Abstract
Free full text

Bacteriophage-driven emergence and expansion of Staphylococcus aureus in rodent populations
Abstract
Human activities such as agriculturalization and domestication have led to the emergence of many new pathogens via host-switching events between humans, domesticated and wild animals. Staphylococcus aureus is a multi-host opportunistic pathogen with a global healthcare and economic burden. Recently, it was discovered that laboratory and wild rodents can be colonised and infected with S. aureus, but the origins and zoonotic potential of rodent S. aureus is unknown. In order to trace their evolutionary history, we employed a dataset of 1249 S. aureus genome sequences including 393 of isolates from rodents and other small mammals (including newly determined sequences for 305 isolates from 7 countries). Among laboratory mouse populations, we identified multiple widespread rodent-specific S. aureus clones that likely originated in humans. Phylogeographic analysis of the most common murine lineage CC88 suggests that it emerged in the 1980s in laboratory mouse facilities most likely in North America, from where it spread to institutions around the world, via the distribution of mice for research. In contrast, wild rodents (mice, voles, squirrels) were colonized with a unique complement of S. aureus lineages that are widely disseminated across Europe. In order to investigate the molecular basis for S. aureus adaptation to rodent hosts, genome-wide association analysis was carried out revealing a unique complement of bacteriophages associated with a rodent host ecology. Of note, we identified novel prophages and pathogenicity islands in rodent-derived S. aureus that conferred the potential for coagulation of rodent plasma, a key phenotype of abscess formation and persistence. Our findings highlight the remarkable capacity of S. aureus to expand into new host populations, driven by the acquisition of genes promoting survival in new host-species.
Author summary
New pathogens typically emerge after host-switching events, threatening public health and food security. Here we used an evolutionary genomic approach to reveal the origins of Staphylococcus aureus in laboratory and wild rodent populations. S. aureus in laboratory rodents appears to have originated in humans and following host jump events has spread around the world via the distribution of mice for research purposes. Distinct clones of S. aureus widely distributed around Europe are associated with wild rodent populations. We found unique types of prophages in rodent S. aureus that have played a central role in their emergence and expansion including some with genes encoding putative virulence factors. In particular, we identified novel prophages and pathogenicity islands that conferred the potential for coagulation of rodent plasma, a key phenotype of abscess formation and persistence. Taken together, our findings highlight the remarkable capacity of S. aureus to expand into new host populations, driven by the acquisition of genes acquired via mobile genetic elements.
Introduction
Domestication and human activities in general have had a profound impact on the emergence of new pathogens [1]. However, our understanding of the drivers for successful host-switching events and subsequent expansion and dissemination is very limited. Staphylococcus aureus is a major human and animal pathogen with a remarkable ability to colonise and infect a broad range of animal hosts including humans, livestock and wildlife [2,3]. The multi-host associations of S. aureus are related to its capacity to adapt to different ecological niches through an array of evolutionary genetic and functional mechanisms, including acquisition of mobile genetic elements (MGE) [4–8], allelic diversification of specific effectors [9,10], recombination [11] or even single point mutations [12]. Recent studies have revealed the existence of high-risk animal-associated clones of S. aureus that may represent an increased public health threat [13].
Laboratory mice are the most commonly employed species for in vivo modelling of S. aureus pathogenesis or for testing the efficacy of new vaccine candidates [14,15]. Typically, mice are challenged with human-adapted S. aureus isolates, such as USA300. However, it has been recently demonstrated that laboratory mice are natural S. aureus hosts, and that even specific-pathogen-free (SPF) mice are frequently colonized with S. aureus [16]. Natural colonization with S. aureus primes the adaptive immune system and thus likely impacts on the outcome of experimental infections or challenge experiments [16].
Multi locus sequence typing (MLST)-based genotyping of S. aureus isolated from laboratory mice in different countries has identified three main clonal complexes (CC) accounting for the majority of isolates: CC88, CC15 and CC1 [17]. While CC1 and CC15 are commonly associated with human infections, CC88 is rarely found in human populations in Western countries [18] and its origin remains unknown. Rodents in urban or human-influenced environments are typically colonised by S. aureus strains also found in humans and other animals such as CC8, CC398, and CC97, and frequently encode methicillin resistance [19–23]. Similarly, companion rodents such as guinea pigs, and companion rabbits are typically colonized by strains commonly associated with humans [24,25]. In contrast, wild rodents including mice, rats and squirrels are typically colonized with unique S. aureus lineages, including CC49, CC1956 and CC890 [26–28]. In particular, S. aureus CC49 (only sporadically found in humans) has been isolated from different species of wild mice in Germany [26], wild rats in Belgium [27] and red squirrels in the UK [29,30]. The existence of CCs that are unique to wild rodents suggests host-specialization but the specific mechanisms that underpin this tropism are largely unknown.
Rodents are a major reservoir for bacterial zoonotic infections of humans [31]. It is imperative that we understand the origins of newly identified animal pathogens, to investigate the drivers of successful expansion into distinct host populations and to assess the public health threat. Here, we combine population genomic, phylodynamic and genome-wide association approaches along with functional analyses to trace the evolutionary history of rodent populations of S. aureus and to investigate the genetic and functional basis for host-adaptation. Our data reveal the time-frame for the emergence and international dissemination of the major rodent clones of S. aureus, and identify a novel family of bacteriophages that are associated with their ecological success.
Methods
Sequence dataset
Whole genome sequencing (WGS) of 340 previously described S. aureus isolates [17,26] was performed by MicrobesNG (Birmingham, UK) using the Nextera XT library prep protocol on a MiSeq platform (Illumina, San Diego, CA, USA). Sequences were deposited in SRA/ENA under the BioProject PRJEB41130. These isolates included i) 241 laboratory mice and 15 laboratory rat isolates from the US, Germany, Canada, France, New Zealand, the UK and Japan; ii) 43 wild mouse and vole isolates from Germany and the Czech Republic, iii) 5 shrew isolates from Germany; iv) 1 guinea pig isolate from Germany; and v) 35 human isolates belonging to CCs found among the rodent isolates (ST88, ST49, ST130 and ST8). No live participants were used in the current study. S. aureus isolates from laboratory mice were originally obtained as part of routine health surveillance, and occasionally from euthanized mice suffering from S. aureus infections (e.g. preputial gland adenitis). To avoid sample bias, we included a maximum of 4 isolates of the same spa type, barrier, mouse strain and time point in the study cohort.
To complement this dataset, S. aureus RefSeq genomes and short-read data deposited in SRA (S1 Table) were queried for sequences from rodents (mice, rats, squirrels, guinea pigs and others) (n = 64) and lagomorphs (rabbits, hares) (n = 25).
These genomes are listed in S2 Table with their available metadata. Short read sequences were adapter trimmed using Trimmomatic v0.36 [32] and de novo assembled using SPAdes v3.11.1 [33]. Genes were annotated using Prokka v1.13 [34]. All assemblies were typed using mlst v2.10 (https://github.com/tseemann/mlst) and sequence types (STs) were grouped into CCs using the eBURST algorithm in PhyloViz [35].
Phylogenetic and phylodynamic analyses
In order to examine the phylogenetic relationships between the rodent isolates, we constructed a core genome phylogenetic tree using as background sequences a dataset of 798 S. aureus genomes representative of the global genetic diversity [5] using Parsnp v1.1.2 [36].
Monophyletic, rodent-specific clades were identified, and CCs with more than one rodent clade were selected for further phylodynamic analysis. Additional sequences belonging to the STs contained in each of these CCs of interest were obtained from SRA queried using Staphopia [37]. These were downloaded and assembled as explained above. CC-specific core genome alignments were created using snippy and snippy-core v4.1 (https://github.com/tseemann/snippy) using as reference for each dataset a closed genome belonging to that CC when available. Recombinant regions in those core genome alignments as detected by Gubbins v2.3.4 [38] were discarded. An initial tree of the recombinant-free single nucleotide polymorphism (SNP) genome alignment was constructed using IQTree v1.6.12 [39] with the GTR+G4 model including an ascertainment bias correction with the number of invariable sites from the core genome alignment, and 1,000 rapid bootstraps. These trees were used to test the temporal signal of the dataset in TempEst v1.5.3 [40].
Bayesian phylogenetic analyses were performed using BEAST v1.9.0 [41], testing different models for nucleotide substitution (HKY and GTR), molecular clock (strict and uncorrelated lognormal relaxed) and demographic growth (constant, exponential and Bayesian skygrid). Each model combination was run for 100 million generations, with sampling every 10,000 and discarding the initial 10% as burn-in. Runs were compared via a marginal likelihood estimation (MLE) using path sampling and stepping stone sampling methods implemented in BEAST. The posterior distribution of trees was summarised into a maximum clade credibility tree. For the largest, CC88-derived dataset a phylogeographic analysis was performed using the Bayesian stochastic search variable selection (BSSVS) model to reconstruct the geographical locations of the ancestral states [42] contained in a sub-sample of the posterior tree distribution of the BEAST runs described above. SpreaD3 v0.9.6 [43] was used to identify statistically well-supported migration routes using the Bayes Factor (BF).
Genome-wide association analysis (GWAS)
We performed a GWAS analysis in order to find genetic determinants associated to rodent isolates compared to a collection of S. aureus whole genome sequences from other hosts. For the latter, we downloaded all RefSeq S. aureus genomes with known host (mostly human, bovine, swine and avian) as well as a collection of S. aureus genomes representative of the worldwide genetic diversity [5]. The sequence dataset for the GWAS included all rodent sequences (n = 357) with the 50 genetically closest (in terms of mash distance [44]) non-rodent sequences to each of them. Identical sequences were removed, and, in order to avoid over-sampling of the CC88 murine cluster, this lineage was further down-sized. This produced a final dataset of 832 sequences (241 rodent and 591 non-rodent controls).
We performed the GWAS analysis using pyseer v1.1.1 [45] from 3 inputs: k-mers, core SNPs and gene presence/absence. For the k-mer-based GWAS analysis, 31 base pair (bp)-long k-mers were generated from the 832 assemblies using dsk [46], and summarised using the pyseer script combineKmers. A variant calling file (VCF) containing core SNPs was inferred using snippy and snippy-core v4.1 (https://github.com/tseemann/snippy) with the strain RF122 (GCA_000009005.1) as reference. The gene presence/absence matrix was generated using roary v3.12.0 [47]. Different runs of pyseer applied two methods for population structure correction (mash and phylogenetic distances) and three association models: fixed effects (SEER), mixed effects (FaST-LMM) and lineage effects (bugwas) (S1 Data). In all cases we applied a filter p-value of 1E-8, a minimum allele frequency of 0.01 and a maximum allele frequency of 0.95.
An initial threshold for the p-value adjusted for population structure in order to consider statistical significance of the k-mers (p < 1.43E-07) was calculated using the script count_patterns.py included in the pyseer pipeline. Significant k-mers were mapped against a collection of full genome S. aureus sequences from a wide variety of clonal complexes in order to identify the gene each k-mer originated from. Genes containing significant k-mers which belonged to the same cluster of orthologous genes (COG), according to roary, were grouped. After examination of the gene areas containing the k-mers, only those which involved non-synonymous substitutions were further considered. Finally, only k-mers found in more than one major rodent lineage were analysed, a step taken in order to avoid overrepresentation of the k-mers enriched in the CC88 lineage only. The function of the proteins encoded by these selected genes was predicted using eggnog [48].
Prophage identification
Prophage sequences in genome assemblies were identified using PHASTER [49] and prophage-associated regions were extracted and annotated using prokka v1.13 [34]. In order to establish the genetic relationships between the putative prophages, we calculated mash distances [44] between prophage sequences and used them to build a cladogram applying a hierarchical clustering method (function hclust) using R [50]. This cladogram was used to explore the presence and distribution of shared prophage sequences across the study population. The identity of these prophages was confirmed by genome-to-genome comparisons using Artemis Comparison Tool (ACT, [51]).
Prothrombin sequence comparison
Prothrombin protein references were obtained from GenBank for common shrew (XP_004619129.1), rat (NP_075213.2), human (NP_000497.1) and laboratory mouse (NP_034298.1). As no references were readily available for bank vole, common vole and yellow-necked field mouse, we used publicly available raw sequence reads stored in the Sequence Read Archive (SRA) in order to determine the prothrombin mRNA sequences of these species. In detail, we downloaded the SRA datasets SRR1010821 (bank vole), SRR1325018 (common vole), and SRR9990624 (yellow-necked field mouse) using parallel-fastq-dump and trimmed the raw reads using Trimgalore v0.6.10 (https://github.com/FelixKrueger/TrimGalore) running in automatic adapter detection mode. The trimmed reads were assembled using rnaSPAdes v3.15.5 [52] and contigs matching prothrombin were selected using diamond blastx v2.1.6 [53].
Cloning, recombinant protein purification, and coagulation assays
The capacity of several coagulase-like proteins encoded by rodent S. aureus isolates to coagulate plasma from different hosts was tested in vitro. Specifically, nine different coagulase-like proteins were selected for cloning of the coding sequences and protein production: core genome-encoded coagulase (Coa) and van Willebrandt factor-binding protein (vWbp) from three strains each (laboratory mouse ST88, bank vole ST49, and human ST8), phage-encoded coagulase-like (Coa’) from two strains each (common vole ST3252 and yellow-necked field mouse ST890), and one SaPI-encoded vWbp (SaPI-vWbp) present only in ST3033 strains from wild common shrews. The six S. aureus strains were grown overnight in TSB and genomic DNA was isolated with the DNeasy Blood & Tissue Kit (Qiagen, Venlo, The Netherlands) according to manufacturer’s instructions, except for the addition of lysostaphin during lysis of the cells. The corresponding coagulase-like genes, omitting the information for the signal peptide, were amplified via PCR and purified with the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Düren, Germany). DNA was inserted into the plasmid pET21b using the NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA, USA) according to manufacturer’s instructions, and transformed into chemically competent Escherichia coli DH5α. The amplified plasmid was isolated using the High Pure Plasmid Isolation Kit (Sigma-Aldrich, Taufenkirchen, Germany) and transformed into chemically competent E. coli BL21 (DE3) cells. For protein expression LB broth containing 100 μg/mL ampicillin was inoculated from a preculture to an optical density (OD) of 0.05 and incubated at 37°C with agitation. Afterwards, protein expression was induced by adding Isopropyl-β-D-thiogalactopyranosid (IPTG) to a final concentration of 1 mmol/L. After 3h of additional incubation at 37°C with agitation cells were harvested and lysed with a sonicator. The His-tagged proteins were purified using a HisTrap HP affinity column on an ÄKTA start protein purification system according to manufacturer’s instructions (GE Healthcare, Chicago, IL, USA). The imidazole-containing elution buffer was changed to PBS using Amicon Ultra Centrifugal Filters (Merck Millipore, Burlington, MA, US) and the protein concentration was determined with a Bradford assay.
To determine the coagulation properties of these proteins, heparinized plasma was isolated from humans, lab rats, lab mice (C57BL/6), and bank voles. Heparinized lab rat plasma (Spraque-Dawley, Genetex, Irvine, CA, USA) and additional heparinized lab mouse plasma (non-Swiss albino, Equitech-Bio, Kerrville, TX, USA) were obtained from a commercial provider. 376 nmol of protein in a volume of 25 μL PBS was added to 300 μL plasma in a 10 mL glass tube and incubated at 37°C without agitation. The coagulation was examined visually at 0.5, 1, 2, 4 and 20 h using a modified coagulation score [54]: 0 = no coagulation; 1 = small clot/flakes; 2 = medium-sized clot; 3 = large clot; 4 = complete coagulation (coagulum sticks to the inverted tube). The scoring was performed in a blinded fashion.
Results
Rodent S. aureus isolates represent an array of lineages spanning the species diversity
In order to examine the genetic diversity of the rodent S. aureus population and their evolutionary origins, we employed a dataset of 1,249 S. aureus sequences from the following host species: humans (n = 571), laboratory mice (n = 245), cattle (n = 108), wild voles and mice (n = 43), birds (n = 44), goats (n = 25), lagomorphs (rabbits, hares) (n = 25), pigs (n = 20), laboratory rats (n = 15), sheep (n = 12), wild rats (n = 12), wild red squirrels (n = 13), shrews (n = 5), and others (n = 111)(S1 Table). In total, 245 isolates from laboratory mice belonged to 16 different CCs, predominantly CC88 (127; 51.8%), CC15 (37; 15.1%), CC1 (30; 12.2%) and CC5 (11; 4.5%). The 15 isolates from laboratory rats belonged to CC7 (5; 33.3%), CC20 (3; 20.0%), CC88 (2; 13.3%), CC1 (2; 13.3%), a novel ST (2; 13.3%) and CC8 (1; 6.7%). In contrast, isolates from wild voles and mice (n = 43), wild rats (n = 12) and wild red squirrels (n = 13), belonged to CC49 (29; 43.9%), CC1956 (15; 22.7%), CC890 (9; 13.6%), CC398 (7; 10.6%), CC8 (3; 4.5%), CC130 (2; 3.0%), and CC80 (1; 1.5%), whereas five isolates from wild common shrews sampled in the same geographic region belonged to a single CC (CC3033) consistent with host-specificity (S2 Table). S. aureus from 28 urban rats and 7 zoo rodents (guinea pigs, maras and capybaras) belonged to STs typically associated with humans or livestock, including CC398 (12; 34.3%), CC8 (10; 28.6%) and CC97 (4; 11.4%), among others. Maximum-likelihood phylogenetic analysis of all 1,249 S. aureus sequences revealed rodent isolates to be interspersed among human and other animal lineages across the tree, consistent with numerous host-switching or spill-over events into rodents during the recent evolutionary history of S. aureus (Fig 1). Of note, some rodent-associated lineages represent monophyletic, host-specific clades (Fig 1), which are examined in more detail in the following sections.
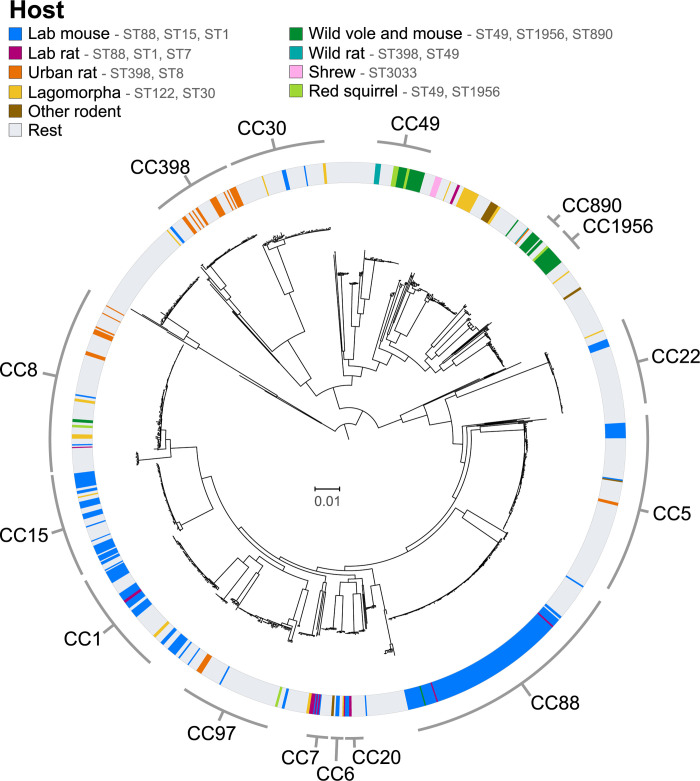
Maximum likelihood tree of 398 S. aureus genomes from rodents and other small mammals together with 851 background sequences from other hosts, mostly humans, representative of the worldwide S. aureus genetic diversity. The main clonal complexes among the study population are highlighted.
CC88 emerged in laboratory mice after a single host switch followed by wide international dissemination
S. aureus CC88 contained the largest rodent-specific phylogenetic cluster observed in our Maximum-Likelihood phylogenetic tree (n = 127), including 124 ST88 isolates and 3 single-locus variants of ST88 (Fig 1). Of the isolates in this clade, 125 were obtained from laboratory mice and 2 from laboratory rats, in 13 different facilities across 6 countries in 4 continents: Canada, France, Germany, Japan, New Zealand and the USA. A CC88-specific Bayesian phylogenetic analysis (Fig 2) included all publicly available sequences belonging to this CC with sampling date and location (n = 209), and allowed us to estimate that the most recent common ancestor (MRCA) of CC88 existed in 1972 (Bayesian Confidence Interval (BCI): 1966.0–1977.5). In addition, phylogeographic reconstruction inferred its most likely origin to have been in South East Asia (Thailand/Cambodia) with a root posterior probability (PP) of 0.32. The CC88 evolutionary rate was determined to be 2.3 (1.92–2.64) × 10−6 substitutions/site/year. The phylogeny indicates that the murine-associated CC88 clade likely resulted from a single introduction of S. aureus into laboratory mice which happened around 1984 (1977.4–1989.4) on the East Coast of North America (Fig 2). The subsequent international spread of murine S. aureus was driven by the distribution of colonized laboratory mice from a limited number of commercial vendors to research institutes with their own breeding facility around the globe. Although the oldest nodes in the murine cluster exhibited a degree of uncertainty (see S1 Fig), the Charles River (CR) facility in Saint-Constant (Canada) was determined to be a possible origin of the lineage (PP = 0.86), after which migration to CR Raleigh (NC, USA) preceded the wider spread to CR facilities and other public and private institutions in North America up to at least 2004 (Fig 2). This is supported by the fact that CR Raleigh isolates exhibited the highest genetic diversity and were distributed in groups across the entire murine CC88 lineage. Isolates from CR Kingston (NY, USA) were largely clustered in a single group, suggesting one or few transmissions from CR Raleigh to CR Kingston (between 1990 and 1995). Murine CC88 were also introduced into three research institute-associated breeding facilities in Germany and the phylogeny suggests that these migrations were closely connected, possibly because CC88-colonized animals were purchased from the same supplier. Moreover, murine CC88 migrated to a university-associated breeding facility in New Zealand in two events between 1990 and 1995, suggestive of the purchase of breeding pairs from different vendors. Most of these migrations involved S. aureus strains that were closely related to strains obtained from USA-based CR customers (vendors, pharmaceutical companies and/or universities), suggesting that CC88-colonized animals originated from the same supplier or were shared between facilities (Fig 2).
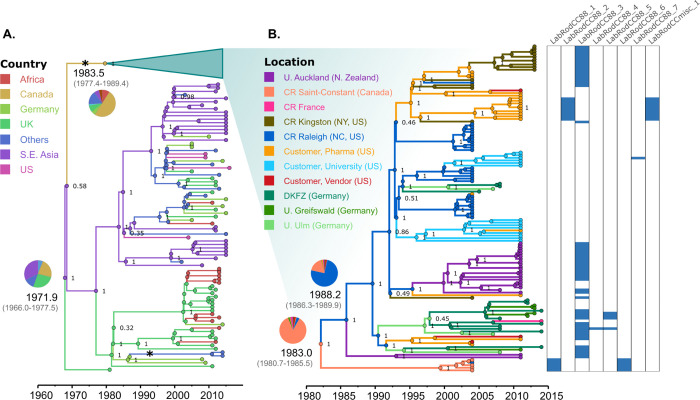
Bayesian phylogenetic tree of the S. aureus CC88 lineage. Branches and node circles are coloured according to the reconstructed location, and the numbers in key nodes represent statistical support. A) Tree of the full lineage, with the main, international murine cluster collapsed. Asterisks represent host jumps into laboratory mice and rat populations. For the root node and the common node to the murine cluster, the reconstructed dates and the probabilities of each possible location are shown. B) Tree of the international murine cluster, with further locations depending on the sampled institution. The columns at the right-hand side indicate the distribution of the prophages found across laboratory isolates. Abbreviations: CR, Charles River, DKFZ, Deutsches Krebsforschungszentrum, U., University.
Of note, we found a second CC88 introduction into the murine population (Fig 2), based on isolates from two French laboratory mice that formed a phylogenetic pair (emerged in 2012 (2010.1–2013.1)) clustered within the human CC88 clade that was independent from the international murine CC88 cluster (Fig 2).
Identification of multiple, independent introductions of S. aureus into laboratory mice and rats from humans with limited onward dissemination
CC1 and CC15 each contained five rodent-specific phylogenetic clusters representing multiple host jumps from humans and expansions into rodent laboratory populations. Phylodynamic analyses (S2 Fig) of these lineages revealed that the oldest of these clusters originated around the year 1980 (1979.2 (1976.3–1982.1) and 1983 (1977.0–1987.8) for the oldest CC15 and CC1 clusters, respectively), in agreement with the reconstructed date of emergence of the international murine CC88 cluster (Fig 2). The oldest CC15 cluster included samples from US, Canada and the UK. Subsequently, other CC15 and CC1 clusters emerged during the 1990s including two that also involved samples from different countries (Germany and France in both cases). In addition, singleton monophyletic clusters were detected for many other S. aureus lineages (CC5, CC22, CC7, CC188, CC20 and CC6), consistent with frequent spillovers into the laboratory mouse and rat population with limited further expansion.
Most S. aureus from wild rodents in Europe belong to a single lineage CC49
Phylodynamic analysis of CC49 indicated an origin of approximately 1840 with an early split into a clade comprised of swine isolates in Switzerland and a second clade comprised of rodent or human isolates in several European countries (Fig 3). The latter clade originated in 1924 (1838.6–1939.5) and comprised of subclades that are largely segregated according to host-species group and geography (Fig 3). For example, CC49 sequences from wild rodents segregated according to sampling location (North East Germany, and South West Germany) interspersed with yellow-necked field mouse and bank vole isolates (Fig 3). S. aureus isolates from red squirrels in the UK Channel Islands were basal to the wild German rodent isolates in the phylogeny (Fig 3). Wild rat isolates from Belgium formed a deep-branching cluster indicating a distinct evolutionary history to S. aureus in wild mice and voles (Fig 3). The remaining S. aureus lineages associated with wild voles and mice (CC890, CC1956) and wild shrews (CC3033) were highly host-specific with limited clonal genetic diversity (Fig 1). Overall, these data indicate distinct evolutionary histories for S. aureus associated with wild and domesticated rodents, respectively.
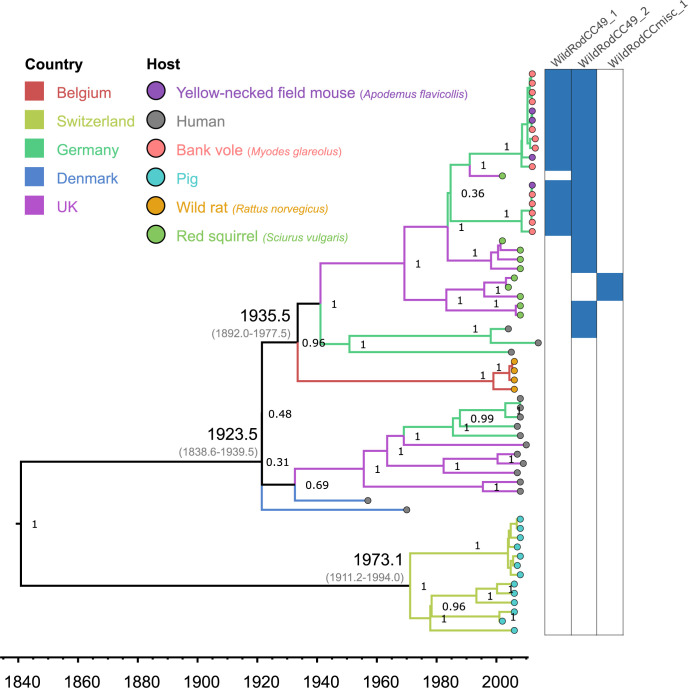
Bayesian phylogenetic tree of the S. aureus CC49 lineage. Branch colours show sampling country, and the circles at the tips indicate the bacterial host. The columns at the right-hand side indicate the distribution of three prophages found across wild rodent isolates. The dates for most recent common ancestors of key nodes are shown.
Genome-wide association analysis identifies prophage genes as key markers of rodent-host-specificity
The identification of S. aureus lineages that have evolved the capacity to colonise and spread in rodent populations prompted us to explore the genetic basis for rodent host-adaptation. Accordingly, we carried out a kmer-based GWAS analysis using pyseer to identify genetic determinants (including whole genes or gene mutations) of S. aureus associated with rodent hosts (see Methods section). A total of 41 unique Clusters of Orthologous Groups of proteins (COGs) contained k-mers significantly enriched in rodent S. aureus sequences. Of these, the majority (n = 26) were located on phage sequences, including 13 representing enriched whole genes, and 13 representing non-synonymous mutations of phage genes. Of note, phage genes accounted for the 18 most significant hits, indicating that specific prophage sequences were the best predictors of rodent host-association, and likely play a key role in host adaptation. The 15 GWAS hits found in the chromosome included non-synonymous mutations in genes encoding putative amino acid synthases, transmembrane transporters, 2 superantigen-like proteins (SSL4 and SSL7), IgG-binding protein, a nuclease, a nucleokinase and a helicase (S3 Table). Finally, a whole gene encoding a putative restriction enzyme belonging to a type II restriction modification system was enriched in the CC890, CC1956 and CC30 clades. Overall, the relatively small number of GWAS hits out-with MGE indicates that convergent adaptations of different S. aureus lineages to the rodent host were rare, implying an important role of lineage-specific traits of host-adaptation, as highlighted previously for bovine S. aureus [13].
Rodent S. aureus isolates contain a diverse complement of novel prophages
The identification of phage genes as markers of rodent S. aureus led us to examine in detail the diversity and distribution of phages of the family Siphoviridae among the rodent S. aureus population. A hierarchical clustering tree constructed from pairwise k-mer-based genetic distances of the prophages identified by PHASTER (S3 Fig) was used to identify a total of 52 prophage lineages that were present in at least 2 rodent S. aureus genomes (see S4 Table for a summary of the features of these 52 phage lineages). Fig 4 depicts the genome maps of the most frequently identified (identified in ≥10 isolates) prophages, and those exhibiting the strongest rodent-specific genetic signatures (see below). S. aureus from wild mice and voles contained on average more prophages than isolates from laboratory mice (mean = 2.2 vs 0.6 prophages/genome, respectively; p<0.01). Most (n = 33) prophages belonged to a single CC, associated with a single host-species. However, the remaining 19 prophages were identified in S. aureus genomes across lineages and/or host-species. A total of 6 different prophages were shared between S. aureus isolates from mice and rats (S4 Table). Of note, prophages were not shared between laboratory and wild mice, consistent with the ecological separation of murine S. aureus populations in laboratory and wild environments. In contrast, we identified prophages that were shared among isolates from laboratory mice and rats, and among urban rats and companion animals, revealing a distinct pool of MGE in human-associated environments.
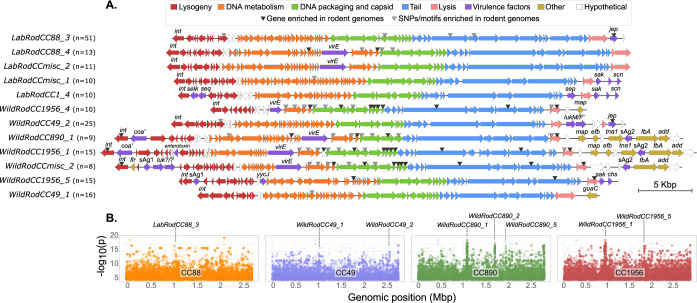
A) Genome maps of the most frequent prophages and those with the highest number of rodent-specific genetic signatures. Genes are coloured according to their function (see legend at the top). Triangles highlight phage genes harbouring rodent-specific genetic signatures (i.e. GWAS hits). Virulence factors and other genes are labelled. B) Manhattan plots showing the GWAS hits across representative rodent S. aureus genomes belonging to the main four lineages found: CC88, CC49, CC890 and CC1956. Peaks with the highest statistical significance correspond to strain-dependent phages as indicated. add: adenosine deaminase; chs: chemotaxis-inhibiting protein (CHIPS); coa’: putative coagulase-like protein; efb: extracellular fibrinogen-binding protein; flr: formyl peptide receptor (FPR)-like 1 inhibitory protein; fba: putative Fg-binding protein; guaC: GMP reductase; int: integrase; jep: JSNZ extracellular protease; luk?/?: undetermined leukocidin; lukM/F’: leukocidin MF’; map: major histocompatibility complex class II analog protein; sAg1: putative superantigen 1; sAg2: putative superantigen 2; sak: staphylokinase; scn: staphylococcal complement inhibitor (SCIN); selk: staphylococcal enterotoxin-like protein K; sep: staphylococcal enterotoxin P; seq: staphylococcal enterotoxin Q; tna1: tryptophanase; virE: virulence-associated protein E; yycJ: Metallo-beta-lactamase family protein.
Among the prophages, we detected 13 different integrase types (the most common being Sa3int (n = 14)), including 5 novel variants in 10 prophage lineages (8 in wild rodents and 2 in laboratory mice), which where phylogenetically distinct from previously described integrase types [55] (S4 Fig), and which exhibited novel chromosomal integration sites. BLAST analysis of the 52 full prophage sequences revealed that 28 phages were novel, i.e., did not have a match in the GenBank sequence database with ≥90% coverage and ≥90% nucleotide sequence identity. All but one (16 of 17, 94.1%) of those identified in wild rodents were novel, whereas only 12 of 35 (34.3%) phages identified in laboratory rodents were novel, highlighting the unique nature of phages associated with wild rodent isolates. Importantly, 32 of the 52 phage lineages contained at least one of the rodent-associated hits identified by GWAS (Fig 4). Most hits were associated with modules for DNA replication, DNA packaging and head morphogenesis and tail. However, a gene encoding a novel serine protease, named jep (JSNZ extracellular protease), was identified in 4 different phages in both laboratory and wild rodents, and in 12.4% of the rodent genomes (and only 0.5% of the non-murine S. aureus genomes). Taken together, we have identified a unique complement of phages associated with rodent S. aureus with distinct pools of phages circulating in wild and laboratory populations, respectively.
Rodent-specific phages encode novel determinants of host-specificity
An array of novel factors predicted to be involved in host-pathogen interactions were encoded among rodent S. aureus phages. Namely, genes for a putative serine protease, superantigens, superantigen-like proteins and coagulase-like proteins were distributed among phages in distinct S. aureus CCs (Figs Figs2B,2B, ,33 and and44). In particular, the jep gene encoding a novel protease is located on a prophage (LabRodCC88_3) distributed across multiple subclades of the globally distributed CC88 laboratory mouse cluster (Fig 2B). Furthermore, the WildRodCC49_2 and WildRodCCmisc_1 phages, which contain their own allelic variant of the jep gene, were identified in all isolates within the wild rodent CC49 lineage (Fig 3) suggesting a role in the adaptation of CC49 to wild rodents.
Another determinant of host-specificity is the pore-forming toxin LukMF’, a member of the family of bicomponent leucocidins that includes the Panton-Valentine leukocidin (PVL) [56]. lukMF’ has recently been implicated in contributing to fulminant skin and soft tissue infections in wild squirrels [30]. LukMF’ was encoded by the phage WildRodCC49_2, common to wild mice (n = 17) and squirrel (n = 5) isolates (S2 Table).
Moreover, phages containing genes encoding allelic variants (72% aa identity) of a putative coagulase-like protein (coa’) were present in all CC890 (WildRodCC890_1, in wild mice) and CC1956 (WildRodCC1956_1, in wild mice and red squirrels) isolates. Unusually for accessory genes, the coagulase-like protein genes were located adjacent to the phage integrase gene (Fig 4). Previous studies have demonstrated that S. aureus produces several secreted proteins with plasma coagulase activity [57]. In particular, S. aureus produces coagulase (Coa) and von Willebrand factor-binding protein (vWbp) which can activate host prothrombin and form fibrin clots, thereby promoting abscess formation [57,58]. The identification of a large array of novel putative coagulase proteins encoded by rodent S. aureus is striking and suggests an important role in host adaptation (S5 Table). Accordingly, we aimed to test the ability of the encoded proteins to coagulate plasma from humans and rodents in vitro. We generated recombinant proteins of the core genome-encoded coagulase (Coa) and vWbp from human-derived ST8, laboratory mouse-derived ST88, and wild rodent-derived ST49 strains, as well as phage-encoded coagulase-like proteins (Coa’) from wild rodent-derived ST3252 and ST890 strains, and one SaPI-encoded vWbp-like protein (vWbp’) identified only in ST3033 strains from wild shrews. While Coa encoded by ST8 and ST88 were closely related (90.4% aa identity), ST49 Coa and the phage-encoded Coa’ variants differed markedly from the Coa ST8 (68.8, 29.3 and 40.1% identity for Coa ST49, Coa’ ST3252 and Coa’ ST890, respectively) (S5 Fig). Similary, vWbp sequences from rodent and shrew-derived S. aureus isolates differed markedly from the vWbp sequence of the human-adapted ST8 lineage (S5 Fig). For both, Coa and vWbp, the greatest sequence variation was observed in the domains required for prothrombin-binding (D1 and D2). All nine recombinant proteins were tested for coagulation activity on plasma obtained from human, laboratory mouse, laboratory rat and bank voles (Fig 5).
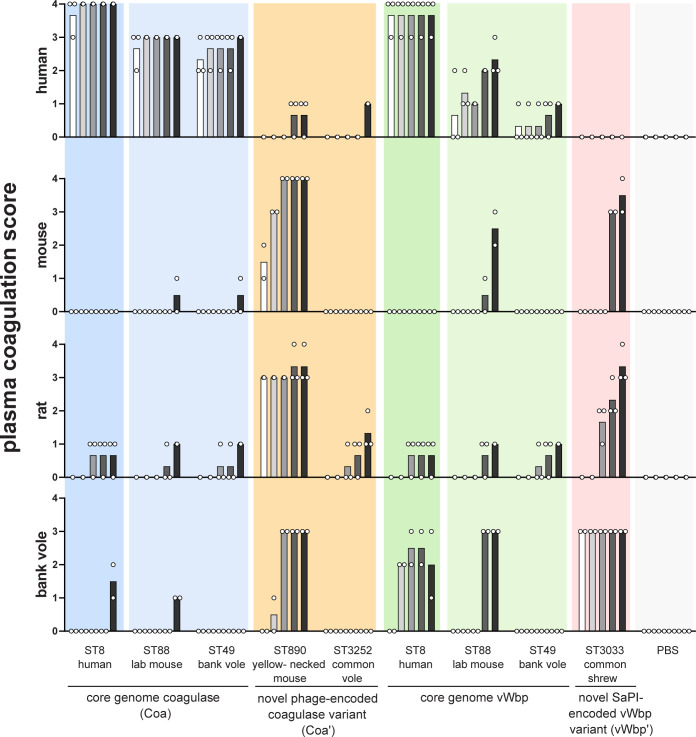
Coagulation of human, rat, mouse and bank vole plasma was induced by different recombinant S. aureus coagulase-like proteins from human, lab mouse, bank vole (Myodes glareolus), yellow-necked field mouse (Apodemus flavicollis), common vole (Microtus arvalis), and common shrew (Sorex araneus). Heparinized plasma from different species was mixed with S. aureus coagulase-like proteins and coagulation was visually assessed after 0.5, 1, 2, 4 and 20 h (indicated by individual bars with increasing grey scale). For the experiments in human plasma, plasma from 3 different donors was tested. For the experiments in animal plasma, plasma pools were used and independently tested two (mouse and bank vole) or three (rat) times. The bars show the respective mean value.
Overall, all proteins tested coagulated plasma from at least one host-species. Notably, coagulation activity was highly species-dependent, suggesting a role for coagulases in host adaptation (Fig 5). The core genome-encoded Coa from human ST8, murine ST88 and ST49 strongly coagulated human plasma, but exhibited no or neglectable activity on mouse, rat and bank vole plasma. In contrast, the phage-encoded Coa’ variant from yellow-necked field mouse-derived ST890 strongly coagulated bank vole, mouse and rat plasma, but exhibited low level activity for human plasma. Similarly, the core genome-encoded vWbp from human ST8 potently coagulated human plasma as well as blank vole plasma, but exhibited negligible activity for lab mouse and lab rat plasma. Core genome-encoded vWbp from ST88 and ST49 had low activity for human plasma and negligible activity for mouse and rat plasma. Strikingly, the SaPI-encoded vWbp variant (vWbp’ ST3033) from shrews potently coagulated bank vole plasma, exhibited moderate activity for mouse and rat plasma and none for human plasma.
The host-dependent functionality of different coagulases likely reflects distinct tropisms for the substrate prothrombin which differs in sequence in different host-species [59–62]. The human prothrombin heavy chain interacts with Coa via two binding sites, the Trp148 binding pocket and exosite 1 [63]. Both Coa and vWbp allosterically activate prothrombin through insertion of the first two N-terminal residues of Coa/vWbp into the activation pocket on prothrombin [63,64]. The prothrombin heavy chain has 81% aa identity with the prothrombin of mice (Mus musculus), 79% with rats (both Rattus rattus and Rattus norvegicus), and less than 75% with common shrews (Sorex araneus) (S6 Fig). The interacting amino acid residues in the Coa binding sites and activation pocket were conserved between human and mouse/vole prothrombin sequences, but the shrew prothrombin displayed a R75→K mutation in the exosite 1. In addition, mouse, vole and shrew prothrombin displayed sequence variations adjacent to the binding pocket and the activation pocket (S6 Fig). These prothrombin modifications likely drive selection for new host-specific coagulase variants.
Overall, these data indicate that S. aureus chromosomally-encoded secreted coagulases had limited functional activity for most rodent plasmas whereas coagulases encoded on rodent-specific MGE could mediate coagulation of rodent plasma suggesting a key role for these elements in murine host-adaptation.
Discussion
Addressing the ongoing threat of emerging infectious diseases affecting humans and animals requires identification of the main potential reservoirs for new pathogens, and an understanding of how they evolve and spread within new host-species populations. Recent bacterial typing studies have reported an array of S. aureus STs that are associated with laboratory rodent populations around the world [16,17]. Here, using a phylogenomic approach, we report that these lineages are broadly representative of the S. aureus species diversity. Many cases of S. aureus isolation from rodents likely involved sporadic human-to-rodent spillovers with limited levels of onward transmission, reflecting the nature of laboratory rodent husbandry. However, we identified several monophyletic, rodent-specific populations which have arisen via single host-switching events presumably from humans, followed by widespread intercontinental dissemination.
In particular, S. aureus CC88 represents a large, internationally spread lineage, found only in laboratory rodents. Our phylogenetic data suggest that it most likely emerged in CR facilities in Canada, from which it spread to facilities in the US East Coast and subsequently (from 1990) throughout the world. Several factors should be considered when interpreting the phylodynamic data for S. aureus from laboratory rodents: hygiene measures and caging effectively prevent horizontal transmission between barrier rooms, but still enable vertical transmission from breeding pairs to offspring [16]. In addition, while vendors are active on a global scale, shipping S. aureus-colonized mice between continents, actual animal movements were not documented and thus intermediate steps (e.g. other vendors or users) may be missing in our study. Thus, the locations suggested to be involved in migration events should be interpreted with caution. Nevertheless, it is likely that there is a human origin for CC88 which has been largely reported in humans in South East Asia and sub-Saharan Africa but not typically in other parts of the world [18]. CC88 emerged in laboratory mice around 1983, within a few years of the oldest CC1 and CC15 murine clusters, and these contemporaneous events may reflect animal handling or biosafety practices that were in place during the late 1970s-80s. At that time, barrier to barrier transfer of laboratory mice was still common practise. Of note, only limited numbers of transmission events into mice have been detected subsequently, likely reflecting stricter regulations regarding biosafety. Finally, the narrow geographic sampling of wild mouse, vole, shrew, and guinea pig isolates limits the scope of the phylogeographic analysis.
Some human S. aureus CCs have a particular propensity to undergo host-switches, adapt, and spread in mouse populations. For example, we observed several spillover events for the S. aureus lineages CC1 and CC15, which are commonly found in humans.
As previously reported [26], wild rodents (mice, voles, red squirrels and rats) were colonized by S. aureus lineages distinct to those found in laboratory mice, dominated by CC49, CC890 and CC1956. In particular, CC49 has been reported to be a cause of fatal exudative dermatitis in UK squirrels and it is speculated that the expression of the lukMF’ toxin may be associated with the lethal infections of squirrels following spillover events from a reservoir species (such as bank voles or rats) with overlapping habitats [29,30]. LukMF’ is commonly found in ruminant and rodent S. aureus isolates [65–67]. It lyses bovine, ovine, goat and mouse neutrophils in vitro by binding to chemokine receptor CCR1, but has no cytolytic effect on human neutrophils, which lack CCR1 [68].
Our analysis did not detect evidence of convergent host-adaptation between lineages, and genetic diversification was largely lineage-dependent, consistent with previous findings for S. aureus associated with host-species [5,13]. However, we identified a large and diverse family of novel S. aureus prophages that are associated with a rodent host ecology. The majority of prophages identified among wild rodent S. aureus were not represented in public sequence repositories, encoded novel integrase variants and proteins putatively involved in host-pathogen interactions. For instance, CC1 and CC15 isolates from laboratory mice were marked by the loss of human-associated prophages and acquisition of murine-associated prophages consistent with adaptation to the murine host. Moreover, the remarkable success of the CC88 lineage has not been observed for other lineages and we hypothesise that the expansion was aided by the acquisition of prophages such as one (LabRodCC88_3) that encoded the novel serine protease JEP. This protease was also encoded by unrelated prophages found in CC49 isolates from wild rodents (WildRodCC49_2 and WildRodCCmisc_1, in wild mice, voles, and red squirrels), indicative of a key role in adaptation of S. aureus to both laboratory and wild rodents. Overall, this suggests that the emergence and expansion of S. aureus in rodent populations is driven by bacteriophage-encoded effectors.
To investigate this further we analyzed phage- and SaPI-encoded coagulase-like proteins and demonstrated that they conferred the ability to mediate coagulation of rodent plasma demonstrating a role for acquisition of MGE in adaptation to rodent hosts. Previously, we reported that novel variants of SaPI-encoded vWbp identified among sheep S. aureus and S. aureus subsp. anaerobius, the causative agent of Morels’ disease, had ruminant-specific functional activity [9,69]. Furthermore, different coagulase-positive staphylococcal species have evolved coagulases with a wide spectrum of host-tropisms [61]. These studies highlight the key importance of plasma coagulation for S. aureus survival in vivo and the strong selective pressure driving the diversification of coagulases to promote abscess formation and persistence in different host-species.
In conclusion, we used a combination of population comparative genomics, phylodynamics and GWAS to reveal the genomic diversity of S. aureus isolates found in rodent populations. S. aureus isolates from laboratory mice resulted from numerous human-to-mouse host jump events, including a CC88 clade that disseminated worldwide via introductions from commercial mouse providers. In contrast, wild rodents contained a distinct set of S. aureus lineages consistent with their independent ecological and evolutionary history. A unique set of diverse prophages were identified to be circulating among S. aureus in rodent populations, some of which encoded novel determinants of host-specificity that likely promote colonization and persistence. Our findings highlight the remarkable promiscuity of S. aureus and its capacity to adapt to different niches and expand into new host-species populations. Our findings reinforce the importance of employing a One Health approach to investigate the dynamics of bacterial pathogens at the interface between humans, and domesticated and wild animals.
Supporting information
S1 Fig
Densitree of Bayesian posterior distribution of CC88 international murine cluster.The plot shows all possible topologies, parts of the tree with a higher topology agreement look sharper, whereas areas with more uncertain topologies look more blurred.
(DOCX)
S2 Fig
CC1 and CC15 murine-specific clusters extracted from the phylogenetic Bayesian trees of both S. aureus lineages.(DOCX)
S3 Fig
Hierarchical clustering tree built from pairwise genetic distances (calculated with Mash) between prophage sequences (as predicted by PHASTER) across the whole study dataset.Tips names and circles are coloured according to the integrase type detected through BLAST.
(DOCX)
S4 Fig
Maximum-likelihood phylogenetic analysis of the integrases in rodent prophages.Reference sequences for each integrase type are coloured in cyan. Putative novel integrase types, genetically different and showing different integration sites, are coloured in orange.
(DOCX)
S5 Fig
Protein sequence variation among core genome- and MGE-encoded coagulases.A) Amino acid-based identity matrix for core-genome encoded coa (human-derived ST8, laboratory mouse-derived ST88, and bank vole-derived ST49 strains) as well as phage-encoded vWbp variants from yellow-necked field mice (Coa’ ST980) and common voles (Coa’ ST3252). B) Alignment of core genome- and phage-encoded coagulases. The prothrombin-binding domains D1 and D2 and the fibrinogen-binding domain are indicated. Amino acid residues interacting with the exosite 1 on prothrombin are highlighted. The highlighted N-terminal amino acids induce allosteric activation of prothrombin by inserting into the prothrombin activation pocket. C) Amino acid-based identity matrix for core-genome encoded vWbp (human-derived ST8, laboratory mouse-derived ST88, and bank vole-derived ST49 strains), as well as SaPI-endoded vWbp’ (common shrew-derived ST3033). D) Alignment of core genome- and SaPI-encoded vWbps.The N-terminal peptide, the prothrombin-binding domains D1 and D2 and the van Willebrand factor-binding domain are indicated.
(DOCX)
S6 Fig
Protein sequence variation among human, rodent and shrew prothrombin.A) Amino acid-based identity matrix for the prothrombin heavy chain derived from humans, laboratory mice, yellow-necked field mice, rats, bank voles and common voles. B) Alignment of human, rodent and shrew prothrombin sequences (heavy chain). The two Coagulase binding sites, Trp148 binding pocket and exosite 1, are indicated. Both Coa and vWbp allosterically activate prothrombin through insertion of their first two N-terminal residues into activation pocket on prothrombin.
(DOCX)
S5 Table
List of identified genes predicted to encode coagulases.(XLSX)
Funding Statement
The study was supported by grants to JRF from the Wellcome Trust (UK) (201531/Z/16/Z/WT) and the Biotechnology and Biological Sciences Research Council, UK (Institute Strategic programme; BBS/E/D/20002173/UKRI) and to SH by the Deutsche Forschungsgemeinschaft (GRK 2719/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Illumina sequence data is available in the European Nucleotide Archive under the BioProject PRJEB41130.
References
Decision Letter 0
13 May 2024
Dear Prof. Fitzgerald,
Thank you very much for submitting your manuscript "Bacteriophage-driven emergence and expansion of Staphylococcus aureus in rodent populations" for consideration at PLOS Pathogens. As with all papers reviewed by the journal, your manuscript was reviewed by members of the editorial board and by several independent reviewers. The reviewers appreciated the attention to an important topic. Based on the reviews, we are likely to accept this manuscript for publication, providing that you modify the manuscript according to the review recommendations.
Please address the issues related to sampling that were raised by both reviewers, and the GWAS issues indicated by reviewer 2. In addition, the posterior probabilites are not high in some of your phylogeographic and phylogenetic analyses, which needs to be better acknowledged. For example, analysis in Figure 2A may best support an origin of CC88 in SE Asia, but it does so at only 32% PP - hardly convincing. When looking at the geographic origin of murine CC88 in Figure 2A the pie-chart seems to only support Canada at about 50% PP, but in the text describing Figure 2B you indicate the support is 86% (probably should also show pie-charts in 2B). The Figure 2B result is better, but why the difference? Is it sampling related? Based on the analysis presented, you should be more circumspect in the Abstract about where this murine lineage may have emerged. In Figure 3, the CC49 node that includes human and rodent isolates has weak support, possibly due to a closely timed radiation of lineages?
Please prepare and submit your revised manuscript within 30 days. If you anticipate any delay, please let us know the expected resubmission date by replying to this email.
When you are ready to resubmit, please upload the following:
[1] A letter containing a detailed list of your responses to all review comments, and a description of the changes you have made in the manuscript.
Please note while forming your response, if your article is accepted, you may have the opportunity to make the peer review history publicly available. The record will include editor decision letters (with reviews) and your responses to reviewer comments. If eligible, we will contact you to opt in or out
[2] Two versions of the revised manuscript: one with either highlights or tracked changes denoting where the text has been changed; the other a clean version (uploaded as the manuscript file).
Important additional instructions are given below your reviewer comments.
Thank you again for your submission to our journal. We hope that our editorial process has been constructive so far, and we welcome your feedback at any time. Please don't hesitate to contact us if you have any questions or comments.
Sincerely,
D. Ashley Robinson, Ph.D.
Academic Editor
PLOS Pathogens
Helena Boshoff
Section Editor
PLOS Pathogens
Michael Malim
Editor-in-Chief
PLOS Pathogens
***********************
Please address the issues related to sampling that were raised by both reviewers, and the GWAS issues indicated by reviewer 2. In addition, the posterior probabilites are not high in some of your phylogeographic and phylogenetic analyses, which needs to be better acknowledged. For example, analysis in Figure 2A may best support an origin of CC88 in SE Asia, but it does so at only 32% PP - hardly convincing. When looking at the geographic origin of murine CC88 in Figure 2A the pie-chart seems to only support Canada at about 50% PP, but in the text describing Figure 2B you indicate the support is 86% (probably should also show pie-charts in 2B). The Figure 2B result is better, but why the difference? Is it sampling related? Based on the analysis presented, you should be more circumspect in the Abstract about where this murine lineage may have emerged. In Figure 3, the CC49 node that includes human and rodent isolates has weak support, possibly due to a closely timed radiation of lineages?
Reviewer Comments (if any, and for reference):
Reviewer's Responses to Questions
Part I - Summary
Please use this section to discuss strengths/weaknesses of study, novelty/significance, general execution and scholarship.
Reviewer #1: Thank you for the opportunity to review “Bacteriophage-driven emergence and expansion of Staphylococcus aureus in rodent populations” by Yebra and colleagues. The authors present a fascinating study that includes population genomics, phylodynamic, and genome-wide association analysis of S. aureus from rodent populations to trace the evolutionary history and investigate for host-adaptation. The results coalesce into an intriguing story of rodent-associated lineages and host-adaptation through bacteriophage acquisition. In particular, the identify ST88 as a rodent-associated lineage that is likely associated with laboratory rodents. They also identify a number of previously-unreported S. aureus phages, which the subsequently found to encode host-specific proteases. This is one of the few examples of where the evolutionary impact of temperate bacteriophages are revealed.
Overall, their analysis was an excellent combination of genomic and molecular approaches. Their methods are clear, comprehensive, and easy to follow. In addition, the use and incorporation of data from Staphopia presents an excellent use case for the repository. The figures are aesthetically pleasing and informative.
In terms of critiques, I have none that are substantial. Usually in similar papers there are shortcomings in the phylodynamic analysis, but here the authors are thorough with model testing and also present their limitations. The only minor comment is that it may be worth mentioning that the limited geographic sampling of wild mouse, vole, shrew, and guinea pig isolates limits the phylogeographic analysis (i.e., you can’t infer ancestral locations from unsampled locations). However, the discussion mentions something to this effect. In supplemental figure 3, you could probably drop the tip labels and just use shapes since you can’t read the text, but again, that is a minor stylistic critique. I really enjoyed reading the publication and think that it will be of broad interest to the microbial genomics, genomic epidemiology, and molecular biology readership.
Reviewer #2: This population genomics study looked at the evolutionary history of rodent-adapted Staphylococcus aureus using a combination of phylodynamics and genome-wide association studies (GWAS). The authors describe the emergence of CC88 as a laboratory mice clone in the ~1980 as well as several limited introductions of S. aureus into laboratory rodents, while distinct clones are found in wild rodents. Finally, they identify an association between mice adaptation and the presence of a novel phage. This association was further investigated by assessing the host-specificity of the recombinant coagulase proteins obtained from the core genome vs. phage-encoded coagulases.
The manuscript is well written, and the genomics methods are state-of-the art. The conclusions are supported by the genomic analysis and the coagulase activity testing, however, no major new findings emerge: the CC88 has been previously identified and the association between presence of a phage and host adaptation has been reported in previous GWAS with a similar design.
**********
Part II – Major Issues: Key Experiments Required for Acceptance
Please use this section to detail the key new experiments or modifications of existing experiments that should be absolutely required to validate study conclusions.
Generally, there should be no more than 3 such required experiments or major modifications for a "Major Revision" recommendation. If more than 3 experiments are necessary to validate the study conclusions, then you are encouraged to recommend "Reject".
Reviewer #1: None
Reviewer #2: 1. Sampling is key in this kind of studies, yet little is said on how the isolates were collected. How were they obtained? Were they selected somehow?
2. Multiple GWAS approaches were used (fixed effect models, linear mixed model, lineage effects), yet it is unclear how the output of the approaches overlaps (or not) and how the final 41 hits were selected (i.e. just one approach? Intersection of multiples approaches?). I recommend providing all GWAS results in the supplementary data, including non-significant hits.
**********
Part III – Minor Issues: Editorial and Data Presentation Modifications
Please use this section for editorial suggestions as well as relatively minor modifications of existing data that would enhance clarity.
Reviewer #1: Minor changes to supplemental figures, but not required
Reviewer #2: 1. Please add line numbers to the manuscript
2. Please provide the heritability estimated calculated by Pyseer
3. From figure 4B it looks like the GWAS was done on a per-CC basis, however this is not clearly said in the results: please clarify.
4. Table S1: please list all genomes included in the analysis
5. “Overall, the relatively small number of GWAS hits out-with MGE indicates that convergent adaptations of different S. aureus lineages to the rodent host were rare”: this sentence is unclear to me
**********
PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: Yes: Taj Azarian
Reviewer #2: No
Figure Files:
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email us at gro.solp@serugif.
Data Requirements:
Please note that, as a condition of publication, PLOS' data policy requires that you make available all data used to draw the conclusions outlined in your manuscript. Data must be deposited in an appropriate repository, included within the body of the manuscript, or uploaded as supporting information. This includes all numerical values that were used to generate graphs, histograms etc.. For an example see here: http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1001908#s5.
Reproducibility:
To enhance the reproducibility of your results, we recommend that you deposit your laboratory protocols in protocols.io, where a protocol can be assigned its own identifier (DOI) such that it can be cited independently in the future. Additionally, PLOS ONE offers an option to publish peer-reviewed clinical study protocols. Read more information on sharing protocols at https://plos.org/protocols?utm_medium=editorial-email&utm_source=authorletters&utm_campaign=protocols
References:
Please review your reference list to ensure that it is complete and correct. If you have cited papers that have been retracted, please include the rationale for doing so in the manuscript text, or remove these references and replace them with relevant current references. Any changes to the reference list should be mentioned in the rebuttal letter that accompanies your revised manuscript. If you need to cite a retracted article, indicate the article’s retracted status in the References list and also include a citation and full reference for the retraction notice.
Author response to Decision Letter 0
23 Jun 2024
Decision Letter 1
27 Jun 2024
Dear Prof. Fitzgerald,
We are pleased to inform you that your manuscript 'Bacteriophage-driven emergence and expansion of Staphylococcus aureus in rodent populations' has been provisionally accepted for publication in PLOS Pathogens.
Before your manuscript can be formally accepted you will need to complete some formatting changes, which you will receive in a follow up email. A member of our team will be in touch with a set of requests.
Please note that your manuscript will not be scheduled for publication until you have made the required changes, so a swift response is appreciated.
IMPORTANT: The editorial review process is now complete. PLOS will only permit corrections to spelling, formatting or significant scientific errors from this point onwards. Requests for major changes, or any which affect the scientific understanding of your work, will cause delays to the publication date of your manuscript.
Should you, your institution's press office or the journal office choose to press release your paper, you will automatically be opted out of early publication. We ask that you notify us now if you or your institution is planning to press release the article. All press must be co-ordinated with PLOS.
Thank you again for supporting Open Access publishing; we are looking forward to publishing your work in PLOS Pathogens.
Best regards,
D. Ashley Robinson, Ph.D.
Academic Editor
PLOS Pathogens
Michael Wessels
Section Editor
PLOS Pathogens
Michael Malim
Editor-in-Chief
PLOS Pathogens
***********************************************************
Reviewer Comments (if any, and for reference):
Acceptance letter
15 Jul 2024
Dear Prof. Fitzgerald,
We are delighted to inform you that your manuscript, "Bacteriophage-driven emergence and expansion of Staphylococcus aureus in rodent populations," has been formally accepted for publication in PLOS Pathogens.
We have now passed your article onto the PLOS Production Department who will complete the rest of the pre-publication process. All authors will receive a confirmation email upon publication.
The corresponding author will soon be receiving a typeset proof for review, to ensure errors have not been introduced during production. Please review the PDF proof of your manuscript carefully, as this is the last chance to correct any scientific or type-setting errors. Please note that major changes, or those which affect the scientific understanding of the work, will likely cause delays to the publication date of your manuscript. Note: Proofs for Front Matter articles (Pearls, Reviews, Opinions, etc...) are generated on a different schedule and may not be made available as quickly.
Soon after your final files are uploaded, the early version of your manuscript, if you opted to have an early version of your article, will be published online. The date of the early version will be your article's publication date. The final article will be published to the same URL, and all versions of the paper will be accessible to readers.
Thank you again for supporting open-access publishing; we are looking forward to publishing your work in PLOS Pathogens.
Best regards,
Michael Malim
Editor-in-Chief
PLOS Pathogens
Articles from PLOS Pathogens are provided here courtesy of PLOS
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/165802941
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (1 citation) BioProject - PRJEB41130
GCA - NCBI genaome assembly
- (1 citation) GCA - GCA_000009005.1
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Laboratory Mice Are Frequently Colonized with Staphylococcus aureus and Mount a Systemic Immune Response-Note of Caution for In vivo Infection Experiments.
Front Cell Infect Microbiol, 7:152, 02 May 2017
Cited by: 26 articles | PMID: 28512627 | PMCID: PMC5411432
Fatal exudative dermatitis in island populations of red squirrels (Sciurus vulgaris): spillover of a virulent Staphylococcus aureus clone (ST49) from reservoir hosts.
Microb Genom, 7(5), 01 May 2021
Cited by: 6 articles | PMID: 34016250 | PMCID: PMC8209723
Wild rodents and shrews are natural hosts of Staphylococcus aureus.
Int J Med Microbiol, 308(6):590-597, 22 Sep 2017
Cited by: 25 articles | PMID: 28967544
Temperate Phages of Staphylococcus aureus.
Microbiol Spectr, 7(5), 01 Sep 2019
Cited by: 31 articles | PMID: 31562736
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (2)
Grant ID: BBS/E/D/20002173/UKRI
Grant ID: BBS/E/D/20002173
Wellcome Trust (2)
Understanding bacterial host adaptation to combat infectious disease
Prof Ross Fitzgerald, University of Edinburgh
Grant ID: 201531/Z/16/Z
Grant ID: 201531/Z/16/Z/WT

 2
,* and
2
,* and 

