Abstract
Free full text

Efficient control of the fungal pathogens Colletotrichum gloeosporioides and Penicillium digitatum infecting citrus fruits by native soilborne Bacillus velezensis strains
Abstract
Destruction of citrus fruits by fungal pathogens during preharvest and postharvest stages can result in severe losses for the citrus industry. Antagonistic microorganisms used as biological agents to control citrus pathogens are considered alternatives to synthetic fungicides. In this study, we aimed to identify fungal pathogens causing dominant diseases on citrus fruits in a specialized citrus cultivation region of Vietnam and inspect soilborne Bacillus isolates with antifungal activity against these pathogens. Two fungal pathogens were characterized as Colletotrichum gloeosporioides and Penicillium digitatum based on morphological characteristics and ribosomal DNA internal transcribed spacer sequence analyses. Reinfection assays of orange fruits confirmed that C. gloeosporioides causes stem-end rot, and P. digitatum triggers green mold disease. By the heterologous expression of the green fluorescent protein (GFP) in C. gloeosporioides using Agrobacterium tumefaciens-mediated transformation, we could observe the fungal infection process of the citrus fruit stem-end rot caused by C. gloeosporioides for the first time. Furthermore, we isolated and selected two soilborne Bacillus strains with strong antagonistic activity for preventing the decay of citrus fruits by these pathogens. Molecular analyses of 16 S rRNA and gyrB genes showed that both isolates belong to B. velezensis. Antifungal activity assays indicated that bacterial culture suspensions could strongly inhibit C. gloeosporioides and P. digitatum, and shield orange fruits from the invasion of the pathogens. Our work provides a highly effective Bacillus-based preservative solution for combating the fungal pathogens C. gloeosporioides and P. digitatum to protect citrus fruits at the postharvest stages.
1. Introduction
Citrus species, including oranges, lemons, grapefruits, pomelos, and limes, are the economically important and preferred fruit crops worldwide. The Food and Agriculture Organization of the United Nations (FAO) reported that citrus crops are commercially cultivated in more than 140 countries, with a global citrus production of 143,755,600 tons in 2019 [1]. However, the citrus production industry may be severely affected by the destruction caused by postharvest pathogens [2]. One of the most common Colletotrichum species infecting tropical and temperate fruit crops is reported as C. gloeosporioides [[3], [4], [5]]. This fungus is a hemibiotrophic pathogen of anthracnose disease in many plant hosts. C. gloeosporioides is the major causative agent of leaf blight, leaf spot, and stem-end rot, followed by premature fruit drop and postharvest anthracnose in citrus [4,6,7]. Morphological characteristics of C. gloeosporioides are highly similar to those of other Colletotrichum species. These features are not reliable enough for fungal identification at the species level because they can be changed under different environmental conditions. Therefore, molecular analysis based on the sequencing of some target genes is usually required to accurately identify Colletotrichum species [3,8]. Another severe disease in citrus at postharvest stages is green mold decay caused by Penicillium digitatum. This necrotrophic fungus infects citrus fruits through injuries during handling, transportation, storage, and marketing [2]. P. digitatum is the most destructive postharvest pathogen on citrus fruits and accounts for approximately 90% of total postharvest losses [9,10].
Synthetic fungicides such as imazalil, thiabendazole, prochloraz, mancozeb, fludioxonil, pyrimethanil, etc., are mainly used to control postharvest diseases, including stem-end rot and green mold in citrus. Applying these fungicides for fruit preservation raises public concerns over their harmful risks to human health and the environment. Additionally, overuse of these chemical agents causes fungicide resistance in numerous fungal strains, especially in the green mold fungus P. digitatum. Therefore, biological control of postharvest diseases exhibits a safer and eco-friendly approach. Several strategies based on microbial antagonists and plant-derived compounds have been developed for controlling postharvest pathogens. Among them, antagonistic microbes appear to be a promising source for developing preservative applications in citrus production at the postharvest stages [6,[11], [12], [13], [14]].
To date, several non-pathogenic Bacillus species with strong antifungal activity, including B. subtilis, B. amyloliquefaciens, and B. velezensis have been exploited as biocontrol agents. These Gram-positive, aerobic, endospore-forming bacteria can inhibit a broad spectrum of fungal plant pathogens [13,15,16]. However, the inhibitory effects of the Bacillus species on the pathogenicity of both citrus pathogens, C. gloeosporioides and P. digitatum, still need to be further investigated for potential preservative applications in the future. In the present study, we identified fungal postharvest pathogens causing citrus stem-end rot and green mold in a specialized citrus cultivation region in Northern Vietnam as C. gloeosporioides and P. digitatum, respectively. Furthermore, two native soilborne B. velezensis isolates with strong antifungal activity against both C. gloeosporioides and P. digitatum were selected as potential bioagents for preserving citrus fruits against postharvest diseases.
2. Materials and methods
2.1. Sample collection
Orange fruits with stem-end rot or green mold disease symptoms and soil samples were collected from a specialized citrus cultivation region of Ha Giang province in Northern Vietnam. Samples were stored in sterile bags and delivered immediately to the laboratory for microbial isolation.
2.2. Isolation of fungal pathogens infecting citrus fruits
Fungal materials were collected from the infected orange fruits of stem-end rot or green mold by sterile toothpicks, diluted in sterile distilled water (SDW), and streaked on potato dextrose agar (PDA) medium. PDA was supplemented with chloramphenicol (100 μg/mL) to prevent bacterial contamination. The plates were incubated at 25 °C for 2−3 days. Fungal isolates were purified by single spore isolation and identified by morphological analyses under a microscope (Krüss, Hamburg, Germany).
To confirm the citrus pathogenicity of fungal strains, orange fruits were cleaned with SDW and 70% ethanol. For the P. digitatum infection, artificial wounds (10 mm long and 2 mm deep) were made on the surface of the oranges using a sterile tool, while smaller wounds (5 mm long and 1 mm deep) were generated on the fruit pedicel for the C. gloeosporioides infection. A volume of 10 μL of fungal spore suspension (106 spores/mL) was inoculated to the respective wounded position on the fruit. SDW was used for the uninfected control fruits (mock) instead of the spore suspensions. The orange fruits were incubated in sterile plastic boxes at 25 °C for 3−10 days.
2.3. Isolation of native soilborne Bacillus strains with antifungal activity
Soil samples were resuspended and successively diluted in SDW before being heated at 100 °C for 10 min. The bacterial suspensions were then spread on Luria Bertani (LB) agar medium, and the plates were incubated at 30 °C for 1 day. Single colonies appearing on the plates were purified for morphological analysis under microscopy. The antifungal activity of the Bacillus isolates against the stem-end rot and green mold pathogens was examined using dual-culture assays.
To examine the antifungal activity of liquid cultures, the Bacillus strains were cultivated in Erlenmeyer flasks containing the LB medium at 30 °C for 1 day. The cultures were centrifuged at 12000 rpm for 10 min at 4 °C, and the supernatants were collected for antifungal activity assays by the agar well diffusion method. A volume of 50 μL of fungal spore suspension (106 spores/mL) was spread on a PDA plate using sterile glass beads. The plate was kept in a sterile cabinet at room temperature for 30 min to dry the agar surface to avoid the spreading growth of residual Bacillus cells in the supernatants after centrifugation. Next, one well was made in the center of the agar plate, and 30 μL of a bacterial supernatant was added to the well. SDW and the LB medium were used for the control plates instead of bacterial culture supernatants. Plates were incubated at 4 °C for 4 h to promote the diffusion of antifungal substances into the agar medium and then at 25 °C for 5 days to facilitate the fungal growth. The diameters of the inhibition zones were measured to calculate the fungal inhibition as previously described [17]. The experiments were performed in triplicate.
2.4. Molecular identification of microbial strains
Fungal strains were cultivated in potato dextrose broth (PDB) medium at 25−28 °C for 3 days using a shaking incubator. Meanwhile, bacterial isolates were grown in the LB liquid medium at 28−30 °C for 1 day. Total DNA samples were prepared from bacterial biomass or fungal mycelial biomass using our in-house chemical protocols or G-spin™ Total DNA Extraction Mini Kit (iNtRON Biotechnology, Gyeonggi-do, South Korea). DNA samples were directly used for PCR amplifications or stored at −20 °C for later use.
PCR amplification of the fungal internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) was performed with the universal primer pair ITS1 (TCCGTAGGTGAACCTGCGG)/ITS4 (TCCTCCGCTTATTGATATGC). Bacterial sequences of 16 S rRNA and gyrB were amplified with the universal primer pairs fD1 (AGAGTTTGATCCTGGCTCAG)/rP1 (ACGGTTACCTTGTTACGACTT) and UP-1 (GAAGTCATCATGACGTTCTGCAYGCNGGNGGNAARTTYGA)/UP-2r (AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRTCNGTCAT), respectively [[18], [19], [20]]. Phusion™ high-fidelity DNA polymerase (Thermo Scientific, Massachusetts, USA) was used for the PCR amplifications. PCR thermal cycle includes 94 °C for 3 min; 35 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 60 s; 72 °C for 3 min. PCR products were purified with MEGAquick-spin™ Plus Total Fragment DNA Purification Kit (iNtRON Biotechnology, Gyeonggi-do, South Korea) for sequencing by the 1st BASE company (Singapore). DNA sequences were analyzed with BioEdit 7.2 and compared to the GenBank database using the BLAST tool. Phylogenetic analyses were performed with MEGA X using the statistical neighbor-joining method and bootstrap values of 1000 replicates.
2.5. Agrobacterium tumefaciens-mediated transformation of the fungal pathogens C. gloeosporioides and P. digitatum
C. gloeosporioides HGC201 and P. digitatum HGP01 were cultivated on PDA plates at 25 °C for 5−7 days to prepare spore suspensions. Fungal spore suspensions as starting materials for genetic transformation were collected from the plates with SDW and adjusted to a concentration of 106 spores/mL. The binary vector pGreen2 [21] was employed for the GFP tagging of both fungal pathogens. The steps of Agrobacterium tumefaciens-mediated transformation (ATMT) were implemented as previously reported [18]. Some parameters were adjusted for the genetic transformation of C. gloeosporioides. Mainly, the co-culture process was performed at 22 °C for 60 h, and the hygromycin concentration of 300 μg/mL was used for selecting transformants.
Fungal colonies appearing on the filter membranes were transferred to the PDA plates containing the hygromycin selection agent (200−300 μg/mL) to confirm the resistance. Hygromycin-resistant transformants were purified by single spore isolation and cultivated in the PDB medium for total DNA extraction. GoTaq® Green Master Mix (Promega, Wisconsin, USA) was used for PCR amplification to confirm fungal transformants. Primer pairs, including HPH-F/HPH-R and GFP-F/GFP-R [18], were recruited for the specific amplification of the hygromycin resistance cassette or the GFP reporter gene from genomes of fungal transformants, respectively. The GFP expression in fungal transformants was examined under the Axioplan fluorescence microscope (Carl Zeiss, Oberkochen, Germany) using the slide culture method [18].
To follow the host colonization process, spores of the GFP-tagged strains were inoculated to wounded sites on orange fruits. The infected fruits were incubated at 25 °C for 3−5 days. After the incubation, thin orange tissue slices of the infected areas were prepared with a sharp blade. Samples were then observed under fluorescence microscopy.
2.6. Evaluating effects of temperature and filtration on antifungal activity of bacterial cultures
Bacterial culture supernatants obtained from centrifugation were incubated at different temperatures, including 30 °C, 40 °C, 60 °C, 80 °C, and 100 °C, for 30 min using a block heater (Corning, Arizona, USA). The heat-treated supernatants were used for antifungal activity assays against C. gloeosporioides and P. digitatum. These supernatants were further filtered through sterile syringe filters with a pore size of 0.22 μm or 0.45 μm to remove bacterial cells completely. Cell-free filtrates were evaluated for antifungal activity by the agar well diffusion method or the microdilution method [17]. For microdilution assays, 1% or 5% of each filtrate was supplemented to PDA before pouring this medium into Petri plates. Spore suspensions were prepared at 104−106 spores/mL by serial dilutions, and 10 μl of each diluted spore suspension was inoculated onto the culture filtrate-containing PDA plates. Filtrate-free PDA plates were used for the controls. Plates were incubated at 25−28 °C for 5 days.
2.7. In vivo assays for antifungal activity against the pathogens on citrus fruits
Two orange varieties were used in the study, including Citrus sinensis and Citrus reticulata × sinensis (a citrus hybrid originating in Vietnam with the name “Cam Sanh”), which were also grown in Ha Giang province, Northern Vietnam. Orange fruits were cleaned with SDW and subsequently with 70% ethanol. The fruits were soaked into the bacterial cultures for 30 min. Afterward, the fruits were air-dried at room temperature before being infected with either C. gloeosporioides or P. digitatum. For uninfected control fruits (mock), SDW was used instead of fungal spore suspensions. Ten orange fruits were used for each treatment, and the experiment was performed in triplicate. The GFP-tagged strains of C. gloeosporioides and P. digitatum were also recruited to verify this assay. The fungal invasion was examined under fluorescence microscopy with the GFP signal. The fungal strains were re-isolated by placing thin slices of infected orange tissue on PDA supplemented with chloramphenicol (100 μg/mL) to inhibit bacterial growth. The inoculated fruits were kept separately in sterile plastic boxes at 25 °C for 3−10 days. Symptomatic progression of fruit decay was initially observed after 3 days.
2.8. Statistical analysis
Experiments were performed in triplicate, and data were expressed as mean ± standard deviation (SD). Statistical analyses were conducted with GraphPad Prism 8 using one-way ANOVA. A statistically significant difference was considered when p < 0.05.
3. Results and discussion
3.1. The virulent strains of C. gloeosporioides and P. digitatum isolated in Northern Vietnam are the main causative agents of stem-end rot and green mold diseases on orange fruits
C. gloeosporioides is the causative agent of anthracnose on a wide range of plant hosts, including mango, papaya, apple, pomegranate, citrus, etc. This pathogenic fungus produces a lot of asexual spores (conidia) for dispersal in nature by rain and wind. The anthracnose disease is symptomatically characterized by dark spots or lesions on different parts of plants, including leaves, shoots, twigs tip, petals, and fruits [3,7,22]. Regarding citrus crops, C. gloeosporioides were reported to cause anthracnose on citrus hosts in Italy, Spain, Portugal, Greece, Malta, Australia, New Zealand, USA, Tunisia, and Mexico [14,[22], [23], [24], [25]]. The moldy fungus was also revealed as the infectious agent of citrus post-bloom fruit drop in Brazil [4]. In this study, we collected orange fruits with the symptoms of stem-end rot and green mold diseases directly at a specialized orange crop cultivation region in Ha Giang, a mountainous province in Vietnam. Fungal materials separated from the infected fruit tissues were diluted with SDW and spread on the PDA plates supplemented with chloramphenicol to obtain single fungal colonies. Dominant fungal colonies were selected for further analyses. From orange fruits with decay symptoms around the fruit pedicel, we isolated two fungal strains designated as HGC201 and HGC286. The morphological characteristics of these strains are similar to those of Colletotrichum species [8]. They possess light grey mycelia after 7 days of cultivation on PDA at 25 °C. Under microscopy, fungal mycelia are branched with single cylindrical conidia at hyphal tips (Fig. 1A). Although the ITS region of rDNA is unreliable for resolving relationships within Colletotrichum genus, it allows the separation of C. gloeosporioides from all other species [8]. Our molecular analyses based on the rDNA ITS sequences classified HGC201 and HGC286 as C. gloeosporioides (Fig. 1B). Reinfection assays confirmed that HGC201 and HGC286 cause the black rot symptom around the pedicel of the infected orange fruits (also called stem-end rot) within 10 days at 25 °C (Fig. 1C).
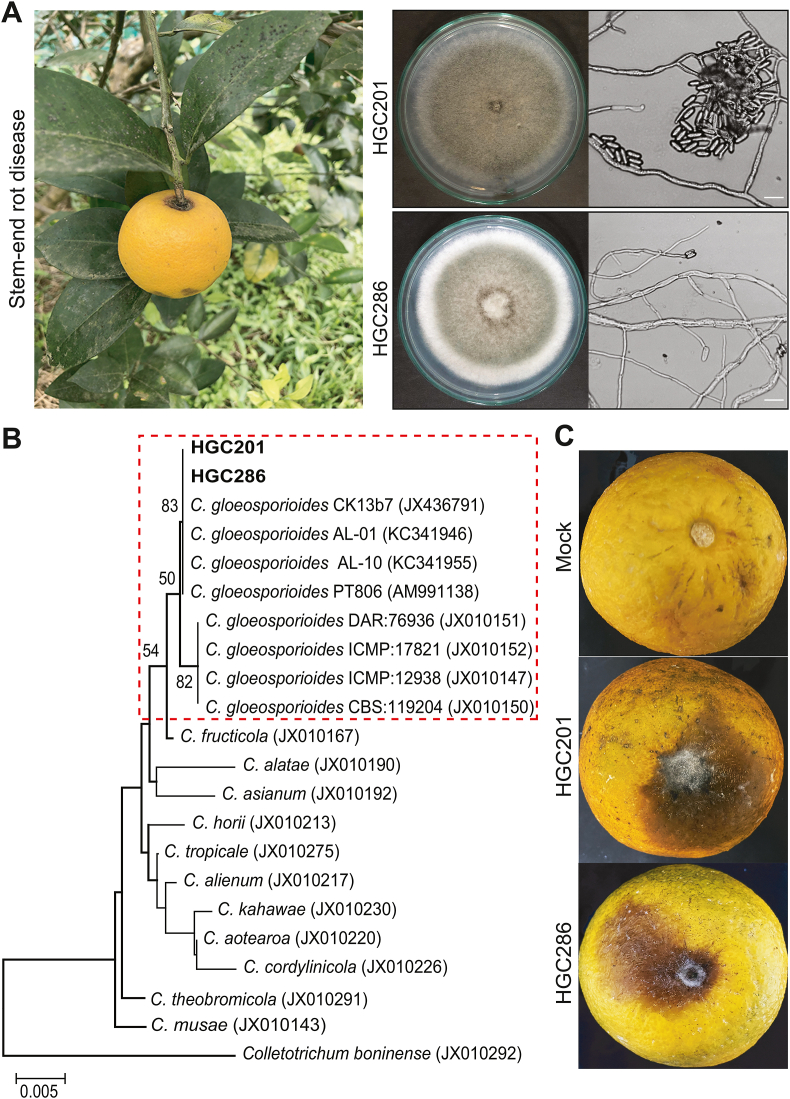
Identification of the stem-end rot pathogen in citrus fruits. (A) Two fungal strains (HGC201, HGC286) resembling Colletotrichum fungi were isolated from stem-end rot orange fruits. Fungal strains were cultivated on PDA at 25 °C for 5 days for morphological analyses. (B) Molecular identification of strains HGC201 and HGC286 by ITS rDNA sequencing. The ITS sequences were compared to GenBank using the BLAST tool. The phylogenetic tree was constructed with MEGA X using the neighbor-joining method and 1000 bootstrap replications. Statistical support values at branch nodes of the tree, genetic distance scale, and accession numbers for the ITS sequences from other fungi are indicated. The ITS sequences for two C. gloeosporioides strains (HGC201 and HGC286) are deposited in GenBank with accession numbers OL739236−OL739237. (C) Reinfection of the isolated strains on citrus fruits to confirm their pathogenicity. Fungal spore suspensions (106 spores/mL) were employed for the fruit infection assays. Negative control fruits (mock) were inoculated with SDW. The fruits were incubated in sterile plastic boxes for 10 days. Scale bars indicate the sizes of the images. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, from decayed oranges with green mold symptoms, three other fungal strains (HGP01, HGP82, HGP116) were isolated. Colonies of these strains are turned from white into olive green after 3 days of growth on PDA at 25 °C. Microscopic examinations revealed that fungal mycelia are irregularly branched, and conidiophores possess terverticillate structures with chains of oval conidia at tips (Fig. S1A). These morphological features resemble P. digitatum PdVN1, which was previously reported [18]. Analyses of the ITS sequences using the universal primer pair ITS1/ITS4 indicated that all three strains, HGP01, HGP82, and HGP116, belong to P. digitatum (Fig. S1B). Pathogenicity assays confirmed that these strains trigger the green mold disease on orange fruits within 7 days at 25 °C (Fig. S1C).
Our reinfection results indicated that the strains of each fungal pathogen are quite similar in their virulence (Fig. 1 & S1). Therefore, we selected just one representative strain of each fungus, including C. gloeosporioides HGC201 and P. digitatum HGP01, for further investigation.
3.2. GFP tagging approach allows following the orange fruit colonization of the stem-end rot pathogen C. gloeosporioides and the green mold pathogen P. digitatum
To inspect the invasive progress of citrus fruits by C. gloeosporioides and P. digitatum, the strain HGC201 of C. gloeosporioides and HGP01 of P. digitatum were employed for the GFP tagging using the ATMT method. Fungal transformants were selected on PDA supplemented with hygromycin. The results showed that the transformants could grow well under the selection pressure of the high hygromycin concentrations (200−300 μg/mL). Molecular investigations by PCR analysis using the specific primer pairs indicated that both the cassettes for the hygromycin resistance (HPH) and the GFP expression were present in the genomes of these transformants. Furthermore, the strong expression of the GFP reporter gene under the control of the Aspergillus nidulans gpdA promoter could be easily detected in the transformants generated from C. gloeosporioides and P. digitatum (Fig. 2A and S2). Fungal colonization of citrus fruits via the GFP expression has been recently reported for P. digitatum [18,26]. In this study, the GFP-tagged C. gloeosporioides (strain C4) with the wild-type like phenotype was employed for visualizing the fungal colonization process and decay of the central cores in the infected orange fruits. Compared to the negative control (mock), the severe disease symptom of stem-end rot was detected by direct observation inside and outside the infected fruits after 10 days of spore inoculation. Under fluorescence microscopy, the invasion of this fungal pathogen could be seen at the earlier stage of 5 days, in which the GFP-tagged C. gloeosporioides infected the host by mycelial colonization through the central core of orange fruit (Fig. 2B). This is the first report describing the disease progression of citrus stem-end rot caused by the fungal pathogen C. gloeosporioides.
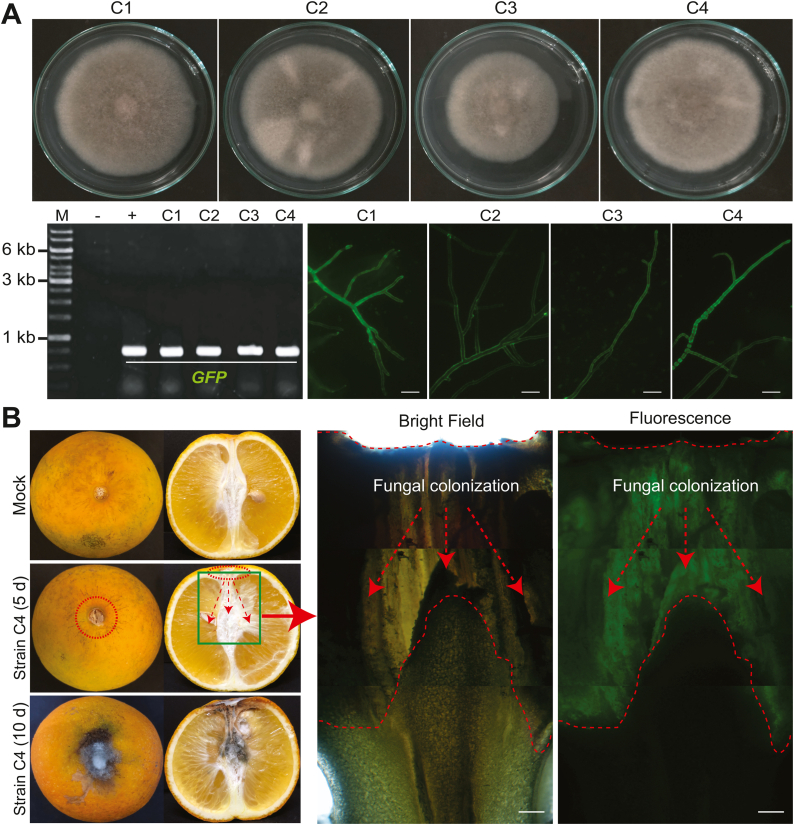
GFP tagging of the fungal pathogens by ATMT for inspecting citrus fruit colonization. (A) The transformants generated from C. gloeosporioides HGC201 were confirmed for the presence of the GFP gene in their genomes by PCR and examined for the GFP expression under fluorescence microscopy. (B) Orange fruits were infected with spores of the transformants of C. gloeosporioides for 5−10 days at 25 °C. Thin slices of the infected fruits were prepared to detect the host colonization by the fungus via the GFP signal. Scale bars indicate the sizes of the images. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. The soilborne B. velezensis isolates possess strong antifungal activity against both fungal pathogens infecting citrus fruits
To prevent the attack of postharvest pathogens, synthetic fungicides are often used to preserve citrus fruits. However, the continuous use of fungicides may pose potential risks to the environment and human health. Additionally, it also promotes fungicide resistance in fungal pathogens. Therefore, biological control is a promising approach to replacing synthetic fungicides in fruit preservation [10,13]; Wang et al., 2020). From native soil samples, we detected three bacterial strains possessing antifungal activity against both the citrus pathogens C. gloeosporioides and P. digitatum. These bacterial isolates were designated as B14, B15, and B16. The results from the assays using the dual-culture method indicated that B14 and B15 exhibited vigorous antifungal activity against the C. gloeosporioides strains, while B16 displayed weak antifungal activity toward this pathogen. However, these bacterial isolates presented robust activity against all tested P. digitatum strains (Fig. 3A). Inspections of culture supernatants using the agar well diffusion method confirmed the strong activity of B14 and B15 against C. gloeosporioides and P. digitatum with inhibition zones in diameters of 21–22 mm and 50−56 mm, respectively (Fig. 3B).
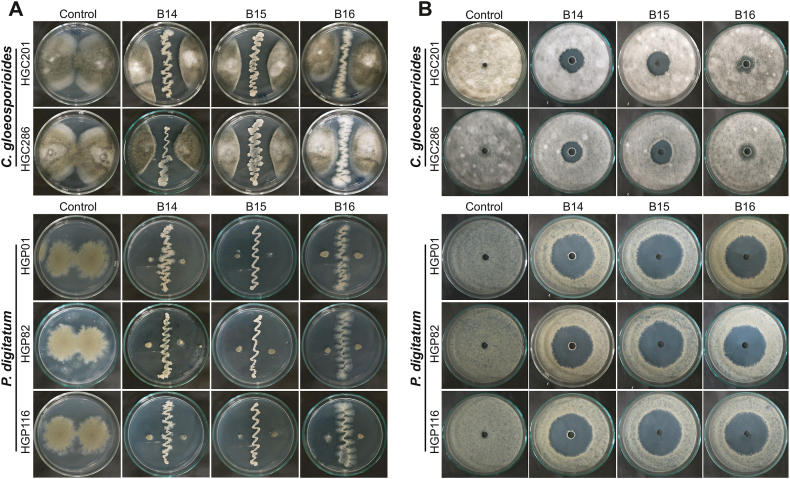
Antifungal activity of the Bacillus isolates. The dual-culture and agar well diffusion methods were used to inspect the antifungal activity of Bacillus isolates against two stem-end rot C. gloeosporioides strains (A) and three green mold P. digitatum strains (B). For the dual-culture method, three Bacillus strains (B14, B15, B16) were cultivated on the PDA plates in the presence of the fungal pathogens. Control plates were inoculated with only fungal strains. For the agar well diffusion method, the Bacillus strains were grown in conical flasks containing the LB medium at 30 °C for 1 day. Culture supernatants collected by centrifugation were added to agar wells on the PDA plates fully covered with fungal spores. For control plates, SDW was used instead of the culture supernatants. The plates were incubated at 25−28 °C for 5 days. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Microscopic examinations indicated that B14 and B15 are Gram-positive bacteria with rod-shaped cell morphology (data not shown). When cultured on the LB agar, colonies of these antagonistic bacteria possess a milky white phenotype with a wrinkled surface (B14) or jagged edges (B15) (Fig. S3A). Molecular analyses showed that the 16 S rRNA sequences from B14 and B15 are almost identical (99.6−100%) to those of B. velezensis, and very closely related to those of B. amyloliquefaciens and B. subtilis deposited in GenBank (Fig. S3B). To ensure the accurate taxonomy of these strains, the gyrB gene encoding DNA gyrase subunit B was recruited as the second signature for the identification. Based on the gyrB sequences, the data confirmed that B14 and B15 belong to the B. velezensis species (Fig. S3C). By DNA sequencing of some target genes, several antagonistic Bacillus strains used as a vital component of commercial bioproducts for plant protection have been recently re-identified as B. velezensis instead of B. subtilis or B. amyloliquefaciens [27]. Therefore, a combination of morphological and DNA analyses is required for discriminating B. velezensis from other closely related Bacillus species.
A prominent role of Bacillus species in biological control against phytopathogens has been recently documented [15,28,29]. Many isolates of B. velezensis possess antimicrobial activity against pathogenic bacteria, fungi, and nematodes [29,30]. The B. velezensis TS3B-45 strain was shown to have inhibitory activity in vitro against the anthracnose pathogen C. gloeosporioides, similar to the synthetic fungicide prochloraz [31]. The volatile organic compounds produced by B. velezensis strains exhibited antifungal activity against postharvest fungal pathogens, including P. digitatum [32]. Two strains of B. velezensis isolated in this study could suppress the citrus pathogens C. gloeosporioides and P. digitatum. These antagonistic isolates may also inhibit the growth of other pathogenic fungi. Further analyses of the strains B. velezensis B14 and B15 still need to be investigated.
3.4. Antifungal activity of the bacterial liquid cultures was decreased by heat treatment or sterile filtration
B. velezensis has emerged as a vital member of plant microbiomes for agricultural applications. Numerous studies have shown that B. velezensis and other closely related Bacillus species possess gene clusters involved in the biosynthesis of secondary metabolites, which play significant roles in suppressing microbial pathogens. These bioactive metabolites include surfactin, iturin A, bacillomycin D, fengycin, bacillibactin, macrolactin, bacillaene, and difficidin [15,[29], [30], [31],33,34]. Furthermore, volatile organic compounds from B. velezensis strains were reported to inhibit the growth of P. digitatum by up to 28% [32]. Antifungal compounds from Bacillus species are usually stable after treating with abiotic stresses [35,36]. To assess the thermostability of bacterial liquid cultures for antifungal activity, B14 and B15 were grown in LB, and culture supernatants were collected by centrifugation. After treatment at different temperatures, the supernatants were tested for antifungal activity. Our results showed that heat-treated supernatants maintained significant ability against C. gloeosporioides and P. digitatum. However, antifungal activity against P. digitatum of the supernatants treated at 100 °C was strongly decreased for B14, and even lost for B15 (Fig. S4).
Removing bacterial cells from a liquid culture by sterile filtration to obtain a cell-free filtrate is essential for conducting antifungal assays [37]. Strains B14 and B15 were cultivated in the liquid medium, and the cultures were filtered through sterile syringe filters with a pore size of 0.22 μm or 0.45 μm. The cell-free culture filtrates were examined for antifungal activity against the pathogens by the agar well diffusion method and the microdilution method. The results showed that the antifungal activity of the filtrates was significantly reduced for P. digitatum and even lost entirely for C. gloeosporioides (Fig. S5). Our data suggest that potential antifungal molecules produced by B. velezensis B14 and B15 might be somehow retained on the membranes of the syringe filters during sterile filtration. Therefore, it is worthwhile to consider the direct use of liquid cultures of the B. velezensis strains to maximize antagonistic activity against the citrus fungal pathogens.
3.5. Bacterial cultures as preservatives to protect orange fruits from the attacks of the pathogens
Like other closely related Bacillus species, including B. subtilis and B. amyloliquefaciens, B. velezensis is also a beneficial and safe microorganism for humans and the environment [30,33]. When mixed with cultivation media, the culture filtrate of B. velezensis CE 100 effectively suppressed C. gloeosporioides. A cyclic tetrapeptide from B. velezensis CE 100 was indicated to inhibit the growth of this pathogen in vitro even more effectively than benzoic acid [38]. In this work, we examined whether bacterial cultures from the B. velezensis strains (B14 and B15) could serve as preservatives to protect citrus fruits from stem-end rot and green mold pathogens. The wounded oranges were soaked in bacterial liquid cultures or SDW before being infected with either C. gloeosporioides HGC201 or P. digitatum HGP01. The uninfected control experiments (mock) were also conducted similarly but without fungal inoculation. The results showed that the cultures prepared with B14 or B15 suppressed symptomatic disease development on the treated fruits that resembled the uninfected control fruits. In contrast, the fruits soaked in SDW and infected with the fungi displayed severe symptoms of stem-end rot or green mold (Fig. 4A and B).
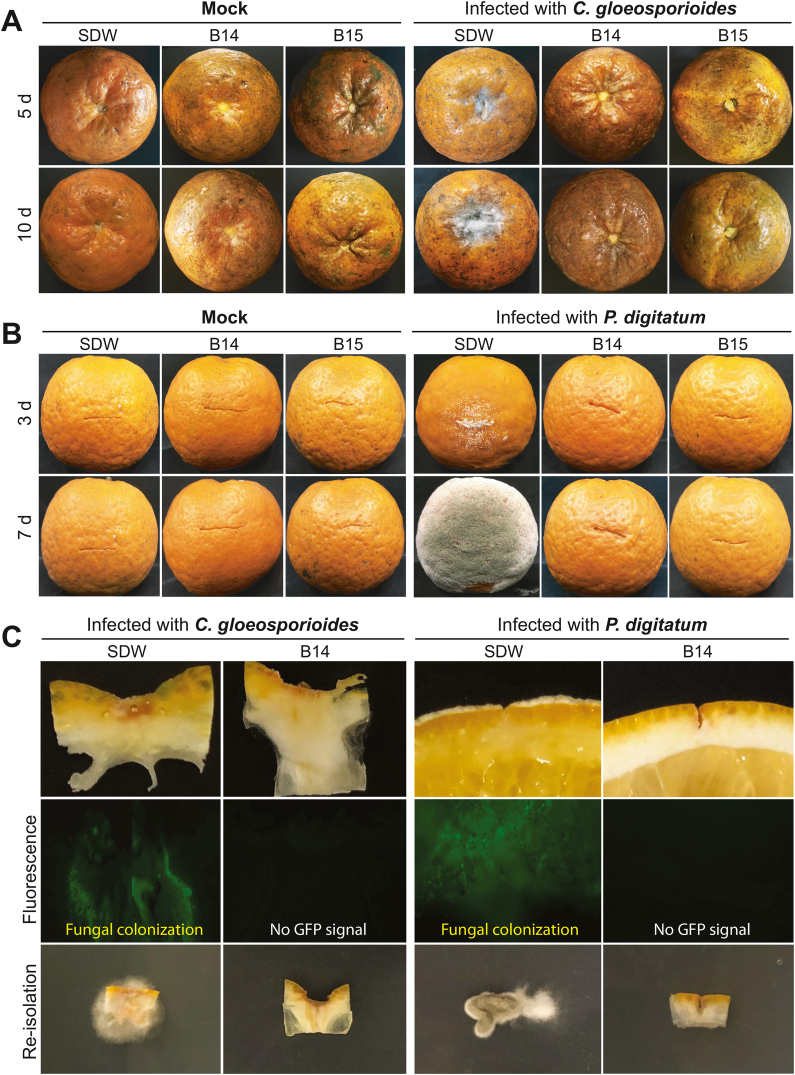
In vivo assays of the antagonistic B. velezensis strains in protecting citrus fruits. Oranges were soaked in bacterial cultures for 30 min and air-dried at room temperature. The fruits were wounded and infected with fungal spores from (A) C. gloeosporioides HGC201 or (B) P. digitatum HGP01. The infected oranges were incubated in sterile boxes at 25 °C for 3−10 days. (C) The antifungal effects of bacterial cultures on citrus fruit infection were evaluated using the GFP-tagged strain of C. gloeosporioides HGC201 or C. gloeosporioides HGC201. The fungal strains were re-isolated from the infected fruit tissues using PDA supplemented with chloramphenicol (100 μg/mL) to eliminate bacterial cells and facilitate fungal growth. The plates were incubated at 25 °C for 5 days.
Further inspections using the GFP-tagged strains of C. gloeosporioides HGC201 or P. digitatum HGP01 also resulted in supportive pieces of evidence. In this assay, the liquid culture of B14 also effectively protected the wounded orange fruits from the invasion of the GFP-tagged strains. Under fluorescence microscopy, no green signal of the GFP reporter protein could be detected in the fruits treated with the B14 culture suspension before fungal infection. Inversely, in the control fruits soaked in SDW, the systemic colonization of fungal mycelia inside the orange tissues could be observed with clear GFP signals. The GFP-tagged strains were easily re-isolated from the infected tissues (Fig. 4C).
We also performed additional antifungal tests to compare the antifungal ability of the B. velezensis strains with the common synthetic fungicide mancozeb. As the commercial name AN-K-ZEB 80WP, this synthetic fungicide contains 80% of the antifungal agent mancozeb and is widely distributed in Vietnam for controlling fungal plant pathogens. Our results revealed that mancozeb strongly inhibited the growth of both fungal pathogens on the agar medium at a low concentration of 0.001%. However, it was not as effective in protecting citrus fruits from fungal infection as the antagonistic B. velezensis strains, even at the recommended fungicide concentration of 0.25% (Fig. S6). Recently, the metabolite bacillomycin D from B. velezensis HN-2 has been demonstrated to possess strong activity against C. gloeosporioides compared to the commonly used fungicide mancozeb [39]. The potential of B. velezensis as a probiotic in animal feeds and bioproducts for agriculture has also been reviewed [29,40]. Our data showed that the soilborne B. velezensis strains, B14 and B15, can serve as biological agents for citrus fruit preservation instead of synthetic fungicides. However, additional analyses still need to be conducted to identify antifungal metabolites, the safety, and other biological characteristics of these antagonistic B. velezensis strains.
4. Conclusions
In this work, we reported two predominant fungal pathogens causing stem-end rot and green mold diseases on citrus fruits cultivated in Ha Giang province of Northern Vietnam. Based on morphological and DNA sequencing analyses, these postharvest pathogens were identified as C. gloeosporioides and P. digitatum. Further, we provided experimental evidence for the citrus fruit infection process by C. gloeosporioides using a GFP-tagged strain to observe fungal colonization directly under fluorescence microscopy. To search for potential bioagents to control these fungal pathogens, two B. velezensis strains with strong antifungal activity were isolated from the native soil samples. These bacterial isolates efficiently antagonized the citrus causal agents C. gloeosporioides and P. digitatum in vitro and in vivo. We also indicated that the direct application of bacterial liquid cultures was more effective in preventing the pathogens from attacking citrus fruits. The present study revealed that the selected B. velezensis strains are promising candidates for developing biofungicides to prevent postharvest losses in citrus crops due to stem-end rot and green mold diseases. These B. velezensis strains may also be exploited as bioagents to replace synthetic fungicides in controlling other phytopathogenic fungi.
Declarations
Author contribution statement
Tao Xuan Vu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Tram Bao Tran, Minh Binh Tran, Trang Thi Kim Do, Linh Mai Do, Mui Thi Dinh, Hanh-Dung Thai, Duc-Ngoc Pham: Performed the experiments; Analyzed and interpreted the data. Van-Tuan Tran: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
The authors are grateful to the Center for Experimental Biology (National Center for Technological Progress, Ministry of Science and Technology of Vietnam) and the University of Science (Vietnam National University, Hanoi) for the excellent infrastructure support.
Footnotes
Appendix ASupplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13663.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
Articles from Heliyon are provided here courtesy of Elsevier
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/142581435
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.heliyon.2023.e13663
Article citations
Combined transcriptomic and metabolomic analysis of the mechanism by which <i>Bacillus velezensis</i> induces resistance to anthracnose in walnut.
Front Microbiol, 15:1420922, 09 Oct 2024
Cited by: 0 articles | PMID: 39444687 | PMCID: PMC11496756
Biocontrol of citrus fungal pathogens by lipopeptides produced by Bacillus velezensis TZ01.
Front Microbiol, 15:1471305, 04 Sep 2024
Cited by: 0 articles | PMID: 39296284 | PMCID: PMC11408202
Ethanolic extract from fruiting bodies of Cordyceps militaris HL8 exhibits cytotoxic activities against cancer cells, skin pathogenic yeasts, and postharvest pathogen Penicillium digitatum.
Arch Microbiol, 206(3):97, 13 Feb 2024
Cited by: 0 articles | PMID: 38349544
Comparative study of antioxidant and antimicrobial activity of berberine-derived Schiff bases, nitro-berberine and amino-berberine.
Heliyon, 9(12):e22783, 25 Nov 2023
Cited by: 1 article | PMID: 38058428 | PMCID: PMC10696212
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Nucleotide Sequences (2)
- (1 citation) ENA - OL739237
- (1 citation) ENA - OL739236
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A highly efficient Agrobacterium tumefaciens-mediated transformation system for the postharvest pathogen Penicillium digitatum using DsRed and GFP to visualize citrus host colonization.
J Microbiol Methods, 144:134-144, 23 Nov 2017
Cited by: 9 articles | PMID: 29175534
Effectiveness of Trichoderma strains isolated from the rhizosphere of citrus tree to control Alternaria alternata, Colletotrichum gloeosporioides and Penicillium digitatum A21 resistant to pyrimethanil in post-harvest oranges (Citrus sinensis L. (Osbeck)).
J Appl Microbiol, 129(3):712-727, 29 Apr 2020
Cited by: 7 articles | PMID: 32249987
Phenylalanine: A Promising Inducer of Fruit Resistance to Postharvest Pathogens.
Foods, 9(5):E646, 18 May 2020
Cited by: 12 articles | PMID: 32443417 | PMCID: PMC7278716
Penicillium digitatum infection mechanisms in citrus: What do we know so far?
Fungal Biol, 123(8):584-593, 13 May 2019
Cited by: 26 articles | PMID: 31345412
Review




