Abstract
Free full text

Egg-adaptive mutations of human influenza H3N2 virus are contingent on natural evolution
Abstract
Egg-adaptive mutations in influenza hemagglutinin (HA) often emerge during the production of egg-based seasonal influenza vaccines, which contribute to the largest share in the global influenza vaccine market. While some egg-adaptive mutations have minimal impact on the HA antigenicity (e.g. G186V), others can alter it (e.g. L194P). Here, we show that the preference of egg-adaptive mutation in human H3N2 HA is strain-dependent. In particular, Thr160 and Asn190, which are found in many recent H3N2 strains, restrict the emergence of L194P but not G186V. Our results further suggest that natural amino acid variants at other HA residues also play a role in determining the preference of egg-adaptive mutation. Consistently, recent human H3N2 strains from different clades acquire different mutations during egg passaging. Overall, these results demonstrate that natural mutations in human H3N2 HA can influence the preference of egg-adaptation mutation, which has important implications in seed strain selection for egg-based influenza vaccine.
Author summary
Growing influenza virus in chicken eggs remains a widely used method for producing seasonal influenza vaccines. However, it is common for influenza virus to acquire mutations on the hemagglutinin (HA) protein to facilitate their growth in eggs. These egg-adaptive mutations may change the antigenic property of HA and thus decrease the vaccine effectiveness, as exemplified by mutation L194P. In this study, we demonstrate that two natural variants Thr160 and Asn190 in recently circulating human H3N2 strains can prevent the emergence of L194P. Consistently, we also show that the preference of egg-adaptive mutation varies from strains to strains, with various previously uncharacterized egg-adaptive mutations in recent human H3N2 clades. These observations indicate that the preference of egg-adaptive mutation changes as human influenza virus evolves over time. Overall, our findings provide valuable insights into the mechanism of egg-adaptation of influenza virus and have important implications for the development of more effective influenza vaccines.
Introduction
Vaccination is considered the most effective approach to prevent influenza infection. Seasonal influenza vaccine offers protection against two influenza A subtypes, namely H1N1 and H3N2, as well as influenza B viruses. However, the vaccine effectiveness against H3N2 is typically lower than H1N1 and influenza B [1–3]. Besides vaccine mismatch in certain influenza seasons, the occurrence of egg-adaptive mutations during vaccine production can also lead to the reduction of vaccine effectiveness [4–6]. When grown in eggs, human H3N2 viruses often acquire mutations in the receptor-binding site (RBS) of hemagglutinin (HA) to facilitate the binding to α2,3-linked sialic acid, which is the prototypical receptor for avian influenza viruses [7–9]. Since the HA RBS partially overlaps with several major antigenic sites [10], some egg-adaptive mutations can alter the antigenicity of HA [4, 5, 9, 11–13].
G186V and L194P are the two most common egg-adaptive mutations in H3N2 HA [14]. Although both mutations are in the RBS, L194P but not G186V significantly alters the antigenicity of HA [15]. Thus, identifying factors that influence the preference of egg-adaptive mutation can help optimize the effectiveness of egg-based influenza vaccine. Our previous study has demonstrated a strong incompatibility between G186V and L194P, in which their co-occurrence is extremely deleterious to the virus [14]. This epistatic interaction suggests that G186V and L194P represent two mutually exclusive egg-adaptive mutations. Given that epistasis is pervasive in the HA RBS and that HA RBS is constantly evolving [13,16–19], it is possible that different strains have different preferences of egg-adaptive mutation. However, it remains to be explored whether such differential preference exists.
In this study, sequence analysis and mutagenesis experiments are used to identify amino acid variants on human H3N2 HA that can affect the preference of egg-adaptive mutation. We discovered that L194P is incompatible with T160 and N190, both of which are prevalent in recent human H3N2 strains. Consistently, recent human H3N2 strains are incompatible with L194P. Our results further indicate that amino acid variants at other residues also contribute to the L194P incompatibility in recent human H3N2 strains. Along with the observation that human H3N2 strains from different clades prefer different egg-adaptive mutations, we conclude that the preference of egg-adaptive mutation changes as human H3N2 virus evolves.
Results
Differential egg-adaptation preferences in historical vaccine strains
To examine whether different human H3N2 strains have different preferences of egg-adaptive mutations, HA sequences from egg-passaged strains were collected from the Global Initiative for Sharing Avian Influenza Data (GISAID) [20] (S1 Table). Specifically, we focused on egg-passaged strains that were derived from WHO-recommended candidate vaccine viruses for influenza seasons between 2008 and 2023. Egg-passaged strains that were derived from different parental viruses have acquired different egg-adaptive mutations (Fig 1A). For example, all egg-passaged strains from A/Victoria/361/2011, A/Texas/50/2012, A/Switzerland/9715293/2013, and A/Hong Kong/2671/2019 have acquired G186V, whereas those from A/Hong Kong/4801/2014, A/Singapore/INFIMH-16-0019/2016 (Sing16) and A/Switzerland/8060/2017 (Switz17) have acquired L194P. This observation is consistent with the hypothesis that the preference of egg-adaptive mutation is strain dependent.
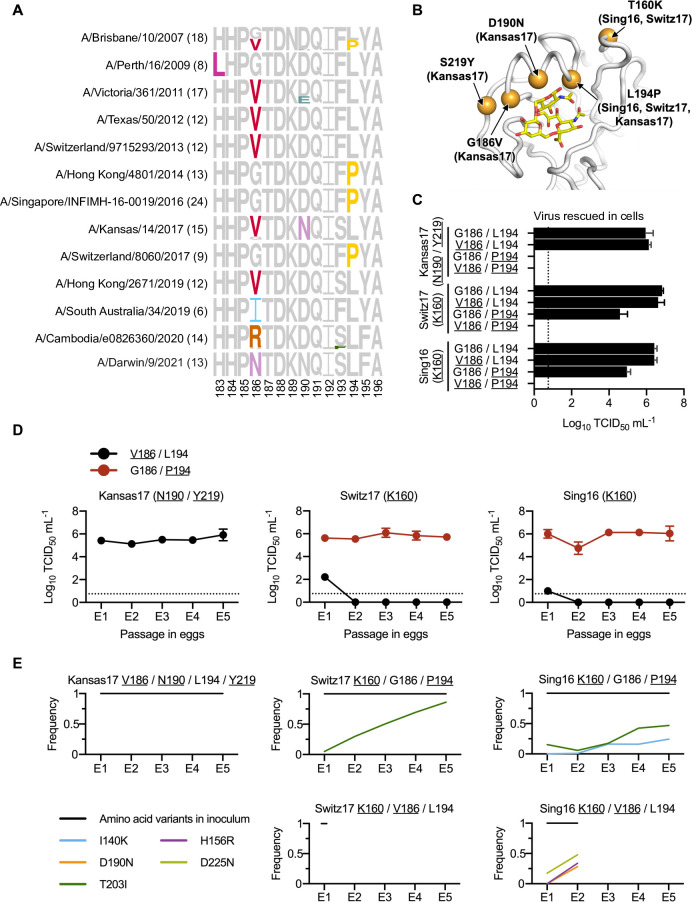
(A) Frequencies of mutations among different egg-passaged seasonal H3N2 vaccine strains are shown as sequence logos. Amino acid variants from residues 183 to 196 are shown (H3 numbering). The number of egg-passaged strains included in this analysis is indicated in the parenthesis. The relative size of each amino acid letter represents its frequency in the sequences. Grey letters represent the amino acid variants that are observed in the corresponding unpassaged parental strains. (B) Key egg-adaptive mutations in this study are shown in orange (PDB 6BKT) [17]. Sialylated glycan receptor is in yellow sticks representation. Strains in parentheses are associated with the corresponding egg-adaptive mutations. Only three strains of interest (Sing16, Switz17, and Kansas17) are included here. (C) Replication fitness of different mutants of human H3N2 vaccine strains was examined in a virus rescue experiment in mammalian cells (293T/hMDCK). Viral titers were measured by TCID50. (D) The viral titer of each human H3N2 vaccine strain with either V186/L194 or G186/P194 was measured during egg-passaging. (C-D) The means of three independent experiments are shown with SD indicated by the error bars. The dashed line represents the lower detection limit. Amino acid variant representing an egg-adaptive mutation is underlined. (E) Frequencies of mutations in the receptor-binding subdomain (residues 117–265) [21] that emerged during serial passaging in eggs. The strain of inoculum for each passaging experiment is indicated above each plot, with those representing egg-adaptive mutations underlined. Frequencies are shown as means of three biological replicates. Only those mutations that reached a minimum average frequency of 10% after the fifth passage are plotted.
To further test our hypothesis, we measured the replication fitness of G186V and L194P in Sing16, Switz17, and A/Kansas/14/2017 (Kansas17) (S2 Table). Our mutagenesis experiments were based on egg-adapted strains. As a result, all the Kansas17 mutants in this experiment were constructed in the presence of D190N and S219Y, which were present in 100% (15/15) and 87% (13/15), respectively, of egg-passaged Kansas17 strains in GISAID. Similarly, all the Sing16 and Switz17 mutants were constructed in the presence of T160K, which was found in all egg-passaged strains derived from Sing16 and Switz17 in GISAID and could alter the antigenicity of the virus (Fig 1B) [5]. The fitness effects of four different combinations of amino acid variants at residues 186 and 194 were tested, namely 1) G186/L194, which represented the amino acid variants in the unpassaged strains, 2) V186/L194, which represented the single egg-adaptive mutation G186V, 3) G186/P194, which represented the single egg-adaptive mutation L194P, and 4) V186/P194, which represented the co-occurrence of two incompatible egg-adaptive mutations [14].
As expected, all variants with G186/L194 could be rescued, whereas all variants with V186/P194 had no detectable titer (Fig 1C). Furthermore, all rescuable variants showed comparable replication kinetics in hMDCK cells, despite G186/P194 had a slightly lower titer in Switz17 and V186/L194 had a slower growth in Kansas17 (S1A Fig). In contrast, the fitness effects of V186/L194 and G186/P194 in eggs differed dramatically among strains. For example, V186/L194 only facilitated the egg-adaptation of Kansas17-N190/Y219, but not Sing16-K160 and Switz17-K160 (Fig 1D). Similarly, G186/P194 could only be rescued in Sing16-K160 and Switz17-K160, but not Kansas17-N190/Y219 (Fig 1C). Next-generation sequencing further showed that during egg-passaging, V186/L194 were stable in Kansas17-N190/Y219, and G186/P194 were stable in both Sing16-K160 and Switz17-K160 (Fig 1E and S3 Table. These results substantiate that there are differential preferences of egg-adaptive mutations among different strains.
Residues 160 and 190 influence the egg-adaptation preference
The inability of rescuing G186/P194 in Kansas17-N190/Y219 suggests that G186V is not the only mutation that is incompatible with L194P. We therefore aimed to identify additional mutations that were incompatible with L194P. The HA sequence of Kansas17 X-327, which was an egg-adapted strain equivalent to the Kansas17-V186/N190/L194/Y219 (see above, Fig 1C), was compared to that of Switz17 NIB-112 and Sing16 NIB-104, which were two egg-adapted strains with K160/P194. Of note, the HA sequences are equivalent between Kansas17 X-327 with V186G/L194P (Fig 2A) and the unrescuable Kansas17-G186/N190/P194/Y219 (see above, Fig 1C). This sequence analysis led us to identify four candidate amino acid variants in the RBS that might be incompatible with L194P, namely S159, N190, S193 and Y219. Subsequently, S159Y, N190D, S193F and Y219S, which represented the RBS mutations from Kansas17 X-327 to Switz17 NIB-112 and Sing16 NIB-104, were introduced individually into the HA of Kansas17 X-327 with V186G/L194P. V186G was included in since V186 and P194 were known to be incompatible [14]. Our virus rescue experiment showed that the fitness defect of V186G/L194P in Kansas17 X-327 could be restored by N190D (Fig 2B). Interestingly, although Kansas17 X-327 with V186G/L194P/N190D had a lower replication fitness in hMDCK cells compared to X-327, it showed comparable replication fitness with X-327 in eggs (Figs Figs2C2C and S1B). Next-generation sequencing further confirmed that Kansas17 X-327 with V186G/L194P/N190D was genetically stable during egg-passaging (Fig 2D and S3 Table). To examine if the incompatibility between N190 and P194 could be generalized to other strains, we further introduced D190N into Switz17-K160/P194 and Sing16-K160/P194, which were equivalent to Switz17 NIB-112 and Sing16 NIB-104, respectively. Consistent with the results for Kansas17 X-327, D190N was deleterious in Switz17-K160/P194 and Sing16-K160/P194 (Fig 2E). Together, these results demonstrate the incompatibility between N190 and P194.
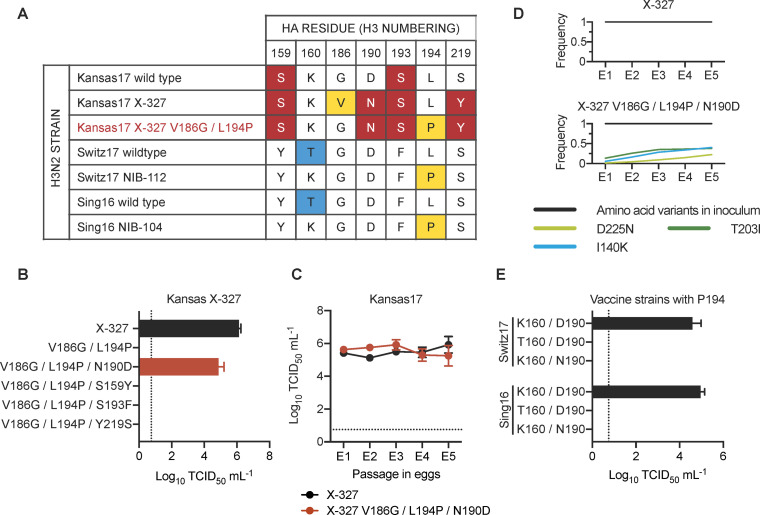
(A) Amino acid variants from residues 150 to 220 among the indicated strains are shown. The rest of the sequences from residues 150 to 220 were completely conserved among these strains. Major egg-adaptive mutations G186V and L194P are in yellow. Amino acid variants that are unique to Kansas17 strains are in red while variants unique to Switz17 and Sing16 are in blue. Of note, Kansas17 X-327, Switz17 NIB-112, and Sing16 NIB-104 are egg-adapted strains, whereas Kansas17 X-327 V186G/L194P (red) was a mutant generated in this study. (B) Replication fitness of Kansas17 X-327 with indicated mutations was examined in a virus rescue experiment in mammalian cells (293T/hMDCK). (C) The replication fitness of Kansas17 X-327 and Kansas17 X-327 with triple mutations (V186G/L194P/N190D) in eggs were examined during serial egg-passaging. (D) Frequencies of mutations in the receptor-binding subdomain (residues 117–265) [21] that emerged during egg-passaging of the indicated strains. Results are shown as means of three independent biological replicates. Only those mutations that reached a minimum average frequency of 10% after the fifth passage are plotted. (E) Replication fitness of Sing16 and Switz17 L194P virus with or without K160T and D190N mutations was assessed in a virus rescue experiment in mammalian cells (293T/hMDCK). (B, C, E) All viral titers were measured by TCID50. The means of three independent experiments are shown with SD indicated by the error bars. The dashed line represents the lower detection limit.
During our sequence analysis, we noticed that the egg-adaptive mutation T160K [5] often co-occurred with L194P, as exemplified by Switz17 NIB-112 and Sing16 NIB-104. Our rescue experiment showed that T160 was incompatible with P194 in both Sing16 and Switz17 (Fig 2E). As a result, in addition to V186 and N190, T160 is another amino acid variant that is incompatible with P194.
Recent human H3N2 strains are incompatible with L194P
Among the amino acid variants that are incompatible with P194, T160 and N190 have reached high occurrence frequency in recently circulating human H3N2 strains. T160, which introduced an N-glycosylation site at N158, was fixed in clade 3C.2a during the 2014–2015 influenza season [5], whereas N190 emerged more recently along with I160 and D186 and reached >90% in 2021 (Fig 3A). Not surprisingly, A/Victoria/22/2020 (Vic20) and A/Italy/11871/2020 (Italy20), which were both from clade 3C.2A1b.1a and had T160/N190, could not be rescued in the presence of L194P (Fig 3B). In contrast, introducing V186 into Italy20 yielded high titers in both virus rescue and egg-passaging experiments (Fig 3C). These observations illustrate that recent human H3N2 strains have a strong preference against L194P as determined in the egg-adapted viruses.
(A) Frequencies of amino acid variants at residues 160, 186 and 190 in human H3N2 HA over time are shown. Only those variants that reached a maximum annual frequency of 35% are plotted. (B) Two wild type (WT) strains in clade 3C.2a1b.1a (Vic20 and Italy20) with or without L194P mutation were examined in a virus rescue experiment in mammalian cells (293T/hMDCK). Of note, most unpassaged strains in clade 3C.2a1b.1a, including both Vic20 and Italy20, naturally contain N190. (C) The replication fitness of Italy20 with indicated mutations was examined in a virus rescue experiment in mammalian cells (293T/hMDCK) as well as in an egg-passaging experiment, in which 104 TCID50 of the rescued viruses were propagated in eggs for 48 hours. (D) The replication fitness of Italy20 with indicated mutations was measured in a virus rescue experiment in mammalian cells (293T/hMDCK). (B-D) Viral titers were measured by TCID50. The means of three independent experiments are shown with SD indicated by the error bars. The dashed line represents the lower detection limit. (E) The locations of mutations between Sing16 and Italy20 in the receptor-binding subdomain (residues 117–265) [21] are shown in blue (PDB 6BKT). The locations of T160 and Y98 are shown in grey [17]. L194P is in orange. Sialylated glycan receptor is in yellow sticks representation. (F) The structural impact of each mutation was modeled by Rosetta [48]. The distance between the phenolic oxygen of Tyr98 (OH98) and the Cα of residue 190 was computed as the height of receptor-binding site (RBS). Three replicates, each with 100 simulations, were performed. Each data point represents the lowest scoring pose in a replicate. Error bars represent the SD. P-value was computed by two-tailed t-test. Only the difference between WT and G186V is statistically significant (P < 0.05).
While we have shown above that T160 and N190 were incompatible with L194P in Kansas17, Switz17, and Sing16 (Fig 2), Italy20 with T160K/N190D was still incompatible with L194P (Fig 3D). This result indicates that besides T160 and N190, additional amino acid variants in Italy20 were also incompatible with L194P. We postulated that those additional amino acid variants could be identified by comparing the HA sequences of Italy20 and Sing16, since Sing16 was compatible with L194P in the presence of K160/D190 (Fig 2E). Eight mutations in the receptor binding subdomain (residues 117–265) [21] were identified between Sing16 and Italy20 (Fig 3E). Based on spatial proximity between these eight mutations and L194P, we hypothesized that the differential fitness effects of L194P in Sing16 and Italy20 could be due to the presence of mutations G186D, F193S, and S198P in Italy20. Since the compatibility with L194P in Kansas17 was not influenced by the presence of S193 as shown in our rescue experiment above (Fig 2A and 2B), we focused on D186 and P198 here. However, even with mutations D186G and P198S, Italy20-T160K/D186G/N190D/P198S/L194P still could not be rescued (Fig 3D). As demonstrated in our previous study, G186V significantly increases the RBS height [14]. In contrast, L194P decreases the RBS height. The co-existence of these two mutations with opposite structural effects destabilizes the 190-helix and abolishes receptor binding [14]. However, structural modeling showed that none of the eight mutations increased the height of the RBS (Fig 3F). Together, these results demonstrate that the L194P incompatibility in recent human H3N2 strains is conferred by multiple amino acid variants in addition to T160 and N190, and likely involves a mechanism that is different from G186V.
Egg-adaptation preference is clade dependent
The recent emergence of L194P incompatibility suggests that the preference of egg-adaptive mutation has been evolving. To understand the preference of egg-adaptation in recent human H3N2 strains, we analyzed the HA sequences of 72 egg-passaged H3N2 strains from years 2019 to 2021, which belong to antigenically distinct clades [22–24]. By comparing the HA sequences between the egg-passaged strains and their non-egg-passaged counterparts, we observed that egg-adaptive mutations often emerged at residues 160, 186, 190, 194, 219 and 225 (Fig 4 and S4 Table). In addition, different clades showed different preferences of egg-adaptive mutation. This differential preference could be exemplified by residues 186 and 225. While all non-egg-passaged strains from both clades 3C.2a1b.1a and 3C.2a1b.2a2 had D186, egg-adaptive mutation D186N was common in 3C.2a1b.2a2 but not observed in 3C.2a1b.1a. Similarly, although D225 was present in all non-egg-passaged strains across all clades, egg-adaptive mutation D225G was prevalent in 3C.2a1b.2a2 but not observed in clades 3C.2a1b.2a, 3C.2a1b.2b, and 3C.2a1b.1b. These observations are consistent with the notion that the preference of egg-adaptive mutation in human H3N2 HA is influenced by natural mutations.
A rooted phylogenetic tree was built using maximum likelihood (ML) method with 100 bootstraps on the HA sequences of 61 human H3N2 strains without egg-passaging (unpassaged strains). Different clades are highlighted in different colors. WHO-recommended vaccine strains are indicated by a red triangle symbol. These unpassaged human H3N2 strains correspond to the parental strains of 72 egg-passaged strains. Since multiple egg-passaged strains with different HA sequences could be generated from a single unpassaged strain, the number of unpassaged strains is less than that of egg-passaged strains. Frequencies of amino acids at HA residues 160, 186, 190, 194, 219 and 225 among egg-passaged strains in different clades are shown as sequence logos. The relative size of each amino acid letter represents its frequency in the sequences. For each clade, amino acid variants that are also observed in the unpassaged strains are in grey and listed below each sequence logo.
Discussion
Egg-adaptation of influenza virus has been central to the production of seasonal influenza vaccine. Our work here shows that the preference of egg-adaptive mutation is strain dependent, due to epistasis between egg-adaptive mutations and natural amino acid variants. Specifically, we identified two natural amino acid variants T160 and N190 that are incompatible with the egg-adaptive mutation L194P. Our results also indicate that additional amino acid variants are responsible for the L194P incompatibility in recent human H3N2 HA. Besides L194P, many other egg-adaptive mutations, albeit at a lower prevalence, were observed in historical vaccine strains, such as H156Q, H156R, H183L and A196T [14,25–27]. The sequence determinants that influence the preference of other egg-adaptive mutations are currently unclear. Comprehending the epistatic interactions that involve egg-adaptive mutations as well as the underlying mechanisms will be key to accurately predict mutations that arise during egg-adaptation.
Our previous studies have shown that epistasis is pervasive between mutations in the HA RBS [16–18]. One interesting observation is that epistasis between two HA RBS mutations can be modulated by a third mutation. For example, epistasis between the engineered mutations L226R and S228E in an H1 HA could only be observed in the presence of G225E [16]. This type of higher-order epistasis may play a role in determining the preference of egg-adaptive mutation. Despite the incompatibility between T160 and P194 in Sing16 and Switz17 (Fig 2E), we noticed the co-occurrence of T160 and P194 in four egg-adapted H3N2 strains (EPI1588456, EPI1584616, EPI1526498 and EPI1440496), which all belong to clade 3C.2a1b.2b (Fig 4 and S4 Table). This observation implies that the incompatibility between T160 and P194 can be masked by other amino acid variants. As shown by a recent study, epistasis can also alter the antigenic effect of a given mutation in the HA RBS [28]. Whether the antigenic effects of egg mutations are influenced by epistasis will need to be explored in future studies.
We anticipate that the preference of egg-adaptive mutation will keep changing as human H3N2 HA continues to evolve. As a result, future studies of egg-adaptive mutations in human H3N2 strains are warranted. Currently, 3C.2a1b.2a2 is the dominant clade of circulating human H3N2 virus. Although our data show that egg-adaptive mutation L194P is unlikely to occur in clade 3C.2a1b.2a2, egg-adaptive mutations D186N and D225G are common in this clade [15]. However, the antigenic effects of D186N and D225G in clade 3C.2a1b.2a2 remain to be explored. Given that different egg-adaptive mutations can have different antigenic effects [5,8,9,12,15,24–32], systematic characterization of individual egg-adaptive mutations is critical for the selection of vaccine seed strains with minimal antigenic change. Nevertheless, egg-based influenza vaccine production is an 80-year-old technology that needs to be replaced by more advanced alternatives. An important step forward was the commercialization of cell-based and recombinant influenza vaccines, which have shown better effectiveness than egg-based influenza vaccines [33,34]. In addition, a Phase 1/2 clinical trial of an mRNA seasonal influenza vaccine candidate has initiated recently [35]. Despite the potentially slow transition, these alternatives will provide an ultimate solution to the problems of egg adaptation.
Materials and methods
Sequence analysis
Full-length HA sequences of WHO-recommended H3N2 candidate vaccine virus strains from 2008 to 2022 were downloaded from the Global Initiative for Sharing Avian Influenza Data (GISAID; http://gisaid.org) [20]. Information for these sequences is shown in S1 Table. Information for the full-length HA sequences of viruses from 2019 to 2021, with and without egg-passaging, is shown in S4 Table. Sequences were aligned by MAFFT version 7 (https://mafft.cbrc.jp/alignment/software/) with default setting [36]. Passaging history was determined by parsing the regular expression in FASTA headers as described [37]. The frequencies of amino acids were plotted using WEBLOGO (https://weblogo.berkeley.edu/logo.cgi) [38]. Phylogenetic tree was built and rooted using maximum likelihood (ML) method by Geneious Prime (Geneious), with 100 bootstraps.
Cell culture
Humanized Madin-Darby canine kidney (hMDCK) cells with higher and stable expression of human 2,6-sialtransferase were kindly provided by Professor Yoshihiro Kawaoka, the University of Tokyo [39]. Both hMDCK cells and human embryonic kidney (HEK) 293T cells were maintained in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 25 mM HEPES, and 100 U mL-1 penicillin-streptomycin (PS).
Virus rescue experiments
All H3N2 viruses generated in this study were based on the influenza eight-plasmid reverse genetics system [40,41]. The HA and neuraminidase (NA) genes of the strains of interest (S2 Table) were synthesized by Sangon Biotech, and cloned into the pHW2000 vector as previously described [14]. For HA, the ectodomain was from the strains of interest, whereas the non-coding region, N-terminal secretion signal, C-terminal transmembrane domain, and cytoplasmic tail were from H1N1 A/PR/8/34 (PR8). For NA, the entire coding region was from the strains of interest, whereas the non-coding region of NA was from PR8. Mutations were introduced into the constructs by polymerase chain reaction (PCR). Primers used in this study were produced by Integrated DNA Technologies. Recombinant chimeric 6:2 reassortant viruses with the six internal genes (PB2, PB1, PA, NP, M, and NS) from PR8 were rescued by transfecting a co-culture of HEK 293T and hMDCK cells at a 6:1 ratio with 70% confluence in 6-well plate. Transfection was performed using 16 μL of TransIT-LT1 (Mirus Bio) and 1 μg each of the eight plasmids encoding the corresponding influenza virus segments (a total amount of 8 μg of DNA). At 6 h post-transfection, medium was replaced with 1 mL of MEM. At 24 h post-transfection, another 1 mL of MEM supplemented with 1 μg mL-1 tosylphenylalanyl chloromethyI ketone (TPCK)-trypsin was added. Supernatant was harvested at 72 h post-transfection, all of which was further inoculated into hMDCK cells at 90% confluence in a T75 flask. Supernatant of the hMDCK cells was harvested when more than 70% of cells had cytopathic effect (CPE). If less than 70% of cells had CPE by 72 h post-infection, cell supernatant was harvested regardless. Virus titer was determined by titration in hMDCK cells as previously described [42,43]. For each mutant, three independent rescue experiments were performed. All our rescue experiments included a positive control that contained wild type (WT) HA and a negative control that contained no HA. Viral RNAs were extracted, reverse transcribed by Superscript III reverse transcriptase (Thermo Fisher Scientific), and sequenced by Sanger sequencing to confirm its sequence integrity before passaging in eggs.
Multicycle viral growth kinetics
To measure the viral multicycle viral growth kinetics, rescuable viruses were inoculated into hMDCK cells at an MOI of 0.01, and supernatants were harvested at 24, 48 and 72 h post-infection [44]. Viral titers in the supernatants were determined by titration in hMDCK cells as previously described [42,43].
Virus passaging in embryonic eggs
Embryonic eggs at 10 days post-fertilization were used for the virus passaging. For each passage, 104 TCID50 of viruses were inoculated into one egg. The only exception was for the next-generation sequencing experiment (see below), 106 TCID50 was used for the first passage of Sing16-K160/V186/L194. The allantoic fluid was harvested at 48 h post-infection. Virus titer was determined by titration in hMDCK cells as previously described [42,43]. Each experiment consisted of five consecutive passaging and was performed in triplicate.
Next-generation sequencing of egg-passaged viruses
The viral RNA of the egg-passaged virus was extracted using QIAamp Viral RNA Mini Kit (QIAGEN). The extracted RNA was then reverse transcribed to cDNA using ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs). Subsequently, PCR was performed using PrimeSTAR Max DNA Polymerase (Takara Bio) with the cDNA as template and virus-specific primers 5’-CAC TCT TTC CCT ACA CGA CGC TCT TCC GAT CT NNN GGT CAC TAG TTG CCT CAT CCG G-3’ and 5’- GAC TGG AGT TCA GAC GTG TGC TCT TCC GAT CTG GTG CAT CTG ATC TCA TTA TTG-3’. A second round of PCR was performed to add the rest of adapter sequence and index to the amplicon using primers 5’- AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT-3’ and 5’- CAA GCA GAA GAC GGC ATA CGA GAT XXX XXX GTG ACT GGA GTT CAG ACG TGT GCT-3’. Nucleotides in primers annotated by an ‘N’ or “X” represented the index sequence for distinguishing PCR products derived from different viruses, passages, and biological replicates. The final PCR products were sequenced by Illumina MiSeq PE300.
Analysis of next-generation sequencing data
Sequencing data was obtained in FASTQ format and analyzed using in-house Python and shell scripts. Briefly, paired-end reads were merged by PEAR [45]. The merged reads were then parsed by SeqIO module in BioPython and translated into protein sequences [46]. For each sample, the frequency of amino acid variant i was computed as follows:
where readcounti represents the number of reads that contain amino acid variant i and sequencingdepth represents the total number of reads in the sample of interest.
Structural modeling
Residues 57 to 261 of a monomer of A/Michigan/15/2014 (H3N2) influenza virus HA (PDB 6BKT) [17] was extracted. The PDB file was renumbered using pdb-tools [47]. Point mutagenesis for T128A, V130I, T135K, A138S, G186D, G186V, D190N, F193S, L194P and S198P (all in original numbering) were performed using the fixed backbone (fixbb) design application in Rosetta (RosettaCommons). For each replicate, one hundred poses were generated and the lowest scoring pose was used for downstream analysis. The fixbb application for each mutation was performed in triplicate. A constraint file for each lowest scoring pose was obtained using the minimize_with_cst application in Rosetta and converting the log file to a constraint (cst) file using the convert_to_cst_file application in Rosetta. Subsequently, fast relax was performed on wild type or mutant using the relax application in Rosetta with the corresponding constraint file [48]. For each replicate, 30 poses were generated and the lowest scoring pose was used to measure the height of the RBS, which is the distance between the side chain oxygen atom of Y98 (H3 numbering) and Cα of amino acid 190 (H3 numbering) using PyMOL (Schrödinger).
Supporting information
S1 Fig
Viral replication kinetics of different H3N2 variants in hMDCK cells.(A) Viral replication kinetics of rescuable mutants of H3N2 vaccine strains were examined in hMDCK cells. (B) Viral replication kinetics of Kansas17 X-327 and Kansas17 X-327 with V186G/L194P/N190D were examined in hMDCK cells. (A, B) hMDCK cells were inoculated with the indicated variant at an MOI of 0.01. Viral titers in supernatants harvested at 24, 48 and 72 h post-infection were measured by TCID50 using hMDCK cells. The means of three independent experiments are shown with SD indicated by the error bars. The dashed line represents the lower detection limit. Amino acid variant representing an egg-adaptive mutation is underlined.
(TIF)
S1 Table
H3N2 components of WHO-recommended influenza vaccines for influenza seasons between 2008 and 2023.(DOCX)
S2 Table
H3N2 genes that were used in this study.(DOCX)
S3 Table
Frequencies of mutations in egg-passaged viruses that showed an average frequency of >10% in the fifth passage.(DOCX)
S4 Table
Information of HA from human H3N2 strains with or without egg-passaging.(DOCX)
Funding Statement
This work was supported by a fellowship from the Pasteur Foundation Asia (W.L.), the Calmette and Yersin scholarship from the Pasteur International Network Association (H.L.), the funding from Centre for Immunology & Infection, InnoHK, an initiative of the Innovation and Technology Commission, the Government of the Hong Kong SAR (R.B.), the startup fund from the University of Illinois at Urbana-Champaign (N.C.W.), the Health and Medical Research Fund (no.19180932) (C.K.P.M), the National Research Foundation of Korea (NRF) grant funded through the Korea government (NRF-2018M3A9H4055203) (C.K.P.M.), Guangdong-Hong Kong-Macau Joint Laboratory of Respiratory Infectious Disease (20191205) (C.K.P.M.) and the visiting scientist scheme from Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore (C.K.P.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Raw sequencing data have been submitted to the NIH Short Read Archive under accession number: BioProject PRJNA800806. Codes for analyzing next-generation sequencing data have been deposited to https://github.com/Wangyiquan95/HA_egg_passage. Code and source files for structural modeling using Rosetta have been deposited to https://github.com/timothyjtan/HA-RBD_height.
References
Decision Letter 0
9 Jun 2022
Dear Dr Wu,
Thank you very much for submitting your manuscript "Egg-adaptation pathway of human influenza H3N2 virus is contingent on natural evolution" for consideration at PLOS Pathogens. As with all papers reviewed by the journal, your manuscript was reviewed by members of the editorial board and by several independent reviewers. In light of the reviews (below this email), we would like to invite the resubmission of a significantly-revised version that takes into account the reviewers' comments.
We cannot make any decision about publication until we have seen the revised manuscript and your response to the reviewers' comments. Your revised manuscript is also likely to be sent to reviewers for further evaluation.
When you are ready to resubmit, please upload the following:
[1] A letter containing a detailed list of your responses to the review comments and a description of the changes you have made in the manuscript. Please note while forming your response, if your article is accepted, you may have the opportunity to make the peer review history publicly available. The record will include editor decision letters (with reviews) and your responses to reviewer comments. If eligible, we will contact you to opt in or out.
[2] Two versions of the revised manuscript: one with either highlights or tracked changes denoting where the text has been changed; the other a clean version (uploaded as the manuscript file).
Important additional instructions are given below your reviewer comments.
Please prepare and submit your revised manuscript within 60 days. If you anticipate any delay, please let us know the expected resubmission date by replying to this email. Please note that revised manuscripts received after the 60-day due date may require evaluation and peer review similar to newly submitted manuscripts.
Thank you again for your submission. We hope that our editorial process has been constructive so far, and we welcome your feedback at any time. Please don't hesitate to contact us if you have any questions or comments.
Sincerely,
Florian Krammer, PhD
Associate Editor
PLOS Pathogens
Mark Heise
Section Editor
PLOS Pathogens
Kasturi Haldar
Editor-in-Chief
PLOS Pathogens
orcid.org/0000-0001-5065-158X
Michael Malim
Editor-in-Chief
PLOS Pathogens
***********************
Reviewer's Responses to Questions
Part I - Summary
Please use this section to discuss strengths/weaknesses of study, novelty/significance, general execution and scholarship.
Reviewer #1: Liang et al. complete a nice set of experiments analyzing the compatibility of different egg-adaptative HA substitutions in different H3N2 backgrounds. Wu and colleagues recently demonstrated that G186V and L194P substitutions in HA are incompatible, and the current study expands this to examine additional substitutions in other HA backgrounds. They first identify different egg-adaptive substitutions in different egg based H3N2 vaccine strains. They then study replicative fitness (or ability to rescue) viruses with different combinations of HA mutations. They ultimately identify epistatic interactions, including those involving key egg/antigenic substitutions such as T160K, that are in these vaccine strains, and conclude that different egg-adaptations are favored in different genetic backgrounds.
Reviewer #2: In this manuscript by Liang et al., the authors sought out to understand how egg adaptations of H3N2 differ across distinct H3N2 viruses. The authors nicely show epistasis of key residues associated with egg mutations and how this relates to distinct H3N2 clades. The manuscript is very well written and the topic is important. The manuscript can be hard to follow at times, largely due to the massive number of mutations being studied. Simplification of the key residues and relationship between these as a panel somewhere in the paper would greatly benefit the presentation of this manuscript. Otherwise, all my comments are quite minor and easily addressable.
Reviewer #3: The authors assess how natural mutations in seasonal H3N2 virus impact on the adaptation of these same viruses when cultured in eggs. H3N2 viruses are frequently associated to a higher mismatch of current seasonal vaccines in comparison to group 1 and influenza B viruses. One important aspect to consider here is that most currently licensed influenza vaccines are manufactured in eggs. Therefore, it is of interest to understand if changes in antigenicity of H3N2 viruses when passaged in eggs are related to specific DNA/protein sequences of circulating H3N2 influenza viruses. The manuscript is well-written and conveys a clear message to the audience. There are some minor points that should be addressed before acceptance.
Reviewer #4: In this manuscript Liang et al, perform a series of experiments to elucidate the trajectory and limitations of the mutation (amino acid variations) that arise in the hemagglutinin (HA) protein of H3N2 human seasonal viruses during egg-adaptation. In previous studies they identified the amino acid change L194P as an important egg-adaptation that often occurs during passaging of human H3N2 in eggs. Herein they identify specific amino acid changes that appear to be compensatory mutations that are generated in the presence of amino acid P194 in in HA. The authors also identify two naturally occurring amino acids (T160 and N190) that are appear to be incompatible with mutation P194, suggesting a potential structural restrictions on accommodating some of these residue variations. The results provide relevant information on egg-adaptations that impact vaccine development. Nonetheless, there are a few issues that need to be addressed to improve the clarity of the manuscript, in particular on how the authors are assessing and interpreting fitness.
**********
Part II – Major Issues: Key Experiments Required for Acceptance
Please use this section to detail the key new experiments or modifications of existing experiments that should be absolutely required to validate study conclusions.
Generally, there should be no more than 3 such required experiments or major modifications for a "Major Revision" recommendation. If more than 3 experiments are necessary to validate the study conclusions, then you are encouraged to recommend "Reject".
Reviewer #1: 1. Overall, the study is very nice and the experiments are well control and the data are solid. There are some parts of the manuscript where I felt ‘a bit in the weeds’ trying to follow along with different combinations of substitutions, but the overall experimental question is important and the data support the idea that different egg adaptations are favored in different genetic backgrounds.
2. The study could be strengthened if there was a better structural/mechanistic explanation for some of the findings. For example, Wu and colleagues showed earlier that RBS height explained the incompatibility of L194P and G186V. The authors of the current study show that is not the case in panel Fig 3F for incompatibility of N190 and P194, but additional structural experiments could address the mechanism further about this incompatibility.
3. The authors conclude that the specific types of egg-adaptations that arise is complicated and dependent on the evolving H3N2 strains. It would be useful to be able to predict specific egg-adaptations based on sequence—is this impossible at this point or could preliminary predictive models be made?
Reviewer #2: N/A
Reviewer #3: (No Response)
Reviewer #4: - It is unclear how the authors assessed replication fitness of the rescued viruses. In the methods they mention that after transfection the viruses were harvested and inoculated into hMDCKs, and then the supernatant were harvested once CPE was observed. These supernatants (containing the viruses) were then titrated by a TCID50 assay. Hence, it seems that the authors are determining fitness as a measure ability to rescue viruses and the generation of progeny virus (titer) after CPE was observed.
Was there any standardization to control the time that each of the viral infections took to generate CPE?; e.g. Do you collect the supernatant at the first sign of CPE? After a few hours of CPE? Until all the cells show CPE?
- To determine differences in replication fitness, one would have to perform a side-by-side growth curve for 48 hrs and plot them as such to determine and compare the kinetics of replication. As this is a major methodology of the paper (shown in Figs 1B, 2B and E, and 3B-D), these experiments need to be explained in more detail to understand how the authors are controlling for time differences that the cultures take to determine CPE, and the time when the sups were collected. It will also be valuable to actually show growth curves in cases where the growth kinetics are in fact slower (or faster).
- Can you comment on the number of rescue attempts that were done for each of the mutant viruses? After how many attempts did the authors determined that a virus could not be rescued? How were the rescue attempts controlled? Please also clarify if all viral stocks were sequenced after rescue and prior to passage in eggs, to confirm the sequence integrity of each rescue. Include that information in detail in the methods section.
- For the figure legends 1B, 2B and E, and 3B-D, provide more information on the experiment? e.g. since the results of the rescue attempts in the 293T/hMDCK… For expalme in Line 356, “…in a virus rescue experiment in mammalian cells (293T/hMDCK) after XX attempts”. Indicate what the error bars are (e.g. Line 359: SD of three experiments represented by the error bars).
- The use of “egg-adaptation pathway” is a bit confusing, as this would imply a unique way in which the mutations might generate. The data shown (e.g. Figs 2, 3 and 4) indicate that different mutations (amino acid variations) in deed occur depending on the genotype of the initial human H3N2 strain. Hence, it appears that alternative amino acids (e.g. more then one residue) can emerge at different positions as compensatory mutations. Therefore, I suggest revising the use of “egg-adaptation pathway” throughout the manuscript, including the tittle, to better reflect the conclusions of the study. For example, a tittle a long these lines: “Egg-adaptation compensatory mutations of human influenza H3N2 virus are dependent on natural evolution”, might be more aligned with the results shown.
**********
Part III – Minor Issues: Editorial and Data Presentation Modifications
Please use this section for editorial suggestions as well as relatively minor modifications of existing data that would enhance clarity.
Reviewer #1: It might be nice to add details about the new 2022-23 Northern Hemisphere H3N2 vaccine strain (and current SH vaccine) which doesn’t have the N158 glycosylation site.
Reviewer #2: 1. At the end of figure 1 please include a phylogenetic tree of the different viruses being tested and the mutations associated with which. This will help clarify how the egg-adaptation and H3N2 strains relate. Alternatively, a structure of HA and which residues are associated with each strain would be useful.
2. Line 150, please clarify this is circulating virus and not egg-grown virus.
3. Line 175, can the authors expand on why RBS height may matter? I’d assume this obstructs receptor binding, but it’s not clear.
4. Line 215 – should read “which all belong to clade…”
5. The authors do a lot of virus rescuing but it’s not explicitly clear how they did so. They seem to use reverse genetics to generate their viruses and to rescue them in MDCK cells and then use eggs to show stability of mutations. Clarity on methods are appreciated.
Reviewer #3: Line 95-97) What do authors mean by background mutations? Are these egg mutations having a deleterious impact on the antigenicity of the virus?
Line 150-151) What do authors mean by “T160, which introduced an N-glycosylation site at N158”? Does this N-glycosylation occur due to a higher accessibility to this site of glycosyltransferases after the T160 mutation takes place?
Line 226-227) Do authors mean that if for instance there is a prevalent circulating H3N2 strain but it is likely that when growing it in eggs there appears remarkable antigenic changes, it is better to select another circulating strain, though less prevalent in humans, but less prone to antigenic changes in eggs? Of course we are not considering here the use of mammalian cell lines to grow these viruses, we assume only that the egg platform is the only possibility.
Discussion point) Is the rationale of this study based on the egg-adapted mutations discussed in this study since they are the most prevalent seen in H3N2 strains? Are there other relevant mutations not mentioned in this study?
Discussion point) according to the data provided by the authors, it seems the deleterious mutation L194P is less prevalent in egg-passaged H3N2 strains. Do authors believe that mismatches to current circulating H3N2 strains due to egg-passaging are less likely to occur? Discussion on this topic would be interesting for the audience.
Reviewer #4: 1. In the model on Figure 3, for orientation it would be helpful to include/highlight the locations of residues 160 and 98.
2. In the Figure 4 legend, include the method and bootstrap used to construct the phylogenetic tree and whether the tree was rooted or not.
3. Line 52; change to “which is the prototypical receptor for avian influenza viruses”
4. Line 52; Change to “Since the HA RBS partial…”
5. Line 62; should be “discovered”
6. Line 153; is this supposed to be 2020 and not 2021??
7. Line 155; it should be “titers”
8. Line 157-158; change to “…as determined in the egg-adapted viruses”
**********
PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: No
Reviewer #3: No
Reviewer #4: No
Figure Files:
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email us at gro.solp@serugif.
Data Requirements:
Please note that, as a condition of publication, PLOS' data policy requires that you make available all data used to draw the conclusions outlined in your manuscript. Data must be deposited in an appropriate repository, included within the body of the manuscript, or uploaded as supporting information. This includes all numerical values that were used to generate graphs, histograms etc.. For an example see here on PLOS Biology: http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1001908#s5.
Reproducibility:
To enhance the reproducibility of your results, we recommend that you deposit your laboratory protocols in protocols.io, where a protocol can be assigned its own identifier (DOI) such that it can be cited independently in the future. Additionally, PLOS ONE offers an option to publish peer-reviewed clinical study protocols. Read more information on sharing protocols at https://plos.org/protocols?utm_medium=editorial-email&utm_source=authorletters&utm_campaign=protocols
Author response to Decision Letter 0
29 Jul 2022
Attachment
Submitted filename: Liang_PLOS Pathg Response_V7_20220728.docx
Decision Letter 1
12 Sep 2022
Dear Dr Wu,
We are pleased to inform you that your manuscript 'Egg-adaptive mutations of human influenza H3N2 virus are contingent on natural evolution' has been provisionally accepted for publication in PLOS Pathogens.
Before your manuscript can be formally accepted you will need to complete some formatting changes, which you will receive in a follow up email. A member of our team will be in touch with a set of requests.
Please note that your manuscript will not be scheduled for publication until you have made the required changes, so a swift response is appreciated.
IMPORTANT: The editorial review process is now complete. PLOS will only permit corrections to spelling, formatting or significant scientific errors from this point onwards. Requests for major changes, or any which affect the scientific understanding of your work, will cause delays to the publication date of your manuscript.
Should you, your institution's press office or the journal office choose to press release your paper, you will automatically be opted out of early publication. We ask that you notify us now if you or your institution is planning to press release the article. All press must be co-ordinated with PLOS.
Thank you again for supporting Open Access publishing; we are looking forward to publishing your work in PLOS Pathogens.
Best regards,
Florian Krammer, PhD
Associate Editor
PLOS Pathogens
Mark Heise
Section Editor
PLOS Pathogens
Kasturi Haldar
Editor-in-Chief
PLOS Pathogens
orcid.org/0000-0001-5065-158X
Michael Malim
Editor-in-Chief
PLOS Pathogens
***********************************************************
Reviewer Comments (if any, and for reference):
Reviewer's Responses to Questions
Part I - Summary
Please use this section to discuss strengths/weaknesses of study, novelty/significance, general execution and scholarship.
Reviewer #1: (No Response)
Reviewer #2: The authors have addressed my concerns.
Reviewer #3: (No Response)
**********
Part II – Major Issues: Key Experiments Required for Acceptance
Please use this section to detail the key new experiments or modifications of existing experiments that should be absolutely required to validate study conclusions.
Generally, there should be no more than 3 such required experiments or major modifications for a "Major Revision" recommendation. If more than 3 experiments are necessary to validate the study conclusions, then you are encouraged to recommend "Reject".
Reviewer #1: (No Response)
Reviewer #2: N/A
Reviewer #3: (No Response)
**********
Part III – Minor Issues: Editorial and Data Presentation Modifications
Please use this section for editorial suggestions as well as relatively minor modifications of existing data that would enhance clarity.
Reviewer #1: (No Response)
Reviewer #2: N/A
Reviewer #3: (No Response)
**********
PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: No
Reviewer #3: No
Acceptance letter
20 Sep 2022
Dear Dr Wu,
We are delighted to inform you that your manuscript, "Egg-adaptive mutations of human influenza H3N2 virus are contingent on natural evolution," has been formally accepted for publication in PLOS Pathogens.
We have now passed your article onto the PLOS Production Department who will complete the rest of the pre-publication process. All authors will receive a confirmation email upon publication.
The corresponding author will soon be receiving a typeset proof for review, to ensure errors have not been introduced during production. Please review the PDF proof of your manuscript carefully, as this is the last chance to correct any scientific or type-setting errors. Please note that major changes, or those which affect the scientific understanding of the work, will likely cause delays to the publication date of your manuscript. Note: Proofs for Front Matter articles (Pearls, Reviews, Opinions, etc...) are generated on a different schedule and may not be made available as quickly.
Soon after your final files are uploaded, the early version of your manuscript, if you opted to have an early version of your article, will be published online. The date of the early version will be your article's publication date. The final article will be published to the same URL, and all versions of the paper will be accessible to readers.
Thank you again for supporting open-access publishing; we are looking forward to publishing your work in PLOS Pathogens.
Best regards,
Kasturi Haldar
Editor-in-Chief
PLOS Pathogens
orcid.org/0000-0001-5065-158X
Michael Malim
Editor-in-Chief
PLOS Pathogens
Articles from PLOS Pathogens are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.ppat.1010875
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1010875&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/136743966
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.ppat.1010875
Article citations
Adaptive truncation of the S gene in IBV during chicken embryo passaging plays a crucial role in its attenuation.
PLoS Pathog, 20(7):e1012415, 30 Jul 2024
Cited by: 0 articles | PMID: 39078847 | PMCID: PMC11315334
Seasonal antigenic prediction of influenza A H3N2 using machine learning.
Nat Commun, 15(1):3833, 07 May 2024
Cited by: 3 articles | PMID: 38714654 | PMCID: PMC11076571
Epistasis mediates the evolution of the receptor binding mode in recent human H3N2 hemagglutinin.
Nat Commun, 15(1):5175, 18 Jun 2024
Cited by: 1 article | PMID: 38890325 | PMCID: PMC11189414
Predictive evolutionary modelling for influenza virus by site-based dynamics of mutations.
Nat Commun, 15(1):2546, 21 Mar 2024
Cited by: 1 article | PMID: 38514647 | PMCID: PMC10958014
Zoonosis and zooanthroponosis of emerging respiratory viruses.
Front Cell Infect Microbiol, 13:1232772, 05 Jan 2024
Cited by: 1 article | PMID: 38249300 | PMCID: PMC10796657
Review Free full text in Europe PMC
Go to all (6) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GISAID - Global Initiative on Sharing All Influenza Data (3)
- (1 citation) GISAID - EPI1440496
- (1 citation) GISAID - EPI1526498
- (1 citation) GISAID - EPI1584616
Protein structures in PDBe
-
(3 citations)
PDBe - 6BKTView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Preventing an Antigenically Disruptive Mutation in Egg-Based H3N2 Seasonal Influenza Vaccines by Mutational Incompatibility.
Cell Host Microbe, 25(6):836-844.e5, 28 May 2019
Cited by: 29 articles | PMID: 31151913 | PMCID: PMC6579542
Comparison of A(H3N2) Neutralizing Antibody Responses Elicited by 2018-2019 Season Quadrivalent Influenza Vaccines Derived from Eggs, Cells, and Recombinant Hemagglutinin.
Clin Infect Dis, 73(11):e4312-e4320, 01 Dec 2021
Cited by: 12 articles | PMID: 32898271
In-depth phylogenetic analysis of the hemagglutinin gene of influenza A(H3N2) viruses circulating during the 2016-2017 season revealed egg-adaptive mutations of vaccine strains.
Expert Rev Vaccines, 19(1):115-122, 19 Jan 2020
Cited by: 9 articles | PMID: 31875483
Nucleoside-Modified mRNA-Based Influenza Vaccines Circumvent Problems Associated with H3N2 Vaccine Strain Egg Adaptation.
J Virol, 97(1):e0172322, 19 Dec 2022
Cited by: 6 articles | PMID: 36533954 | PMCID: PMC9888232
Funding
Funders who supported this work.
Calmette and Yersin scholarship from the Pasteur International Network Association
Guangdong-Hong Kong-Macau Joint Laboratory of Respiratory Infectious Disease (1)
Grant ID: 20191205
Lee Kong Chian School of Medicine, Nanyang Technological University
Pasteur Foundation Asia
The Health and Medical Research Fund (1)
Grant ID: No.19180932
University of Illinois at Urbana-Champaign
the National Research Foundation of Korea (1)
Grant ID: NRF-2018M3A9H4055203

 4
,
5
,* and
4
,
5
,* and 


