Abstract
Free full text

Understanding the divergent evolution and epidemiology of H3N8 influenza viruses in dogs and horses
Abstract
Cross-species virus transmission events can lead to dire public health emergencies in the form of epidemics and pandemics. One example in animals is the emergence of the H3N8 equine influenza virus (EIV), first isolated in 1963 in Miami, FL, USA, after emerging among horses in South America. In the early 21st century, the American lineage of EIV diverged into two ‘Florida’ clades that persist today, while an EIV transferred to dogs around 1999 and gave rise to the H3N8 canine influenza virus (CIV), first reported in 2004. Here, we compare CIV in dogs and EIV in horses to reveal their host-specific evolution, to determine the sources and connections between significant outbreaks, and to gain insight into the factors controlling their different evolutionary fates. H3N8 CIV only circulated in North America, was geographically restricted after the first few years, and went extinct in 2016. Of the two EIV Florida clades, clade 1 circulates widely and shows frequent transfers between the USA and South America, Europe and elsewhere, while clade 2 was globally distributed early after it emerged, but since about 2018 has only been detected in Central Asia. Any potential zoonotic threat of these viruses to humans can only be determined with an understanding of its natural history and evolution. Our comparative analysis of these three viral lineages reveals distinct patterns and rates of sequence variation yet with similar overall evolution between clades, suggesting epidemiological intervention strategies for possible eradication of H3N8 EIV.
Introduction
The processes driving virus outbreak epidemiology and cross-species virus transmission are topics of great importance. Studying viruses involved in emerging outbreaks in animals can reveal the fundamental processes that mediate successful transfer between hosts and the subsequent establishment of viral variants as epizootic or even zoonotic and pandemic pathogens. Such studies also suggest approaches to protecting humans and other animals from potential zoonotic viruses by preventing cross-species epizootic outbreaks, as well as new methods for blocking the emergence of transferred viruses or reducing the adaptation of viruses to new hosts, thereby reducing their pandemic potential (Antia et al. 2003; Parrish et al. 2008; Plowright et al. 2017; Wasik et al. 2019a).
Most subtypes of influenza A virus (IAV) are well established in their reservoir hosts, mainly birds living in aquatic environments (Yoon, Webby, and Webster 2014). However, several subtypes appear to have a relatively high propensity for infecting other animal hosts, including domestic poultry, humans, swine, ferrets, seals, horses, and most recently dogs (Yoon, Webby, and Webster 2014; Parrish, Murcia, and Holmes 2015). Some mammals occasionally suffer outbreaks from avian origin influenza, including seals and mink notably during recent spread of highly pathogenic avian influenza (HPAI) H5N1, that may suggest new subtype risks (Agüero et al. 2023; Gamarra-Toledo et al. 2023). Most spillovers cause short-term outbreaks that rapidly die out, while some have transitioned from emergent to epidemic or pandemic pathogens in their mammalian hosts. In human populations, some IAV subtypes (primarily H1N1 and H3N2 in the modern era) have become endemic diseases (Krammer et al. 2018). IAV are members of the family Orthyomyxoviridae, and have a negative-sense RNA genome arranged as eight segments: PB2, PB1, PA, HA, NP, NA, M, and NS (Krammer et al. 2018).
Horses have historically been an important mammalian host for IAV, likely including epidemics in the 1870s in North America that posed a significant burden on animal health, as well as on a human society that was highly dependent on horses (Morens and Taubenberger 2010). In the 20th century, horses were infected by an H7N7 virus that was first recognized in 1956 and became extinct in the late 1970s (Webster 1993). H3N8 equine influenza virus (EIV) was first isolated in 1963 in Florida, likely after being introduced along with infected horses from South America, and descendants of that virus continue to circulate in horses today (Waddell, Teigland, and Sigel 1963; Scholtens et al. 1964; Murcia, Wood, and Holmes 2011; Chambers 2022). The appearance and circulation of reassortant H7N7 and H3N8 EIVs in the 1970s suggests that the H3N8 contributed to displacement and extinction of the H7N7 subtype. Vaccines against H3N8 EIV were first introduced in the late 1960s, and the period of the 1970s and early 1980s resulted in several subclade divergences and increasing H3N8 diversity (Murcia, Wood, and Holmes 2011; Murcia et al. 2013). Around the year 2000, the H3N8 EIV population divided into different lineages: the Florida Clade 1 (FC1) was primarily in North America but also observed in outbreak in other regions of the world, while the Florida Clade 2 (FC2) was found primarily in Europe, yet both virus clades have spread to many different countries and continents (Daly et al. 1996; Oxburgh and Klingeborn 1999; Karamendin et al. 2014; Miño et al. 2019; Diallo et al. 2021; Nemoto et al. 2021). However, these clades appear to have kept some degree of geographic separation, and they do not appear to have been in direct competition (Bryant et al. 2009).
EIV is a common respiratory tract infection in horses and closely related equids, causing influenza-like signs similar to those seen in humans (Dionísio et al. 2021). Current estimations of EIV incidence in US horses comes from voluntary surveillance, and are frequently under 10 per cent with seasonal variability (Pusterla et al. 2022; Chappell et al. 2023). Vaccines for EIV have been available since the 1970s and include antigens from the most commonly circulating strains, and those are widely used in domesticated horses (Oladunni et al. 2021). While human influenza vaccines are updated frequently with the most widespread antigenic variants, EIV vaccines are updated less often. In the USA, roughly 60
per cent with seasonal variability (Pusterla et al. 2022; Chappell et al. 2023). Vaccines for EIV have been available since the 1970s and include antigens from the most commonly circulating strains, and those are widely used in domesticated horses (Oladunni et al. 2021). While human influenza vaccines are updated frequently with the most widespread antigenic variants, EIV vaccines are updated less often. In the USA, roughly 60 per cent of equine operations vaccinate for EIV (66.4
per cent of equine operations vaccinate for EIV (66.4 per cent in 1998, 54.1
per cent in 1998, 54.1 per cent in 2005, and 60.4
per cent in 2005, and 60.4 per cent in 2015) (USDA 2016). It is unclear to what degree low vaccine effectiveness or antigenic mismatch have contributed to recently observed ‘breakthrough’ outbreaks in multiple countries (Gildea et al. 2018; Castro et al. 2019; Olguin-Perglione et al. 2020; Daly et al. 2021; Nemoto et al. 2021). Recommendations for vaccine formulations are regularly proposed by the World Organization for Animal Health (formerly Office International des Epizooties, OIE) expert surveillance panel (OIE 2020; WOAH 2022), but strain inclusion varies in different vaccine formulations (Pavulraj et al. 2021). The potential for horses to be sources of zoonotic disease given their close association with humans makes EIV an identified neglected disease threat of concern (Sack et al. 2019, 2020). Experimental inoculation of human volunteers in the 1960s showed that the virus was able to replicate in humans with sufficient exposure (Kasel et al. 1965; Kasel and Couch 1969). Only through proper surveillance and sequencing of circulating EIV in different populations will a clearer picture of the virus variation be obtained. In particular, this would enable targeted antigenic testing to justify the production of vaccines with a better antigenic match and greater protective efficacy (WOAH 2022; Nemoto et al. 2023). Determining the relationships between viruses causing different outbreaks through surveillance will also reveal epidemiological pathways that arise during epidemics, allowing better strategies for control and potentially eradication.
per cent in 2015) (USDA 2016). It is unclear to what degree low vaccine effectiveness or antigenic mismatch have contributed to recently observed ‘breakthrough’ outbreaks in multiple countries (Gildea et al. 2018; Castro et al. 2019; Olguin-Perglione et al. 2020; Daly et al. 2021; Nemoto et al. 2021). Recommendations for vaccine formulations are regularly proposed by the World Organization for Animal Health (formerly Office International des Epizooties, OIE) expert surveillance panel (OIE 2020; WOAH 2022), but strain inclusion varies in different vaccine formulations (Pavulraj et al. 2021). The potential for horses to be sources of zoonotic disease given their close association with humans makes EIV an identified neglected disease threat of concern (Sack et al. 2019, 2020). Experimental inoculation of human volunteers in the 1960s showed that the virus was able to replicate in humans with sufficient exposure (Kasel et al. 1965; Kasel and Couch 1969). Only through proper surveillance and sequencing of circulating EIV in different populations will a clearer picture of the virus variation be obtained. In particular, this would enable targeted antigenic testing to justify the production of vaccines with a better antigenic match and greater protective efficacy (WOAH 2022; Nemoto et al. 2023). Determining the relationships between viruses causing different outbreaks through surveillance will also reveal epidemiological pathways that arise during epidemics, allowing better strategies for control and potentially eradication.
A single H3N8 EIV transferred to dogs around 1999 to create the first known epidemic of canine influenza virus (CIV). Molecular and epidemiological evidence suggests the epidemic originated from a single, cross-species transmission event of a non-reassortant H3N8 EIV, with cases being initially identified in greyhounds in a training facility in Florida in 2003 and 2004 (Crawford et al. 2005; Payungporn et al. 2008; Anderson et al. 2012; Collins et al. 2014). The H3N8 CIV spread to several regions of the USA around this period, likely due to transport of infected greyhounds, although the virus only achieved prolonged transmission in dogs within larger dense populations, including animal shelters, kennels, and canine day cares (Dalziel et al. 2014). The virus appears to have persisted in large metropolitan areas (e.g. Denver and Colorado Springs, New York City, and Philadelphia) with large shelter dog populations that had high standing populations and high rates of turnover (Hayward et al. 2010). While the emergence of CIV H3N8 led to an epidemic, other observed spillovers of EIV H3N8 into dogs (in 2002 in the UK and 2007 in Australia) did not result in onward canine transmission (Daly et al. 2008; Crispe et al. 2011). No human infections with H3N8 CIV were observed, despite frequent interactions (Krueger et al. 2014). A second CIV arose around 2005 in Asia, and that H3N2 strain appears to have been derived directly from an avian influenza virus that transferred to dogs (Li et al. 2010). H3N2 CIV appears to have circulated widely in Korea and China for a number of years, and has been repeatedly introduced into the USA and Canada where it has mostly caused local and self-limiting outbreaks (Voorhees et al. 2018). Risk assessment of H3N2 CIV transmission to humans was found to be minimal (Martinez-Sobrido et al. 2020).
Host transfers underpin many infectious disease epidemics or pandemics, and in recent years, such events have given rise to new emerging viruses in humans and other animals, including the 2009 pandemic swine influenza (pdmH1N1), Ebolavirus, SARS-CoV-1 and -2, and the H3N8 and H3N2 CIV (Parrish et al. 2008; Morens et al. 2020). Influenza surveillance networks (such as the World Health Organization Global Influenza Surveillance & Response System, WHO GISRS) have made considerable leaps in the past decade, with rapid identification of human cases of HPAI (Hay and McCauley 2018; Ziegler, Mamahit, and Cox 2018). Despite this, gaps remain in our ability to predict changes in host-range that result in virus emergence, or to create early warning systems to detect and mitigate outbreaks before they get underway (Pepin et al. 2010; Plowright et al. 2017; Fagre et al. 2022). Strategies for controlling emerging viruses primarily involve aggressively managing outbreaks after they first occur. Yet, this requires a clear understanding of the routes of transmission, ongoing circulation, and likely host adaptation leading to increased spread or immune evasion, and such knowledge is often lacking in the early stages of outbreaks (Wasik et al. 2019a). It is also often unclear whether and how virus evolution in the original host facilitates the initial successful spillover infection, or how the viruses later evolve and adapt to their new hosts compared to evolution that occurs in their original hosts. Understanding these processes in more detail may allow us to anticipate and forestall the emergence of other viruses in the future.
As well as local outbreaks, a defining feature of the most threatening virus emergence events is longer distance and international transfers that allow viral outbreaks to sustain over time and develop into pandemics, particularly within human host populations (Lemey et al. 2014; Worobey et al. 2020). Long-distance transfers are also a feature of H3N8 EIV epidemiology, likely associated with the movement of infected racing and sport horses (Murcia and Wood 2011; Gildea, Fitzpatrick, and Cullinane 2013; Gildea et al. 2018). It is estimated that just 0.7 per cent of operations transport horses outside North America, yet these international transfers of horses have made EIV a global pathogen of concern (Cullinane and Newton 2013; Equine 2015 2015; WOAH 2022). Among dogs, H3N2 CIV has similarly transferred between continents (Voorhees et al. 2018). In contrast, the H3N8 CIV outbreaks were more confined, circulating within and between animal shelters and kennels within the central and eastern regions in the USA (Hayward et al. 2010; Dalziel et al. 2014). Notably, that virus lineage did not appear to spread outside the USA.
per cent of operations transport horses outside North America, yet these international transfers of horses have made EIV a global pathogen of concern (Cullinane and Newton 2013; Equine 2015 2015; WOAH 2022). Among dogs, H3N2 CIV has similarly transferred between continents (Voorhees et al. 2018). In contrast, the H3N8 CIV outbreaks were more confined, circulating within and between animal shelters and kennels within the central and eastern regions in the USA (Hayward et al. 2010; Dalziel et al. 2014). Notably, that virus lineage did not appear to spread outside the USA.
Modern genomic epidemiology has allowed the detailed examination of viral outbreaks, including the spread and replacement of viral variants (Antia et al. 2003; Hadfield et al. 2018; Grubaugh et al. 2019; Rochman, Wolf, and Koonin 2022). For example, the emergence of more transmissible or antigenic variant viruses that displace earlier lineages is commonly seen in evolving virus populations, including 2009 pdmH1N1, SARS-CoV-2 (B.1.1.7 ‘Alpha’ or B.1.1.529 ‘Omicron’), and the canine parvovirus type-2a variant that emerged and efficiently replaced the earlier strains of that virus (Parrish et al. 1988; Elderfield et al. 2014; su et al. 2015; Harari et al. 2022; Hill et al. 2022; Viana et al. 2022; Carabelli et al. 2023). These events highlight the importance of continued surveillance and analysis of viruses; both in new hosts and in their original hosts, as such comparative analysis allows us to identify mechanisms of true adaptive evolution following emergence.
To resolve the evolution of H3N8 in dogs and horses we collected genomic data from our own sequencing and public repositories of H3N8 beginning around 2001. Complete genome sequences of all segments enable phylogenetic relationships and host-specific evolution to be determined. We further employed use of the much larger collection of HA1 sequences (HA1 being the hemagglutinin ‘head’ containing the receptor binding domain and dominant antigenic epitopes, H3 numbering 1–329). Here, we investigate the emergence the H3N8 CIV and its origin from the H3N8 EIV, describe the later extinction of H3N8 in dogs while also following the continuing evolution and epidemiology of EIV among horses in several geographic regions of the world.
Results
H3N8 experienced major clade divergence in the early 2000s, including the emergence of CIV in dogs
Despite years of spread and evolution in horses since its first emergence around 1963, H3N8 EIV diversity had largely collapsed to a single North American clade by the end of the 20th century (Fig. 1, inset) (Daly et al. 1996; Lai et al. 2001). This ‘Florida’ clade then gave rise to the FC1 and FC2 subclades. A full genome phylogeny since 2001 shows the presence of pre-clade divergence viruses (preC), and viruses within the monophyletic FC2, FC1, and CIV groups. Divergence occurred roughly after the appearance of Kentucky/5/2002, California/8560/2002, and Newmarket/5/2003 preC (Fig. 1), so that the first full-genome samples from the FC2, FC1, and CIV clades are Richmond/1/2007, Wisconsin/1/2003, and Florida/242/2003, respectively. These FC2 and FC1 sequences are currently (or closely match) the EIV formulations frequently manufactured and used for vaccination (Pavulraj et al. 2021).
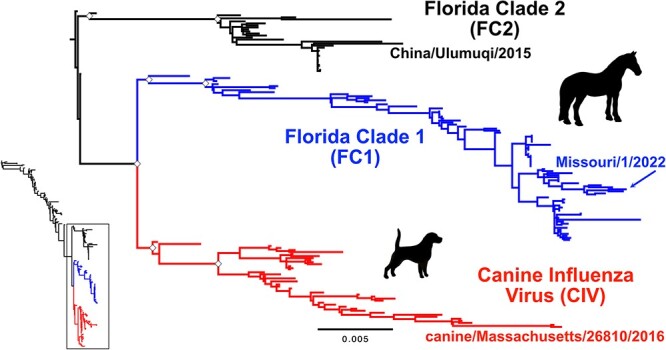
Recent evolution of H3N8 influenza virus. Equine influenza virus (EIV) H3N8 evolution from 1963 to present has resulted in three distinct clades post 2002 (inset). ML tree of the whole genome data set revealing three distinct clades: the Florida Clade 1 (FC1) and Florida Clade 2 (FC2) in horses, and canine influenza virus (CIV) in dogs. Tree was rooted to pre-clade (preC) genome Kentucky/5/2002. Scale denotes nucleotide divergence. White diamonds at major early nodes represent bootstrap support >99 per cent. Most recent genome of each clade is noted.
per cent. Most recent genome of each clade is noted.
We further gathered all detailed collection dates at best accuracy (YYYY-MM-DD) to review the temporal nature of the data set. A root-to-tip regression analysis of the full genome data set revealed strong clock-like structure (R2 =
= 0.92) and an evolutionary rate of 1.5 × 10−3 nucleotide substitutions/site/year (Figure S1A). It was obtained a reliable root-to-tip regression for the expanded HA1 data set from 2001 to present, yet with rooting divergence from the common ancestor between FC1/CIV and FC2 lineages (Figure S1B). The establishment and persistence of these lineages allows a direct comparison of H3N8 influenza virus evolution in novel (dog) and original (horse) hosts following the host transfer and emergence event giving rise to CIV. To determine what genomic characteristics may have allowed FC1 (and FC2) to persist in nature, while CIV went extinct, we analyzed the CIV, FC1, and FC2 clades since their divergence in parallel (Fig. 1).
0.92) and an evolutionary rate of 1.5 × 10−3 nucleotide substitutions/site/year (Figure S1A). It was obtained a reliable root-to-tip regression for the expanded HA1 data set from 2001 to present, yet with rooting divergence from the common ancestor between FC1/CIV and FC2 lineages (Figure S1B). The establishment and persistence of these lineages allows a direct comparison of H3N8 influenza virus evolution in novel (dog) and original (horse) hosts following the host transfer and emergence event giving rise to CIV. To determine what genomic characteristics may have allowed FC1 (and FC2) to persist in nature, while CIV went extinct, we analyzed the CIV, FC1, and FC2 clades since their divergence in parallel (Fig. 1).
CIV was restricted to the USA and went extinct around 2016
The last known positive cases of H3N8 CIV were reported in the Northeast USA in 2016, two samples of which (Maine and Massachusetts) were sequenced shortly thereafter by our group. No further H3N8 positive cases in the USA have been reported, even though this was a time of frequent canine influenza testing due to the emergence and spread of H3N2 CIV in the USA in 2015 and 2016 (Voorhees et al. 2018).
We gathered a data set of 43 complete CIV genomes (and an expanded 88 HA1 reading frame sequences) and employed the same methods as for the full H3N8 data set. Our full genome CIV phylogeny (Fig. 2A) closely matches the monophyletic group seen in the larger H3N8 full genome data set (Fig. 1) and previous analyses (He et al. 2019). The full genomes of CIV showed a strong clock-like evolution (R2 =
= 0.94) at approximately the same rate (1.8 × 10−3 substitutions/site/year, Figure S1C) as observed with the larger H3N8 full genome data set. We used the expanded HA1 data set to connect the phylogenetic tree to the geographic data (Fig. 2B). Subclades of CIV were highly structured by regional introduction and spread, mostly localizing to outbreak centers in Florida, New York City, Philadelphia, and the cities of Colorado. Secondary geographic sites appear to have involved secondary outbreaks that were related by phylogenetics back to their source locations.
0.94) at approximately the same rate (1.8 × 10−3 substitutions/site/year, Figure S1C) as observed with the larger H3N8 full genome data set. We used the expanded HA1 data set to connect the phylogenetic tree to the geographic data (Fig. 2B). Subclades of CIV were highly structured by regional introduction and spread, mostly localizing to outbreak centers in Florida, New York City, Philadelphia, and the cities of Colorado. Secondary geographic sites appear to have involved secondary outbreaks that were related by phylogenetics back to their source locations.
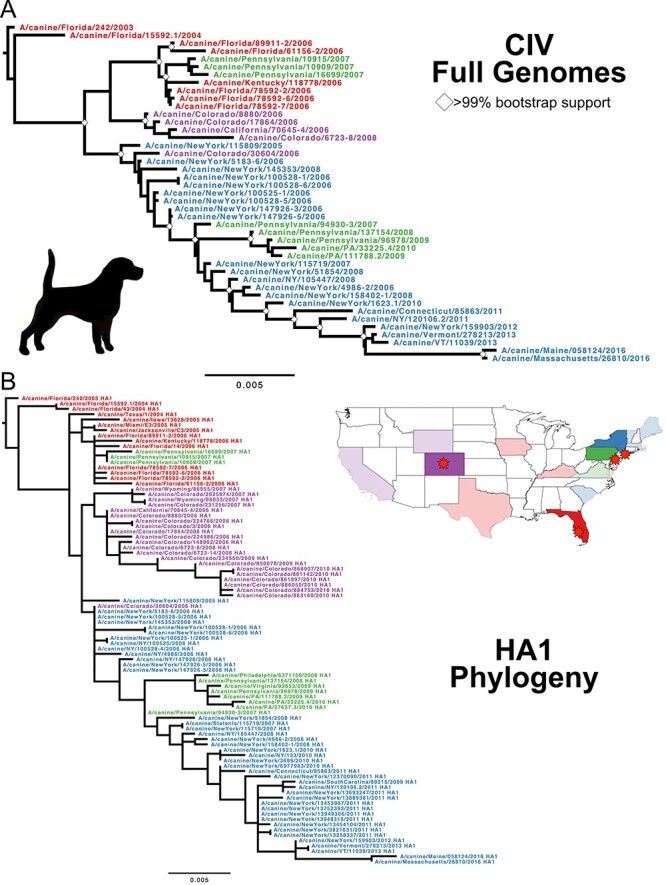
Evolution of the CIV H3N8 clade. (A) ML tree of the full genome data set of CIV, with taxa color defined by outbreak source region: Red, Florida/SE; Blue, New York City; Green, Philadelphia; and Purple, Colorado. Tree was rooted to earliest sample, Florida/242/2003. Scale denotes nucleotide divergence. White diamonds at major nodes represent bootstrap support >99 per cent. (B) ML tree of expanded HA1 data set reveals the strong geographic organization of subclades.
per cent. (B) ML tree of expanded HA1 data set reveals the strong geographic organization of subclades.
We further inferred the demographic history of these populations using Bayesian analysis with a Bayesian Skyline Plot (BSP) tree prior, enabling us to estimate changes in genetic diversity—a measure of effective population size (Ne)—through time. For both data sets, we found that Ne plots correspond to virus natural history and epidemiology: large population and diversity growth occurring during the initial US-wide outbreaks of 2005–2007, followed by regional die out and the eventual complete extinction (Figures S2A and B).
EIV FC1 is a US-centric virus that contributes to international transfers and outbreaks
We compiled 82 FC1 full genome and 252 HA1 sequence data sets including those newly generated here. The FC1 full genome phylogeny shows a fairly linear clade divergence (Fig. 3). The trees show strong signatures of geographic clustering, suggesting frequent single-introduction outbreaks in particular locations including South America, Europe, and other regions. There is a strong clock-like evolution (R2 =
= 0.99) at approximately the same rate (1.7 × 10−3 substitutions/site/year, Figure S1E) seen in the larger H3N8 full genome data set and the CIV clade. We again estimated Ne changes through time, and while sampling bias and poor admixture limits making broad conclusions, we were able to identify key demographic transitions and data gaps (Figures S2A, S6, and S7).
0.99) at approximately the same rate (1.7 × 10−3 substitutions/site/year, Figure S1E) seen in the larger H3N8 full genome data set and the CIV clade. We again estimated Ne changes through time, and while sampling bias and poor admixture limits making broad conclusions, we were able to identify key demographic transitions and data gaps (Figures S2A, S6, and S7).
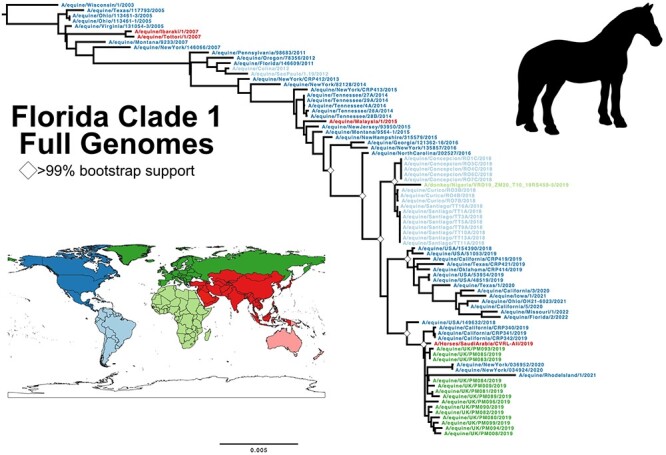
Evolution of the H3N8 EIV clade FC1. ML tree of full genome data set of FC1, with taxa color defined by worldwide region. Tree was rooted to earliest sample, Wisconsin/1/2003. Scale denotes nucleotide divergence. White diamonds at major nodes represent bootstrap support >99 per cent.
per cent.
The phylogenetic tree of our expanded HA1 sequences largely paralleled the full-length genome tree, further clarifying the larger geographic movements of FC1 since its divergence. The overall phylogenetic structure reveals an apparent US-based continuous circulation that seeds multiple international transfers and outbreaks in different regions of the world (Fig. 4). Some of these historical transfers have been reported and analyzed previously and confirmed here (Moloney 2011; Motoshima et al. 2011; Gahan et al. 2019; Diallo et al. 2021; Olguin-Perglione and Barrandeguy 2021; Lee et al. 2022a). Our Bayesian demographic analysis of the FC1 HA1 data set (Figure S8) also shows that the effective population size largely mirrors the full-length genome data set, yet with more points of transition and changes in diversity, some of which are likely revealed by the increased sampling resolution (Figure S2B).

Evolution of the H3N8 EIV clade FC1. ML tree of the expanded HA1 data set (n =
= 251), also colored by regional isolate. Tree was rooted to earliest sample, Wisconsin/1/2003. Scale denotes nucleotide divergence. Dates of and geography of regional subclades shows the recurring impact of US circulation on international transfers. Country ISO codes used: AE, United Arab Emirates; AR, Argentina; AU, Australia; BR, Brazil; CL, Chile; DE, Germany; EG, Egypt; FR, France; GB, United Kingdom; JP, Japan; MY, Malaysia; NE, Niger; NG, Nigeria; SA, Saudi Arabia; SN, Senegal; US, United States; and UY, Uruguay.
251), also colored by regional isolate. Tree was rooted to earliest sample, Wisconsin/1/2003. Scale denotes nucleotide divergence. Dates of and geography of regional subclades shows the recurring impact of US circulation on international transfers. Country ISO codes used: AE, United Arab Emirates; AR, Argentina; AU, Australia; BR, Brazil; CL, Chile; DE, Germany; EG, Egypt; FR, France; GB, United Kingdom; JP, Japan; MY, Malaysia; NE, Niger; NG, Nigeria; SA, Saudi Arabia; SN, Senegal; US, United States; and UY, Uruguay.
An analysis of recent FC1 phylogenetic structure (in both full genome and HA1 data sets) reveals two closely related subclades at this point in time: the viruses circulating in the USA, and those collected from the 2019 European outbreak. The directions of transmission appear complex, but since the virus in the USA was circulating for a few years prior to 2019, that was likely transferred from the USA to the UK and elsewhere in Europe. A few viruses collected in California in 2019 clustered with the European subclade, noted in horses that had been imported from Europe (metadata of samples collected by UC-Davis for this study). Some 2020–2021 US cases in the Northeastern states of New York and Rhode Island also more closely match the UK samples, suggesting likely transfers from Europe back to the USA. Further sampling in the USA, particularly from the Northeast region, is needed to clarify the relatedness of the North American and European FC1 viruses at this time.
FC2 represents an under sampled and likely limited clade of EIV with current circulation in some regions of Asia
The public repositories have limited numbers of full genomes from the FC2 clade, although more HA1 sequences are available. Sampling appears to be relatively sparce, with multiple samples and sequences from notable outbreaks, so that there is extensive genetic redundancy among the samples available. Our FC2 full genome maximum likelihood (ML) tree showed no variation from the full H3N8 data set analysis (Figs 1 and 5A). The more substantial HA1 data set (n =
= 254, Fig. 5B) clearly revealed geographic distribution of recent FC2 samples: while FC2 were centered in Europe between around 2000 and 2014, the clade appears to have been transferred to Asia around 2009. Europe has seen fewer FC2 cases, with last reports in 2017, while since 2018 only a single subclade has been present in mainland Asia. The small number of full FC2 genomes (Figure S1G–H) resulted in Bayesian analyses characterized by high statistical uncertainty (Figures S2A–B, S9–11).
254, Fig. 5B) clearly revealed geographic distribution of recent FC2 samples: while FC2 were centered in Europe between around 2000 and 2014, the clade appears to have been transferred to Asia around 2009. Europe has seen fewer FC2 cases, with last reports in 2017, while since 2018 only a single subclade has been present in mainland Asia. The small number of full FC2 genomes (Figure S1G–H) resulted in Bayesian analyses characterized by high statistical uncertainty (Figures S2A–B, S9–11).

Evolution of the H3N8 EIV clade FC2. (A) ML tree of full genome data set of FC2, with taxa color defined by worldwide region. Tree was rooted to earliest full genome sample, Richmond/1/2007. Scale denotes nucleotide divergence. White diamonds at major nodes represent bootstrap support >99 per cent. (B) ML tree of the expanded HA1 data set (n
per cent. (B) ML tree of the expanded HA1 data set (n =
= 254), also colored by regional origin. Tree was rooted to earliest HA1 FC2 sample, NewMarket/5/2003. Dates and geography of regional subclades show that FC2 is now centered in mainland Asia.
254), also colored by regional origin. Tree was rooted to earliest HA1 FC2 sample, NewMarket/5/2003. Dates and geography of regional subclades show that FC2 is now centered in mainland Asia.
We also note that FC2 isolates have been identified in various equid species (Mongolian horses, Arabian horses, and donkeys), as well as dogs, although their epidemiological relevance is unknown. Horses in Asia appear to be regularly exposed to avian influenza but have generally failed to support a new emergent strain due to a lack of adaptation (Zhu et al. 2019).
H3N8 clades show distinct mutational signatures reflecting host biology and unique epidemiology
We next compared H3N8 evolution in different hosts to see if there were obvious signatures of host-specific evolution after transfer to, and sustained transmission in, dogs. The rate of genome evolution (i.e. nucleotide substitutions per site per year for each segment) for each clade showed that substitution rates in CIV and FC1 were essentially the same in all segments except HA (Fig. 6A), suggesting that if new-host adaptation was present it was masked in the general background rate of genetic drift (McCrone and Lauring 2018; Sigal, Reid, and Wahl 2018). In contrast, the HA, NA, and M1 substitution rates are significantly greater within the FC1 clade than the FC2. All H3N8 clades within horses and dogs exhibit lower substitution rates than reported for influenza circulating in human host populations (huH3N2, Fig. 6A), particularly in HA and NA where antigenic selection would be most apparent.
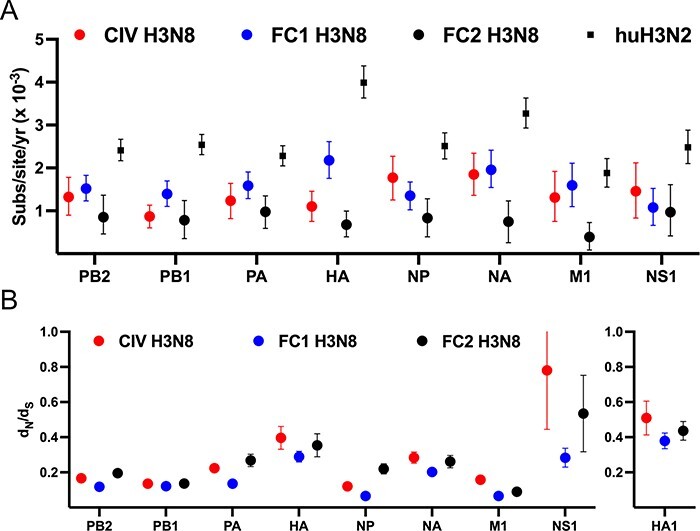
Molecular evolution of the H3N8 clades. (A) The nucleotide substitution rate for each segment major ORF in each clade are compared to known huH3N2 reference data set (Westgeest et al. 2014). Datapoints are mean values and error bars represent 95 per cent highest posterior density (HPD) values. (B) The mean dN/dS for each segment major ORF was determined using the SLAC program from the Datamonkey platform. Datapoints are mean values and error bars represent 95
per cent highest posterior density (HPD) values. (B) The mean dN/dS for each segment major ORF was determined using the SLAC program from the Datamonkey platform. Datapoints are mean values and error bars represent 95 per cent confidence intervals (CI).
per cent confidence intervals (CI).
We have previously reported comparative analyses of CIV and FC1 through 2015 (Feng et al. 2015). Only two CIV isolates have been generated since this time, while FC1 has continued to circulate widely in different regions of the world. Here, to better identify any residues subject to natural selection, we have updated that analysis based on the many additional the full genome sequences of the different clades. To do this we utilized several approaches in the Datamonkey analytical toolset (https://datamonkey.org/; Weaver et al. 2018). Using SLAC (Single-Likelihood Ancestor Counting), we measured the mean dN/dS ratio of each segment for each clade (Fig. 6B). An elevated ratio is seen in the HA ORF (particularly in the HA1 antigenic domain), but with no significant difference between clades. The divergent selection acting on the CIV and EIV H3N8 NS segment, reflected in variant segment length, has previously been analyzed in detail (Na et al. 2015). In sum, it appears that FC1 is undergoing greater evolutionary change in horses than the FC2 clade, or than the H3N8 CIV in dogs up until 2016.
CIV early mutations suggest host adaptation to dogs, yet with no change in epidemiology and avoidance of extinction
The sequences of viruses spilling over into new hosts will reflect the composition of the single virus that makes the initial transfer, and then will likely undergo adaptation to the properties and physiology of the new host animal, including optimizing onward transmission (Wasik et al. 2019a). We therefore compared the sequences and evolution of the viruses that were closely related to the common ancestor of CIV (preC), the evolution of H3N8 CIV in its new host, and parallel continued evolution of the early FC1 H3N8 EIV in horses (Fig. 7A). This revealed multiple transitional mutations in CIV (Fig. 7B), some of which have been previously analyzed (Collins et al. 2014; Feng et al. 2015; Na et al. 2015; Feng et al. 2016). However, these genetic changes appear not to have allowed CIV to increase its spread among the general dog populations, such that the virus continued to be maintained as shelter-driven outbreaks which resulted in viral extinction, likely through stochastic fade out (Hayward et al. 2010). It is also likely that selection on the virus for spread within dense and high turn-over shelter or kennel populations did not result in the selection of mutations enabling enhanced spread in the household dog population required to develop a true epidemic or pandemic (Dalziel et al. 2014). Despite this successful transfer of the H3N8 EIV to dogs, all other spillovers have resulted in only single canine infections or short-lived local outbreaks (Crispe et al. 2011).
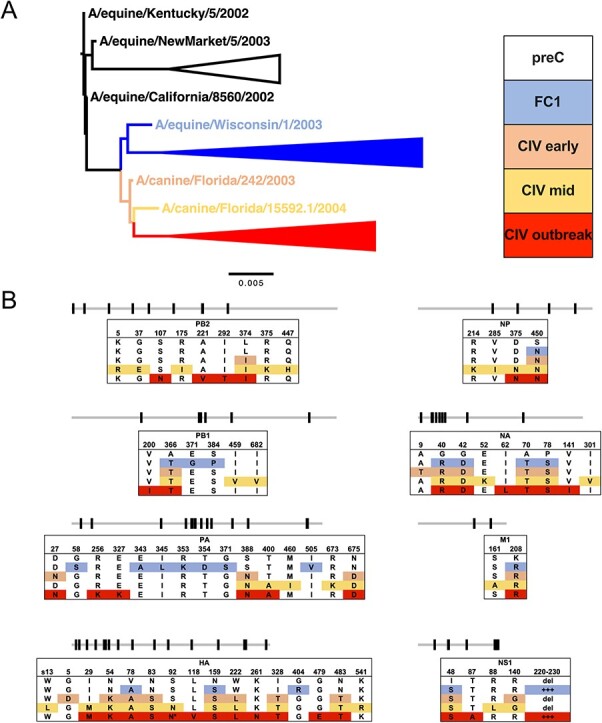
Early molecular evolution of H3N8 clade divergence and CIV emergence. (A) The H3N8 whole genome ML tree focused on the earliest preC, FC1, and CIV taxa for comparative alignment. (B) Key nonsynonymous changes in the earliest diverging taxa are identified by genome segment and position. Some late CIV mutations (*) are not present in all outbreak subclades.
It is also noteworthy that the H3N8 EIV FC1 acquired additional mutations during its parallel spread in horses. There is some evidence that H3N8 may have undergone further host adaptation in horses over time (Amat et al. 2021). This change in evolutionary potential of the EIV may suggest that the preC consensus genome was particularly suited to clade divergence and host-switching in 1999. While some antigenic selection may occur (Nemoto et al. 2021; Lee et al. 2022b; Nemoto et al. 2023), most mutations are not in the antigenic sites of the HA or NA, and likely represent genetic drift during the outbreak-driven circulation of FC1 (Fig. 4). Discerning true antigenic evolution from background genetic drift will require greater surveillance and testing of immunity in horses.
Discussion
A detailed understanding of the replication and spread of infectious diseases allows effective control strategies to be devised. While endemic human influenza outbreaks appear to involve common global movement and widespread infections, IAVs in other hosts involves more complex epidemiology, often with constrained geographic spread, and many outbreaks dying out without obvious human interventions. The recent apparent extinction of the Influenza B Yamagata lineage in humans suggests that influenza viruses may have a more tenuous foothold in mammalian host population than has sometimes been assumed (Dhanasekaran et al. 2022).
Equine and canine H3N8 have different evolutionary fates
The analysis presented here focuses on the avian-mammalian emergent H3N8 viruses: first in their original equine hosts in which they have been circulating for 60 years, and then with comparisons to the same virus in dogs, to understand the divergent evolution of the same virus in different hosts. While EIV continues to circulate and infect horses around the world, we report here that CIV was last seen in 2016 in the Northeast USA, and is apparently now extinct.
years, and then with comparisons to the same virus in dogs, to understand the divergent evolution of the same virus in different hosts. While EIV continues to circulate and infect horses around the world, we report here that CIV was last seen in 2016 in the Northeast USA, and is apparently now extinct.
H3N8 CIV was likely entirely contained to the USA during its ~17 years of transmission in dogs, as no cases of this lineage have been reported in other countries. Our new complete genome sequencing of the H3N8 CIV isolates from throughout the course epidemic show that its geographic structure arose from an initial burst of widespread disease that may have been associated with the movement of infected greyhounds. After 2007, only local transmission was seen in Colorado and close by in Wyoming, up to around 2010 (Rivailler et al. 2010). In the Northeast of the USA, the virus was first reported in New York in 2005 and in Pennsylvania in 2007. The virus continued to circulate in New York, where it was isolated from short-lived outbreaks that occurred after introduction of virus (likely due to transport of infected dogs from the shelters where the virus was endemic), and those were seen in Connecticut, Pennsylvania, Vermont, Maine, and Massachusetts up until 2016 (Hayward et al. 2010; Dalziel et al. 2014). Our analysis confirms that the outbreaks in Colorado and the Northeast were distinct, and also that after 2012 only a single New York-centered lineage of virus continuing to circulate in dogs.
years of transmission in dogs, as no cases of this lineage have been reported in other countries. Our new complete genome sequencing of the H3N8 CIV isolates from throughout the course epidemic show that its geographic structure arose from an initial burst of widespread disease that may have been associated with the movement of infected greyhounds. After 2007, only local transmission was seen in Colorado and close by in Wyoming, up to around 2010 (Rivailler et al. 2010). In the Northeast of the USA, the virus was first reported in New York in 2005 and in Pennsylvania in 2007. The virus continued to circulate in New York, where it was isolated from short-lived outbreaks that occurred after introduction of virus (likely due to transport of infected dogs from the shelters where the virus was endemic), and those were seen in Connecticut, Pennsylvania, Vermont, Maine, and Massachusetts up until 2016 (Hayward et al. 2010; Dalziel et al. 2014). Our analysis confirms that the outbreaks in Colorado and the Northeast were distinct, and also that after 2012 only a single New York-centered lineage of virus continuing to circulate in dogs.
Equine influenza virus circulates widely in the USA and in different regions of the world
In contrast, the H3N8 EIV has been found throughout North America for the 60 years since it was first isolated. Ongoing passive surveillance systems confirm US circulation, with some observed seasonality (Chappell et al. 2023). However, frequent international transfers of the virus are also a feature of its epidemiology. We confirm here that outbreaks in several countries around the world were likely directly, or indirectly, caused by viruses from North America. Those included widespread outbreaks in South America that started in 2018, and in the UK and elsewhere in Europe starting in 2019. Similarly, viruses from Japan, Australia, and Malaysia have been closely related to viruses from the USA. Recent outbreaks in Western Africa also fit this pattern, as viruses from Senegal, Nigeria, and Niger were closely related to those from South America, which were due to viruses originating in the USA (Olguin-Perglione and Barrandeguy 2021). SARS-CoV-2 drastically changed human behavior during 2020 and 2021, with international travel spending being reduced by 76 per cent in 2020 compared with 2019 following international travel bans imposed by many countries (US Travel Association 2022). Equine sport activities in the USA and around the world were consequently impacted by varied economic curtailment, including the international transfer of horses. Yet, we see continued FC1 infections in the USA between 2020 and 2022, and a potential inter-relatedness between US and European samples in that time, suggesting a potential return to the norm. Overall, it appears that the FC1 lineage of EIV is maintained in the horse population of the USA, and that provides viruses causing outbreaks in other regions of the world (Lee et al. 2022a).
years since it was first isolated. Ongoing passive surveillance systems confirm US circulation, with some observed seasonality (Chappell et al. 2023). However, frequent international transfers of the virus are also a feature of its epidemiology. We confirm here that outbreaks in several countries around the world were likely directly, or indirectly, caused by viruses from North America. Those included widespread outbreaks in South America that started in 2018, and in the UK and elsewhere in Europe starting in 2019. Similarly, viruses from Japan, Australia, and Malaysia have been closely related to viruses from the USA. Recent outbreaks in Western Africa also fit this pattern, as viruses from Senegal, Nigeria, and Niger were closely related to those from South America, which were due to viruses originating in the USA (Olguin-Perglione and Barrandeguy 2021). SARS-CoV-2 drastically changed human behavior during 2020 and 2021, with international travel spending being reduced by 76 per cent in 2020 compared with 2019 following international travel bans imposed by many countries (US Travel Association 2022). Equine sport activities in the USA and around the world were consequently impacted by varied economic curtailment, including the international transfer of horses. Yet, we see continued FC1 infections in the USA between 2020 and 2022, and a potential inter-relatedness between US and European samples in that time, suggesting a potential return to the norm. Overall, it appears that the FC1 lineage of EIV is maintained in the horse population of the USA, and that provides viruses causing outbreaks in other regions of the world (Lee et al. 2022a).
What host differences explain the different viral evolutionary fates?
As dogs usually live in households as single animals or in small groups as their population structure and movement patterns likely differ from that of horses. Larger numbers of susceptible dogs may be found in animal shelters, kennels, or dog day cares, and these will allow increased transmission. CIVs will spread rapidly within most shelters, and then the virus will die out once the population has all been infected and become immune, or transmission may be stopped within the shelter by quarantining affected dogs (Hayward et al. 2010; Dalziel et al. 2014). Horses are often found in groups or stables, yet its impact on transmission is unclear. The transfer of horses for sport (including internationally) is likely a large driver of EIV dissemination. Recent estimations by USDA-APHIS report that 57.8 per cent of equine operations in the USA transported any resident equid by vehicle off the operation and back in the preceding 12 months; 14.1
months; 14.1 per cent were >500
per cent were >500 miles, 29.7
miles, 29.7 per cent of operations transported equids to an adjacent state, and 11.8
per cent of operations transported equids to an adjacent state, and 11.8 per cent transported equids beyond adjacent states; while only 0.7
per cent transported equids beyond adjacent states; while only 0.7 per cent of operations transferred horses internationally (2015). Additionally, there appear to have been no international transfers of dogs infected with H3N8 CIV. Given the short lifespan of dogs, limited circulation of H3N8, and the absence of a licensed vaccine until 2009, we assume very little immune pressure on the CIV H3N8. Equids may be similar to humans as long-lived vertebrates where relatively high levels of anti-EIV antibody immunity are induced due to either infection or vaccination but wane in time between lifetime exposures (Hannant, Mumford, and Jessett 1988).
per cent of operations transferred horses internationally (2015). Additionally, there appear to have been no international transfers of dogs infected with H3N8 CIV. Given the short lifespan of dogs, limited circulation of H3N8, and the absence of a licensed vaccine until 2009, we assume very little immune pressure on the CIV H3N8. Equids may be similar to humans as long-lived vertebrates where relatively high levels of anti-EIV antibody immunity are induced due to either infection or vaccination but wane in time between lifetime exposures (Hannant, Mumford, and Jessett 1988).
Interestingly, the reservoir of H3N2 CIV in China and Korea appears to have been dogs being bred in large numbers, with infected dogs then transported to the USA or Canada to cause outbreaks among different dog populations. However, once again there has been no onward transmission of that H3N2 CIV to other countries from North America, suggesting that the recent US dog population structure may represent a ‘dead-end’ for endemic IAV circulation (Voorhees et al. 2018).
Conclusion
Our analysis of H3N8 evolution in the horse and canine clades show similar levels of variation that are lower than that observed for influenza in humans. The data therefore suggest that host population structure and epidemiology rather than specific adaptive evolution are driving the sustained transmission of FC1 in horses. Given the extinction of CIV H3N8, this suggests that well targeted interventions could in turn result in extinction of EIV H3N8 FC1 in horses. This likely requires a more refined epidemiological understanding of circulation in the USA, updated vaccination formulation for FC1 and greater uptake. It may further require screening of international transfers of horses for influenza to reduce worldwide transmission, particularly the export of US-borne FC1 lineages, as well as the ‘return’ of viruses to the USA from Europe. Rational human intervention may be necessary to ‘break’ the host-structure epidemiology that is sustaining this virus.
Materials and Methods
H3N8 Sample Collection
Samples were obtained from various diagnostic centers following routine passive diagnostic testing for respiratory disease (listed in Table S1, red). Samples were received as either nasal or pharyngeal swab material, virus isolated in culture/egg passages, or extracted total nucleic acid (tNA) first utilized for quantitative reverse transcription-PCR (qRT-PCR) positivity. vRNA was extracted from clinical swab samples and isolated using the QIAamp Viral RNA Mini kit. Purified vRNA or tNA was then either directly used for influenza multi-segment RT-PCR or stored at −80˚C.
Generation of Influenza Virus Sequences
Genomes from samples were generated as cDNAs using a whole genome multi-segment RT-PCR protocol as described previously (Wasik et al. 2019b; Mitchell et al. 2021). A common set of primers (5′ to 3′, uni12a, GTTACGCGCCAGCAAAAGCAGG; uni12b, GTTACGCGCCAGCGAAAGCAGG; uni13, GTTACGCGCCAGTAGAAACAAGG) that recognize the terminal sequences of the influenza A segments were used in a single reaction with SuperScript III OneStep RT-PCR with Platinum Taq DNA polymerase (Invitrogen). Following confirmation by gel electrophoresis, viral cDNA was purified either by standard PCR reaction desalting columns or with a 0.45X volume of AMPure XP beads (Beckman Coulter). Nextera XT libraries were generated with 1 ng of cDNA material, while Nextera FLEX libraries were generated with 150–200 ng of cDNA (Invitrogen). Libraries were multiplexed, pooled, and sequenced using Ilumina MiSeq 2× 250 sequencing, and consensus sequences were determined for each genome segment after assuring full genome coverage.
Consensus sequence editing was performed using Geneious Prime 2022.1.1. Paired reads were trimmed using BBduk script (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbduk-guide/) and merged. Each sequence was assembled by mapping to a reference sequence of a previously annotated equine or canine H3N8 sequence. Consensus positions had read depth >300 and >75 per cent identity. We did not observe unresolved positions under these conditions from the samples generated in this study.
per cent identity. We did not observe unresolved positions under these conditions from the samples generated in this study.
Phylogenetic Analysis
H3N8 nucleotide sequences were downloaded from the NCBI Influenza Virus Database and combined with the consensus genomes generated in this study (Bao et al. 2008). We focused on genomes from 2001 to present, encompassing the formation of the major virus clades (FC2, FC1, and CIV). Both inter- and intra-subtype reassortants were identified using RDP4 (seven methods: RDP, GENECONV, Bootscan, MaxChi, Chimaera, SiScan, and 3seq) and excluded if found to be a statistically significant outlier in two or more methods (Martin et al. 2015). Our total data set comprised 157 full genomes (preC: 3, FC2: 29, FC1: 82, and CIV: 43), with an expanded 611 HA1 reading frames (preC: 17, FC2: 254, FC1: 252, and CIV: 88) (Tables S1 and S2). Sequences were manually trimmed to their major open reading frames (PB2: 2280nt, PB1: 2274nt, PA: 2151nt, HA: 1695–1701nt, NP: 1497nt, NA: 1410–1413nt, M1: 759nt, and NS1: 654–693nt) and either analyzed separately or concatenated with all other genome segments from the same virus sample. HA1 nucleotide sequences were trimmed to cover amino acids 1–329 (H3 numbering). Nucleotide sequences were aligned using MUSCLE (Edgar 2004). ML phylogenetic analysis was performed by PhyML (Guindon et al. 2010), employing a generalized time-reversable (GTR) substitution model, gamma-distributed (Γ4) rate variation among sites, and bootstrap resampling. Both MUSCLE and PhyML were run under the Geneious Prime platform. ML trees were visualized and annotated using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
The temporal signal (i.e. molecular clock-like structure) of the data was assessed by a regression of root-to-tip genetic distance against date of sampling using our ML tree and the TempEst v.1.5.3 software (Rambaut et al. 2016). Accurate collection dating to the day (YYYY-MM-DD) was utilized for all samples where this information was available. The sampling dates used for all isolates are listed in Tables S1 and S2.
Bayesian Phylogeny and Analysis
We used a Bayesian Markov chain Monte Carlo (MCMC) approach to better estimate virus divergence times and population dynamics. Analyses were performed in BEAST v.1.10.4 (Suchard et al. 2018), where MCMC sampling was performed with a strict clock, a generalized time-reversable (GTR) substitution model with gamma distribution in four categories (Γ4), and assuming a BSP prior demographic model. Analyses were performed for a minimum of 100 million runs, with duplicates combined in Log Combiner v1.10.4. All output results were examined for statistical convergence in Tracer v1.7.2 (effective sample size [ESS] ≥200, consistent traces, and removed burn-in at 10–15 per cent) (Rambaut et al. 2018). Maximum clade credibility (MCC) phylogenetic trees were generated by Tree Annotator v1.10.4 and visualized by FigTree v1.4.4.
per cent) (Rambaut et al. 2018). Maximum clade credibility (MCC) phylogenetic trees were generated by Tree Annotator v1.10.4 and visualized by FigTree v1.4.4.
Analysis of Selection Pressures
The relative numbers of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per site in each ORF of each segment were analyzed for the signature of positive selection (adaptive evolution, p <
< 0.05 or >0.95 posterior) using a variety of method packages available in the Datamonkey package (https://datamonkey.org/, (Weaver et al. 2018)): BUSTED (Branch-site Unrestricted Statistical Test for Episodic Diversification), FEL (Fixed Effects Likelihood), FUBAR (Fast, Unconstrained Bayesian AppRoximation), MEME (Mixed Effects Model of Evolution), and SLAC (Single-Likelihood Ancestor Counting).
0.05 or >0.95 posterior) using a variety of method packages available in the Datamonkey package (https://datamonkey.org/, (Weaver et al. 2018)): BUSTED (Branch-site Unrestricted Statistical Test for Episodic Diversification), FEL (Fixed Effects Likelihood), FUBAR (Fast, Unconstrained Bayesian AppRoximation), MEME (Mixed Effects Model of Evolution), and SLAC (Single-Likelihood Ancestor Counting).
Acknowledgements
We thank the sources of the new samples used in this study, including diagnostic labs and individual private veterinarians. We wish to thank members of the Parrish Lab for critical feedback and support on this manuscript, particularly Wendy Weichert. S.E.R. and T.M.C. were supported by a project of the Kentucky Agricultural Experiment Station, Project No. KY-014067. P.R.M. was supported by the Medical Research Council of the United Kingdom (Grant No. MC_UU_12014/9), the Horserace Betting Levy Board (Grant No. 797), and the Biotechnology and Biological Sciences Research Council (Grant Nos. BB/V002821/1 and BB/V004697/1). C.R.P. (and by extension B.R.W., E.R., and I.E.H.V.) were supported by National Institutes of Health (Grant No. R01-GM080533). This work was partially funded by the Influenza Division of the Centers for Disease Control, Animal-Human Interface program, under Contract No. 75D30121P12812 to C.R.P. and L.B.G.
Contributor Information
Brian R Wasik, Baker Institute for Animal Health, Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA.
Evin Rothschild, Baker Institute for Animal Health, Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA.
Ian E H Voorhees, Baker Institute for Animal Health, Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA.
Stephanie E Reedy, Department of Veterinary Science, Gluck Equine Research Center, University of Kentucky, Lexington, KY 40546, USA.
Pablo R Murcia, MRC-University of Glasgow Centre for Virus Research, School of Infection and Immunity, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G61 1QH, Scotland.
Nicola Pusterla, Department of Medicine & Epidemiology, School Veterinary Medicine, University of California, Davis, CA 95616, USA.
Thomas M Chambers, Department of Veterinary Science, Gluck Equine Research Center, University of Kentucky, Lexington, KY 40546, USA.
Laura B Goodman, Baker Institute for Animal Health, Department of Public and Ecosystems Health, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA.
Edward C Holmes, Sydney Institute for Infectious Diseases, School of Medical Sciences, University of Sydney, Sydney, NSW 2006, Australia.
James C Kile, Influenza Division, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, GA 30329, USA.
Colin R Parrish, Baker Institute for Animal Health, Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA.
Data availability
Virus genome consensus sequences have been submitted to GenBank, and accession numbers of the sequences used (and generated) for this study are provided in Tables S1 and S2.
IMPORTANCE
The emergence of viruses in new hosts is a threat to human and animal health. The H3N8 equine influenza virus (EIV) emerged in 1963 by transfer of an avian influenza virus and the H3N8 canine influenza virus (CIV) subsequently emerged in 1999 when EIV transferred to dogs. H3N8 CIV persistently circulated in only a few locations in the USA, and has not been detected since 2016. In the same period, H3N8 EIV has circulated as two separate clades, one in North America and other regions of the world, while the other currently appears to be found only in Central Asia. By comparing the hosts, epidemiology, and evolution of these influenza viruses, we explain how these lineages had different evolutionary fates and show why elucidating these evolutionary processes is key to understanding zoonotic disease and viral emergence.
References
- Agüero M. et al. (2023) ‘Highly Pathogenic Avian Influenza A(H5N1) Virus Infection in Farmed Minks, Spain, October 2022’, Eurosurveillance, 28: 2300001. [Europe PMC free article] [Abstract] [Google Scholar]
- Amat J. A. R. et al. (2021) ‘Long-term Adaptation following Influenza A Virus Host Shifts Results in Increased Within-Host Viral Fitness Due to Higher Replication Rates, Broader Dissemination within the Respiratory Epithelium and Reduced Tissue Damage’, PLOS Pathogens, 17: e1010174. [Europe PMC free article] [Abstract] [Google Scholar]
- Anderson T. C. et al. (2012) ‘Serological Evidence of H3N8 Canine Influenza-like Virus Circulation in USA Dogs Prior to 2004’, The Veterinary Journal, 191: 312–6. [Abstract] [Google Scholar]
- Antia R. et al. (2003) ‘The Role of Evolution in the Emergence of Infectious Diseases’, Nature, 426: 658–61. [Europe PMC free article] [Abstract] [Google Scholar]
- Bao Y. et al. (2008) ‘The Influenza Virus Resource at the National Center for Biotechnology Information’, Journal of Virology, 82: 596–601. [Europe PMC free article] [Abstract] [Google Scholar]
- Bryant N. A. et al. (2009) ‘Antigenic and Genetic Variations in European and North American Equine Influenza Virus Strains (H3N8) Isolated from 2006 to 2007’, Veterinary Microbiology, 138: 41–52. [Abstract] [Google Scholar]
- Carabelli A. M. et al. (2023) ‘SARS-CoV-2 Variant Biology: Immune Escape, Transmission and Fitness’, Nature Reviews Microbiology, 23: 162–77. [Europe PMC free article] [Abstract] [Google Scholar]
- Castro E. R. et al. (2019) ‘Epidemiological and Virological Findings during an Outbreak of Equine Influenza in Uruguay in 2018’, Revue Scientifique Et Technique de l’OIE, 38: 737–49. [Abstract] [Google Scholar]
- Chambers T. M. (2022) ‘Equine Influenza’, Cold Spring Harbor Perspectives in Medicine, 12: a038331. [Europe PMC free article] [Abstract] [Google Scholar]
- Chappell D. E. et al. (2023) ‘Voluntary Surveillance Program for Equine Influenza Virus in the United States during 2008–2021’, Pathogens, 12: 192. [Europe PMC free article] [Abstract] [Google Scholar]
- Collins P. J. et al. (2014) ‘Recent Evolution of Equine Influenza and the Origin of Canine Influenza’, Proceedings of the National Academy of Sciences, 111: 11175–80. [Europe PMC free article] [Abstract] [Google Scholar]
- Crawford P. C. et al. (2005) ‘Transmission of Equine Influenza Virus to Dogs’, Science, 310: 482–5. [Abstract] [Google Scholar]
- Crispe E. et al. (2011) ‘Infection of Dogs with Equine Influenza Virus: Evidence for Transmission from Horses during the Australian Outbreak’, Australian Veterinary Journal, 89: 27–8. [Abstract] [Google Scholar]
- Cullinane A., and Newton J. R. (2013) ‘Equine Influenza—A Global Perspective’, Veterinary Microbiology, 167: 205–14. [Abstract] [Google Scholar]
- Daly J. M. et al. (2008) ‘Transmission of Equine Influenza Virus to English Foxhounds’, Emerging Infectious Diseases, 14: 461–4. [Europe PMC free article] [Abstract] [Google Scholar]
- ——— et al. (1996) ‘Antigenic and Genetic Evolution of Equine H3N8 Influenza A Viruses’, Journal of General Virology, 77: 661–71. [Abstract] [Google Scholar]
- ——— et al. (2021) ‘Retrospective Serological Survey for Influenza in Horses from Brazil’, Brazilian Journal of Microbiology, 52: 461–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Dalziel B. D. et al. (2014) ‘Contact Heterogeneity, Rather than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus’, PLoS Pathogens, 10: e1004455. [Europe PMC free article] [Abstract] [Google Scholar]
- Dhanasekaran V. et al. (2022) ‘Human Seasonal Influenza under COVID-19 and the Potential Consequences of Influenza Lineage Elimination’, Nature Communications, 13: 1721. [Europe PMC free article] [Abstract] [Google Scholar]
- Diallo A. A. et al. (2021) ‘Transboundary Spread of Equine Influenza Viruses (H3N8) in West and Central Africa: Molecular Characterization of Identified Viruses during Outbreaks in Niger and Senegal, in 2019’, Transboundary and Emerging Diseases, 68: 1253–62. [Abstract] [Google Scholar]
- Dionísio L. et al. (2021) ‘Equine Influenza: A Comprehensive Review from Etiology to Treatment’, Animal Health Research Reviews, 22: 56–71. [Abstract] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Elderfield R. A. et al. (2014) ‘Accumulation of Human-adapting Mutations during Circulation of A(H1N1)pdm09 Influenza Virus in Humans in the United Kingdom’, Journal of Virology, 88: 13269–83. [Europe PMC free article] [Abstract] [Google Scholar]
- USDA . (2016). ‘Equine 2015, “Baseline Reference of Equine Health and Management in the United States, 2015”’, Fort Collins: USDA–APHIS–VS–CEAH–NAHMS.
- Fagre A. C. et al. (2022) ‘Assessing the Risk of Human-to-wildlife Pathogen Transmission for Conservation and Public Health’, Ecology Letters, 25: 1534–49. [Europe PMC free article] [Abstract] [Google Scholar]
- Feng K. H. et al. (2015) ‘Equine and Canine Influenza H3N8 Viruses Show Minimal Biological Differences despite Phylogenetic Divergence’, Journal of Virology, 89: 6860–73. [Europe PMC free article] [Abstract] [Google Scholar]
- ——— et al. (2016) ‘Comparing the Functions of Equine and Canine Influenza H3N8 Virus PA-X Proteins: Suppression of Reporter Gene Expression and Modulation of Global Host Gene Expression’, Virology, 496: 138–46. [Europe PMC free article] [Abstract] [Google Scholar]
- Gahan J. et al. (2019) ‘Whole Genome Sequencing of the First H3N8 Equine Influenza Virus Identified in Malaysia’, Pathogens, 8: E62. [Europe PMC free article] [Abstract] [Google Scholar]
- Gamarra-Toledo V. et al. (2023) First Mass Mortality of Marine Mammals Caused by Highly Pathogenic Influenza Virus (H5N1) in South America. <10.1101/2023.02.08.527769>. [CrossRef] [Google Scholar]
- Gildea S. et al. (2018) ‘Multifocal Equine Influenza Outbreak with Vaccination Breakdown in Thoroughbred Racehorses’, Pathogens, 7: 43. [Europe PMC free article] [Abstract] [Google Scholar]
- Gildea S., Fitzpatrick D. A., and Cullinane A. (2013) ‘Epidemiological and Virological Investigations of Equine Influenza Outbreaks in Ireland (2010–2012)’, Influenza and Other Respiratory Viruses, 7: 61–72. [Europe PMC free article] [Abstract] [Google Scholar]
- Grubaugh N. D. et al. (2019) ‘Tracking Virus Outbreaks in the Twenty-first Century’, Nature Microbiology, 4: 10–9. [Europe PMC free article] [Abstract] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [Abstract] [Google Scholar]
- Hadfield J. et al. (2018) ‘Nextstrain: Real-time Tracking of Pathogen Evolution’, Bioinformatics, 34: 4121–3. [Europe PMC free article] [Abstract] [Google Scholar]
- Hannant D., Mumford J. A., and Jessett D. M. (1988) ‘Duration of Circulating Antibody and Immunity following Infection with Equine Influenza Virus’, Veterinary Record, 122: 125–8. [Abstract] [Google Scholar]
- Harari S. et al. (2022) ‘Drivers of Adaptive Evolution during Chronic SARS-CoV-2 Infections’, Nature Medicine, 28: 1501–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Hay A. J., and McCauley J. W. (2018) ‘The WHO Global Influenza Surveillance and Response System (GISRS) – A Future Perspective’, Influenza and Other Respiratory Viruses, 12: 551–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Hayward J. J. et al. (2010) ‘Microevolution of Canine Influenza Virus in Shelters and Its Molecular Epidemiology in the United States’, Journal of Virology, 84: 12636–45. [Europe PMC free article] [Abstract] [Google Scholar]
- He W. et al. (2019) ‘Host-Range Shift of H3N8 Canine Influenza Virus: A Phylodynamic Analysis of its Origin and Adaptation from Equine to Canine Host’, Veterinary Research, 50: 87. [Europe PMC free article] [Abstract] [Google Scholar]
- Hill V. et al. (2022) ‘The Origins and Molecular Evolution of SARS-CoV-2 Lineage B.1.1.7 in the UK’, Virus Evolution, 8: veac080. [Europe PMC free article] [Abstract] [Google Scholar]
- Karamendin K. et al. (2014) ‘Continuing Evolution of Equine Influenza Virus in Central Asia, 2007–2012’, Archives of Virology, 159: 2321–7. [Abstract] [Google Scholar]
- Kasel J. A. et al. (1965) ‘Experimental Infection of Human Volunteers with Equine Influenza Virus’, Nature, 206: 41–3. [Abstract] [Google Scholar]
- Kasel J. A., and Couch R. B. (1969) ‘Experimental Infection in Man and Horses with Influenza A Viruses’, Bulletin of the World Health Organization, 41: 447–52. [Europe PMC free article] [Abstract] [Google Scholar]
- Krammer F. et al. (2018) ‘Influenza’, Nature Reviews. Disease Primers, 4: 3. [Europe PMC free article] [Abstract] [Google Scholar]
- Krueger W. S. et al. (2014) ‘No Evidence for Zoonotic Transmission of H3N8 Canine Influenza Virus among US Adults Occupationally Exposed to Dogs’, Influenza and Other Respiratory Viruses, 8: 99–106. [Europe PMC free article] [Abstract] [Google Scholar]
- Lai A. C. et al. (2001) ‘Diverged Evolution of Recent Equine-2 Influenza (H3N8) Viruses in the Western Hemisphere’, Archives of Virology, 146: 1063–74. [Abstract] [Google Scholar]
- Lee K. et al. (2022a) ‘Investigation of Cross-regional Spread and Evolution of Equine Influenza H3N8 at US and Global Scales Using Bayesian Phylogeography Based on Balanced Subsampling’, Transboundary and Emerging Diseases, 69: e1734–e1748. [Abstract] [Google Scholar]
- ——— et al. (2022b) ‘Genome-informed characterisation of antigenic drift in the haemagglutinin gene of equine influenza strains circulating in the United States from 2012 to 2017’, Transboundary and Emerging Diseases, 69: e52–e63. [Abstract] [Google Scholar]
- Lemey P. et al. (2014) ‘Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2’, PLoS Pathogens, 10: e1003932. [Europe PMC free article] [Abstract] [Google Scholar]
- Li S. et al. (2010) ‘Avian-origin H3N2 Canine Influenza A Viruses in Southern China’, Infection, Genetics and Evolution, 10: 1286–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: vev003. [Europe PMC free article] [Abstract] [Google Scholar]
- Martinez-Sobrido L. et al. (2020) ‘Characterizing Emerging Canine H3 Influenza Viruses’, PLOS Pathogens, 16: e1008409. [Europe PMC free article] [Abstract] [Google Scholar]
- McCrone J. T., and Lauring A. S. (2018) ‘Genetic Bottlenecks in Intraspecies Virus Transmission’, Current Opinion in Virology, 28: 20–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Miño S. et al. (2019) ‘Equine Influenza Virus in Asia: Phylogeographic Pattern and Molecular Features Revealed the Circulation of an Autochthonous Lineage’, Journal of Virology, 93: e00116–19. [Europe PMC free article] [Abstract] [Google Scholar]
- Mitchell P. K. et al. (2021) ‘Method Comparison of Targeted Influenza A Virus Typing and Whole-genome Sequencing from Respiratory Specimens of Companion Animals’, Journal of Veterinary Diagnostic Investigation, 33: 191–201. [Europe PMC free article] [Abstract] [Google Scholar]
- Moloney B. (2011) ‘Overview of the Epidemiology of Equine Influenza in the Australian Outbreak’, Australian Veterinary Journal, 89: 50–6. [Abstract] [Google Scholar]
- Morens D. M. et al. (2020) ‘Pandemic COVID-19 Joins History’s Pandemic Legion’, mBio, 11: e00812–20. [Europe PMC free article] [Abstract] [Google Scholar]
- Morens D. M., and Taubenberger J. K. (2010) ‘An Avian Outbreak Associated with Panzootic Equine Influenza in 1872: An Early Example of Highly Pathogenic Avian Influenza?’, Influenza and Other Respiratory Viruses, 4: 373–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Motoshima M. et al. (2011) ‘Antigenic and Genetic Analysis of H3N8 Influenza Viruses Isolated from Horses in Japan and Mongolia, and Imported from Canada and Belgium during 2007-2010’, Archives of Virology, 156: 1379–85. [Abstract] [Google Scholar]
- Murcia P. R. et al. (2013) ‘Evolution of Equine Influenza Virus in Vaccinated Horses’, Journal of Virology, 87: 4768–71. [Europe PMC free article] [Abstract] [Google Scholar]
- Murcia P. R., and Wood J. L. N. (2011) ‘Equine Influenza Virus: A Jumping Virus that Races with Thoroughbred Horses and Greyhounds’, The Veterinary Journal, 189: 3–4. [Abstract] [Google Scholar]
- Murcia P. R., Wood J. L. N., and Holmes E. C. (2011) ‘Genome-scale Evolution and Phylodynamics of Equine H3N8 Influenza A Virus’, Journal of Virology, 85: 5312–22. [Europe PMC free article] [Abstract] [Google Scholar]
- Na W. et al. (2015) ‘Inefficient Transmissibility of NS-truncated H3N8 Equine Influenza Virus in Dogs’, Journal of Microbiology and Biotechnology, 25: 317–20. [Abstract] [Google Scholar]
- Nemoto M. et al. (2021) ‘Antigenic Differences between Equine Influenza Virus Vaccine Strains and Florida Sublineage Clade 1 Strains Isolated in Europe in 2019’, The Veterinary Journal, 272: 105674. [Abstract] [Google Scholar]
- ——— et al. (2023) ‘Antigenic Comparison of H3N8 Equine Influenza Viruses Belonging to Florida Sublineage Clade 1 between Vaccine Strains and North American Strains Isolated in 2021-2022’, Archives of Virology, 168: 94. [Abstract] [Google Scholar]
- OIE (2020) ‘OIE Expert Surveillance Panel on Equine Influenza Vaccine Composition’, Official 2020-1.
- Oladunni F. S. et al. (2021) ‘Equine Influenza Virus and Vaccines’, Viruses, 13: 1657. [Europe PMC free article] [Abstract] [Google Scholar]
- Olguin-Perglione C. et al. (2020) ‘Multifocal Outbreak of Equine Influenza in Vaccinated Horses in Argentina in 2018: Epidemiological Aspects and Molecular Characterisation of the Involved Virus Strains’, Equine Veterinary Journal, 52: 420–7. [Abstract] [Google Scholar]
- Olguin-Perglione C., and Barrandeguy M. E. (2021) ‘An Overview of Equine Influenza in South America’, Viruses, 13: 888. [Europe PMC free article] [Abstract] [Google Scholar]
- Oxburgh L., and Klingeborn B. (1999) ‘Cocirculation of Two Distinct Lineages of Equine Influenza Virus Subtype H3N8’, Journal of Clinical Microbiology, 37: 3005–9. [Europe PMC free article] [Abstract] [Google Scholar]
- Parrish C. R. et al. (1988) ‘The Global Spread and Replacement of Canine Parvovirus Strains’, Journal of General Virology, 69: 1111–6. [Abstract] [Google Scholar]
- ——— et al. (2008) ‘Cross-species Virus Transmission and the Emergence of New Epidemic Diseases’, Microbiology and Molecular Biology Reviews, 72: 457–70. [Europe PMC free article] [Abstract] [Google Scholar]
- Parrish C. R., Murcia P. R., and Holmes E. C. (2015) ‘Influenza Virus Reservoirs and Intermediate Hosts: Dogs, Horses, and New Possibilities for Influenza Virus Exposure of Humans’, Journal of Virology, 89: 2990–4. [Europe PMC free article] [Abstract] [Google Scholar]
- Pavulraj S. et al. (2021) ‘Immunogenicity of Calvenza-03 EIV/EHV® Vaccine in Horses: Comparative in Vivo Study’, Vaccines (Basel), 9: 166. [Europe PMC free article] [Abstract] [Google Scholar]
- Payungporn S. et al. (2008) ‘Influenza A Virus (H3N8) in Dogs with Respiratory Disease, Florida’, Emerging Infectious Diseases, 14: 902–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Pepin K. M. et al. (2010) ‘Identifying Genetic Markers of Adaptation for Surveillance of Viral Host Jumps’, Nature Reviews. Microbiology, 8: 802–13. [Europe PMC free article] [Abstract] [Google Scholar]
- Plowright R. K. et al. (2017) ‘Pathways to Zoonotic Spillover’, Nature Reviews. Microbiology, 15: 502–10. [Europe PMC free article] [Abstract] [Google Scholar]
- Pusterla N. et al. (2022) ‘Frequency of Detection and Prevalence Factors Associated with Common Respiratory Pathogens in Equids with Acute Onset of Fever And/or Respiratory Signs (2008–2021)’, Pathogens, 11: 759. [Europe PMC free article] [Abstract] [Google Scholar]
- Rambaut A. et al. (2018) ‘Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7’, Systematic Biology, 67: 901–4. [Europe PMC free article] [Abstract] [Google Scholar]
- ——— et al. (2016) ‘Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen)’, Virus Evolution, 2: vew007. [Europe PMC free article] [Abstract] [Google Scholar]
- Rivailler P. et al. (2010) ‘Evolution of Canine and Equine Influenza (H3N8) Viruses Co-circulating between 2005 and 2008’, Virology, 408: 71–9. [Abstract] [Google Scholar]
- Rochman N. D., Wolf Y. I., and Koonin E. V. (2022) ‘Molecular Adaptations during Viral Epidemics’, EMBO Reports, 23: e55393. [Europe PMC free article] [Abstract] [Google Scholar]
- Sack A. et al. (2019) ‘Equine Influenza Virus—A Neglected, Reemergent Disease Threat’, Emerging Infectious Diseases, 25: 1185–91. [Google Scholar]
- ——— et al. (2020) ‘Zoonotic Diseases from Horses: A Systematic Review’, Vector-Borne and Zoonotic Diseases, 20: 484–95. [Europe PMC free article] [Abstract] [Google Scholar]
- Scholtens R. G. et al. (1964) ‘U.S. Epizootic of Equine Influenza, 1963’, Public Health Reports (1896-1970), 79: 393–402. [Europe PMC free article] [Abstract] [Google Scholar]
- Sigal D., Reid J. N. S., and Wahl L. M. (2018) ‘Effects of Transmission Bottlenecks on the Diversity of Influenza A Virus’, Genetics, 210: 1075–88. [Europe PMC free article] [Abstract] [Google Scholar]
- Su Y.C.F. et al. (2015) ‘Phylodynamics of H1N1/2009 Influenza Reveals the Transition from Host Adaptation to Immune-driven Selection’, Nature Communications, 6: 7952. [Europe PMC free article] [Abstract] [Google Scholar]
- Suchard M. A. et al. (2018) ‘Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10’, Virus Evolution, 4: vey016. [Europe PMC free article] [Abstract] [Google Scholar]
- US Travel Association (2022). International Travel Spending in the United States from 2019 to 2021, with a Forecast until 2026 (In Billion U.S. Dollars) [Graph] In Statista <https://www.statista.com/statistics/207083/international-travel-spending-in-the-us-since-2002/> Accessed February 16, 2023.
- Viana R. et al. (2022) ‘Rapid Epidemic Expansion of the SARS-CoV-2 Omicron Variant in Southern Africa’, Nature, 603: 679–86. [Europe PMC free article] [Abstract] [Google Scholar]
- Voorhees I. E. H. et al. (2018) ‘Multiple Incursions and Recurrent Epidemic Fade-Out of H3N2 Canine Influenza A Virus in the United States’, Journal of Virology, 92: e00323–18. [Europe PMC free article] [Abstract] [Google Scholar]
- Waddell G. H., Teigland M. B., and Sigel M. M. (1963) ‘A New Influenza Virus Associated with Equine Respiratory Disease’, Journal of the American Veterinary Medical Association, 143: 587–90. [Abstract] [Google Scholar]
- Wasik B. R. et al. (2019a) ‘Onward Transmission of Viruses: How Do Viruses Emerge to Cause Epidemics after Spillover?’ Philosophical Transactions of the Royal Society B: Biological Sciences, 374: 20190017. [Europe PMC free article] [Abstract] [Google Scholar]
- ——— et al. (2019b) ‘Influenza Viruses in Mice: Deep Sequencing Analysis of Serial Passage and Effects of Sialic Acid Structural Variation’, Journal of Virology, 93: e01039–19. [Europe PMC free article] [Abstract] [Google Scholar]
- Weaver S. et al. (2018) ‘Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes’, Molecular Biology and Evolution, 35: 773–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Webster R. G. (1993) ‘Are Equine 1 Influenza Viruses Still Present in Horses?’, Equine Veterinary Journal, 25: 537–8. [Abstract] [Google Scholar]
- Westgeest K. B. et al. (2014) ‘Genomewide Analysis of Reassortment and Evolution of Human Influenza A(H3N2) Viruses Circulating between 1968 and 2011’, Journal of Virology, 88: 2844–57. [Europe PMC free article] [Abstract] [Google Scholar]
- WOAF (2022) ‘Expert surveillance panel on equine influenza vaccine composition’, Paris: World Organization for Animal Health. <https://www.woah.org/en/home/>. [Google Scholar]
- Worobey M. et al. (2020) ‘The Emergence of SARS-CoV-2 in Europe and North America’, Science, 370: 564–70. [Europe PMC free article] [Abstract] [Google Scholar]
- Yoon S.-W., Webby R. J., and Webster R. G. (2014) ‘Evolution and Ecology of Influenza A Viruses’, Current Topics in Microbiology and Immunology, 385: 359–75. [Abstract] [Google Scholar]
- Zhu H. et al. (2019) ‘Absence of Adaptive Evolution is the Main Barrier against Influenza Emergence in Horses in Asia despite Frequent Virus Interspecies Transmission from Wild Birds’, PLOS Pathogens, 15: e1007531. [Europe PMC free article] [Abstract] [Google Scholar]
- Ziegler T., Mamahit A., and Cox N. J. (2018) ‘65 Years of Influenza Surveillance by a World Health Organization-coordinated Global Network’, Influenza and Other Respiratory Viruses, 12: 558–65. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Virus Evolution are provided here courtesy of Oxford University Press
Citations & impact
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/153990305
Article citations
Upper Respiratory Tract Disease in a Dog Infected by a Highly Pathogenic Avian A/H5N1 Virus.
Microorganisms, 12(4):689, 29 Mar 2024
Cited by: 4 articles | PMID: 38674633 | PMCID: PMC11051868
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Host-range shift of H3N8 canine influenza virus: a phylodynamic analysis of its origin and adaptation from equine to canine host.
Vet Res, 50(1):87, 30 Oct 2019
Cited by: 6 articles | PMID: 31666126 | PMCID: PMC6822366
Equine Influenza Virus in Asia: Phylogeographic Pattern and Molecular Features Reveal Circulation of an Autochthonous Lineage.
J Virol, 93(13):e00116-19, 14 Jun 2019
Cited by: 13 articles | PMID: 31019053 | PMCID: PMC6580976
Equine and Canine Influenza H3N8 Viruses Show Minimal Biological Differences Despite Phylogenetic Divergence.
J Virol, 89(13):6860-6873, 22 Apr 2015
Cited by: 28 articles | PMID: 25903329 | PMCID: PMC4468500
Equine Influenza Virus and Vaccines.
Viruses, 13(8):1657, 20 Aug 2021
Cited by: 22 articles | PMID: 34452521 | PMCID: PMC8402878
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (3)
Grant ID: BB/V002821/1 BB/V004697/1
Grant ID: BB/V002821/1
Grant ID: BB/V004697/1
Horserace Betting Levy Board (1)
Grant ID: Grants 797
Kentucky Agricultural Experiment Station (1)
Grant ID: KY-014067
Medical Research Council (1)
Respiratory infections
Professor Pablo Murcia, University of Glasgow
Grant ID: MC_UU_12014/9
NIGMS NIH HHS (1)
Grant ID: R01 GM080533
National Institute of General Medical Sciences (1)
Grant ID: R01-GM080533




