Abstract
Free full text

Altered CXCR4 dynamics at the cell membrane impairs directed cell migration in WHIM syndrome patients
Significance
New imaging-based approaches are incorporating new concepts to our knowledge of biological processes. The analysis of receptor dynamics involved in cell movement using single-particle tracking demonstrates that cells require chemokine-mediated receptor clustering to sense appropriately chemoattractant gradients. Here, we report that this process does not occur in T cells expressing CXCR4R334X, a mutant form of CXCR4 linked to WHIM syndrome (warts, hypogammaglobulinemia, infections, myelokathexis). The underlaying molecular mechanism involves inappropriate actin cytoskeleton remodeling due to the inadequate β-arrestin1 activation by CXCR4R334X, which alters its lateral mobility and spatial organization. These defects, associated to CXCR4R334X expression, contribute to the retention of hematopoietic precursors in bone marrow niches and explain the severe immunological symptoms associated with WHIM syndrome.
Abstract
Chemokine receptor nanoscale organization at the cell membrane is orchestrated by the actin cytoskeleton and influences cell responses. Using single-particle tracking analysis we show that CXCR4R334X, a truncated mutant chemokine receptor linked to WHIM syndrome (warts, hypogammaglobulinemia, infections, myelokathexis), fails to nanoclusterize after CXCL12 stimulation, and alters the lateral mobility and spatial organization of CXCR4 when coexpressed. These findings correlate with multiple phalloidin-positive protrusions in cells expressing CXCR4R334X, and their inability to correctly sense chemokine gradients. The underlying mechanisms involve inappropriate actin cytoskeleton remodeling due to the inadequate β-arrestin1 activation by CXCR4R334X, which disrupts the equilibrium between activated and deactivated cofilin. Overall, we provide insights into the molecular mechanisms governing CXCR4 nanoclustering, signaling and cell function, and highlight the essential scaffold role of β-arrestin1 to support CXCL12-mediated actin reorganization and receptor clustering. These defects associated with CXCR4R334X expression might contribute to the severe immunological symptoms associated with WHIM syndrome.
WHIM syndrome (an acronym for warts, hypogammaglobulinemia, recurrent bacterial infections, and myelokathexis) is a rare combined immunodeficiency disorder that is linked to inherited, heterozygous autosomal-dominant mutations in the gene for the chemokine receptor CXCR4 (1). Typically, mutations cause premature termination or a frameshift in the region that encodes the cytoplasmic tail of the protein, which is important for signaling (2, 3). Indeed, elevated G protein-dependent signaling has been reported in leukocytes from patients with WHIM syndrome and in cell lines expressing CXCR4WHIM mutants, which correlates with impaired CXCR4 desensitization and internalization in response to the receptor ligand CXCL12 (2). This can be mechanistically linked to an inability of mutant CXCR4 to activate negative feedback loops, including the induction of G protein-coupled receptor kinase and protein kinase C-mediated phosphorylation of the C-terminus domain of the receptor for coupling to β-arrestins (4, 5). Accordingly, CXCR4WHIM mutant receptors show enhanced signaling activation in the presence of CXCL12, and are therefore considered as gain-of-function mutants (6). The most common and best-studied WHIM mutation is CXCR4R334X, which lacks the C-terminal 19 amino acids (7). Mice and zebrafish expressing CXCR4R334X recapitulate the neutropenia and myelokathexis observed in patients with WHIM syndrome (8, 9). In addition to explaining the loss of receptor internalization (10), the partial deletion of the cytoplasmic tail could also lead to changes in its interactions with the actin cytoskeleton (11). Chemokines are known to regulate actin dynamics (12), which coordinate cell responses (13). Among its physiological roles, the actin cytoskeleton regulates membrane diffusion, protein compartmentalization and clustering, and controls receptor signaling (14). Recent data suggest that the actin cytoskeleton is essential for ligand-mediated CXCR4 nanoclustering, which is necessary for achieving complete receptor functions (15).
The C-terminal end of CXCR4 is implicated in the recruitment of β-arrestin adaptor proteins, which link CXCR4 to a clathrin lattice and participate in coupling the receptor to the actin cytoskeleton (11). Previous studies have shown that G protein-dependent actin reorganization is mediated by mechanisms involving members of the Rho family of small GTPases and cofilin activation (16, 17). This signaling cascade contributes to the Rac1-mediated generation of free actin barbed ends at the leading edge of motile cells (18). β-Arrestins have been postulated to spatially regulate actin polymerization by scaffolding cofilin at the leading edge (19, 20). Complexes between β-arrestins and the actin-binding protein filamin A have been identified by proteomics analysis (21) and have been shown to functionally cooperate to regulate ERK activation and actin cytoskeleton reorganization (22).
Using quantitative single-molecule spatio-dynamic imaging, we show here that CXCR4R334X is unable to form large nanoclusters in response to CXCL12. Similar behavior was observed for CXCR4 in cells lacking β-arrestin1. These results suggest that—due to the lack of negative feedback loops—CXCR4R334X triggers continuous G protein activation in the presence of CXCL12, which in turn distorts the balance between active and inactive cofilin, perturbing actin dynamics and ligand-mediated receptor nanoclustering in migrating cells. This phenotype concours with the formation of multiple lamellae in CXCR4R334X-expressing cells through mechanisms involving sustained cofilin activation, which might also contribute to the inability of these cells to correctly sense chemokine gradients. Our results also indicate that by altering the activity of the actin-binding protein cofilin, β-arrestin–mediated signaling pathways influence the lateral partitioning of CXCR4 and the ability of cells to correctly sense chemoattractant gradients.
Results
Gain-of-Function CXCR4R334X Does Not Enhance Cell Migration in a Lipid-Bilayer System.
CXCR4R334X behaves as a gain-of-function mutant because of its inability to be desensitized (6). We tested the capacity of CXCR4R334X to promote cell migration in transwell assays using transiently transfected Jurkat (JK) cells (cells expressing CXCR4 endogenously) or JKX4−/− cells (CXCR4 deficient cells) (SI Appendix, Fig. S1 A and B). As expected, cell migration of GFP+ cells to CXCL12 was higher in JKX4−/−-R334X cells (CXCR4-deficient cells that transiently express CXCR4R334X-AcGFP) than in JKX4−/−-X4 cells (CXCR4-deficient cells that transiently express CXCR4-AcGFP), at all concentrations tested (Fig. 1A). In agreement with the dominant effect of mutant CXCR4 receptors when coexpressed in heterozygosis with wild-type CXCR4 (1), we found that ligand-mediated cell migration was higher in JK-R334X cells than in JK-X4 cells (Fig. 1A). We also found that the WHIM mutation promoted a significant reduction of CXCL12-induced receptor internalization in both JKX4−/−-R334X and JK-R334X cells (Fig. 1 B and C), confirming previous work (6).
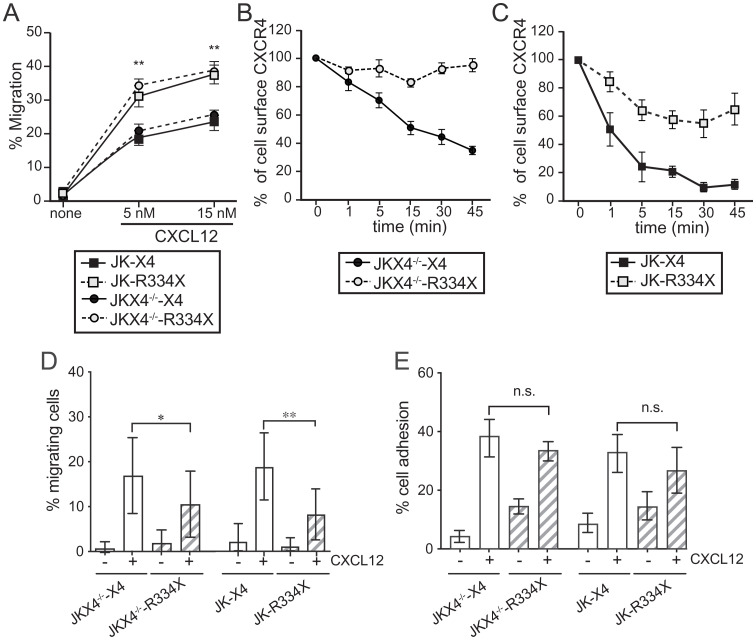
CXCR4R334X expressed in both homo- and heterozygosis alters CXCL12-mediated responses. (A) JK-X4, JK-R334X, JKX4−/−-X4, and JKX4−/−-R334X cell migration in Boyden chambers in response to CXCL12. Data are shown as the mean percentage (plus SD) of input cells that migrate (n = 5; **P ≤ 0.01). (B) Cell surface expression of CXCR4 and CXCR4R334X in JK-X4 and JK-R334X cells after stimulation with CXCL12 (40 nM) at different time points and analyzed by flow cytometry using an anti-CXCR4 antibody in nonpermeabilized cells. Results show mean ± SEM of the percentage of CXCR4 expression at the cell surface (n = 4). (C) Surface receptor expression in JKX4−/−-X4 and JKX4−/−-R334X cells was analyzed as in B. Results show mean ± SEM of the percentage of CXCR4 expression at the cell surface (n = 3). (D) Migration frequency of JKX4−/− or JK cells transiently transfected with CXCR4 wild-type (-X4) or with CXCR4R334X (-R334X) on ICAM-1-containing lipid bilayers alone or together with CXCL12 (mean ± SD, n = 3; *P ≤ 0.05, **P ≤ 0.01). (E) Cell adhesion frequency of cells as in D on ICAM-1–containing lipid bilayers alone or together with CXCL12 (mean ± SD, n = 3; n.s., not significant).
As transwell experiments reflect the contribution of only some molecular components involved in migration, we tested the behavior of cells using another experimental model where cell migration depends not only on the expression of chemokine receptors, but also on integrin activation. Using a two-dimensional lipid bilayer system with embedded intercellular adhesion molecule 1 (ICAM-1) and CXCL12, we found that the percentage of migrating (GFP+) cells was significantly lower for JKX4−/−-R334X and JK-R334X cells than for JKX4−/−-X4 or JK-X4 cells (Fig. 1D). Although many factors can affect cell migration (23), we discarded significant differences in CXCL12-mediated cell adhesion to ICAM-1 between the different cells used (Fig. 1E). The reduction in cell migration in the absence of differences in cell adhesion strongly suggests that CXCR4R334X functions less efficiently than CXCR4 in experiments mimicking physiological cell migration.
CXCL12 Fails to Promote Larger CXCR4R334X Nanoclusters.
By acting as a physical barrier, the actin cytoskeleton modulates plasma membrane compartmentalization and membrane protein dynamics (24, 25). Hence, actin dynamics not only define the cell shape during migration, but also have an essential role in coordinating chemokine receptor signaling (26). We previously found that abrogating actin polymerization with the actin monomer sequestering drug latrunculin A abolishes CXCL12-mediated CXCR4 nanoclustering (15). We next used single-particle tracking (SPT) in total internal reflection fluorescence (TIRF) mode to examine CXCR4 and CXCR4R334X dynamics in JKX4−/− cells. (Movies S1–S4). This technique allows the detection of individual molecules close to the plasma membrane with great signal-to-background ratio, and has been used to capture the dynamics of individual receptors or complexes as well as determining their stoichiometries (see SI Appendix, Supplementary Methods for more discussion) (27).
Using cells expressing the receptors in homozygosis, we observed that CXCR4 and CXCR4R334X dynamics were very similar in steady state; in both cases the highest proportion of CXCR4 particles corresponded to mobile particles (~93% vs. ~92%) (Fig. 2A). The median value of the short time-lag diffusion coefficient (D1–4) for both CXCR4 and the mutant CXCR4R334X was also similar (0.027 μm2/s for CXCR4 and 0.023 μm2/s for the mutant) (Fig. 2B). CXCL12 promoted a significant reduction in overall receptor diffusivity (basal, median D1–4 = 0.027 μm2 s−1; CXCL12, median D1–4 = 0.011 μm2 s−1) and increased the percentage of immobile particles from ~7% (basal) to ~20% (CXCL12) in JKX4−/−-X4 cells but not in JKX4−/−-R334X cells (basal, median D1–4 = 0.023 μm2 s−1; CXCL12, median D1–4 = 0.037 μm2 s−1). In the latter case, we also detected a similar percentage of immobile particles independently of ligand activation (~8% basal vs. ~6% CXCL12) (Fig. 2 A and B).
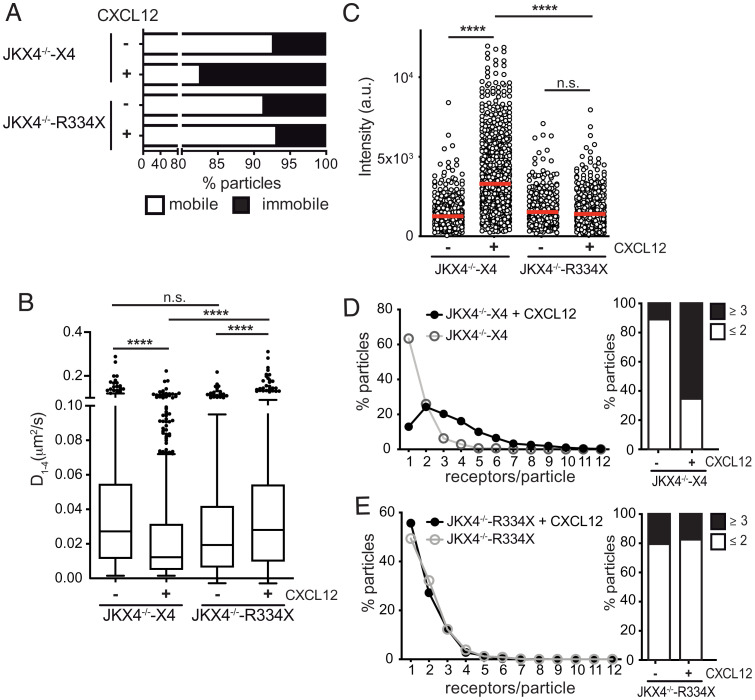
CXCL12 does not influence CXCR4R334X dynamics and nanoclustering. SPT analysis of CXCR4-AcGFP and CXCR4R334X-AcGFP in JKX4−/− cells on fibronectin (FN) or FN+CXCL12-coated coverslips (697 particles in 64 cells on FN; 1,221 in 66 cells on FN+CXCL12 in JKX4−/−-X4 cells; 461 in 54 cells on FN; 775 in 72 cells on FN+CXCL12 in JK ×4−/−-R334X cells; n = 4). (A) Percentage of mobile and immobile CXCR4- and CXCR4R334X-AcGFP particles at the cell membrane. (B) Diffusion coefficients (D1–4) of mobile single trajectories, with median (black line) corresponding to JKX4−/−-X4 and JKX4−/−-R334X cells as in A. (n.s., not significant, ****P ≤ 0.0001). (C) Intensity distribution (arbitrary units, a.u.) from individual CXCR4- and CXCR4R334X-AcGFP trajectories on unstimulated and CXCL12-stimulated JKX4−/− transfected cells, mean is indicated (red) (n = 3; n.s., not significant; ****P ≤ 0.0001). (D and E) Frequency of CXCR4-AcGFP particles containing different number of receptors expressed as a histogram of unstimulated and CXCL12-stimulated JKX4−/−-X4 (D) and JKX4−/−-R334X (E) cells, calculated from MSI values of each particle as compared with the MSI value of monomeric CD86-AcGFP. The frequency of particles expressing monomers plus dimers (≤2) or nanoclusters (≥3) in both cell types is also shown (Right).
Both CXCR4 and CXCR4R334X were found as predominantly monomers and dimers in steady state (~90% for CXCR4 vs. ~81% for CXCR4R334X) (Fig. 2 C–E). Accordingly, basal intensity distribution was comparable for both receptor types (1,255 arbitrary units for CXCR4 vs. 1,521 arbitrary units for CXCR4R334X) (Fig. 2C). We observed an increase in the number of larger CXCR4 nanoclusters at the membrane of JKX4−/−-X4 cells upon CXCL12 activation (~64% of nanoclusters of ≥3 receptors), but this was not evident for CXCR4R334X in CXCL12-activated JKX4−/−-R334X cells (~17%) (Fig. 2 D and E).
Essentially similar results were obtained when CXCR4R334X dynamics were evaluated in heterozygosis (JK-R334X) (Fig. 3 and SI Appendix, Fig. S2), although in this case the endogenous expression of CXCR4 was associated with a reduction of CXCR4R334X nanoclusters both in steady state and after CXCL12 stimulation (SI Appendix, Fig. S2). In addition, the expression of CXCR4R334X altered the typical behavior of CXCR4 in response to CXCL12 when analyzed in JKX4−/− cells stably expressing CXCR4R334X (JKX4−/−R334X+) and transiently transfected with CXCR4-AcGFP (35.1% of nanoclusters in JKX4−/−R334X+-X4 vs. 51.2% in JK-X4) (Fig. 3B). These results confirm a dominant effect of CXCR4R334X on wild-type CXCR4 and concur with the ability of the mutant receptor to heterodimerize with CXCR4 even in the absence of ligand stimulation (28) (SI Appendix, Fig. S3).
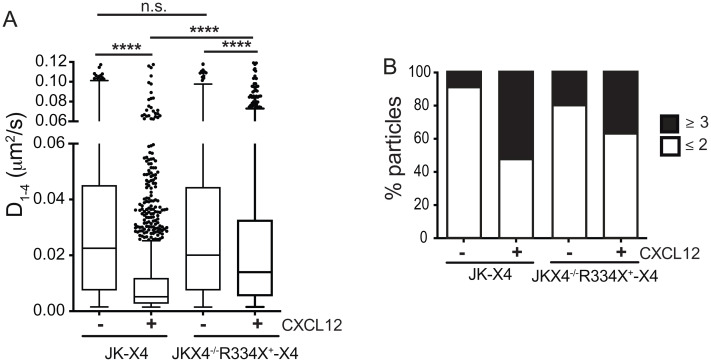
Heterozygous expression of CXCR4R334X abrogates CXCL12-mediated CXCR4 dynamics and nanoclustering. SPT analysis of CXCR4-AcGFP in JKX4−/−-X4 and JKX4−/−-R334X+-X4 cells on fibronectin (FN) or FN+CXCL12-coated coverslips (1,176 particles in 60 cells on FN; 3,037 in 66 cells on FN+CXCL12 in JK-X4 cells; 867 in 72 cells on FN; 1,525 in 75 cells on FN+CXCL12 in JKP ×4−/−R334X+-X4 cells; n = 3). (A) Diffusion coefficients (D1–4) of mobile single trajectories, with median (black line) corresponding to JK-X4 and JKX4−/−-R334X+X4 cells (n.s., not significant; ****P ≤ 0.0001). (B) Frequency of CXCR4-AcGFP particles containing the same number of receptors [monomers plus dimers (≤2) or nanoclusters (≥3) in JK and JKX4−/−R334X+ cells, calculated from MSI values of each particle as compared with the MSI value of monomeric CD86-AcGFP.
Altogether, the data indicate that CXCL12 does not trigger CXCR4R334X nanoclustering nor does it increase the percentage of immobile particles; instead, it increases the diffusion of the mutant receptor at the cell membrane. These observations might correlate with the defects observed in the migration of JK-R334X and JKX4−/−-R334X cells on lipid bilayers.
CXCR4R334X Abrogates Directed Cell Migration.
Receptor nanoclustering influences several CXCL12-mediated responses, including ligand-induced directed cell migration (15). We utilized fibronectin-coated chemotaxis chambers to assess the ability of JKX4−/−-R334X cells to migrate toward CXCL12 gradients. Results showed that whereas JKX4−/−-X4 cells sensed the gradient, JKX4−/−-R334X cells did not (Fig. 4A and Movies S5–S8). Quantitation of the results indicated that, compared with JKX4−/−-X4 cells, CXCL12 exposure failed to increase the forward migration index and track straightness in JKX4−/−-R334X cells (Fig. 4 B and C). Similarly, JK cells coexpressing both receptors, JKX4−/−R334X+-X4 (heterozygosis), were unable to sense CXCL12 gradients (Fig. 4 D–F). These data show that the ability to sense CXCL12 gradients is blocked in cells expressing CXCR4R334X.
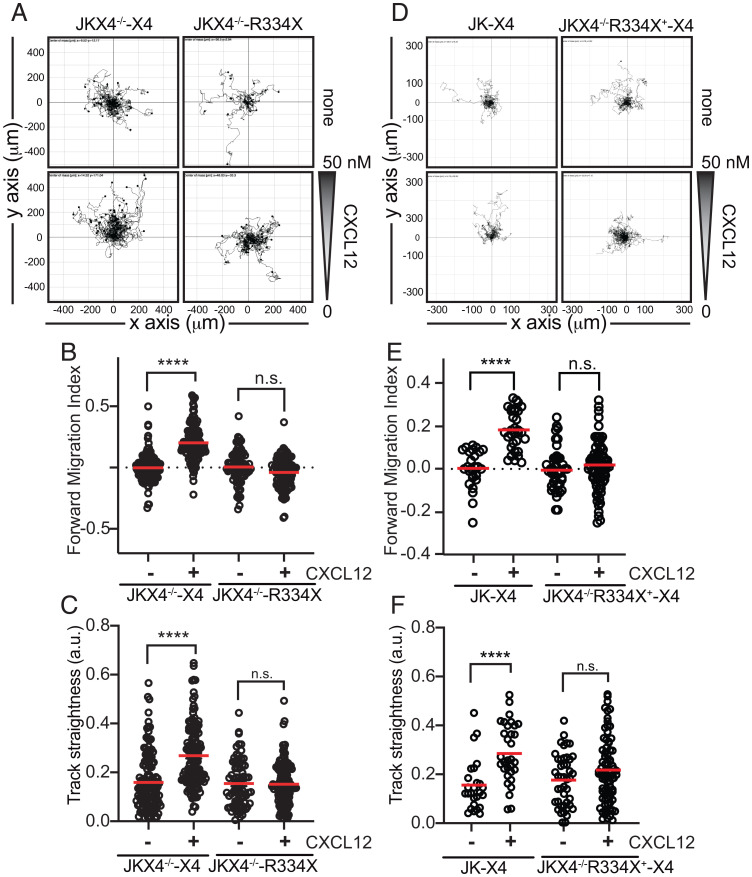
CXCL12 does not trigger directed migration of cells expressing CXCR4R334X both in homo- or heterozygosis. Migration of JKX4−/− transiently transfected with CXCR4-AcGFP (JKX4−/−-X4) or CXCR4R334X-AcGFP (JKX4−/−-R334X) cells (homozygosis) (A–C) or JK-expressing CXCR4-AcGFP (JK-X4) or JKX4−/−R334X+ cells transiently transfected with CXCR4-AcGFP (JKX4−/−-R334X+-X4) (heterozygosis) (D and E) on fibronectin-coated μ-slide chemotaxis chambers in response to the indicated gradient of CXCL12 concentration for 18 h. (A and D) Representative spider plots showing the trajectories of tracked cells migrating along the gradient. Dots in the plots represent the final position of each single tracked cell. Gray triangle indicates CXCL12 gradient. (B, C, E, and F) Quantitative evaluation of forward migration index (B and E) and track straightness (C and F). These graphs show the data of individual cells, with the mean indicated (red) (n = 3; n.s., not significant; ****P ≤ 0.0001).
Actin cytoskeleton dynamics not only regulate receptor compartmentalization, but are also important for maintaining directional migration by forming and stabilizing protrusions or lamellipodia at the leading edge of motile cells (29), a key element in the response to chemoattractant gradients. We thus tested the ability of CXCL12 to promote lamellipodia formation. JKX4−/−-X4 and JKX4−/−-R334X cells were activated with CXCL12, and phalloidin staining was evaluated in fixed cells by confocal microscopy. Quantitation of the number of protrusions demonstrated that both types of cells showed a spherical phenotype in steady-state conditions, with a weak phalloidin-staining pattern around the cell. CXCL12 promoted the rapid polarization of JKX4−/−-X4 cells and phalloidin staining concentrated mainly in a unique protrusion (lamellipodium) (Fig. 5 A–C). In contrast, phalloidin staining in CXCL12-stimulated JKX4−/−-R334X cells was randomly distributed in the cell in multiple protrusions (Fig. 5 A–C). Comparable results were obtained when we compared primary CD3+ T cells from WHIM patients (CXCR4R334X) with those of healthy donors. Whereas CXCL12 triggered multiple phalloidin-enriched protrusions in cells from WHIM patients, a unique polarized lamellipodium was evident in cells from healthy donors (Fig. 5 D–F). These results suggest that defects in actin cytoskeleton reorganization might not only affect the migration of cells expressing CXCR4R334X but might also affect CXCL12-mediated receptor nanoclustering.
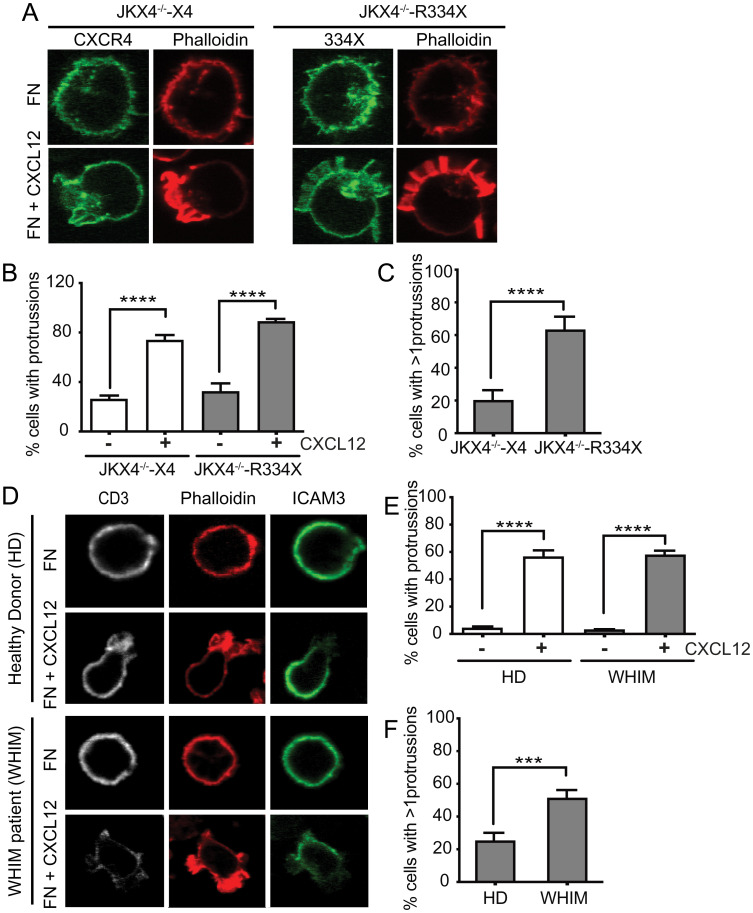
CXCL12 induces multiple lamellipodia through interaction with mutant CXCR4R334X. (A–C) F-actin (phalloidin-TRITC, red) and CXCR4 (AcGFP, green) visualized by confocal microscopy in JKX4−/−-X4 and JKX4−/−-R334X cells adhered to fibronectin and treated or not with CXCL12 (100 nM), as indicated (n = 2, more than 150 cells analyzed). (A) A representative cell type of each condition is shown. Original magnification 63×. (B) Percentage of cells with positive phalloidin staining (mean ± SEM, ****P ≤ 0.0001), and (C) percentage of cells showing >1 phalloidin+ protussion/cell (mean ± SEM, ****P ≤ 0.0001). (D–F) F-actin (phalloidin-TRITC, red), anti-CD3 (white) and anti-ICAM3 (green) visualized by confocal microscopy in PBMCs isolated from blood of patients with WHIM (R334X) and of healthy controls, adhered to fibronectin and treated or not with CXCL12 (100 nM), as indicated (n = 2, more than 150 cells analyzed of each condition). (D) A representative cell type of each condition is shown. Original magnification 63×. (E) Percentage of PBMCs with positive phalloidin staining (mean ± SEM, ****P ≤ 0.0001) and (F) percentage of PBMCs showing more than one phalloidin+ protrusion per cell (mean ± SEM, ***P ≤ 0.001). Images were acquired using a plan-apochromat 63 × /1.4 NA Oil DIC objective and without zoom. Resolution of acquired images is 1024 × 1024, although, for the figure, cells were cropped.
The absence of negative feedback mechanisms for CXCR4R334X facilitates the continuous activation of Gαi-mediated signaling. To determine whether this hyperactivated pathway might be linked to actin dynamics, we studied the actin-binding protein cofilin, which is activated in response to CXCL12 (30). Cofilin is inactivated by LIM kinase (LIMK)-mediated phosphorylation, which inhibits its binding to actin, and is reactivated by the phosphatase slingshot homolog 1 (SSH1), which enables actin filament depolymerization (31). We found that whereas CXCL12 mediated the rapid dephosphorylation/phosphorylation cycle of cofilin in JKX4−/−-X4 cells, dephosphorylation of cofilin was sustained in JKX4−/−-R334X cells (Fig. 6A). However, other ligand-mediated signaling pathways, such as AKT phosphorylation, were normally activated (Fig. 6A). Again, heterozygous expression of CXCR4R334X mimics the results obtained in JKX4−/−-R334X. In this heterozygous model, we detected prolonged cofilin activation without a significant effect on AKT phosphorylation (Fig. 6B).
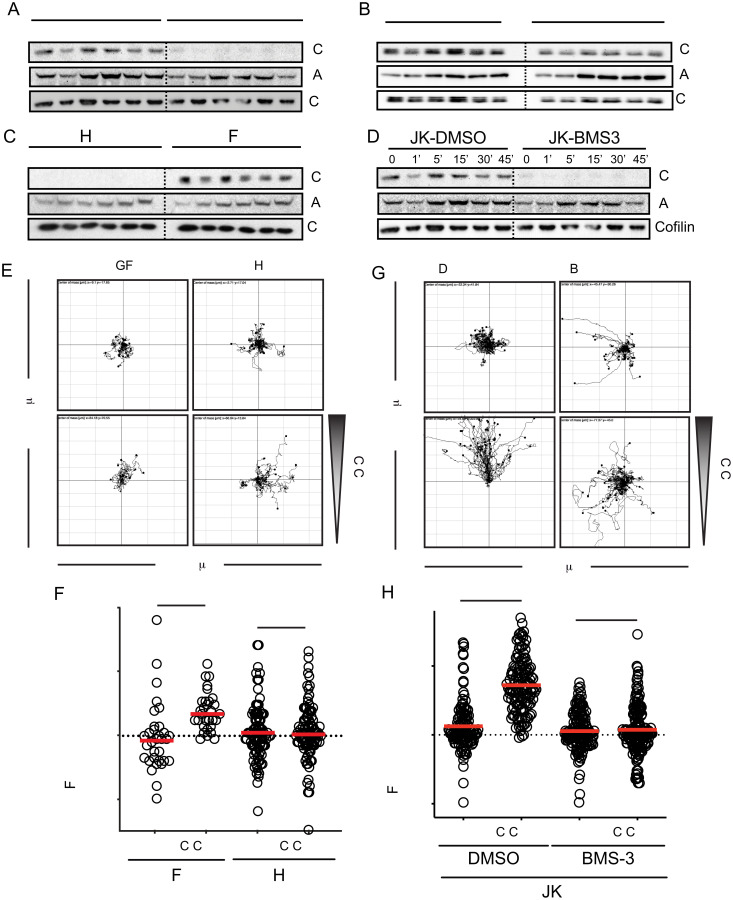
CXCR4R334X triggers sustained cofilin activation in response to CXCL12. Western blot analysis of cofilin phosphorylation in (A) JKX4−/−-X4 or JKX4−/−-R334X cells (homozygosis), (B) JK-X4 or JKX4−/−R334X+-X4 cells (heterozygosis), (C) JK cells transiently transfected with GFP-SSH1 (JK-SSH1) or with GFP (JK-GFP) and (D) JK cells treated with BMS-3 or DMSO. As loading control, the membrane was reblotted with an anticofilin antibody. As control of cell activation, phospho-Akt was assessed in all cells used in A–D, which were treated with CXCL12 at the indicated time points (n = 3). (E–H) Cells in C and D were plated on fibronectin-coated μ-slide chemotaxis chambers and allowed to migrate in response to the indicated gradient of CXCL12 concentration for 18 h. (E and G) Representative spider plots showing the trajectories of tracked JK-GFP–, JK-SSH1–, BMS-3–treated JK or DMSO-treated JK migrating along the gradient (n = 3, in duplicate). Dots in the plots represent the final position of each single tracked cell. Gray triangle indicates CXCL12 gradient. (F and H) Quantitative evaluation of the forward migration index of experiments performed in E and G, respectively. Figures show the data of individual cells, with the mean indicated (red) (n = 3; n.s., not significant; ***P ≤ 0.001; ****P ≤ 0.0001).
To determine whether a disequilibrium in the dynamics of active/inactive cofilin might explain the inability of CXCL12 to mediate directed cell migration in JKX4−/−-R334X cells, we blocked cofilin in its dephosphorylated state using BMS-3, a chemical inhibitor of LIMK1/2 activity (32), or in JK cells transiently transfected with GFP-SSH1 or GFP alone (as a control). As expected, cofilin was constitutively active independently of ligand stimulus in both JK cells treated with BMS-3 and in SSH1-transfected cells (Fig. 6 C and D). Neither BMS-3 treatment nor SSH1 overexpression affected other signaling pathways, such as Akt phosphorylation (Fig. 6 C and D). In both cases, CXCL12 failed to trigger direct cell migration (Fig. 6 E–H). These data support a role for cofilin in promoting directed cell migration and underscore the importance of CXCL12 controlling the function of this actin-binding protein through regulation of the kinase LIMK1 and the phosphatase SSH1.
Our results thus far led us to hypothesize that in the absence of negative feedback, due to impaired β-arrestin activation mediated by CXCR4R334X, CXCL12 promotes continuous Gαi-mediated signaling and alters the equilibrium of active/inactive cofilin and, consequently, the actin dynamics. In turn, this limits the ability of CXCL12 to promote correct cell polarization and to trigger large receptor nanoclusters.
β-Arrestin1 Links CXCR4 with Actin Cytoskeleton Dynamics and Regulates CXCL12-Mediated CXCR4 Nanoclustering.
The scaffolding role of the β-arrestins has been implicated both in receptor desensitization and in the actin assembly events needed for the formation of gradient-sensing filopodia and lamellipodia at the leading edge of motile cells (20). Impaired chemokine receptor desensitization has been also associated with random rather than directed migration (33). Additionally, although some reports implicate β-arrestins in chemokine-mediated cell migration, only β-arrestin1 has been associated to CXCL12-induced directed cell migration (34, 35). We thus questioned whether defects in β-arrestin activation might explain the lack of ligand-mediated CXCR4R334X nanoclustering and directed cell migration. To do this, we transiently transfected CXCR4-AcGFP into JKX4−/−β-arrestin1 knockout cells (JKX4−/−βarr−/−) (SI Appendix, Fig. S1C) to produce JKX4−/−βarr−/−-X4 cells. We found that CXCL12 failed to induce cofilin phosphorylation (Fig. 7A) and to trigger directed cell migration (Fig. 7B and Movies S9 and S10), as demonstrated by forward migration index and track straightness quantitation (Fig. 7 C and D). In contrast, other CXCL12-mediated signaling pathways, such as Akt phosphorylation, were unaffected (Fig. 7A).
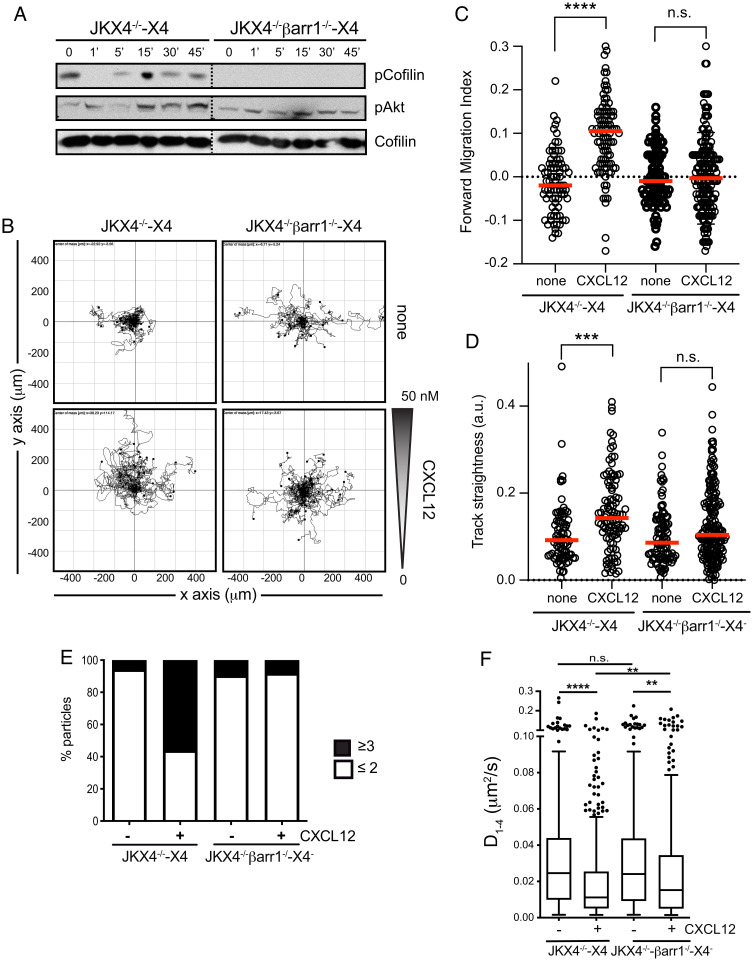
β-Arrestin1 regulates the formation of large CXCR4 nanoclusters and directional cell migration in response to CXCL12. (A) Western blot analysis of cofilin phosphorylation in JKX4−/−-X4 and JKX4−/−βarr−/−-X4 cells treated with CXCL12 at the indicated time points (Top). As loading control, the membrane was reblotted with an anti-cofilin antibody (Bottom). As control of cell activation, phospho-Akt was assessed in JKX4−/−-X4 and JKX4−/−βarr−/−-X4 cells treated with CXCL12 at the indicated time points (Middle) (n = 3). (B–D) JKX4−/−-X4 and JKX4−/−βarr−/−-X4 cells were plated on fibronectin-coated μ-slide chemotaxis chambers and allowed to migrate in response to the indicated gradient of CXCL12 concentration for 18 h. (B) Representative spider plots showing the trajectories of tracked JKX4−/−-X4 and JKX4−/−βarr−/−-X4 cells migrating along the gradient (n = 3, in duplicate). Dots in the plots represent the final position of each single tracked cell. Gray triangle indicates CXCL12 gradient. (C) Quantitative evaluation of the forward migration index and (D) track straightness of experiments performed as in B. Figures show the data of individual cells, with the mean indicated (red) (n.s., not significant; ***P ≤ 0.001; ****P ≤ 0.0001). (E and F) SPT analysis of CXCR4-AcGFP in JKX4−/−-X4 and JKX4−/−βarr−/−-X4 cells on CXCL12-coated coverslips with or without coated FN (JKX4−/−-X4, 531 trajectories in 50 cells plated on FN, 634 trajectories in 50 cells on FN+CXCL12; JKX4−/−βarr−/−-X4, 551 trajectories in 78 cells plated on FN, 587 trajectories in 70 cells on FN+CXCL12, n = 3). (E) Frequency of CXCR4-AcGFP particles containing the same number of receptors (monomers plus dimers [≤2] or nanoclusters [≥3]), calculated from MSI values of each particle as compared with the MSI value of monomeric CD86-AcGFP. (F) Diffusion coefficients (D1–4) of mobile single trajectories, with median (black line), corresponding to JKX4−/−-X4 and JKX4−/−βarr−/−-X4 cells (n.s., not significant; **P ≤ 0.01; ****P ≤ 0.0001).
To evaluate whether defects in β-arrestin1 activation might be involved in the lack of CXCL12-mediated CXCR4R334X nanoclustering, we next analyzed the consequences of β-arrestin1 deficiency for CXCR4-AcGFP dynamics. In steady state, CXCR4 behaved similarly in both JKX4−/−-X4 and JKX4−/−βarr−/−-X4 cells with respect to receptor nanocluster distribution (~4% in JKX4−/−-X4 vs. ~9% JKX4−/−βarr−/−-X4 of complexes of ≥3 receptors) (Fig. 7E and Movies S11 and S12) and the diffusion coefficient (D1–4 values of 0.024 μm2 s−1 and 0.025 μm2 s−1, respectively) (Fig. 7F). In contrast, β-arrestin1 deficiency abrogated ligand-mediated receptor nanoclustering (~54% in JKX4−/−-X4 vs. ~6% in JKX4−/−βarr−/−-X4 of complexes of ≥3 receptors) (Fig. 7E). We also detected a 2-fold decrease in the diffusion in JKX4−/−-X4 cells, while in JKX4−/−βarr−/−-X4 the decrease was only 1.5-fold (0.011 μm2 s−1 in JKX4−/−-X4 vs. 0.015 μm2 s−1 in JKX4−/−βarr−/−-X4) (Fig. 7F).
Among other signaling pathways that can be involved, our data implicate β-arrestin1 in CXCL12-mediated CXCR4 nanoclustering and dynamics and indicate that defects in its activation through CXCR4R334X promote deficiencies in lateral partitioning of this mutant receptor. The mechanism might involve, through regulation of LIMK/SSH1 balance, the spatial control of cofilin activity, which was lost in cells lacking β-arrestin1. Indeed, we detected a continuous activation of cofilin, which in turn might affect actin reorganization in these cells. As actin polymerization also regulates CXCR4 nanoclustering, our data might also explain the evident differences in the lateral partitioning of CXCR4 and CXCR4R334X receptors after ligand activation.
Discussion
Cell migration is a complex process involving myriad signaling proteins and receptors that act coordinately to activate intracellular pathways and promote polarized cell states and directional migration. To migrate directionally in response to external stimuli, the internal machinery of cells needs to be spatially organized, which involves the integration of biochemical and mechanical factors to generate force in a specific direction to move the cell forward. The classic image of migration on substrates is one of actin-driven protrusions occurring primarily at the front, and myosin-driven contractile forces generated at the rear, causing the cell to detach and move forward (36). Recent evidence indicates that the actin cytoskeleton also governs the nanoscale organization and lateral dynamics of chemokine receptors, which is essential for correct receptor function to allow cells to sense chemoattractant gradients (15).
Due to the absence of desensitization mechanisms, CXCR4R334X and other mutant CXCR4 proteins causing WHIM syndrome have been considered as gain-of-function receptors (1, 37). But, in agreement with the observed retention of mature neutrophils in the bone marrow of patients with WHIM, resulting in neutropenia and panleukopenia (7), we found that JK cells expressing CXCR4R334X failed to exhibit enhanced migration on lipid bilayers with embedded ICAM-1 and CXCL12, and were unable to sense CXCL12 gradients in fibronectin-coated μ-chambers. The results argue against differences in CXCL12-mediated adhesion to ICAM-1 of these cells compared with controls, although the cells showed higher adhesion in steady state.
Chemokine receptors are found concentrated at the leading edge of motile cells, on the flattened cell-substratum contact area (38). There, they act as a sensor mechanism for the directed migration of leukocytes through a chemoattractant gradient, as was elegantly shown using a chimeric receptor between rhodopsin and CXCR4 in T cells (39). Using SPT in TIRF-M mode, we observed that, at steady state, CXCR4 and CXCR4R334X exhibited comparable spatial organization at the cell membrane, preferentially forming nonclustered entities (monomers and dimers) and a small fraction of nanoclusters (groups of ≥3 receptors) that are CXCR4 aggregates present at the cell membrane that facilitate some chemokine-mediated signaling events (15). However, we observed some variability in the percentage of nanoclusters between the cell types used in this study, specifically when CXCR4R334X was expressed in heterozygosis versus homozygosis. Although further work is needed to clarify this effect, differences in the CXCR4/CXCR4R334X ratio at the cell surface might affect dynamic determinations, as unlabeled receptors are not detected in our technical approach. Surprisingly, whereas CXCL12 stimulated the formation of large receptor nanoclusters on CXCR4-expressing cells, reduced their diffusion coefficient, and increased the percentage of immobile particles at the cell membrane, this did not occur in cells expressing CXCR4R334X alone or coexpressed with CXCR4. We also observed that CXCR4R334X coexpression altered CXCL12-mediated CXCR4 nanoclustering, consistent with the known dominant characteristics of the mutant receptor detected in patients. In primary cells, WHIM receptors are always expressed in heterozygosis and are therefore coexpressed with wild-type CXCR4 (40). Analysis of FRET data confirmed the heterodimerization between CXCR4R334X and CXCR4 (28), which is a possible mechanism to explain the observed dominant effect of WHIM mutant receptors.
Receptor clustering increases cell sensitivity to external stimuli (41), and is also a means for efficient cell signal propagation (42), increasing the robustness of signaling systems (43). The present data suggest that the structural differences between CXCR4 and CXCR4R334X affect the dynamics of these receptors at the cell surface and, consequently, the migratory behavior of the cells expressing them. The results also point to the relevance of the C-terminal tail of CXCR4 and of its coupling to β-arrestin1 for these processes.
The actin cytoskeleton has a key role in regulating membrane diffusion, protein compartmentalization and clustering, and in controlling receptor signaling (14). Previous data from our group indicate that the actin cytoskeleton is also essential for ligand-mediated CXCR4 nanoclustering, as latrunculin A treatment interferes with CXCR4 nanoclustering and abrogates CXCL12-mediated cell migration (15). Analysis of phalloidin immunofluorescence images revealed clear differences between JKX4−/−-R334X and control (JKX4−/−-X4) cells in actin dynamics. In cells expressing CXCR4, CXCL12 treatment concentrated phalloidin staining in a unique cellular localization, the lamellipodium, whereas phalloidin staining was randomly distributed in cells expressing CXCR4R334X, and several protrusions were detected around the cell. Notably, a comparable phenotype was observed when we compared phalloidin staining on CXCL12-stimulated CD3+ T cells from healthy donors and from patients with WHIM.
β-Arrestins are involved in chemokine receptor desensitization and internalization (44), but they also have a scaffolding role for a number of signaling molecules (45, 46) and participate in actin reorganization and chemotaxis processes (28, 47, 48). In vivo studies have demonstrated the involvement of β-arrestins in tumor cell migration and metastasis (49), and in the recruitment of immune cells to sites of inflammation (50), although only β-arrestin1 participates in chemokine-mediated directed cell migration (35). There is also evidence supporting a role for β-arrestins in the spatial control of actin assembly events at the leading edge of primary leukocytes and cultured cells (19). β-Arrestins are required for PAR-2–dependent activation of the actin-binding protein cofilin, which binds and destabilizes actin filaments, promoting actin severing (51). Through a process involving Ser/Thr phosphorylation of specific residues by G protein-coupled receptor kinase, the C-tail of CXCR4 is essential for β-arrestin association and activation (52). Interestingly, a recent study reported that the phosphorylation pattern of the C-terminal end of G protein-coupled receptors defines not only the binding of β-arrestins, but also their spatial conformation and scaffolding role (53). CXCR4R334X has a truncated C terminal, and cells expressing this receptor show defects not only in desensitization processes (54), but also in those pathways related to the scaffold role of β-arrestin. Our findings show that in cells expressing CXCR4, CXCL12 triggered rapid activation/inactivation of cofilin, allowing a correct balance to regulate actin dynamics.
In contrast, in cells expressing CXCR4R334X, both in homo- and heterozygosis, CXCL12 triggered sustained cofilin dephosphorylation, indicating a permanent activated status of the actin binding protein. This observation correlated with the presence of multiple foci of polymerized actin after CXCL12 activation. Cofilin is temporally and spatially regulated by the kinase LIMK1 and the phosphatase SSH1. LIMK1 knockdown suppresses chemokine-induced lamellipodia formation and directed cell migration, and SSH1 is critically involved in directional cell migration by restricting the membrane protrusion to one direction during the early stages of cell responses (55). Although the lack of suitable reagents prevented us from evaluating the activation status of LIMK and SSH1 in our cells, the direct interaction between β-arrestins and LIMK has been previously demonstrated (19, 56). In JK cells treated with BMS-3, a drug that inhibits LIMK1/2 activity or in JK cells overexpressing SSH1, cofilin was permanently activated and CXCL12-mediated directed migration was abolished. A similar result was obtained when we evaluated CXCL12-directed cell migration of β-arrestin1–deficient JK cells, which showed permanent cofilin activation and directed cell migration was abolished.
Correct lamella formation requires appropriated actin polymerization dynamics, a process that also depends on the recruitment of several scaffold proteins to ensure the proper localization of actin polymerization (57). Among these scaffold proteins, Arp2/3 is involved in actin filament branching (58) and VASP plays a role in actin assembly (59). Also, two members of the small GTPases of the Rho family, Rac1 and Cdc42 (60) and their guanine nucleotide-exchange factors, such as Vav1 (61), are key elements. β-Arrestins have been reported to play both positive and negative roles in Rac1 signaling (62, 63); for example, knockdown of β-arrestin1 blocked β2 adrenergic receptor-dependent activation of Rac1 (64). Additionally, β-arrestins have been reported to negatively regulate PAR2 receptor-mediated Cdc42 activation (65), and a recent study found that β-arrestins and Src kinase conjointly traffic to endomembranes, where Src phosphorylates CCR7 to form an endomembrane-residing signaling complex comprising the chemokine receptor, Vav1, and its effector Rac1, to promote lamellipodia formation at the leading edge (66). The direct interaction between β-arrestin and the actin-binding protein filamin A (FLNa) was reported in a proteomics study (21), and the role of a β-arrestin–FLNa–receptor complex in membrane ruffling has also been studied (22).
We suggest that β-arrestin1, by regulating the LIMK/SSH1 equilibrium, also modulates cofilin activity. Our results indicate that CXCR4R334X internalization and desensitization is impaired due to the defective recruitment of β-arrestin1, a process that also affects the scaffolding role of this protein. As a consequence, an imbalance between LIMK1/2 and SSH1 occurs that disturbs the active/inactive cycle of cofilin and, therefore, cells show defects in the focalization of actin polymerization in a unique cellular pole. Cells expressing CXCR4R334X have multiple foci of polymerized actin randomly distributed, defective ligand-mediated receptor nanoclustering, and impaired ligand-mediated directed cell migration.
Although we cannot discard other differences between CXCR4 and CXCR4R334X that affect cell migration, including distinct regulation of integrin dynamics or additional β-arrestin–independent mechanisms mediated by the C-terminal region of CXCR4, our data indicate that the C-terminal end of chemokine receptors and β-arrestins are key elements for the modulation of actin cytoskeleton dynamics, cell polarization, and receptor nanoclustering. The process involves the interaction of β-arrestin1 with CXCR4 and a precise regulation of cofilin dynamics. These signaling pathways are altered in CXCR4R334X-expressing cells, resulting in a loss of cell movement directionality. This might contribute to the retention of immune cells in the bone marrow of patients with WHIM, and also might mechanistically explain (as yet undefined) the hypogammaglobulinemia that affects many patients with defects in one or more subclasses of antibodies (67). Detailed B cell functional studies performed in a few patients have documented restricted immunoglobulin heavy chain variable region diversity, impaired class switching, and poor or unsustainable responses to vaccines (68, 69). In addition, reduced B cell immunoglobulin gene class switching was observed after immunization in some WHIM patients (69). Similarly, WHIM mice also show distorted germinal centers (70). The organization of germinal centers depends on sorting of centroblasts into the dark zone by CXCR4 and into the light zone by CXCR5 (71). Therefore, the ability of B cells to sense chemoattractant gradients is essential for their correct activation, the generation of plasma cells and memory B cells, immunoglobulin secretion, and the ability to generate proper class switching.
Materials and Methods
Cells and Reagents.
HEK-293T cells were obtained from the American Type Culture Collection (CRL-11268 and CRL-10915, respectively). JK CD4+ cells were kindly donated by J. Alcamí, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid, Spain. We employed the CRISPR/Cas9 system to generate a JK cell line lacking CXCR4, using the pIFA1 plasmid, kindly donated by R. M. Rios, Department of Cell Signaling, Centro Andaluz de Biología Molecular y Medicina Regenerativa/CSIC, E-41092, Sevilla, Spain (72). CXCR4 expression was analyzed by flow cytometry using an anti-human CXCR4-PE (12G5, BioLegend) (SI Appendix, Fig. S1A). Finally, CXCR4-deficient cells (JKX4−/−) were isolated and cloned by limiting dilution and flow cytometry. The absence of CXCR4 in the selected clones was confirmed by analyzing their ability to migrate toward CXCL12 gradients in Boyden (Transwell) chambers (SI Appendix, Fig. S1B).
To generate JKX4−/− cells lacking β-arrestin1 (JKX4−/−βarr1−/−), crRNA, and tracrRNA olignucleotides were chemically synthesized and annealed to form guide RNAs according to the manufacturer’s guidelines (Integrated DNA Technologies). crRNA/tracrRNA duplexes were then mixed with recombinant TrueCut Cas9 Protein v2 (cat. no. A36499; Thermo Fisher Scientific) to form ribonucleoproteins (RNPs), which were electroporated (2,200 mV, 20 ms, 1 pulse) after complexing using the Neon transfection system from Thermo Fisher Scientific. After transfection, cells were cultured in complete medium (RPMI 1640 10% FCS; 37 °C, 5% CO2). Single-cell clones were isolated by limiting dilution, and antibiotic-resistant cells were analyzed for gene silencing by PCR and for protein expression by Western blotting (SI Appendix, Fig. S1C).
°C, 5% CO2). Single-cell clones were isolated by limiting dilution, and antibiotic-resistant cells were analyzed for gene silencing by PCR and for protein expression by Western blotting (SI Appendix, Fig. S1C).
Stable JKX4−/−R334X+ cells were generated by electroporation (Bio-Rad platform; as described above) of JKX4−/− cells with CXCR4R334X (20 μg) and antibiotic selection. Receptor expression was determined by flow cytometry using a specific anti-CXCR4 monoclonal antibody (SI Appendix, Fig. S1D).
When needed, JK cells were transiently transfected by electroporation with CXCR4-AcGFP or CXCR4R334X-AcGFP (20 μg) (Bio-Rad platform; as described). Cells were analyzed for GFP expression by flow cytometry 24 h after transfection.
When needed, JK cells were transfected with GFP-SSH1 (kindly donated by P. Roda, Universidad Complutense de Madrid, Spain) or GFP, and GFP expression was analyzed by flow cytometry (SI Appendix, Fig. S1E).
Human peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats or, when required, from blood of WHIM patients carrying CXCR4R334X or healthy donors, by centrifugation through Percoll density gradients (760 × g, 45 min, room temperature). The study was approved by the Institutional Review Board of the 12 de Octubre Health Research Institute, and was conducted according to the Declaration of Helsinki principles. Informed consent was obtained from all patients.
Antibodies used were as follows: monoclonal anti-CXCR4 (clone 44717, R&D Systems), anti-human CXCR4-biotin (12G5, R&D Systems), and CD3-APC (clone UCHT1, Beckman Coulter #IM2467); antiphospho-Akt (Ser473), anti-Akt (#9272), phospho-cofilin (Ser3, #3313), and phospho-cofilin (#5175) (all from Cell Signaling Technology); anti–β-arrestin1 (Cell Signaling Technology) and anti–β-arrestin2 (Santa Cruz Biotechnology); phalloidin-TRITC (#P1951, Sigma-Merck). The LIMK inhibitor (BMS-3) was obtained from MedChemExpress (#HY-18304). Human CXCL12 was obtained from PeproTech. Human CXCR4 was cloned into pECFP-N1, pEYFP-N1, and pAcGFPm-N1 (Clontech Laboratories), as described previously (15). Human CXCR4R334X was constructed from pcDNA3.1-CXCR4 using specific primers to delete the last 19 amino acids by PCR. The primers used were: Forward-5′ATGGAGGGGATCAGTATATACAC-3′’ and Reverse-5′TTACCGGTTTTCCTTTGGAG AGGATCTTGAG-3′’. Human CXCR4R334X was cloned into pECFP-N1, pEYFP-N1 and pAcGFPm-N1.
Western Blotting.
Cells (3 × 106) were activated with CXCL12 (40 nM) at the indicated time points. Cells were lysed in detergent buffer (20 mM triethanolamine, 1% digitionin, 1 mM PMSF, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 10 μM sodium orthovanadate) for 30 min at 4 °C, and extracts were analyzed by Western blotting using specific antibodies.
°C, and extracts were analyzed by Western blotting using specific antibodies.
Transwell Migration Assay.
Cells (3 × 105 in 0.1 mL of RPMI medium containing 10 mM Hepes and 0.1% BSA) were placed in the upper wells of uncoated 24-well transmigration chambers (5-μm pore; Transwell, Costar). CXCL12 (20 nM) in 0.6 mL of the same medium was added to the lower well. Plates were incubated for 180 min (37 °C, 5% CO2) and cells that migrated to the lower chamber were counted by flow cytometry (Gallios flow cytometer, Beckman Coulter), corrected for variations in input concentrations, and expressed as the mean (SD) percentage of cell migration.
°C, 5% CO2) and cells that migrated to the lower chamber were counted by flow cytometry (Gallios flow cytometer, Beckman Coulter), corrected for variations in input concentrations, and expressed as the mean (SD) percentage of cell migration.
Cell Adhesion/Migration on Planar Lipid Bilayers.
Planar lipid bilayers were prepared as described previously (73). Briefly, unlabeled GPI-linked ICAM-1 liposomes were mixed with 1,2-dioleoyl-PC. Membranes were assembled in FCS2 chambers (Bioptechs), blocked with PBS containing 2% FCS for 1 h at room temperature, and coated with CXCL12 (200 nM, 30 min, room temperature). Cells (3 × 106 cells/mL) in PBS containing 0.5% FCS, 0.5 g/l D-glucose, 2 mM MgCl2, and 0.5 mM CaCl2 were then injected into the prewarmed chamber (37 °C). Confocal fluorescence, differential interference contrast (DIC) and interference reflection microscopy (IRM) images were acquired on a Zeiss Axiovert LSM 510-META inverted microscope with a 40× oil-immersion objective. Imaris 7.0 software (Bitplane) and ImageJ 1.49v were used for qualitative and quantitative analysis of cell dynamics parameters, fluorescence and IRM signals. The fluorescence signal of the planar bilayer in each case was set up as the background fluorescence intensity. The frequency of adhesion (IRM+ cells) per image field was estimated as [number of cells showing IRM contact/total number of cells (estimated by DIC)] × 100; similarly, we calculated the frequency of migration (cells showing and IRM contact and moving over time).
°C). Confocal fluorescence, differential interference contrast (DIC) and interference reflection microscopy (IRM) images were acquired on a Zeiss Axiovert LSM 510-META inverted microscope with a 40× oil-immersion objective. Imaris 7.0 software (Bitplane) and ImageJ 1.49v were used for qualitative and quantitative analysis of cell dynamics parameters, fluorescence and IRM signals. The fluorescence signal of the planar bilayer in each case was set up as the background fluorescence intensity. The frequency of adhesion (IRM+ cells) per image field was estimated as [number of cells showing IRM contact/total number of cells (estimated by DIC)] × 100; similarly, we calculated the frequency of migration (cells showing and IRM contact and moving over time).
Single-Molecule TIRF Imaging and Analysis.
Transfected cells expressing 8,500 to 22,000 receptors per cell, which equates to a density of <4.5 particles/μm2, were selected for detection and tracking analysis.
Experiments were performed at 37 °C with 5% CO2 using a TIRF microscope (Leica AM TIRF inverted). Image sequences of individual particles (500 frames) were then acquired at 49% laser (488-nm diode laser) power with a frame rate of 10 Hz (100 ms per frame). Penetration depth of the evanescent field was 90 nm.
°C with 5% CO2 using a TIRF microscope (Leica AM TIRF inverted). Image sequences of individual particles (500 frames) were then acquired at 49% laser (488-nm diode laser) power with a frame rate of 10 Hz (100 ms per frame). Penetration depth of the evanescent field was 90 nm.
Particles were detected and tracked using previously described algorithms (U-Track2 25 implemented in MATLAB, as described previously (15). Mean spot intensity (MSI), number of mobile and immobile particles and diffusion coefficients (D1–4) were calculated from the analysis of thousands of single trajectories over multiple cells (statistics provided in the respective figure legends), using described routines (74). Receptor number along individual trajectories was determined as described previously (15). We measured the average fluorescence intensity for the first 20 frames of each trajectory and used the intensity of the monomeric protein CD86-AcGFP as a reference. Values were confirmed using single-step photobleaching analysis. For more detailed information about TIRF experiments and the analysis of their results, see SI Appendix.
FRET Saturation Curves by Sensitized Emission.
FRET50 and FRETmax values were determined in transfected HEK-293T cells, as described previously (15).
Directional Cell Migration.
Cells were seeded into fibronectin-coated chambers (Ibidi μSlide Chemotaxis System; 80326) and 50 nM CXCL12 was added to the upper reservoir, following the manufacturer’s instructions. Migration toward CXCL12 was analyzed over 6 h (5% CO2, 37 °C) with a time lapse of 2 min using a Microfluor inverted microscope (Leica). Single-cell tracking was evaluated using the manual tracking plug-in tool in ImageJ. Forward migration index and straightness values were obtained using the Manual Tracking and Chemotaxis Tool plugins for Fiji software (NIH).
°C) with a time lapse of 2 min using a Microfluor inverted microscope (Leica). Single-cell tracking was evaluated using the manual tracking plug-in tool in ImageJ. Forward migration index and straightness values were obtained using the Manual Tracking and Chemotaxis Tool plugins for Fiji software (NIH).
Immunofluorescence Analyses.
Cells on fibronectin (20 μg/mL, Sigma) -coated glass slides were stimulated or not with 100 nM CXCL12 (5 min at 37 °C), fixed with 4% paraformaldehyde (10 min, room temperature), permeabilized with 0.25% saponin (10 min, room temperature), and stained with phalloidin-TRITC (Sigma-Merck; 30 min, room temperature). Preparations were analyzed using a Leica TCS SP8 confocal multispectral microscope. For primary cells, preparations were blocked with PBS containing 150 mM NaCl, 0.1% goat serum, and 1% BSA (60 min, room temperature) before staining with anti-human ICAM-3 plus AlexaFluor 488 goat anti-mouse IgG (30 min, room temperature; Thermo Fisher Scientific) and with mouse anti-human CD3 APC IgG1 (30 min, room temperature), prior to cell permeabilization and staining with phalloidin-TRITC.
°C), fixed with 4% paraformaldehyde (10 min, room temperature), permeabilized with 0.25% saponin (10 min, room temperature), and stained with phalloidin-TRITC (Sigma-Merck; 30 min, room temperature). Preparations were analyzed using a Leica TCS SP8 confocal multispectral microscope. For primary cells, preparations were blocked with PBS containing 150 mM NaCl, 0.1% goat serum, and 1% BSA (60 min, room temperature) before staining with anti-human ICAM-3 plus AlexaFluor 488 goat anti-mouse IgG (30 min, room temperature; Thermo Fisher Scientific) and with mouse anti-human CD3 APC IgG1 (30 min, room temperature), prior to cell permeabilization and staining with phalloidin-TRITC.
Flow Cytometry Studies.
Cells were incubated with specific antibodies (30 min, 4 °C) and mean fluorescence intensity was determined on a Gallios or a FC500 flow cytometer (Beckman Coulter).
°C) and mean fluorescence intensity was determined on a Gallios or a FC500 flow cytometer (Beckman Coulter).
Receptor internalization was determined by flow cytometry after activation with CXCL12 (20 nM) at the indicated times. Results are expressed as a percentage of the mean fluorescence intensity of treated cells relative to that of unstimulated cells.
Statistical Analyses.
All results were analyzed using GraphPad PRISM (n.s. = nonsignificance, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001). Cell migration in transwells and in planar lipid bilayers, directional cell migration assays, and cell polarization under the various conditions were analyzed to determine significant differences between means using one-way ANOVA followed by Tukey’s multiple comparison test. A two-tailed Mann–Whitney nonparametric test was used to analyze the diffusion coefficient (D1–4) of single particles. We used contingency tables to compare two or more groups of categorical variables, such as the percentages of mobile or immobile particles, and these were compared using a χ2 test with a two-tailed P value.
Supplementary Material
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Acknowledgments
We thank Dr. P. M. Murphy from the Laboratory of Molecular Immunology, National Institute of Allergy and Infectious Diseases, Bethesda, MD, for critical review of the manuscript; and the Advance Light Microscopy Unit at the Centro Nacional de Biotecnología/Consejo Superior de Investigaciones Cientificas for technical help. This work was supported by grants from the Spanish Ministry of Science and Innovation (SAF 2017-82940-R and PID2020-114980RB-I00) and the Redes Temáticas de Investigación Cooperativa en Salud Program (RD12/0009/009; Red de inflamación y Enfermedades Reumáticas [RIER]). L.I.G.-G. is supported by a grant of Fondo de Investigaciones Sanitarias PI21/01642; F.S. and the Institute for Research in Biomedicine are supported by the Helmut Horten Foundation; L.M.-M. is supported by the JIN program from the Spanish Ministry of Science and Innovation (RT12018-101789-J-100). N.M. and M.F.G.-P. acknowledge funding from the European Union H2020 under the Marie Sklodowska-Curie Grant 754558-PREBIST (to N.M.) and European Research Council Grant 788546-NANO-MEMEC (to M.F.G.-P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/10.1073/pnas.2119483119/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/128771856
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.2119483119
Article citations
Allosteric modulation of the CXCR4:CXCL12 axis by targeting receptor nanoclustering via the TMV-TMVI domain.
Elife, 13:RP93968, 09 Sep 2024
Cited by: 0 articles | PMID: 39248648 | PMCID: PMC11383527
Expanding CXCR4 variant landscape in WHIM syndrome: integrating clinical and functional data for variant interpretation.
Front Immunol, 15:1411141, 08 Jul 2024
Cited by: 0 articles | PMID: 39040098 | PMCID: PMC11260667
Review Free full text in Europe PMC
The complex nature of CXCR4 mutations in WHIM syndrome.
Front Immunol, 15:1406532, 05 Jul 2024
Cited by: 0 articles | PMID: 39035006 | PMCID: PMC11257845
Review Free full text in Europe PMC
TrackAnalyzer: A Fiji/ImageJ toolbox for a holistic analysis of tracks.
Biol Imaging, 3:e18, 11 Oct 2023
Cited by: 0 articles | PMID: 38510172 | PMCID: PMC10951927
The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention.
Cell Mol Immunol, 20(3):217-251, 01 Feb 2023
Cited by: 85 articles | PMID: 36725964 | PMCID: PMC9890491
Review Free full text in Europe PMC
Go to all (7) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of a new WHIM syndrome mutant reveals mechanistic differences in regulation of the chemokine receptor CXCR4.
J Biol Chem, 298(2):101551, 30 Dec 2021
Cited by: 7 articles | PMID: 34973340 | PMCID: PMC8802859
The negative charge of the 343 site is essential for maintaining physiological functions of CXCR4.
BMC Mol Cell Biol, 22(1):8, 23 Jan 2021
Cited by: 3 articles | PMID: 33485325 | PMCID: PMC7825245
The complex nature of CXCR4 mutations in WHIM syndrome.
Front Immunol, 15:1406532, 05 Jul 2024
Cited by: 0 articles | PMID: 39035006 | PMCID: PMC11257845
Review Free full text in Europe PMC
Filamin A interaction with the CXCR4 third intracellular loop regulates endocytosis and signaling of WT and WHIM-like receptors.
Blood, 125(7):1116-1125, 29 Oct 2014
Cited by: 16 articles | PMID: 25355818 | PMCID: PMC4347305
Funding
Funders who supported this work.
European Research Council (1)
Grant ID: 788546

 a
,
1
a
,
1


