAm findings in an independent (American) cohort of youth with PTSD and a non-traumatized comparison group.
In addition to our MWAS, we investigated whether altered methylation was further related to brain structure in regions involved in emotion regulation. This combined approach has two major advantages. First, the use of independent cohorts for discovery and replication of DNAm findings should reduce the likelihood of false positive findings and enhance generalizability of results. Second, identifying which peripherally-derived methylation differences are linked to structural brain differences in trauma-exposed and PTSD youth may point to genes with the most biological relevance for pediatric trauma and PTSD through their link to central nervous system abnormalities. Given prior work implicating altered methylation in glucocorticoid pathway genes in childhood trauma and adult PTSD, we hypothesized that youth with PTSD would show altered methylation in genes annotated to the HPA axis, which would then be further associated with prefrontal and hippocampal gray matter volume abnormalities previously identified in pediatric PTSD (15-19). We aimed to minimize DNAm findings that were either likely to occur by chance (by requiring replication) and that may not have an impact on neural systems (by requiring association with gray matter volume). Complementing these specific hypotheses, we aimed to identify novel methylation abnormalities and methylation-brain relationships in pediatric PTSD through our MWAS.
METHODS
Participants
In the present study, two independent MWAS were performed. The first cohort (N=224) was recruited at the Specialized Centre for Trauma and Family Treatment of the Bascule/Department of Child and Adolescent Psychiatry of the Amsterdam University Medical Center(The Netherlands). The second cohort (N=44) was recruited from area mental health facilities in Madison, Wisconsin, USA. In both cohorts, medication-free youth with PTSD were included (Partial or Full PTSD diagnosis on CAPS-CA interview). They were matched for age and sex with non-traumatized, typically-developing healthy comparison (NTC) youth. In the Dutch cohort an additional control group of traumatized comparison youth without PTSD (TC) was recruited. These cohorts have been previously described (20, 21)with additional information in supplementary methods and materials. All procedures were approved by the Medical Ethical Committee of the University Medical Center and the University of Wisconsin Health Sciences institutional review board.
Clinical and Behavioral Assessments
Assessments in these cohorts have been previously described. Briefly, participants and their caregivers completed structured psychiatric and trauma screening including the CAPS-CA. Additional self- and parent-report of youth symptoms of depression and anxiety were obtained using standardized questionnaires. Please see supplementary methods and materials for additional detail. To facilitate comparisons between study samples, Table 1 provides the rates of children that score above clinical cut-off of internalizing and externalizing symptoms based on these questionnaires.
Table 1:
Demographic information, trauma history and clinical characteristics in the Dutch cohort
| PTSD youth (n=74) | Traumatized Controls (n=75) | Healthy non-traumatized | |
|---|---|---|---|
| Sex | |||
 Boys Boys | 31 (41.9 %) | 37 (49.3 %) | 36 (48.0 %) |
 Girls Girls | 43 (58.1 %) | 38 (50.7 %) | 39 (52.0 %) |
| Age | 12.13 (3.44) | 11.95 (2.28) | 10.77 (2.15)a** |
| Ethnicity | |||
 Caucasian Caucasian | 44 (59.5 %) | 61 (81.3 %) c** | |
 Other Other | 30 (40.5 %) | 14 (18.7 %) | 64 (85.3 %)c** |
| Index trauma | |||
 Interpersonal violence Interpersonal violence | 38 (54.4%) | 21 (35.6%) | X |
 Sexual abuse Sexual abuse | 11 (14.9%) | 0 (0%) | X |
 Accidents/medical Accidents/medical | 9 (12.2%) | 37 (49.3%) | X |
| Comorbid diagnosis | |||
| Internalizing problems | 23 (31.1 %) | 2 (2.7 %)c** | 0 (0 %)c** |
| Externalizing problems | 18 (24.3 %) | 3 (4.0 %)c** | 0 (0 %)c** |
| CAPS-CA Severity Score | 52.91 (26.07) | 14.45 (12.88)b** | X |
| CRIES-13 Severity Score | 37.44 (15.35) | 11.04 (15.07)b** | X |
Continuous variables presented as mean (standard deviation); categorical variables presented as frequency (percentage).
DNA Acquisition and Extraction and generation of methylation signal
In both cohorts three milliliters of saliva was collected and stored in Oragene DNA sample collection kits (DNA Genotek, Canada). DNA was extracted using a Gentra autopure LS system following manufacturers protocol. For additional detail about the bisulfite conversion, see supplementary file. Methylation signal was generated using the HumanMethylation EPIC/850 BeadChip (Dutch cohort) and 450 BeadChip (USA cohort) following the manufacturer’s guidelines. See for details, supplementary file.
Quality Control and Data Processing
In both cohorts, prior to hybridization, cases and controls were randomized across the 96 well plates. Technical replicates (n = 8) were included for quality control of array, monitoring potential batch effects. Identical analysis pipelines were implemented across cohorts using R (v.3.4.2). In the supplementary file, each step is briefly described, followed by the R package and functions used during implementation. According to these steps, plate batch, ethnicity, sex and age were selected to be included as covariates in the statistical model. Ethnicity was based on genetic information from 27523 SNPs, thus we decided to keep these SNPs for genetic information, however without the X and Y genes, and cross hybridization probes. The second component of our prinicipal component analysis (PCA2) correlated with ethnicity (.03). We therefore added PCA2 as a covariate in our analysis. Next, we identified group-related differentially methylated positions (DMPs) and regions (DMRs), using a general linear model (lmfit) that accounts for the main effects between groups: (1) PTSD vs NTC in both cohorts and (2) PTSD vs TC in the Dutch cohort. To correct for inflation of p-values the BACON package was used (22), the Lambda’s after using BACON are reported in Supplementary Table 6. A false discovery rate (FDR) for DMPs and a family wise error rate (FWER) for DMRs was used to adjust for multiple testing (p. < .05). These corrections were applied to the results from the Dutch correct, but not the American cohort. Because the American cohort was used as a replication sample, stringent corrections would risk unduly and artificially increasing the rate of false negative findings. Therefore in the American cohort a Bonferroni correction (p < 0.007) was applied.The Bonferroni threshold was the by-product of dividing critical alpha = 0.05 by the number of GOIs identified in the Dutch cohort. Bonferroni corrections were combined with Fisher’s method for combining p values from the independent tests across CpGs to approximate the DMR p-value (p. <0.5) (23, 24).DMRs were analyzed with use of the Minfi function bumphunter. DMRs were defined to include at least 2 probes in the cluster. For each significantly associated DMR beta-values of each of the individual CpGs were extracted, which were used in subsequent post-hoc and sensitivity analyses. CpGs were visualized using the R package coMET (v.1.14.0) (25). We used M-values for the analysis (also for calculating the Bacon P values), and Beta values for visualizing our data in the figures.
MRI Acquisition
For the Dutch cohort all scans were acquired using a 3.0T Philips Achieva scanner (Philips Healthcare, Best, The Netherlands) equipped with a SENSE eight-channel receiver head coil. For each participant, a T1-weighted structural MRI image was acquired with the following parameters: TE: 3.527 ms, TR: 9 ms, slice thickness: 1 mm, 170 slices, flip angle: 8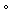 and image matrix 256 x 256 that covert the entire brain. For the American cohort, MRI acquisition parameters are similar and have been reported elsewhere (Keding & Herringa 2015).
and image matrix 256 x 256 that covert the entire brain. For the American cohort, MRI acquisition parameters are similar and have been reported elsewhere (Keding & Herringa 2015).
Image Preprocessing and Voxel-Based Morphometry
The Dutch and American cohorts used the same image preprocessing and voxel-based morphometry (VBM) procedures, which have been reported elsewhere (26), see supplement for additional information. Gray matter volume (GMV) from previously identified regions of interest in youth with PTSD (ROIs - hippocampus, mPFC, fusiform gyrus) (21) were extracted for focused comparisons with DNA methylation. See supplemental methods and materials for additional information.
Post Hoc Analyses
Peak DNA methylation at DMPs and DMRs and their associations with GMV were assessed using partial correlation and regression analyses in R (ppcor) in a subset of the Dutch cohort (see Supplementary Table 4) and in the entire American cohort. Due to regulatory restrictions in the Dutch cohort it was not possible to collect neuroimaging in the non-traumatized control group.
We selected one significant DMP (significant in the Dutch cohort after multiple testing correction) and nine DMRs that showed an overlap within the two cohorts for our post-hoc analysis. MWAS replication analysis was defined as overlap for at least one DMP within a DMR for both cohorts, comparing the top 1000 DMRs. For post hoc analysis we performed both partial correlation analyses between GMV available for each cohort and the information from the original DMR. Analyses were covaried for age, sex, trauma type and PTSD duration. Additionally, these results were further corrected for multiple comparisons using an FDR correction and a Bonferroni correction for the number of post-hoc analyses conducted. Post-hoc results with a corrected p < 0.007 (7 sets of partial correlation analyses) were considered statistically significant.
RESULTS
Demographic and mental health measures of participating youth are shown in Table 1 and and22 and further described in supplement.
Table 2:
Demographic Information, Trauma History and Clinical Characteristics in the USA cohort
| PTSD Youth (n=22) | Healthy non-traumatized | |
|---|---|---|
| Sex | ||
 Boys Boys | 8 | 7 |
 Girls Girls | 14 | 13 |
| Age | 14.22 (2.70) | 14.21 (2.80) |
| Ethnicity | ||
 Caucasian Caucasian | 18 | 17 |
 Other Other | 4 | 3 |
| IQ | 103.2 (13.0) | 110.5 (12.5) |
| Left Handed | 0 | 0 |
| Index Trauma | ||
 Interpersonal Violence Interpersonal Violence | 3 (13.6%) | - |
 Sexual Abuse Sexual Abuse | 11 (50%) | - |
 Severe Severe | 3 (13.6%) | - |
Accident/medical trauma Other (Traumatic News, Natural Disaster) Other (Traumatic News, Natural Disaster) | 5 (22.7%) | - |
| Comorbid Diagnoses | ||
 Depression Depression | 19 (86.4%) | - |
 Anxiety Anxiety | 14 (63.6%) | - |
 ADHD ADHD | 6 (27.3%) | - |
| Previous Psychotropic Medication-Use | ||
 Depression Depression | 8 (36.4%) | - |
 Anxiety Anxiety | 3 (13.6%) | - |
 ADHD ADHD | 7 (31.8%) | - |
| CAPS-CA Severity Score | 77.11 (16.75) | - |
| PTSD-RI Severity Score | 55.36 (11.14) | - |
Continuous variables presented as mean (standard deviation); categorical variables presented as frequency (percentage). There were no significant group differences (PTSD Youth vs. Healthy Youth; p > 0.05) in sex, ethnicity, or handedness distribution, age, or IQ (based on χ2 tests of independence and independent samples t-tests respectively). PTSD = Post-Traumatic Stress Disorder; ADHD = Attention Deficit, Hyperactivity Disorder; CAPS-CA = Clinician-Administered PTSD Scale for Children and Adolescents; PTSD-RI = UCLA PTSD Reaction Index. Comorbid diagnosis were based on the Kiddie Schedule for Affective Disorders and Schizophrenia (KSADS), Mood and Feelings Questionnaire (MFQ), and Screen for Child Anxiety Related Emotional Disorders (SCARED).
EWAS analysis
A set of MWAS (differentially-methylated regions [DMRs] and differentially-methylated positions [DMPs]) contrasting the PTSD and comparison groups identified statistically significant CpG sites. DMR analysis comparing the groups identified significant CpG regions at FWER < 0.05. In the Dutch cohort comparing PTSD with the NTC, the most significant effect was reported on the TNXB gene (FWER = 0.03) and the PM20D1 gene (FWER = 0.04). At the TNXB gene PTSD youth showed hyper methylation compared to NTC, at the PM20D1 gene PTSD youth showed hypomethylation compared to NTC. The list of the top 10 CpG regions in both cohorts are provided in Supplementary Table 1 and 2. The findings annotated to the TNXB and PM20D1 gene (PTSD vs. NTC youth) were replicated with the same set of MWAS in the American cohort. Results are shown in Table 3.
Table 3:
Overlapping DMRs identified in the PTSD vs Non-Traumatized Control group in both cohorts.
| Gene | Chr | p-value Dutch Cohort | FWER Dutch Cohort | p-value USA Cohort | p-value Fisher’s method | Direction | |
|---|---|---|---|---|---|---|---|
| PTSD Youth vs. NTC Youth | TNXB | 6 | < 0.001 | 0.030 | 0.00033 | 0.02334** | PTSD > NTC |
| PM20D1 | 1 | < 0.001 | 0.040 | 6.58E-05 | 0.01104** | NTC > PTSD | |
| TNXB | 6 | < 0.001 | 0.100 | 0.00169 | 0.0992 | PTSD > NTC | |
| DUSP22 | 6 | < 0.001 | 0.320 | 0.00119 | 0.13888 | NTC > PTSD | |
| GDF7 | 2 | 0.002 | 0.958 | 0.00119 | 0.415772 | NTC > PTSD | |
| SLC1A4 | 2 | 0.004 | 0.990 | 0.00550 | 0.99 | NTC > PTSD | |
| KLHL35 | 11 | 0.004 | 0.998 | 0.00018 | 0.596804 | PTSD > NTC | |
| ZNF714 | 19 | 0.009 | 1 | 0.00206 | 0.996 | NTC > PTSD | |
| OLFM3 | 1 | 0.016 | 1 | 0.02906 | 1 | PTSD > NTC |
Detected DMRs (L>2) using minfi’s “bumphunter” function; comparison based on the top 1000 hits in both cohorts. All findings met threshold of a combined FWER calculated with Fisher’s method of < 0.05.
In the Dutch cohort, the most significant DMP was on the corticotrophin-releasing hormone binding protein (CRHBP) gene, located at cg26196496 (FDR < 0.02). Here, PTSD youth showed hyper methylation at this CpG relative to NTC youth. A plot showing greater detail on the CpGs in or near CRHBP is provided in Supplementary Figure 1. Furthermore, PTSD youth showed hypomethylation at a CpG site located on cg21972431 relative to trauma comparison (TC) youth (FDR < 0.01). These DMP findings did not replicate in the American cohort. The lists of the top 5 CpG sites, derived from the DMP analysis in both cohorts, are shown in Supplementary Table 3 and 4..
Exploratory Analysis of DMR/DMP Conjunction in the Dutch and American Cohorts
Next, we compared the top 1000 DMRs between the two cohorts to assess potentially important DNAm abnormalities not detected by the more stringent multiple comparison correction threshold in the Dutch cohort EWAS analysis. This revealed nine DMRs that were associated with PTSD (relative to NTC youth) in both cohorts. These DMRs are shown in Table 3. In both cohorts, we identified overlap on two very large DMRs showing hypermethylation in PTSD youth relative to NTC youth (63 CpGs in the Dutch cohort, 68 CpGs in the American cohort) annotated on the TNXB gene. An overview of the DMR outcomes per cohort are shown in Supplementary Table 1. The DMP conjunction analysis did not reveal overlap between the two cohorts. The results of the DMP analyses by cohort are reported in Supplementary Table 3 and 4..
Associations Between DNA Methylation and Gray Matter Volume
We next assessed potential relationships between DNAm findings with gray matter volume in regions of interest (ROIs) previously identified in pediatric PTSD including the ventromedial prefrontal cortex, hippocampus, and amygdala (13). We performed partial correlations to associate regional grey matter volume with peak methylation on the DMPs/DMRs that survived our multiple testing correction in the Dutch cohort or that were replicated in the American cohort. Only a subgroup of the youth participated in this post hoc analyses (see online methods).
These analyses revealed that a DMR annotated to Olfactomedin 3 (OLFM3) showed a significant negative correlation with right anterior hippocampus GMV in both cohorts (Supplementary Figure 2). Additional findings only detected in one cohort are shown in Table 4 and Supplementary Figure 2.
Table 4:
Significant post-hoc partial correlation results comparing peak methylation on DMPs/DMRs with regional grey matter volume estimates
| Gene | Chr | Region | r Dutch Cohort | p-value Dutch Cohort | FDR p-value Dutch Cohort | r USA Cohort | p-value USA Cohort | FDR p-value USA Cohort | |
|---|---|---|---|---|---|---|---|---|---|
| PTSD Youth and NTC Youth | CRHBP | 5 | R Fusiform Gyrus | 0.328 | 0.002 | 0.018 | - | - | |
| CRHBP | 5 | L Fusiform Gyrus | 0.350 | < 0.001 | 0.016 | - | - | ||
| GDF7 | 2 | R Anterior Hippocampus | 0.288 | 0.006 | 0.037 | - | - | ||
| GDF7 | 2 | R vmPFC | 0.294 | 0.005 | 0.037 | - | - | ||
| GDF7 | 2 | R Fusiform Gyrus | - | - | - | 0.455 | < 0.001 | 0.003 | |
| GDF7 | 2 | L Fusiform Gyrus | - | - | - | 0.375 | 0.003 | 0.018 | |
| OLFM3 | 1 | R Anterior Hippocampus | −0.323 | 0.002 | 0.018** | −0.453 | < 0.001 | 0.003** | |
| OLFM3 | 1 | R vmPFC | −0.369 | < 0.001 | 0.014 | - | - | - | |
| PM20D1 | 1 | R Fusiform Gyrus | - | - | - | 0.384 | 0.003 | 0.017 | |
| SLC1A4 | 2 | R Fusiform Gyrus | - | - | - | 0.511 | < 0.001 | 0.001 | |
| SLC1A4 | 2 | L Fusiform Gyrus | - | - | - | 0.335 | 0.010 | 0.043 | |
| TNXB | 6 | R Anterior Hippocampus | - | - | - | −0.406 | 0.001 | 0.012 |
Partial correlations were controlled for age and sex of participant. All findings met threshold of FDR p-value < 0.05.
All results presented above remained significant after sensitivity analyses controlling for trauma-related variables, suggesting the difference is specific to the development of pediatric psychopathology and not a consequence of trauma type, time elapsed since the trauma, and trauma load. For an overview of the outcomes see Table 4.
DISCUSSION
To our knowledge, this is the first study in which biological pathways of altered gene expression and brain structures implicated in pediatric PTSD have been investigated. Using two independent, international cohorts, the results of this study confirm for the first time the hypothesis that pediatric PTSD is associated with epigenetic modifications which, in turn, are associated with structural neurophenotypes implicated in pediatric PTSD. Most notably, altered DNA methylation on genes annotated to PM20D1, TNXB and OLFM3 genes were replicated across cohorts and exhibited a relationship to neural structure in at least one of the cohorts. Because this is the first genome wide methylation study in youth with PTSD to date, we cannot compare these results with other cohorts (15). It is notable, however, that our results do not overlap with recent findings in adult PTSD (Smith, Ratanatharathorn (6). This is perhaps not surprising, given that the current study focused on a developmental sample, in which trauma load, comorbidities, substance use, and many other factors markedly differ from adult PTSD samples. However, our findings do thematically overlap with studies of childhood trauma and adult PTSD, suggesting abnormal methylation in genes annotated to the inflammation and stress response systems (15).
First, changes observed on OLFM3 gene were related to hippocampal volume in both cohorts. This may represent an interaction between methylation and neuronal change related to early exposure to traumatic events and the development of PTSD at an early stage in life. OLFM3 is a neuronal protein, found throughout the brain and related to the development of microglia (27, 28). Microglia serve as key immune cells of the brain, and are activated in response to signals they receive or detect in their microenvironment. For example, they respond to injuries and infections and are able to modify the structure and function of a cell. They can for example lead to neuronal degeneration, impaired microglia activation has been linked to the development of brain disorders, such as neurodegenerative diseases (29). Epigenetic mechanisms have emerged as important regulators in this process (30). In addition to their essential role in the development of the central nervous system (CNS), microglial dysfunction is suggested to be involved in stress vulnerability and depression recurrence (31). These findings are in line with our results that suggest that hyper methylation of OLFM3 may be a molecular mechanism by which hippocampal volume is decreased in youth with PTSD, potentially via enhanced CNS inflammation in affected youth.
Secondly, methylation changes in the TNXB gene were observed in both cohorts. TNXB encodes an extracellular-matrix associated glycoprotein that has been implicated in cell migration processes (ProteinAtlas), and was shown to be hypermethylated in PTSD youth relative to NTC youth. Furthermore, an additional significant effect was reported comparing PTSD with NTC youth in the Dutch cohort. This suggests that there is an effect specific to pediatric PTSD (and/or its comorbid disorders) rather than trauma-exposure generally. We also observed that in the American cohort across PTSD youth and NTC youth, TNXB methylation is negatively associated with gray matter volume in the anterior hippocampus, originally identified in a group by age interaction in Keding & Herringa (2015). This relationship was not observed in the Dutch cohort, however this could be due to the missing NTC in the MRI group. Interestingly, TNXB has been shown to have protein-protein interactions with the protein products of VEGFA, VEGFB, NEURL1, and NEURL 1B (https://string-db.org/), all four of which have been heavily implicated in cellular processes in the anterior hippocampus. More specifically, these proteins are functionally related to hippocampal-dependent synaptic plasticity, learning, and memory (https://www.genecards.org/). This suggests that methylation of TNXB may act as a molecular mechanism of altered synaptic plasticity or growth affecting hippocampal volume in pediatric PTSD, which may then have implication for multiple hippocampal functions such as context discrimination, fear and extinction learning, and emotion regulation implicated in pediatric PTSD (13).
In addition to the results of the DMR analysis, we detected DNAm abnormalities on the CRHBP gene in the Dutch cohort. This gene is known for its critical role in the regulation of corticotrophin-releasing hormone binding protein (CRH-BP), which in turn plays a modulatory role in the function of the HPA axis as well as CRH signaling within the brain. The HPA-axis is thought to play an important role in the etiology of PTSD, and the regulation of emotions and behavior (15). In addition, numerous studies in rodents have shown that levels of CRH-BP in various brain regions are highly responsive to psychological stressors (32). Furthermore, studies comparing post mortem brain tissue found that altered CRH-BP expression has been associated with adult psychopathology and suicide completion (33, 34). In the case of pediatric PTSD, it is possible that altered CRH-BP methylation, and thus altered expression, may contribute to abnormal CRH signalizing both peripherally and centrally, contributing to heightened physiological and stress responses associated with PTSD. In line with earlier studies, these findings further support the involvement of abnormal gene regulation associated with HPA axis dysregulation.
While this study highlights potentially novel genetic and molecular mechanisms contributing to pediatric PTSD, several limitations should be acknowledged. First, we used data generated on different arrays between the two cohorts (HumanMethylation450 in the Dutch cohort and EPIC bead chip array in the US cohort). Data generated with these arrays are comparable (the EPIC bead chip covers the whole 450k array), but only capture a fraction of the CpG sites in the genome. Second, this study has a limited sample size in both cohorts, creating risk for both false negative and false positive findings particularly in a genetic study. To mitigate the latter, we included youth with carefully diagnosed, phenotypes to increase the probability that important, and true, group differences would be identified. However, replication is a strength that in part helps to overcome the small sample size. Another limitation regarding our cohorts are the comorbid problems that have been reported, such as ADHD and depression which may limit specificity of findings to PTSD.
Additionally, we applied strict corrections in the Dutch (MWAS)search, with replication in the American cohort at an uncorrected level, in order to reduce the overall risk of false positive results while balancing the risk of false negatives. Third, our study examined gene methylation using only saliva samples. Though this approach likely captures part of the underlying pathophysiology of pediatric PTSD, it is unlikely to represent exact DNA methylation in brain regions most relevant for PTSD. To mitigate this possibility, we have emphasized DNAm abnormalities that also map onto known structural brain abnormalities in pediatric PTSD.
Future studies would be warranted to collect longitudinal data in youth with and without trauma-exposure and PTSD to determine the temporal course of methylation-neural structure relationships in relation to childhood trauma exposure and the emergence of PTSD. Given the implication of inflammatory and glucocorticoid pathways in this initial study, future studies would also benefit from including other peripheral or central markers such as cortisol and cytokine levels, and determine whether these represent mediating factors between altered peripheral methylation and structural brain changes leading to pediatric PTSD. Finally, future studies would benefit from collecting additional tissue sources (blood, saliva and eventually post-mortem brain tissue), as well as information about RNA and protein expression.
In conclusion, our findings provide new insights into the underlying biology and potential biomarkers for PTSD in youth. To our knowledge, this is the first investigation into differential DNA methylation in pediatric PTSD and its association to structural brain abnormalities. While youth with PTSD are an inherently difficult population to recruit, we utilized a combination of two independent, international, and well-phenotyped cohorts, a common data analytic pipeline, strict statistical corrections, and linkage to neural phenotypes, to increase the likelihood of important and reproducible findings in pediatric PTSD. These findings offer future targets for hypothesis driven studies in larger samples and statistical rigor for early identification of candidate gene pathways. If replicated in subsequent work, the epigenetic markers identified here could serve as novel therapeutic targets in the prevention and treatment of pediatric PTSD.
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Commercial Assay Or Kit | Human: Saliva | DNA Genotek, Canada | ||
| Other | EPIC/850 BeadChip | Illumina, Inc. | ||
| Other | 450 BeadChip | Illumina, Inc. | ||
| Other | EZ DNA Methylation-Gold kit | Zymo Research | ||
| Other | Qubit | Qiagen, USA | ||
| Software; Algorithm | R version 3.5.0 | R Project for Statistical Computing | RRID:SCR_001905 | |
| Software; Algorithm | Methylaid package | R Project for Statistical Computing | RRID:SCR_002659 | |
| Software; Algorithm | Minfi package | R Project for Statistical Computing | RRID:SCR_012830 | |
| Software; Algorithm | CoMet package | R Project for Statistical Computing | RRID:SCR:011925 | |
| Software; Algorithm | Statistical Parametric Mapping | Wellcome Department of Imaging Neuroscience, London, UK | RRID:SCR:007037 | |
| Software; Algorithm | VBM8 toolbox | http://dbm.neuro.uni-jena.de/vbm/) | ||
| Software; Algorithm | MATLAB v9.1 | Mathworks | RRID:SCR_001622 |
Acknowledgements
We would like to thank Rosanne op den Kelder (Msc), psychologist at the Bascule, Academic Centre for Child an Adolescent psychiatry, for her help with the collection of the clinical data. This project is partly funded by the Augeo Foundation. The funder has no role in the design and conduct of the study, the collection, management, analysis, or interpretation of data, or article preparation. A preliminary print of this paper was posted on Research Square (2020).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures All authors report no biomedical financial interests or potential conflicts of interest.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.bpsc.2021.04.016
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8568739
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/105556717
Article citations
Methylome-wide and meQTL analysis helps to distinguish treatment response from non-response and pathogenesis markers in schizophrenia.
Front Psychiatry, 15:1297760, 07 Mar 2024
Cited by: 0 articles | PMID: 38516266 | PMCID: PMC10954811
Through a Developmental Lens: Emerging Insights to Understand and Treat Pediatric PTSD.
Am J Psychiatry, 180(9):636-644, 01 Sep 2023
Cited by: 1 article | PMID: 37654114 | PMCID: PMC10636806
Review Free full text in Europe PMC
Genome-wide association and replication studies for handedness in a Korean community-based cohort.
Brain Behav, 13(9):e3121, 20 Jun 2023
Cited by: 2 articles | PMID: 37337823 | PMCID: PMC10498080
The shared mother-child epigenetic signature of neglect is related to maternal adverse events.
Front Physiol, 13:966740, 24 Aug 2022
Cited by: 4 articles | PMID: 36091392 | PMCID: PMC9448913
Sex-based variations of prefrontal structure and longitudinal symptoms in pediatric posttraumatic stress disorder.
Depress Anxiety, 39(12):902-912, 09 Nov 2022
Cited by: 1 article | PMID: 36349877 | PMCID: PMC9762118
Go to all (8) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Integration of neural and epigenetic contributions to posttraumatic stress symptoms: The role of hippocampal volume and glucocorticoid receptor gene methylation.
PLoS One, 13(2):e0192222, 07 Feb 2018
Cited by: 11 articles | PMID: 29415058 | PMCID: PMC5802910
Understanding posttraumatic stress disorder: insights from the methylome.
Genes Brain Behav, 13(1):52-68, 28 Nov 2013
Cited by: 25 articles | PMID: 24286388
Review
Genome-wide differentially methylated genes associated with posttraumatic stress disorder and longitudinal change in methylation in rape survivors.
Transl Psychiatry, 11(1):594, 19 Nov 2021
Cited by: 9 articles | PMID: 34799556 | PMCID: PMC8604994
Abnormal Prefrontal Development in Pediatric Posttraumatic Stress Disorder: A Longitudinal Structural and Functional Magnetic Resonance Imaging Study.
Biol Psychiatry Cogn Neurosci Neuroimaging, 4(2):171-179, 16 Aug 2018
Cited by: 32 articles | PMID: 30343133 | PMCID: PMC6371792
Funding
Funders who supported this work.
NIMH
NIMH NIH HHS (3)
Grant ID: R01 MH115910
Grant ID: R01 MH117141
Grant ID: K08 MH100267




