Abstract
Free full text

Loss of myosin light chain kinase induces the cellular senescence associated secretory phenotype to promote breast epithelial cell migration
Abstract
Overexpression or activation of oncogenes or loss of tumor-suppressor genes can induce cellular senescence as a defense mechanism against tumor development, thereby maintaining cellular homeostasis. However, cancer cells can circumvent this senescent state and continue to spread. Myosin light chain kinase (MLCK) is downregulated in many breast cancers. Here we report that downregulation of MLCK in normal breast epithelial cells induces a senescence-associated secretory phenotype and stimulates migration. The reduction of MLCK results in increased p21Cip1 expression, dependent on p53 and the AKT-mammalian target of rapamycin pathway. Subsequently, p21Cip1 promotes the secretion of soluble ICAM-1, IL-1α, IL-6 and IL-8, thereby enhancing collective cell migration in a non-cell-autonomous manner. These findings provide new mechanistic insights into the role of MLCK in cellular senescence and cancer progression.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76868-y.
Introduction
Cellular senescence is considered a barrier against tumorigenesis via cell-cycle arrest1,2. Overexpression or activation of oncogenes or loss of tumor-suppressor genes triggers senescence in several tumor models, restraining tumor growth and progression3,4. However, the role of senescence in tumor development is still controversial because senescent cells secrete several cytokines, growth factors and matrix metalloproteinase, and these secretomes provoke tumor-promoting as well as tumor-suppressing responses5,6. For example, secretory factors such as interleukin (IL)-1, IL-6 and IL-8, can have potent pro-tumorigenic properties7,8. Additionally, senescent tumor cells exhibit greater invasiveness than corresponding non-senescent tumor cells by expressing higher levels of cytokines5,9.
Senescent cells undergo continuous cell-cycle arrest, leading to changes in morphology and a decline in cell proliferation10. Recent evidence has demonstrated that decreased cell proliferation is linked to cancer cell invasion11. Specifically, loss of the cell-cycle inhibitor p21Cip1 (hereafter referred to as p21) decreases the ability of breast cancer to invade and metastasize despite hyperproliferation12,13. Invasive breast tumors collected from xenografted human cancer cells or mouse tumors showed increased expression of genes associated with cell-cycle arrest and decreased proliferation14–16. Furthermore, cancer cells at the invasive front are less proliferative than trailing cells17,18. Together, these studies suggest that efficient tumor invasion requires decreased proliferation, but the underlying mechanism is unclear.
Myosin light chain kinase (MLCK) is a regulator of the actin cytoskeleton, which is involved in fundamental cellular processes such as cell adhesion, migration and survival. MLCK regulates actomyosin contractility by phosphorylation of myosin light chain at Thr18 and Ser1919. MLCK also acts as a molecular scaffold, complexing with ABL and Cortactin, and regulates intracellular signaling pathways in endothelial cells20–22. Inhibition of MLCK kinase activity induces decreased cell proliferation and growth in breast cancer23–25. However, MLCK (MYLK) expression is lower in 73% of cases of invasive breast cancer when compared with normal tissue26. Downregulation of MLCK is known to affect cancer development through increased cell migration and invasion, and cytokinesis failure27–29. In the context of breast cancer development, MLCK plays opposite roles in cell proliferation and migration.
In this study, we show that downregulation of MLCK in MCF10A breast epithelial cells leads to cellular senescence, which causes decreased cell proliferation but increased cell migration. We found that downregulation of MLCK in MCF10A breast epithelial cells causes p53 (TP53)-and AKT-mammalian target of rapamycin (mTOR)-dependent p21 upregulation and mediates cellular senescence. p21 upregulates senescent-associated secretory cytokines resulting in increased collective cell migration in MLCK-depleted MCF10A cells. These findings suggest that MLCK might mediate reciprocal switching between proliferation and migration during breast cancer progression.
Results
Downregulation of MLCK induces cellular senescence
The expression of MLCK is reduced in breast cancer cells compared to normal breast epithelial cells, and its downregulation is associated with increased breast cancer cell migration and metastasis26,28,30. This implies a potential tumor suppressor role for MLCK in breast cancer progression. Downregulation of MLCK promotes cell migration and activates growth signaling pathways in breast epithelial cells28. However, MLCK depletion using siRNAs slowed cell division and reduced the number of Ki-67 positive proliferating cells in MCF10A breast epithelial cells (Fig. 1a-c), suggesting that MLCK regulates cell proliferation and migration through distinct mechanisms.
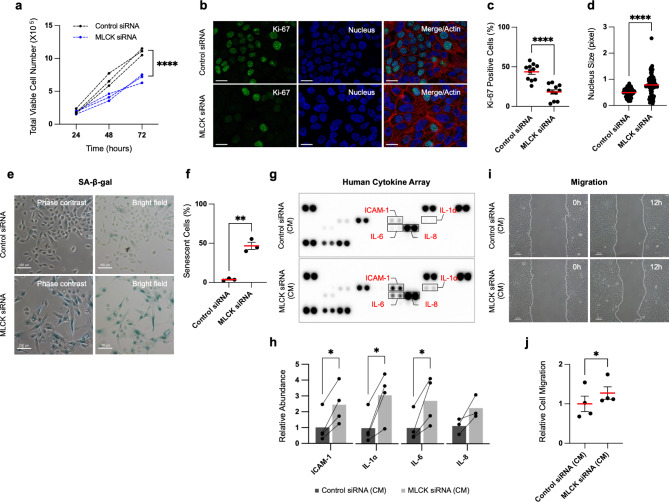
Downregulation of MLCK decreases cell growth and induces cellular senescence-associated secretory phenotypes. (a) MCF10A cells were transfected with MLCK siRNAs for 72 h and seeded the same number of cells. Total cell numbers were counted every 24 h. n =
= 3 independent experiments. Two-way ANOVA with Sidak’s multiple comparisons test. (b, c) Representative images and quantification of Ki-67 positive cells. Scale bars, 20 μm. n
3 independent experiments. Two-way ANOVA with Sidak’s multiple comparisons test. (b, c) Representative images and quantification of Ki-67 positive cells. Scale bars, 20 μm. n =
= 11 fields, 2 independent experiments. Unpaired t-test. (d) The nucleus size of individual cells was measured with DAPI staining. n
11 fields, 2 independent experiments. Unpaired t-test. (d) The nucleus size of individual cells was measured with DAPI staining. n =
= 67 nuclei, 3 independent experiments. Unpaired t-test. (e, f) Representative images and quantification of SA-β-gal staining. Scale bars, 100 μm. n
67 nuclei, 3 independent experiments. Unpaired t-test. (e, f) Representative images and quantification of SA-β-gal staining. Scale bars, 100 μm. n =
= 3 independent experiments, Paired t-test. (g) Antibody-based analysis of human cytokines. Selected 36 cytokines were measured with conditioned media from control or MLCK siRNA-treated cells. (h) Quantification of ICAM-1, IL-1α, IL-6 and IL-8 in control and MLCK siRNA-treated cells. Fold changes were normalized to control cells. n
3 independent experiments, Paired t-test. (g) Antibody-based analysis of human cytokines. Selected 36 cytokines were measured with conditioned media from control or MLCK siRNA-treated cells. (h) Quantification of ICAM-1, IL-1α, IL-6 and IL-8 in control and MLCK siRNA-treated cells. Fold changes were normalized to control cells. n =
= 3 independent experiments. Ratio paired t-test. (i, j) Representative images and quantification of scratch wound migration assays performed under conditioned media from control or MLCK siRNA-treated cells. Scale bars, 100 μm. n
3 independent experiments. Ratio paired t-test. (i, j) Representative images and quantification of scratch wound migration assays performed under conditioned media from control or MLCK siRNA-treated cells. Scale bars, 100 μm. n =
= 4 independent experiments. Paired t-test. mean
4 independent experiments. Paired t-test. mean ±
± s.e.m. *P
s.e.m. *P <
< 0.05, **P
0.05, **P <
< 0.01, ****P
0.01, ****P <
< 0.0001.
0.0001.
In addition to reducing cell proliferation, MLCK depletion with siRNA induced morphological alterations, including enlarged cell size, flattened cell bodies and increased nucleus size (Fig. 1d-e), which are known to be major characteristics of senescent cells31,32. Analysis of senescence-associated β-galactosidase (SA-β-gal) activity revealed an increase in SA-β-gal-positive cells upon MLCK depletion (Fig. 1e-f). Decreased cell proliferation and increased SA-β-gal activity were also observed following MLCK knockdown with a different siRNA, specifically targeting the 3′ UTR of MLCK, suggesting that the siRNA effects are specific (Supplementary Fig. 1a-d). Differences in growth rates between control and MLCK-depleted cells can affect cell density and potentially lead to contact inhibition-induced SA-β-gal activity33. However, SA-β-gal activity remained consistent across varying cell densities in MCF10A cells (Supplementary Fig. 1f). This suggests that MLCK depletion increases SA-β-gal activity independently of cell-cell contact effects, indicating that MLCK downregulation specifically induces cellular senescence.
Downregulation of MLCK stimulates the senescence-associated secretory phenotype
Given that MLCK depletion induces cellular senescence, we questioned whether MLCK affects the senescence-associated secretory phenotype (SASP) in MCF10A cells. To address this question, we collected conditioned media (CM) from both control and MLCK siRNA-treated cells and measured secreted cytokines. Using an antibody-based cytokine array, we assessed levels of 36 different secreted cytokines. MLCK depletion significantly increased the accumulation of soluble Intercellular adhesion molecule-1 (ICAM-1), IL-1α, IL-6 and IL-8 in the CM (Fig. 1g-h).
Secretory factors produced by senescent cells can contribute to collective migration and invasion in various tumors9,34. We asked whether cytokines secreted from MLCK-depleted cells promote breast epithelial cell migration. Confluent monolayers of MCF10A cells were wounded, and migration measured in the presence of CM from control or MLCK-depleted cells. Cell migration was increased by CM from MLCK-depleted cells (Fig. 1i-j, Supplementary Fig. 1e). Live cell imaging showed that cells exposed to CM from MLCK-depleted cells migrate as a collective, with few individually dispersed cells (Supplementary Video 1 and 2).
Our previous work demonstrated that decreased MLCK levels cooperate with HER2 to promote cell migration and are associated with poor survival in HER2-positive breast cancer patients28. We investigated whether downregulation of MLCK in HER2-positive breast cancer cell lines, SK-BR-3 and BT-474, affects cellular senescence. MLCK downregulation slightly reduced growth of both SK-BR-3 and BT-474 cells (Supplementary Fig. 2a-c) and increased the proportion of senescent cells in SK-BR-3 (Supplementary Fig. 2d-e), but did not affect BT-474 cells. The more subtle effects of MLCK depletion on cellular senescence in breast cancer cells than normal MCF10A cells may be due to their already low MLCK levels28. We also explored whether paracrine factors from MLCK-depleted MCF10A cells influence breast cancer cell migration. CM from MLCK-depleted MCF10A cells were incubated with SK-BR-3 or BT-474 during scratch wound assays. Interestingly, CM from MLCK-depleted cells enhanced breast cancer cell migration (Supplementary Fig. 2f-j). Together, these findings suggest that MLCK depletion from epithelial cells promotes the secretion of cytokines that increase cell migration in a non-cell-autonomous manner.
Downregulation of MLCK promotes p21-induced cellular senescence
At the molecular level, pro-senescent stresses are triggered by the DNA damage response, leading to increased expression of cyclin-dependent kinase inhibitors and cell-cycle arrest35,36. Using a reverse-phase protein array, we analyzed 285 phosphorylated or total proteins in control or MLCK-depleted cells (Supplementary Table 1). Notably, MLCK-depleted cells showed downregulation of DNA damage response genes, including phosphorylated ATM (S1981) and CHK (S296 and S345), MSH2, MSH6 and PMS2 (Fig. 2a). No significant changes were observed in DNA damage markers such as RAD51, cleaved PARP1 or phosphorylated H2AX (S139 and S140) (Supplementary Table 1), suggesting that cellular senescence induced by MLCK depletion is not a result of DNA damage. Moreover, MLCK-depleted cells showed significant upregulation of the cyclin-dependent kinase inhibitor p21 (Fig. 2b). These cells also showed upregulation of cyclin D1 and cyclin E1, but downregulation of cyclin B1 and CDK1 (Fig. 2b), indicating cell-cycle arrest at the mitotic entry rather than at the G1 phase.
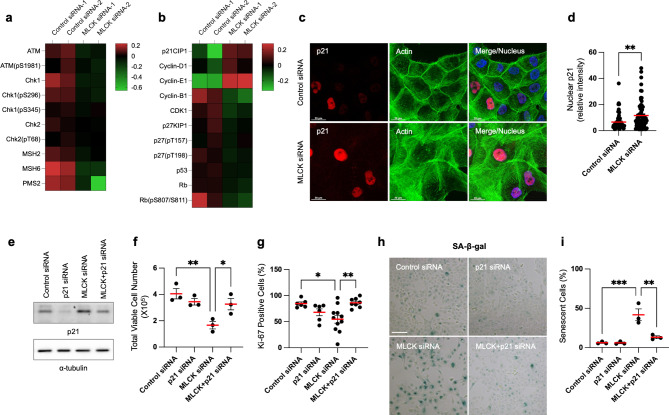
Downregulation of MLCK increases p21 expression and p21-induced cellular senescence. (a, b) MCF10A cells were transfected with MLCK siRNAs for 72 h and protein lysates were subsequently harvested for a reverse-phase protein array. The heatmaps display selected DNA damage response and cell-cycle regulation genes from the reverse-phase protein array. Data represent two independent experiments. (c, d) Representative images and quantification of p21 in the nucleus. Scale bars, 20 μm. n =
= 67 nuclei, 3 independent experiments. Unpaired t-test. (e) Western blot analysis of control, p21, MLCK, MLCK/p21 siRNAs in MCF10A cells. α-tubulin is shown as a loading control. Original blots are presented in Supplementary Fig. 4. (f) Cells were initially seeded at an identical quantity and subsequently counted after 24 h. Dead cells were excluded using trypan blue staining. n
67 nuclei, 3 independent experiments. Unpaired t-test. (e) Western blot analysis of control, p21, MLCK, MLCK/p21 siRNAs in MCF10A cells. α-tubulin is shown as a loading control. Original blots are presented in Supplementary Fig. 4. (f) Cells were initially seeded at an identical quantity and subsequently counted after 24 h. Dead cells were excluded using trypan blue staining. n =
= 3 independent experiments. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (g) Quantification of Ki-67-positive proliferating cells. n
3 independent experiments. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (g) Quantification of Ki-67-positive proliferating cells. n =
= 6–11 fields, 2 independent experiments. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (h, i) Representative images and quantification of SA-β-gal staining. Scale bars, 100 μm. n
6–11 fields, 2 independent experiments. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (h, i) Representative images and quantification of SA-β-gal staining. Scale bars, 100 μm. n =
= 3 independent experiments. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test. mean
3 independent experiments. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test. mean ±
± s.e.m. *P
s.e.m. *P <
< 0.05, **P
0.05, **P <
< 0.01, ***P
0.01, ***P <
< 0.001.
0.001.
Among these proteins, p21 emerged as the most upregulated cyclin-dependent kinase inhibitor in MLCK-depleted cells. With its well-established role in inhibiting cell proliferation, p21 regulates cellular senescence37,38. Western blot analysis confirmed increased expression of p21 in MLCK-depleted cells (Supplementary Fig. 3a). By immunofluorescence, MLCK depletion increased the p21 intensity in the nucleus (Fig. 2c-d). The upregulated p21 was highly localized in the nucleus and undetectable in the cytoplasm in both control and MLCK-depleted cells (Fig. 2c).
We then tested whether p21 regulates cell proliferation and senescence in MLCK-depleted cells. To test this, we inhibited both MLCK and p21 expression by siRNAs (Fig. 2e). Silencing MLCK alone resulted in reduced cell growth and a decreased number of Ki-67-positive proliferating cells. However, co-silencing both p21 and MLCK restored cell growth and proliferation (Fig. 2f-g). Furthermore, p21 depletion prevented the increase of SA-β-gal activity, indicating an inhibition of cellular senescence (Fig. 2h-i). These findings demonstrate that increased p21 expression inhibits cell proliferation and promotes cellular senescence in MLCK-depleted cells.
Downregulation of MLCK increases p21-induced senescence-associated cytokine secretion that is required for increased cell motility and changes in actin filaments
Given that p21 is required to induce cellular senescence in MLCK-depleted cells, we investigated its role in regulating the SASP and increased expression of ICAM-1, IL-1α, IL-6 and IL-8 in MLCK-depleted cells (Fig. 1g-h). The secretion of these cytokines was suppressed upon inhibition of p21 (Fig. 3a-b). mRNA transcripts of ICAM-1, IL-1α, IL-6 and IL-8 were also found to depend on p21 expression (Fig. 3c).
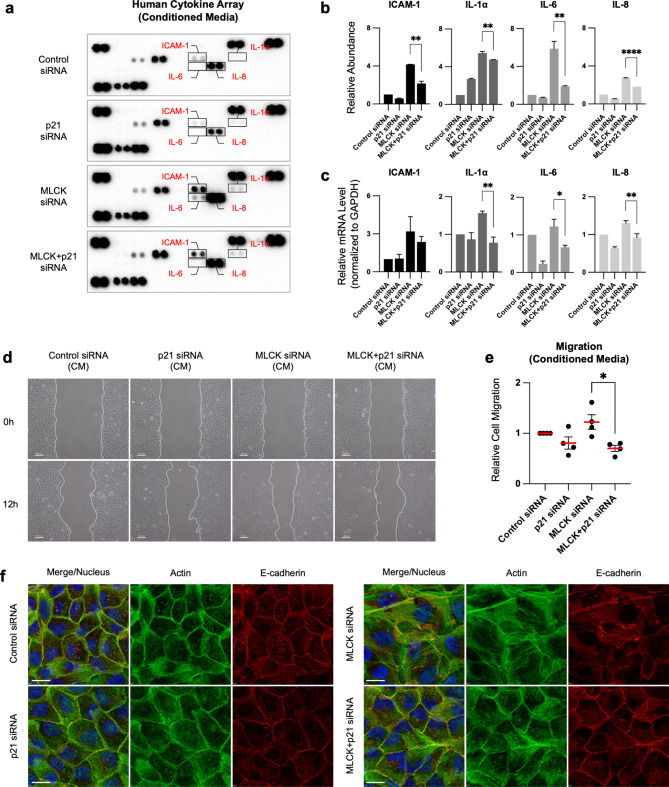
Downregulation of MLCK increases p21-induced senescence-associated cytokine secretion that is required for increased cell motility and changes in actin filaments. (a, b) Human cytokine array analysis of conditioned media from control, p21, MLCK and MLCK/p21 siRNA-treated MCF10A cells. Quantification of ICAM-1, IL-1α, IL-6 and IL-8. Fold changes were normalized to control cells. n =
= 2 independent experiments. One-way ANOVA with Holm-Šídák’s multiple comparisons test. (c) Relative mRNA transcripts of ICAM-1, IL-1a, IL-6 and IL-8 in control, p21, MLCK and MLCK/p21 siRNA-treated cells. n
2 independent experiments. One-way ANOVA with Holm-Šídák’s multiple comparisons test. (c) Relative mRNA transcripts of ICAM-1, IL-1a, IL-6 and IL-8 in control, p21, MLCK and MLCK/p21 siRNA-treated cells. n =
= 3 independent experiments. One-way ANOVA with Holm-Šídák’s multiple comparisons test. (d, e) Representative images and quantification of scratch wound migration assays performed under conditioned media from each siRNA-treated cell. Scale bars, 100 μm. n
3 independent experiments. One-way ANOVA with Holm-Šídák’s multiple comparisons test. (d, e) Representative images and quantification of scratch wound migration assays performed under conditioned media from each siRNA-treated cell. Scale bars, 100 μm. n =
= 3 independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. (f) Representative immunofluorescence images of actin (green), E-cadherin (red) and nucleus (blue). Scale bars, 20 μm. mean
3 independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. (f) Representative immunofluorescence images of actin (green), E-cadherin (red) and nucleus (blue). Scale bars, 20 μm. mean ±
± s.e.m. *P
s.e.m. *P <
< 0.05, **P
0.05, **P <
< 0.01, ****P
0.01, ****P <
< 0.0001.
0.0001.
We further investigated the role of p21-dependent SASP secretion in regulating cell migration by culturing cells with CM from control, p21, MLCK and MLCK/p21 siRNA-treated cells. CM from MLCK siRNA-treated cells increased migration, whereas CM from cells treated with both MLCK and p21 siRNAs restored migration rate (Fig. 3d-e). Interestingly, MLCK depletion increased the formation of actin filaments and impaired cell-cell adhesions, which are potential regulators of cell migration. Inhibition of p21 expression in MLCK-depleted cells restored both actin structure and cell-cell adhesions (Fig. 3f). Additionally, we found that oxidative stress-induced senescent cells exhibited similar phenotypic changes to those observed in MLCK-depleted cells. Short-term treatment with H2O2 in MCF10A cells induced oxidative stress, leading to decreased cell growth and increased SA-β-gal activity (Supplementary Fig. 1g-h). These H2O2-induced senescent cells exhibited increased actin filaments and impaired cell-cell adhesions (Supplementary Fig. 1i). These findings suggest that the phenotypic changes in MLCK-depleted cells may be caused by cellular senescence. Together, these findings demonstrate that p21 expression, stimulated by MLCK depletion, promotes SASP, reorganizes the actin cytoskeleton, and enhances cell migration in a non-cell-autonomous manner.
Downregulation of MLCK increases p21 expression via p53 and protein stability
The regulation of p21 is tightly controlled by p53 in most cancer types39,40. We questioned whether p53 is required for p21 induction in MLCK-depleted cells by silencing p53 together with MLCK and analyzing p21 expression. p53 depletion inhibited both p21 protein and mRNA upregulation in MLCK-depleted cells (Supplementary Fig. 3b-c). These data show that MLCK depletion stimulates p53-dependent p21 expression.
We also investigated whether MLCK affects p21 protein stability and degradation. To test this, we inhibited protein synthesis using cycloheximide. The protein level of p21 decreased over time, and MLCK depletion slowed p21 protein degradation compared to the control (Supplementary Fig. 3d-e). These findings suggest that MLCK regulates both p21 transcription and protein stability.
Downregulation of MLCK activates the AKT-mTOR signaling pathway
AKT-mTOR is often upregulated in senescent cells33,41. Specifically, p21 phosphorylation by AKT enhances p21 protein stability, thereby promoting tumor cell survival42. Moreover, AKT-mTOR signaling is known to elevate IL-1α and IL-6 levels in senescent cells43–45. Inhibition of the mTOR pathway using the rapamycin derivative RAD001 decreases p21 translation and increases the sensitivity of solid tumors to DNA damage-induced apoptosis46. We found that the AKT-mTOR signaling pathway, along with its downstream targets, S6 ribosomal protein kinase (S6K) and S6, are significantly activated in MLCK-depleted cells (Fig. 4a-b). Additionally, we found an increase in the phosphorylation of the NF-κB subunit p65, known as a master regulator of SASP genes, in MLCK-depleted cells (Fig. 4b). Western blot analysis validated that MLCK depletion increased the phosphorylation of AKT, mTOR, S6K and S6 (Fig. 4c). We used specific inhibitors to test whether AKT-mTOR signaling regulates p21 in MLCK-depleted cells. Inhibition of PI3K or AKT with LY294002 or MK-2206, respectively, reduced p21 induction in MLCK-depleted cells (Fig. 4d). Similarly, inhibition of mTOR signaling with Rapamycin or AZD8055 also decreased p21 induction (Fig. 4d-e). We also tested collective migration by treating control or MLCK-depleted cells with AZD8055 and observed both cell-autonomous and non-cell-autonomous effects of mTOR signaling. Importantly, MLCK depletion did not increase migration speed when AKT-mTOR signaling was inhibited (Fig. 4f). These findings suggest that AKT-mTOR signaling facilitates p21 expression and enhances migration in MLCK-depleted cells (Fig. 4g). In addition, and unexpectedly, we also noted that inhibition of p21 expression normalized AKT-mTOR signaling in MLCK-deficient cells (Fig. 4c). This suggests that not only does AKT-mTOR signaling increase p21 expression but p21 also activates AKT-mTOR (Fig. 4g). Combined, both pathways lead to increased SASP and cell migration.
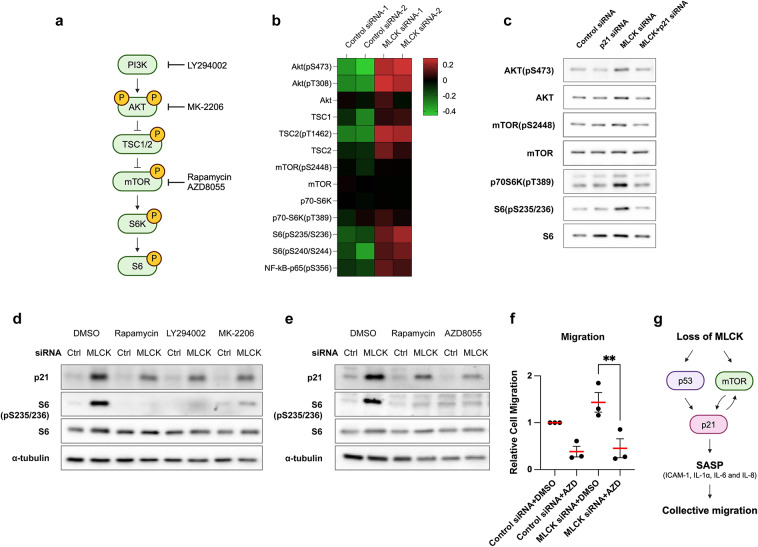
Downregulation of MLCK promotes cell migration via the AKT-mTOR signaling pathway. (a) Schematic diagram illustrating the AKT-mTOR signaling pathway and its selective inhibitors. (b) Heatmap showing the total and phosphorylated levels of AKT-mTOR signaling proteins from a reverse-phase protein array in control and MLCK-siRNA treated MCF10A cells. n =
= 2 independent experiments. (c) Western blot analysis of control, p21, MLCK, MLCK/p21 siRNA. Original blots are presented in Supplementary Fig. 4. (d, e) Western blot analysis following treatment with selective inhibitors for 24 h. α-tubulin is shown as a loading control. Original blots are presented in Supplementary Fig. 4. (f) Scratch wound assays were performed with selective inhibitors. Quantification of wound closed areas 12 h post-wounding. n
2 independent experiments. (c) Western blot analysis of control, p21, MLCK, MLCK/p21 siRNA. Original blots are presented in Supplementary Fig. 4. (d, e) Western blot analysis following treatment with selective inhibitors for 24 h. α-tubulin is shown as a loading control. Original blots are presented in Supplementary Fig. 4. (f) Scratch wound assays were performed with selective inhibitors. Quantification of wound closed areas 12 h post-wounding. n =
= 3 independent experiments. Ordinary one-way with ANOVA Tukey’s multiple comparisons test. mean
3 independent experiments. Ordinary one-way with ANOVA Tukey’s multiple comparisons test. mean ±
± s.e.m. **P < 0.01.
s.e.m. **P < 0.01. (g) Model diagram: Loss of MLCK enhances p21-induced SASP through p53 and AKT-mTOR signaling, thereby stimulating collective cell migration.
(g) Model diagram: Loss of MLCK enhances p21-induced SASP through p53 and AKT-mTOR signaling, thereby stimulating collective cell migration.
Discussion
The cytoskeleton regulates fundamental cellular processes, including survival, proliferation, differentiation and migration. However, the role of the cytoskeleton in senescent cells is poorly understood. Recent studies have indicated that multiple senescence-related alterations affect the reorganization of microtubules and actin cytoskeleton47,48. The cytoskeleton can in turn regulate senescence and cytokine secretion49,50. Our research showed that depletion of MLCK from MCF10A breast epithelial cells increases actin stress fibers and cellular senescence. The increase in stress fibers was unexpected since MLCK catalytic function stimulates actomyosin contractility, so MLCK depletion was expected to reduce stress fibers28. However, an association between increased stress fibers and premature senescence was also noted in AKAP12 knockout mouse embryo fibroblasts51 but the mechanism was unclear. In our system, both the increased stress fibers and senescence phenotypes required p21, which is induced when MLCK is absent. Thus, it is unlikely that the increased actin stress fibers are a direct result of reduced MLCK activity on actomyosin contractility but are secondary to gene expression changes.
Our previous study reported that MLCK expression is downregulated in various breast cancer cell lines compared to non-transformed breast epithelial cells28. MLCK depletion activates oncogenic signaling and migration in MCF10A cells, suggesting that MLCK may function as a tumor suppressor28. Another study also highlights the potential tumor suppressor role of MLCK by screening kinases that regulate oncogene-induced senescence in retinal pigmented epithelial cells52. Our study further supports the idea that MLCK depletion promotes pro-tumorigenic effects on migration by inducing cellular senescence. Importantly, the secretome from MLCK-depleted cells stimulates the releases of cytokines, including ICAM-1, IL-1α, IL-6 and IL-8, and promotes the migration of MCF10A cells and HER2-positive breast cancer cells, suggesting that MLCK depletion in a subset of cells could influence the migratory behavior of the entire tumor microenvironment and promote tumorigenesis in the neighboring cell.
Among the 36 cytokines that we tested, ICAM-1, IL-1α, IL-6 and IL-8 were selectively increased, while the levels of 32 SASP proteins remained unchanged upon MLCK depletion, suggesting that MLCK may be a selective SASP modulator. Increased levels of ICAM-1, IL-1α, IL-6 and IL-8 could potentially regulate the collective cell migration of MCF10A, SK-BR-3 and BT-474 cells in a non-cell-autonomous manner. ICAM-1 is a cytokine-inducible adhesion molecule, which showed the highest upregulation among the proteins secreted by MLCK-depleted cells. ICAM-1 is known to mediate the adhesion of breast cancer cells to bone marrow stromal cells, reducing the retention of hematopoietic stem and progenitor cells in the bone marrow niche53, and it affects matrix attachment, as seen in T cells. High serum levels of ICAM-1 are associated with malignant progression in breast cancer patients54. Decreased level of MLCK in bone or lung metastatic breast cancer30might be associated with their survival in metastatic sites and secretory factors like ICAM-1 are potentially involved. Additionally, IL-1α, IL-6 and IL-8 are well known cytokines that regulate inflammatory responses in tumor microenvironments7,55. Clinical studies have shown that breast cancers express high concentrations of IL-1α, IL-6 and IL-8 compared to normal tissue, strongly correlating with a malignant phenotype56–58. The paracrine secretion of IL-1α, IL-6 and IL-8 promotes breast cancer growth and invasion. For example, senescent skin fibroblasts secrete IL-6, which stimulates MCF-7 breast cancer cells to form tumors and invade in a xenograft mouse model59. Induction of IL-6 or IL-8 promotes epithelial-mesenchymal transition (EMT) phenotypes in breast cancer cells60,61. Interestingly, IL-6 and IL-8 synergize to promote tumor cell migration and metastasis through increased expression of WASF3 and the Arp2/3 complex, which are involved in actin cytoskeleton dynamics62. IL-1α, IL-6 and IL-8 levels are significantly upregulated in high CD44+/CD24- breast cancer cells, which exhibit high invasive properties63. Besides, inhibition of IL-1α has been shown to reduce IL-6 and IL-8 expression in breast cancer cells, suggesting that IL-1α, IL-6 and IL-8 have cooperative effects on breast cancer migration64. Thus, the secretion of IL-1α, IL-6 and IL-8 in MLCK-depleted breast epithelial cells potentially initiates cell migration and promotes breast cancer metastasis. While we analyzed major pro-inflammatory molecules known to mediate the SASP65, it remains to be determined whether MLCK depletion leads to the release of growth factors and proteases, and what the systemic effects of these secretions are in an in vivo model.
P21, upregulated by MLCK depletion, was required for the observed senescence-associated phenotypes. Interestingly, we observed no increase in other genes associated with the G1 cell-cycle arrest, which are typical markers of senescent cells. Although p21 is widely regarded as a tumor suppressor and a negative regulator of cell-cycle progression, high levels of p21 in breast cancer patients correlate with poor overall or metastasis-free survival and are implicated in tumor-promoting roles across several cancer types13,66. Thus, the role of p21 as either a promoter or suppressor of cancer depends on cell context. In our study, p21 not only enhanced cytokine secretion and disrupted actin organization but also facilitated collective cell migration while suppressing cell proliferation in MLCK-depleted breast cells. These distinct actions of p21 are consistent with findings that invasive cancer cells often show lower proliferation than those within the main tumor mass11. We also demonstrated that p21 accumulation is regulated by both p53 and AKT-mTOR signaling in MLCK-depleted cells. Activation of AKT-mTOR signaling impacts protein levels by regulating translational initiation, protein synthesis and degradation67–69, potentially influencing p21 level in MLCK-depleted cells. In addition, crosstalk between mTOR and p53 signaling regulates cellular senescence70,71. For example, mTOR directly binds to p53, interfering with p53-MDM2 binding and increasing p21 accumulation in PTEN-loss-induced senescent cells72, indicating that mTOR can act upstream of p53 activation and p21 accumulation in senescent cells. AKT-mTOR inhibition may be a possible new therapeutic target for breast cancers with low MLCK expression.
Cellular senescence is thought to act as a distinct tumor-suppressing mechanism, initiated by cell-cycle arrest. However, the loss of tumor suppressor genes such as MLCK in breast epithelial cells can trigger the release of SASP, potentially fostering conditions in neighboring cells leading to increased migration. Since cancer treatments can induce senescence, enabling cells to evade conventional anti-cancer therapies, understanding the molecular mechanisms behind senescence and SASP are crucial for enhancing cancer treatment strategies.
Methods
Cell lines and culture
MCF10A cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium/F12 (Welgene, LM002-04; Gibco, 11320082) supplemented with 5% horse serum (Gibco, 16050), 20 ng/ml EGF (Peprotech, AF-100-15), 10 µg/ml bovine insulin (Roche, 11 376 497 001), 0.5 µg/ml hydrocortisone (Sigma, H0888), 0.1 µg/ml cholera toxin (Sigma, C8052) and 1% penicillin/streptomycin (Welgene, LS202-02). SK-BR-3 and BT-474 breast cancer cells were obtained from the ATCC and cultured in RPMI-1640 medium (Gibco, 22400-089) containing 10% fetal bovine serum (Gibco, 16000-044) and 2 mM L-glutamine (Gibco, 25030-081). Cells were incubated at 37 °C in a humidified 5% CO₂ incubator.
siRNAs
For siRNA transfection, 1.0 ×
× 10⁵ cells were plated per well in a six-well plate and allowed to adhere for 24 hours. Transfections were performed using 25 nM siRNA with 3.5 µl of Lipofectamine RNAiMAX reagent (Invitrogen, 13778150) or Lipofectamine 2000 reagent (Invitrogen, 11668019), following the manufacturer’s protocol. Human negative control siRNA (ON-TARGET non-targeting control siRNA, Horizon) was used as an independent control. We employed a mixture of human MYLK siRNAs targeting four different sites within the open reading frame and the 3′ untranslated region (siGENOME SMARTpool, Horizon, M-005351-05-005) or a human MYLK siRNA targeting the 3′ untranslated region (Bioneer, 110543). Additionally, siRNAs targeting human CDKN1A (Bioneer, 1026-1 and 1026-2) and human TP53 (Bioneer, 1155302) were used. The sequences of the siRNAs are provided in Table 1. Subsequent analyses were conducted 72 h after the initial transfection.
10⁵ cells were plated per well in a six-well plate and allowed to adhere for 24 hours. Transfections were performed using 25 nM siRNA with 3.5 µl of Lipofectamine RNAiMAX reagent (Invitrogen, 13778150) or Lipofectamine 2000 reagent (Invitrogen, 11668019), following the manufacturer’s protocol. Human negative control siRNA (ON-TARGET non-targeting control siRNA, Horizon) was used as an independent control. We employed a mixture of human MYLK siRNAs targeting four different sites within the open reading frame and the 3′ untranslated region (siGENOME SMARTpool, Horizon, M-005351-05-005) or a human MYLK siRNA targeting the 3′ untranslated region (Bioneer, 110543). Additionally, siRNAs targeting human CDKN1A (Bioneer, 1026-1 and 1026-2) and human TP53 (Bioneer, 1155302) were used. The sequences of the siRNAs are provided in Table 1. Subsequent analyses were conducted 72 h after the initial transfection.
Table 1
siRNAs used in this study.
| siRNA | Catalog | Target sequence |
|---|---|---|
| Human MYLK siRNA #1 | siGENOME D-005351-05 | UAAGACCAUUCGCGAUUUA |
| Human MYLK siRNA #2 | siGENOME D-005351-06 | AGAUUUGACUGCAAGAUUG |
| Human MYLK siRNA #3 | siGENOME D-005351-07 | GGGAUGACGAUGCCAAGUA |
| Human MYLK siRNA #4 | siGENOME D-005351-17 | CUGCAAGAUUGAAGGAUAC |
| Human MYLK siRNA #5 | Bioneer 1100543 | GAUUAUCGGUUAGGUCAUU |
| Human CDKN1A siRNA | Bioneer 1026-1 | CUGUACUGUUCUGUGUCUU |
| Human CDKN1A siRNA | Bioneer 1026-2 | CAGUUCAUUGCACUUUGAU |
| Human TP53 siRNA | Bioneer 1155302 | CACUACAACUACAUGUGUA |
Senescence assessment
Cell proliferation: 1.0 ×
× 105 cells were plated and incubated at 37 °C, 5% CO2. Every 24 h, the cells were trypsinized and incubated with Trypan blue solution (Invitrogen, 15250061) for 3 min. Live and dead cells were then counted using a hemocytometer.
105 cells were plated and incubated at 37 °C, 5% CO2. Every 24 h, the cells were trypsinized and incubated with Trypan blue solution (Invitrogen, 15250061) for 3 min. Live and dead cells were then counted using a hemocytometer.
Ki-67 staining and nuclear size: Fixed and permeabilized cells were incubated with a rabbit anti-Ki-67 antibody (Abcam, ab1667), followed by incubation with a secondary goat anti-mouse Alexa 488-conjugated antibody (Invitrogen, A11001). Cells were then incubated with 4’,6-diamidino-2-phenylindole (DAPI, D-8417) for 5 min. Ki-67 positive cells were counted using the ZEISS AxioVision 4.8 AutoMeasure module (Zeiss, Germany). Nuclear sizes were measured using Image J.
SA-β-gal staining: Cells were fixed with 4% formaldehyde for 5 min at room temperature. After washing the cells twice with PBS, they were incubated with SA-β-gal staining solution in the dark for 12–16 h at 37 °C73. For SA-β-gal positivity, random fields are shown. SA-β-gal staining was photographed using a Nikon Eclipse TS100. Quantification was performed using ImageJ.
Immunofluorescence
For immunofluorescence, cells were grown on 12 mm round coverglass and co-fixed and permeabilized with 4% formaldehyde and 0.1% Triton X-100 in PBS for 2 min, followed by an additional 15-minute incubation in 4% formaldehyde. Primary antibodies in 1% BSA /PBS were incubated overnight at 4 °C. The following primary antibodies were used: rabbit anti-Ki-67 (Abcam, ab1667, 1:500), rabbit anti-p21 Waf1/Cip1 (Cell Signaling Technology, 2947, 1:200), mouse anti-E-cadherin (BD Bioscience, 610181, 1:200) and mouse anti-β-catenin (BD Bioscience, 610153, 1:200). Oregon Green 488 phalloidin (Invitrogen, O7466) was mixed with goat anti-rabbit Alexa 594 (Invitrogen, A11012) or goat anti-mouse Alexa 594 (Invitrogen, A11005) and incubated for 30 min at room temperature. After a 5-minute incubation with DAPI, the cover glasses were mounted with ProLong Gold Antifade Mountant (Invitrogen, P36934). Cells were observed using a Zeiss Observer Z1 microscope with Apotome 2 or a Leica Stellaris 5 confocal microscope.
Generation of conditioned medium
To collect conditioned medium from control, MLCK, and/or p21 siRNA-transfected cells, the cells were initially transfected with siRNAs. After a 72-hour incubation, the medium from each transfected culture dish was collected and centrifuged at 900 rpm for 3 min. The cell culture supernatant was then filtered through a 0.45 μm syringe filter to remove any remaining cell debris.
Human cytokines assay
To measure secretory proteins in the collected conditioned media, we used the Proteome Profiler Human Cytokine Array Kit (R&D Systems, ARY005B). The array included 36 different cytokine antibodies, each spotted in duplicate on nitrocellulose membranes. Conditioned media were incubated with the antibody mixtures and detected with streptavidin-horseradish peroxidase. The chemiluminescent signals were captured and processed with the Chemi-DOC MP imaging system (Bio-Rad). Quantification was performed using ImageJ.
Scratch wound assay
Cells were cultured on a six-well plate until a confluent monolayer was formed. A wound was scratched across the cell monolayer using 200 µl tips, followed by a change to conditioned medium or culture medium containing DMSO (Sigma, D8418) or AZD8055 (Selleckchem, S1555). For BT-474 cells, they were plated on a three-well silicone insert (Ibidi, 80369) until confluent, and the insert was then removed for migration assays. Phase-contrast images were captured every 6 h for 24 h using a Nikon Eclipse TS100. The area covered by migrating cells was measured at 12 h (MCF10A), 24 h (SK-BR-3) or 6 h (BT-474) post-wounding using ImageJ. For time-lapse imaging, images were taken every 10 min for 16 h on an Incucyte S3 Live-Cell Analysis System (Sartorius).
Reverse phase protein array
Cultured MCF10A cell lysates were denatured by sodium dodecyl sulfate and analyzed at the RPPA Core facility at MD Anderson Cancer Center (Houston, TX, USA). Protein lysates were serially diluted and arrayed on nitrocellulose-coated slides. Each slide was probed with validated primary antibodies, and signals were visualized using a 3,3’-diaminobenzidine (DAB) colorimetric reaction.
Western blot
Protein lysates were collected using ice-cold 2X Laemmli sample buffer (0.125 M Tris-HCl, 20% glycerol, 4% sodium dodecyl sulfate and 0.004% Bromophenol blue) with an additional protease inhibitor cocktail and PhosSTOP (Roche, 4906845001). The total protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The membrane was blocked with 5% skim milk for 30 min and then incubated overnight at 4 °C with the following primary antibodies: MLCK (K-36) (Sigma, M7905), p21 (Cell Signaling Technology, 2947), p53 (BD Transduction Lab, P21020), AKT (Cell Signaling Technology, 9272), Phospho-AKT (Ser473) (Cell Signaling Technology, 9271), mTOR (Cell Signaling Technology, 2983), Phospho-mTOR (Ser2448) (Cell Signaling Technology, 5536), Phospho-p70 S6 Kinase (Thr389) (Cell Signaling Technology, 9234), S6 Ribosomal Protein (Cell Signaling Technology, 2217), Phospho-S6 Ribosomal Protein (Ser235/236) (Cell Signaling Technology, 4858), Integrin β1 (Cell Signaling Technology, 4706), β-actin (Sigma, A5441) and α-tubulin (Sigma, T5168). The membrane was then incubated with peroxidase-conjugated AffiniPure goat anti-mouse IgG or goat anti-rabbit IgG (Jackson ImmunoResearch) for 1 h at room temperature. Peroxidase substrate was applied, and chemiluminescence images were captured and quantified using the Chemi-DOC MP imaging system (Bio-Rad).
RNA isolation and reverse transcription-PCR
Total RNA was prepared using the Hybrid-R RNA isolation kit (GeneAll, 305 −
− 101), according to the manufacturer’s instructions. An equal amount of RNA was used for each sample, and mRNA transcripts were measured using the PrimeScript One Step RT-PCR Kit (Takara, RR055A/B). The primer sets for p21 (CDKN1A), MLCK (MYLK), ICAM-1 (ICAM1), IL-1α (IL1A), IL-6 (IL6), IL-8 (CXCL8) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are listed in Table 2.
101), according to the manufacturer’s instructions. An equal amount of RNA was used for each sample, and mRNA transcripts were measured using the PrimeScript One Step RT-PCR Kit (Takara, RR055A/B). The primer sets for p21 (CDKN1A), MLCK (MYLK), ICAM-1 (ICAM1), IL-1α (IL1A), IL-6 (IL6), IL-8 (CXCL8) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are listed in Table 2.
Table 2
Primer used for reverse transcription-PCR.
| Gene name | Primer | Sequence |
|---|---|---|
| CDKN1A | Forward | CCAAGAGGAAGCCCTAATCC |
| CDKN1A | Reverse | CCCTTCAAAGTGCCATCTGT |
| MYLK | Forward | ATGCTGTCCATGAGGAGGAC |
| MYLK | Reverse | ACGTGTACACTCCCACGTCA |
| ICAM1 | Forward | CAGAGGTTGAACCCCACAGT |
| ICAM1 | Reverse | CATTGGAGTCTGCTGGGAAT |
| IL1A | Forward | GTAAGCTATGGCCCACTCCA |
| IL1A | Reverse | CTTCATCTTGGGCAGTCACA |
| IL6 | Forward | AAAGAGGCACTGGCAGAAAA |
| IL6 | Reverse | GAGGTGCCCATGCTACATTT |
| CXCL8 | Forward | TATAAAAAGCCACCGGAGCA |
| CXCL8 | Reverse | AAATTTGGGGTGGAAAGGTT |
| GAPDH | Forward | GTCAGTGGTGGACCTGACCT |
| GAPDH | Reverse | AGGGGAGATTCAGTGTGGTG |
Hydrogen peroxide-induced senescence
MCF10A cells were seeded at a density of 5 ×
× 104 cells/ml for 24 h. The next day, cells were treated with 250 µM H₂O₂ (Fisher Scientific, H325-500) diluted in DMEM/F12 for 1 h. After treatment, the cells were switched to normal growth medium and cultured for an additional 3 days. To validate cellular senescence, SA-β-gal staining was performed. For comparison at a similar cell density, an equal number of normal or H₂O₂-induced senescent cells were plated onto 12 mm coverglass for 24 h.
104 cells/ml for 24 h. The next day, cells were treated with 250 µM H₂O₂ (Fisher Scientific, H325-500) diluted in DMEM/F12 for 1 h. After treatment, the cells were switched to normal growth medium and cultured for an additional 3 days. To validate cellular senescence, SA-β-gal staining was performed. For comparison at a similar cell density, an equal number of normal or H₂O₂-induced senescent cells were plated onto 12 mm coverglass for 24 h.
p21 protein stability and expression
For p21 protein stability, cells were treated with either DMSO (Sigma, D8418) or 20 µg/ml cycloheximide (Sigma, C7698) at various time points. Protein lysates were collected at 0, 10, 30, 60 and 90 min after the addition of cycloheximide, and p21 levels were analyzed by Western blot. For p21 expression studies, cells were treated with DMSO, LY294002 (Calbiochem, 440202), MK-2206 (Selleckchem, S1078), Rapamycin (Selleckchem, S1039) and AZD8055 (Selleckchem, S1555) for 24 h.
Statistical analysis and graphics
GraphPad Prism v.10 was used for statistical analysis and data graphing. P values were determined using unpaired t-test, paired t-test, ratio paired t-test, ordinary one-way ANOVA with Dunnett’s multiple comparisons test, one-way ANOVA with Holm-Šídák’s multiple comparisons test, ordinary one-way ANOVA with Tukey’s multiple comparisons test or two-way ANOVA with Sidak’s multiple comparisons test. The number of replicates is described in the figure legends. Schematic figures were prepared using Biorender.com.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was financially supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. NFR 2018R1A2A2A05021262), by High Risk High Return Project funds from Korea Advanced Institute of Science and Technology and institutional funds from Fred Hutchinson Cancer Center.
Author contributions
Conceptualization, Formal Analysis, Investigation, Methodology, Validation and Visualization, D.K and D.M.H.; Writing – Original Draft Preparation, Writing – Review & Editing, Resources and Funding acquisition, D.K., J.A.C., and D.M.H; Supervision, D.M.H.
Data availability
The data are available from the corresponding author upon reasonable request.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Articles from Scientific Reports are provided here courtesy of Nature Publishing Group
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/169804528
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cooperation between p21 and Akt is required for p53-dependent cellular senescence.
Aging Cell, 16(5):1094-1103, 09 Jul 2017
Cited by: 61 articles | PMID: 28691365 | PMCID: PMC5595696
IGFBP-rP1 induces p21 expression through a p53-independent pathway, leading to cellular senescence of MCF-7 breast cancer cells.
J Cancer Res Clin Oncol, 138(6):1045-1055, 06 Mar 2012
Cited by: 34 articles | PMID: 22392074
Human pituitary tumor-transforming gene 1 overexpression reinforces oncogene-induced senescence through CXCR2/p21 signaling in breast cancer cells.
Breast Cancer Res, 14(4):R106, 12 Jul 2012
Cited by: 12 articles | PMID: 22789011 | PMCID: PMC3680924
Expression of hepaCAM is downregulated in cancers and induces senescence-like growth arrest via a p53/p21-dependent pathway in human breast cancer cells.
Carcinogenesis, 29(12):2298-2305, 08 Oct 2008
Cited by: 35 articles | PMID: 18845560
Funding
Funders who supported this work.
Fred Hutchinson Cancer Center
National Research Foundation of Korea (1)
Grant ID: NFR 2018R1A2A2A05021262

 1,2
1,2 

