Abstract
Free full text

GPRASP protein deficiency triggers lymphoproliferative disease by affecting B‐cell differentiation
Abstract
Gprasp1 and Gprasp2 encode proteins that control the stability and cellular trafficking of CXCR4, a master regulator of hematopoiesis whose dynamic regulation is required for appropriate trafficking of B‐cells in the germinal center (GC). Here, we report that Gprasp1 and Gprasp2‐deficient B‐cells accumulate in the GC and show transcriptional abnormalities, affecting the mechanisms controlling Aicda expression and exposing them to excessive somatic hypermutation. Consequently, about 30% of mice transplanted with Gprasp‐deficient hematopoietic stem and progenitor cells developed a biologically aggressive and fatal B‐cell hyperproliferative disease by 20–50 weeks posttransplant. Histological and molecular profiling reveal that Gprasp1‐ and Gprasp2‐deficient neoplasms morphologically resemble human high‐grade B‐cell lymphomas of germinal center origin with shared morphologic features of both Burkitt Lymphoma (BL) and diffuse large B‐cell lymphoma (DLBCL), and molecular features consistent with DLBCL, as well as elevated mutational burden and heterogenous transcriptional and mutational signature. Thus, reduced Gprasp1 and Gprasp2 gene expression perturbs B‐cell maturation and increases the risk of B‐cell neoplasms of germinal center origin. As this model recapitulates the essential features of the heterogenous group of human hematopoietic malignancies, it could be a powerful tool to interrogate the mechanisms of lymphomagenesis for these cancers.
INTRODUCTION
The mechanisms that drive normal B‐cell differentiation and activation are frequently subverted in B‐cell lymphomas, which promotes uncontrolled growth and survival. 1 Germinal center (GC) B‐cells are at a particularly high risk for malignant transformation due to attenuation of specific DNA damage and cell proliferation checkpoints essential for immunoglobulin affinity maturation. 2 Although the GC stage is tightly regulated, somatic hypermutation (SHM) can disrupt this equilibrium by generating off‐target mutations that could potentially impose a selective advantage on maturing B‐cells. 2 , 3 Indeed, some healthy individuals harbor premalignant populations of mutant B‐cells, 4 , 5 although it is currently not possible to discern who is at risk for transformation to overt disease. 2 Revelations in the regulation of GC epigenetics, metabolism, signaling, and immune synapses have revealed that these regulatory processes can be hijacked to facilitate lymphomagenesis. 2 In the GC, B‐cells acquire the molecular profiles that determine their fate—proliferation, apoptosis, export—depending on the affinity of their surface receptors for antigen. 6 CXCR4 has a crucial role in the regulation of the dynamic transitions between the light zone and the dark zone of the GC, which is critical for B‐cell differentiation. 7 Indeed, dysregulation of CXCR4, among other lesions identified, contributes to the progression and survival of neoplastic B‐cells, conferring prognostic value to CXCR4. 2 , 3 , 8 , 9 Mature B‐cell malignancies, such as high‐grade B‐cell lymphomas, represent a medical challenge that is only partly met by current therapy, justifying concerted investigation into their molecular circuitry and pathogenesis. Although studies of normal B‐cell biology have yielded insight into the pathogenesis of these B‐cell lymphomas, 1 as mechanisms of oncogenesis are revealed, gaps in our understanding of normal B‐cell function remain.
We recently showed that Gprasp1 and Gprasp2 encode proteins that control the stability and cellular trafficking of CXCR4. 10 Little is known about the function of GPRASP (G protein‐coupled receptors [GPCR]‐associated sorting protein) family members, although several have been implicated in GPCR trafficking and signaling and mitochondria trafficking. 11 Deficiency of either Gprasp1 or Gprasp2 slows the rate of CXCR4 degradation, allowing for its accumulation at the cell surface of hematopoietic progenitors. 10 We report that about 30% of mice transplanted with Gprasp1‐ or Gprasp2‐deficient hematopoietic stem and progenitor cells (HSPCs) develop a fatal, high‐grade B‐cell malignancy 20–50 weeks posttransplant. Prior to the onset of overt malignancy, Gprasp1‐ and Gprasp2‐deficient B‐cells accumulate in the GC, and mature B‐cells display increased CXCR4 expression and transcriptional changes in key regulatory factors of SHM. These data support a model where Gprasp1‐ and Gprasp2‐deficient B‐cells arrest in the GC, increasing the risk of malignant transformation. Our study is the first to implicate Gprasp genes in B‐cell differentiation and lymphomagenesis.
METHODS
Transplants
Lineage−Sca‐1+c‐Kit+ (LSK) cells were transplanted into CD45.1+/CD45.2+ recipient mice preirradiated with 11 Gy (two doses of 5.5
Gy (two doses of 5.5 Gy separated by 3
Gy separated by 3 h). CD45.2+mCherry+ peripheral blood (PB) cell frequency was assessed by flow cytometry every 4 weeks posttransplant for at least 20 weeks. CD45.2+mCherry+ BM HSPC frequency was examined at 20 weeks posttransplant.
h). CD45.2+mCherry+ peripheral blood (PB) cell frequency was assessed by flow cytometry every 4 weeks posttransplant for at least 20 weeks. CD45.2+mCherry+ BM HSPC frequency was examined at 20 weeks posttransplant.
HSPC culture and viral transduction
HSPCs (LSK cells) were isolated and transduced with lentivirus as previously described.
12
To collect cells for transplantation 48 h after transduction, media was slowly removed, and cells washed and resuspended in phosphate‐buffered saline
h after transduction, media was slowly removed, and cells washed and resuspended in phosphate‐buffered saline +
+ 1.5% fetal bovine serum. CD45.1+ competitor cells were processed as describe in this section, but never put in contact with lentiviral particles.
1.5% fetal bovine serum. CD45.1+ competitor cells were processed as describe in this section, but never put in contact with lentiviral particles.
Transcriptional profiling (RNAseq)
Specific populations were sorted directly into lysis buffer and total RNA was isolated (RNeasy Micro kit; QIAGEN). Sequencing was performed at Vantage (Vanderbilt University Medical Center). Sequencing was performed at Paired‐End 150 bp on the Illumina NovaSeq 6000 targeting an average of 50
bp on the Illumina NovaSeq 6000 targeting an average of 50 M reads/sample. For samples with replicates, standard differential gene expression was performed. For the malignant samples, due to their uniqueness, single‐subject RNA‐Seq analysis was performed, using iDEG. Gene set enrichment analysis was performed using the online tool WebGestalt
13
and with Ingenuity Pathway Analysis software (QIAGEN).
M reads/sample. For samples with replicates, standard differential gene expression was performed. For the malignant samples, due to their uniqueness, single‐subject RNA‐Seq analysis was performed, using iDEG. Gene set enrichment analysis was performed using the online tool WebGestalt
13
and with Ingenuity Pathway Analysis software (QIAGEN).
Mutational profile
Control cells and tumor cells were sorted directly into lysis buffer and genomic DNA was isolated (DNeasy Blood and Tissue kit; QIAGEN). Sequencing was performed at Vantage (Vanderbilt University Medical Center). Library preparation and capture were done utilizing the XGen Research Panel probes from IDT. Whole Exome Sequencing (WES) was performed at Paired‐End 150 bp on the Illumina NovaSeq 6000, targeting an average of 40
bp on the Illumina NovaSeq 6000, targeting an average of 40 M reads/sample (×100 coverage).
M reads/sample (×100 coverage).
Additional details for methods regarding mice, pathology analyses, flow cytometry, HSPC isolation, transplant, gene expression analysis, shRNAs, lentivirus production and transduction, colony assays, genotyping, immunoglobulin rearrangement, sample preparation for pathology analysis, and statistics are provided in the File S1. All experiments involving animals were carried out according to procedures approved by the St. Jude Children's Research Hospital Institutional Animal Care and Use Committee.
RESULTS
Gprasp1‐ or Gprasp2‐deficiency increases the risk of mature, high‐grade B‐cell lymphoma
We previously described Gprasp1 and Gprasp2 as negative regulators of acute HSPC repopulating activity.
10
To better understand the long‐term consequences of Gprasp1 and Gprasp2 loss in HSPCs, we isolated HSPCs (defined as Lineage−Sca1+cKit+ [LSK] cells) from CD45.2+ C57Bl/6J mice and then transduced them with control, Gprasp1‐ or Gprasp2‐shRNAs. Transduced cells also express a mCherry reporter (Supporting Information S1: Figure 1A). Forty‐eight hours posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+) (Figure 1A). Knockdown efficiency for each gene was verified by RT‐qPCR (Supporting Information S1: Figure 1B). Recipients transplanted with HSPCs transduced with Gprasp1‐ or Gprasp2‐shRNAs displayed significantly increased CD45.2+mCherry+ peripheral blood (PB) through 20 weeks posttransplant, as previously reported (Figure 1B).
14
Here, we continued to monitor recipient PB for nearly 1‐year posttransplant (Figure 1B). CD45.2+mCherry+ PB reconstitution trended up over time in recipients of Gprasp1‐ or Gprasp2‐deficient HSPCs, relative to controls. This repopulating advantage was unbiased with respect to lineage output and maintained for 48 weeks (Figure 1B, Supporting Information S1: Figure 1C). Strikingly, about 30% of recipients transplanted with Gprasp1‐ (17/48) or Gprasp2‐ (11/38) deficient HSPCs displayed symptoms of illness beginning around 20 weeks post‐transplant (e.g., hunched, scruffy, lethargic, lower limbs paralysis, Figure 1C) and loss of immunophenotypic B cells (B220+ cells) in the peripheral blood (Supporting Information S1: Figure 1D). Pathological examination revealed a widely disseminated hematopoietic neoplasm in these mice that was accompanied by splenomegaly, regional lymphadenopathy, multi‐organ invasion, and organomegaly, that led to eventual death or necessitated humane euthanasia (Figure 1D and Supporting Information S1: Figure 1E–G). In contrast, none of the mice transplanted with control shRNA‐treated HSPCs developed disease (0/23) (Figure 1C–E and Supporting Information S1: Figure 1E–G). The diffusely infiltrative neoplastic population consisted of medium‐sized neoplastic lymphoid cells in a background of small benign appearing CD3‐positive lymphocytes with condensed chromatin pattern that were not immunolabeled for mCherry. Neoplastic cells had a rounded nucleus with open/vesicular chromatin pattern and 1–2 prominent nucleoli associated with the nuclear membrane. The histologic features of neoplastic lymphoid cells were most consistent with a centroblastic morphology. Greater than 50% of the neoplastic populations in all cases (17/17 for Gprasp1‐shRNA and 11/11 for Gprasp2‐shRNA) had this morphology, as determined by light microscopy. In all cases, mitoses were variable and ranged from 0 to 4 based on random assessment of 10 high‐powered fields with an area calculated as 2.37 mm2 (Figure 1E and Supporting Information S1: Figure 1E,F). There were scattered tingible body macrophages consistent with a high proliferation rate and enhanced apoptosis in neoplastic cells arising from the primary transplants. Neither enhanced fibrosis nor stromal deposition were observed (Figure 1E). The morphologic features of the tumor cells were consistent with BL or DLBCL. All high‐grade B‐cell neoplastic cells expressed mCherry, suggesting that the cell‐intrinsic effect of Gprasp1 or Gprasp2 knockdown was causal for malignancy and excluding the possibility of age‐related background B‐cell lymphomas that may arise in laboratory mice. Histopathologic analysis for specific molecular markers revealed neoplastic cells to be PAX5+CD3−TDT−IgM+IRF4+/−KLC+CD43−CD25−LMO2+, an immunophenotype which is highly suggestive of a B‐cell lymphoma of GC origin (Figure 1E and Supporting Information S1: Figure 1H,I).
15
,
16
,
17
mm2 (Figure 1E and Supporting Information S1: Figure 1E,F). There were scattered tingible body macrophages consistent with a high proliferation rate and enhanced apoptosis in neoplastic cells arising from the primary transplants. Neither enhanced fibrosis nor stromal deposition were observed (Figure 1E). The morphologic features of the tumor cells were consistent with BL or DLBCL. All high‐grade B‐cell neoplastic cells expressed mCherry, suggesting that the cell‐intrinsic effect of Gprasp1 or Gprasp2 knockdown was causal for malignancy and excluding the possibility of age‐related background B‐cell lymphomas that may arise in laboratory mice. Histopathologic analysis for specific molecular markers revealed neoplastic cells to be PAX5+CD3−TDT−IgM+IRF4+/−KLC+CD43−CD25−LMO2+, an immunophenotype which is highly suggestive of a B‐cell lymphoma of GC origin (Figure 1E and Supporting Information S1: Figure 1H,I).
15
,
16
,
17
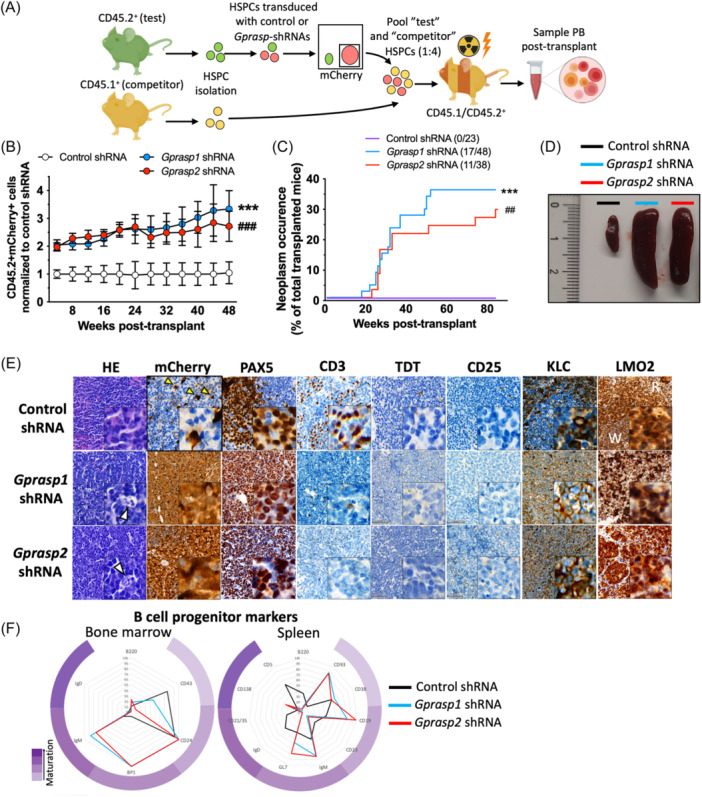
Silencing of
Gprasp1
or
Gprasp2
enhances HSPC in vivo hematopoietic repopulating activity and increases the risk for B‐cell lymphoma. (A) Schematic for transplantation assay. CD45.2+ “test” LSK cells were transduced with control or Gprasp‐shRNAs and then transplanted along with CD45.1+ “competitor” LSK cells into lethally irradiated recipients. Two different shRNAs were used for each gene (Gprasp1‐shRNA‐A =
= 38 recipients, Gprasp1‐shRNA‐B
38 recipients, Gprasp1‐shRNA‐B =
= 10 recipients, Gprasp2‐shRNA‐A
10 recipients, Gprasp2‐shRNA‐A =
= 28 recipients and Gprasp2‐shRNA‐B
28 recipients and Gprasp2‐shRNA‐B =
= 10 recipients). Recipient PB was analyzed for CD45.2+ mCherry+ cells. (B)
Gprasp‐deficient HSPCs display enhanced PB repopulating activity relative to HPSCs treated with control‐shRNA. All mice were included in this data, except for those that died and could not be recovered for analysis. (C) Recipients of Gprasp‐deficient HSPCs show increased frequency of neoplasms. (D) Recipients of Gprasp‐deficient HSPCs suffering neoplasm experience splenomegaly. Representative pictures of the spleen of control and mice where neoplasms were found. (E) Representative images from analysis of histopathologic markers in healthy (control‐shRNA) and neoplastic (Gprasp1 and Gprasp2‐shRNA) samples in the spleen. White arrows in HE highlights tingible body macrophages and the starry sky pattern. Yellow arrows in mCherry indicate donor‐derived cells in control sample. White (W) and red (R) pulp indicated in LMO2 control staining. (F) Expression pattern of B‐cell maturation markers in the BM and spleens from Control (healthy samples), Gprasp1 and Gprasp2‐shRNA recipients (only including those mice where neoplasms were confirmed). Data in (B) from ≥5 independent transplants with N
10 recipients). Recipient PB was analyzed for CD45.2+ mCherry+ cells. (B)
Gprasp‐deficient HSPCs display enhanced PB repopulating activity relative to HPSCs treated with control‐shRNA. All mice were included in this data, except for those that died and could not be recovered for analysis. (C) Recipients of Gprasp‐deficient HSPCs show increased frequency of neoplasms. (D) Recipients of Gprasp‐deficient HSPCs suffering neoplasm experience splenomegaly. Representative pictures of the spleen of control and mice where neoplasms were found. (E) Representative images from analysis of histopathologic markers in healthy (control‐shRNA) and neoplastic (Gprasp1 and Gprasp2‐shRNA) samples in the spleen. White arrows in HE highlights tingible body macrophages and the starry sky pattern. Yellow arrows in mCherry indicate donor‐derived cells in control sample. White (W) and red (R) pulp indicated in LMO2 control staining. (F) Expression pattern of B‐cell maturation markers in the BM and spleens from Control (healthy samples), Gprasp1 and Gprasp2‐shRNA recipients (only including those mice where neoplasms were confirmed). Data in (B) from ≥5 independent transplants with N =
= 5 recipients/condition/transplant. Data in (B) represented as mean
5 recipients/condition/transplant. Data in (B) represented as mean ±
± SEM. Data in (F) as mean from 5 Control‐shRNA, 10 Gprasp1‐shRNA mice and 7 Gprasp2‐shRNA mice. **/##
p
SEM. Data in (F) as mean from 5 Control‐shRNA, 10 Gprasp1‐shRNA mice and 7 Gprasp2‐shRNA mice. **/##
p <
< 0.01 ***/###
p
0.01 ***/###
p <
< 0.001 relative to control. * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
0.001 relative to control. * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
We isolated mCherry+ whole BM (WBM) cells from mice that developed tumors and performed secondary and tertiary transplants into sub‐lethally irradiated recipients (CD45.1+/CD45.2+) (Figure 2A). Disease onset was accelerated and nearly fully penetrant in secondary and tertiary transplant recipients, relative to primary transplants (Figure 2B). Splenomegaly was exacerbated by enhanced neoplastic engraftment (Figure 2C).
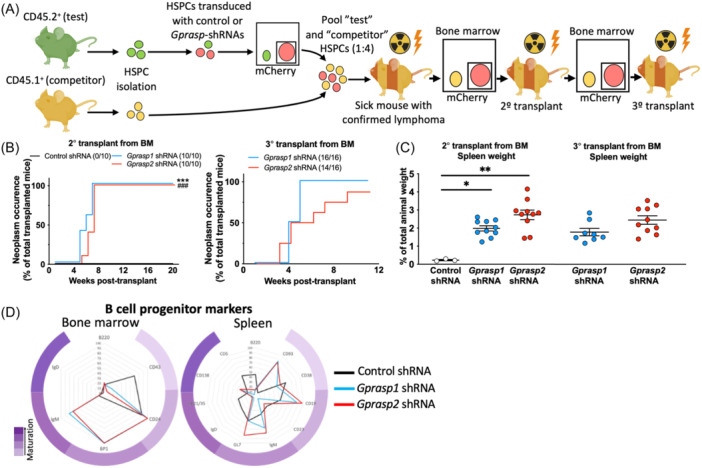
Neoplasms derived from
Gprasp1
or
Gprasp2‐
deficient progenitors are transplantable and increase their aggressiveness overtime. (A) Schematic for serial transplantation assay. Whole bone marrow mCherry+ cells from the mice confirmed of suffering B‐cell lymphoma from Figure 1C were serially transplanted into sublethally irradiated (2 ×
× 485
485 cGy) recipients. Whole bone mCherry+ cells from control mice were serially transplanted for control condition. (B) Neoplasm occurrence in secondary and tertiary recipients. (C) Spleen size in mice from (A). (D) Expression pattern of B‐cell maturation markers in the BM and spleens from Control (healthy samples), Gprasp1 and Gprasp2‐shRNA recipients (only including those mice where neoplasms were confirmed) after secondary transplant. Data in (B, C) from two independent transplants with N
cGy) recipients. Whole bone mCherry+ cells from control mice were serially transplanted for control condition. (B) Neoplasm occurrence in secondary and tertiary recipients. (C) Spleen size in mice from (A). (D) Expression pattern of B‐cell maturation markers in the BM and spleens from Control (healthy samples), Gprasp1 and Gprasp2‐shRNA recipients (only including those mice where neoplasms were confirmed) after secondary transplant. Data in (B, C) from two independent transplants with N =
= 5 recipients/condition/transplant. Data in (C) represented as mean
5 recipients/condition/transplant. Data in (C) represented as mean ±
± SEM. Data in (D) as mean from five Control‐shRNA, eight Gprasp1‐shRNA mice, and eight Gprasp2‐shRNA mice. */#
p
SEM. Data in (D) as mean from five Control‐shRNA, eight Gprasp1‐shRNA mice, and eight Gprasp2‐shRNA mice. */#
p <
< 0.05, **/##
p
0.05, **/##
p <
< 0.01 ***/###
p
0.01 ***/###
p <
< 0.001 relative to control. * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
0.001 relative to control. * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
Lymphoma cell‐of‐origin is defined by light microscopy, histology, immunophenotype, and genetic profiling, reflecting lymphoid cell differentiation and maturation state. 18 , 19 Our data suggests that GPRASP1 and GPRASP2 may regulate B‐cell development, such that perturbing their expression results in malignancy. To further define the phenotypes of these tumors, we investigated the developmental stage of Gprasp‐deficient neoplastic cells by characterizing BM and splenocytes from mice with tumors. We used flow cytometry to examine the frequencies of all BM Hardy fractions 20 [Pro‐B‐cells: Fraction A (B220+CD43+BP1−CD24−), Fraction B (B220+CD43+BP1−CD24+), Fraction C (B220+CD43+BP1+CD24+); pre‐B‐cells: Fraction D (B220+CD43−IgD−IgM−); immature B‐cells: Fraction E (B220+CD43−IgD−IgM+) and mature recirculated B‐cells (B220+CD43−IgD+IgM−/+). We also examined the frequencies of follicular (B220+CD93−CD12/35mid/highCD23mid/high), marginal (B220+CD93−CD12/35+CD23−), transitional (T1: B220+CD93+IgM+CD23− and T2: B220+CD93+IgM+CD23+), B1 (B1a: CD19+,B220low,IgDlowCD23−,CD5+ and B1b: CD19+,B220low,IgDlowCD23−,CD5−), GC B‐cells (dark zone/centroblast: B220+CD138−GL7+CD95+CXCR4+CD86− and light zone/centrocytes: B220+CD138−GL7+CD95+CXCR4−CD86+), mature B‐cells (B220+CD138−GL7−CD38+) and plasma cells (B220low/−CD138+) in the spleen. While expected frequencies were observed at all steps of B‐cell maturation in controls, tumor samples from both Gprasp1 and Gprasp2‐deficient HSPCs displayed a cell surface phenotype that was not reminiscent of any specific stage of B‐cell development, suggesting massively disrupted developmental programs (Figure 1F and Supporting Information S1: Figure 2). A similar aberrant immunophenotype was observed in secondary recipients (Figure 2D).
To further characterize the Gprasp1‐ and Gprasp2‐deficient tumor samples at the molecular level, we performed bulk RNA‐seq on mCherry+ cells from the BM and spleen of primary and secondary recipients. The transcriptional profiles of the tumors showed significant differences when compared to controls and were highly heterogeneous among themselves (Supporting Information S1: Figure 3A,B and Supporting Information S1: Table 1). Despite heterogeneity, commonalities amongst tumor samples that recapitulate transcriptional features of B‐cell lymphomas were readily apparent (Supporting Information S1: Figure 3C,D). For example, multiple cell death pathways were downregulated, potentially promoting tumor cell survival (Supporting Information S1: Figure 3E). Also, downregulation of signaling pathways linked to IL‐7, TH1, RHO GTPases, mTOR, HIF1α, as well as upregulation of RHOGDI and CTLA4 signaling, have previously been reported in B‐cell lymphoma 21 , 22 , 23 , 24 , 25 , 26 , 27 (Supporting Information S1: Figure 3E). Since GPRASP function is mostly associated with GPCR regulation, observed perturbations in GPCR signaling are consistent with Gprasp1 and Gprasp2 gene knockdown (Supporting Information S1: Figure 3E). Interestingly, secondary Gprasp1 and Gprasp2‐deficient tumors clustered after principal component analysis (PCA) (Supporting Information S1: Figure 3A), suggesting a common selective evolution for expression patterns that promote malignancy.
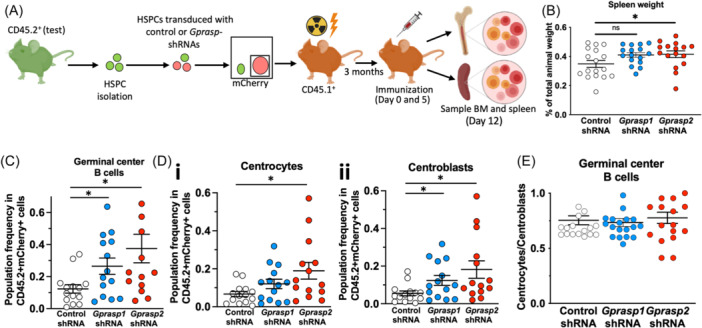
Gprasp1
and
Gprasp2
deficiency perturbs B‐cell normal development. (A) Schematic for transplantation assay. CD45.2+ “test” LSK cells were transduced with control or Gprasp‐shRNAs and then transplanted into lethally irradiated (2 ×
× 550
550 cGy) recipients. Recipient were immunized with SRBC 3 months after transplant. BM and spleen were analyzed for the frequency of B‐cell developmental stages. (B) Spleen size of recipient mice from (A). FACS analysis of the frequency of germinal center B‐cells (C), centrocytes (Di), and centroblasts (Dii). (E) Centrocytes/centroblasts ratio. Data in (B–E) from three independent transplants with N
cGy) recipients. Recipient were immunized with SRBC 3 months after transplant. BM and spleen were analyzed for the frequency of B‐cell developmental stages. (B) Spleen size of recipient mice from (A). FACS analysis of the frequency of germinal center B‐cells (C), centrocytes (Di), and centroblasts (Dii). (E) Centrocytes/centroblasts ratio. Data in (B–E) from three independent transplants with N =
= 5 recipients/condition/transplant. Data represented as mean
5 recipients/condition/transplant. Data represented as mean ±
± SEM and individual values. *p
SEM and individual values. *p <
< 0.05; **p
0.05; **p <
< 0.005; ***p
0.005; ***p <
< 0.001.
0.001.
In sum, knockdown of Gprasp1 or Gprasp2 in transplanted HSPCs results in a serially transplantable hematopoietic malignancy whose histology and molecular phenotype resembles known B‐cell lymphoma biology and demonstrate developmental stage heterogeneity.
Gprasp1‐ or Gprasp2‐deficiency perturbs B‐cell dynamics
Gprasp1 or Gprasp2 knockdown in HSPCs is sufficient to promote B‐cell tumor development following transplantation (Figure 1), suggesting a role for GPRASP1 and GPRASP2 in B‐cell development. To explore this further, we investigated the consequences of Gprasp1 or Gprasp2 loss on B‐cell development prior to the emergence of malignancy. Towards this, CD45.2+ HSPCs were transduced with control, Gprasp1‐ or Gprasp2‐shRNAs. 8000 CD45.2+mCherry+ cells were then transplanted into lethally irradiated recipients (CD45.1+) (Figure 3A). Three months posttransplant, recipients were immunized with SRBC (sheep red blood cells) to trigger the development of GCs and B‐cell differentiation and maturation. 28 Twelve days later, BM and spleen cells were immunophenotyped by flow cytometry for B‐cell stage. Recipients of Gprasp1‐ or Gprasp2‐deficient HSPCs displayed significantly larger spleens than control recipients (Figure 3B). Consistently, GC B‐cell frequency was significantly increased by Gprasp1‐ or Gprasp2‐deficiency relative to controls (Figure 3C and Supporting Information S1: Figure 4A,B). Specifically, centroblasts were elevated with reduced Gprasp1 and Gprasp2, while centrocyte abundance only changed significantly when Gprasp2 was reduced (Figure 3D,E and Supporting Information S1: Figure 4C–F). Histological analysis of the spleens from these mice revealed enlarged follicles and GCs, and more proliferating cells in the GC when Gprasp1 or Gprasp2 expression is reduced (Figure 4A–C). These data suggest that Gprasp1 and Gprasp2 are necessary as B‐cells transition through the GC and its zones during T‐cell‐dependent responses, and that Gprasp1 and Gprasp2 may participate at different levels and steps of that process. Indeed, Gprasp1‐ or Gprasp2‐deficient follicles and GCs lose their circular shape (Figure 4D–F), a phenotype linked to malignant predisposition. 29
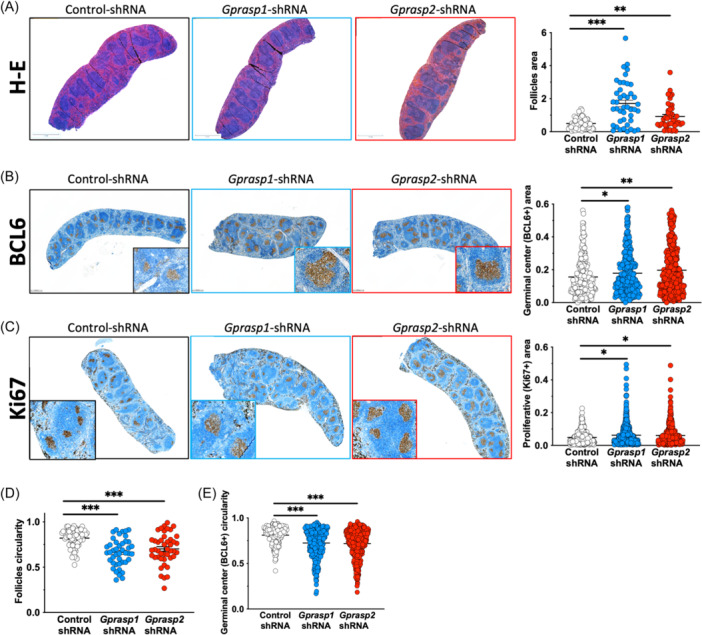
Gprasp1
and
Gprasp2
deficiency perturbs germinal center architecture. Representative images of the histologic analysis of spleens from recipients from recipients from Figure 3A, and scoring for follicle (hematoxylin‐eosin staining) and germinal center size (BCL6 expression) (A, B), proliferation (Ki67 expression) (C) and circularity (as 4p(area/perimeter2) (D, E). Data in (A–E) from three independent transplants with N =
= 5 recipients/condition/transplant. Data represented as mean
5 recipients/condition/transplant. Data represented as mean ±
± SEM and individual values. *p
SEM and individual values. *p <
< 0.05; **p
0.05; **p <
< 0.005; ***p
0.005; ***p <
< 0.001. Size bar in A, B
0.001. Size bar in A, B =
= 1
1 mm and C
mm and C =
= 0.5
0.5 mm.
mm.
Gprasp1‐ or Gprasp2‐deficient malignancy development and progression depend on CXCR4
Our results suggest that Gprasp1 or Gprasp2 deficiency perturbs B‐cell development in the GC and GC organization, which may ultimately result in malignant transformation. We previously reported that Gprasp1 and Gprasp2 regulate CXCR4 turnover in HSPCs. 10 CXCR4 is a master regulator of hematopoietic stem cells. 30 , 31 , 32 , 33 , 34 It also regulates the movement of B‐cells in and out the GC, as well as between the light and dark zones of the GC. 35 We examined Gprasp1 and Gprasp2 expression in pre‐ (follicular), post (plasma and memory), and GC (centrocytes and centroblasts) B‐cells and observed downregulation of both genes in GC B‐cells compared to pre‐ and post‐GC B‐cells (Figure 5A and Supporting Information S1: Figure 5A). Consistently, CXCR4 expression in GC B‐cells increased relative to pre‐ and post‐GC B‐cells (Figure 5A and Supporting Information S1: Figure 5A). Further, when we examined CXCR4 cell surface expression in CD45.2+mCherry+ follicular B‐cells, centrocytes, centroblasts, and mature B‐cells in recipients 12 weeks‐post‐transplant (Figure 3A), CXCR4 was slightly, but significantly, increased in mature B‐cells derived from Gprasp1‐ and Gprasp2‐deficient HSPCs compared to controls (Figure 5B and Supporting Information S1: Figure 5B). Since CXCR4 permits the light zone/dark zone compartmentalization in the GC, 36 the disruption of Gprasp1 or Gprasp2 expression could affect CXCR4 content in these populations, modifying their migratory dynamics relative to the GC. Thus, we hypothesize that in premalignant conditions, Gprasp1‐ and Gprasp2‐deficiency perturbs either pre‐ or post‐GC populations, forcing them to maintain a transcriptional state conducive to ongoing SHM and increasing their susceptibility to malignant transformation. To test this, we transcriptionally profiled follicular cells, centrocytes, centroblasts, and mature cells from the recipient mice described in Figure 3A. Although we found hundreds of differentially expressed genes, when comparing Gprasp1‐ and Gprasp2‐deficient and control populations (Supporting Information S1: Figure 5C and Supporting Information S1: Table 2), these differences did not dramatically alter the cellular identity of these populations (Supporting Information S1: Figure 5D). Gene set enrichment analysis revealed significant upregulation of genes affecting leukocyte differentiation and proliferation (Rag1, Rag2, Lef1, Vpreb1) in centroblasts derived from Gprasp1 and Gprasp2‐deficient progenitors (Supporting Information S1: Figure 5E). We observed that both Gprasp1 and Gprasp2‐deficient derived mature cells express higher levels of Aicda than their control counterparts (Figure 5C). Aicda encodes the enzyme, Activation Induced Cytidine Deaminase, which is required for SHM. 3 Interestingly, Gprasp1‐ and Gprasp2‐deficient HSPC‐derived centrocytes and centroblasts express lower levels of Id2 and Id3 than controls. ID2 and ID3 have been implicated in Aicda transcriptional suppression 37 (Figure 5C). Perturbation of their expression at earlier stages could explain the presence of higher Aicda in mature cells upon suppression of Gprasp1 or Gprasp2. Although further exploration is needed, these results suggest that the absence of Gprasp1 or Gprasp2 influences molecular components of SHM in the populations transitioning the GC. Using these transcription data, we verified the specific knockdown effects of Gprasp1 and Gprasp2‐shRNA in all B cell subsets (Supporting Information S1: Figure 5F).
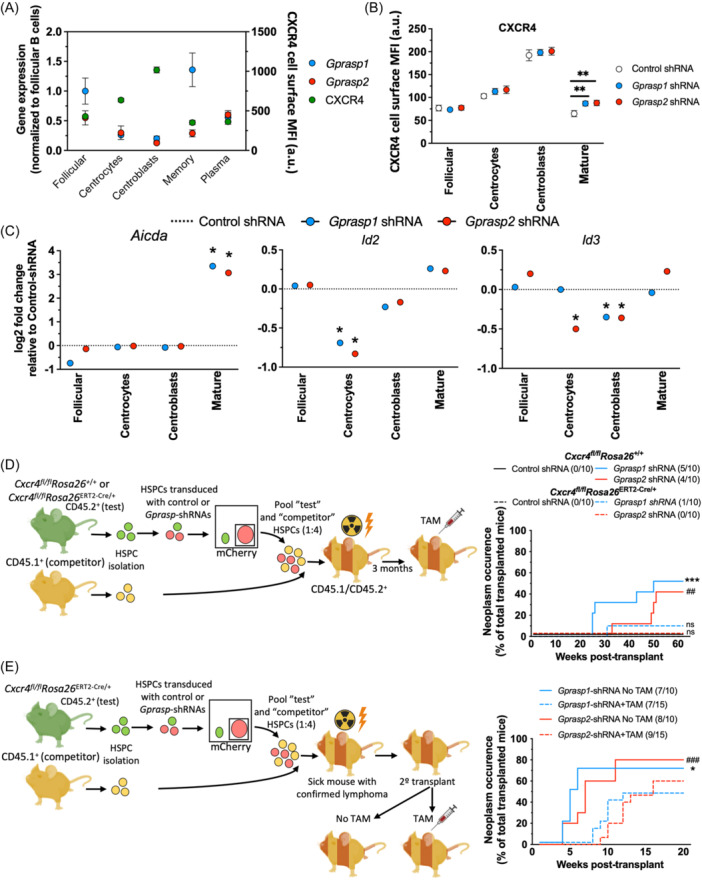
The malignant phenotype derived from
Gprasp
deficiency is affected by CXCR4. (A) Expression profile for Gprasp1, Gprasp2 (gene expression), and CXCR4 (surface protein) in pre‐, post, and germinal center B‐cell subpopulations. (B) FACS analysis for CXCR4 expression (surface protein) in control and Gprasp‐deficient pre‐, post‐, and germinal center B‐cell subpopulations. (C) Aicda, Id2, and Id3 expression (gene expression) in control and Gprasp‐deficient pre‐, post‐, and germinal center B‐cell subpopulations. (D) Schematic for transplantation assay. CD45.2+ “test” LSK cells from Cxcr4
fl/fl
Rosa26
+/+ or Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ mice were transduced with control or Gprasp‐shRNAs and then transplanted along with CD45.1+ “competitor” LSK cells into lethally irradiated (2 ×
× 550
550 cGy) recipients. CXCR4 deletion was induced in the recipient mice three months after transplant. Cxcr4 deletion reduces the occurrence of neoplasms derived from Gprasp‐deficient progenitors. (E) Schematic for transplantation assay. CD45.2+ “test” LSK cells from Cxcr4
fl/fl
Rosa26
+/+ or Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ mice were transduced with control or Gprasp‐shRNAs and then transplanted along with CD45.1+ “competitor” LSK cells into lethally irradiated (2
cGy) recipients. CXCR4 deletion was induced in the recipient mice three months after transplant. Cxcr4 deletion reduces the occurrence of neoplasms derived from Gprasp‐deficient progenitors. (E) Schematic for transplantation assay. CD45.2+ “test” LSK cells from Cxcr4
fl/fl
Rosa26
+/+ or Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ mice were transduced with control or Gprasp‐shRNAs and then transplanted along with CD45.1+ “competitor” LSK cells into lethally irradiated (2 ×
× 550
550 cGy) recipients. Once B‐cell lymphoma was detected and confirmed, the neoplastic population (mCherry+) was transplanted into sublethally irradiated (2
cGy) recipients. Once B‐cell lymphoma was detected and confirmed, the neoplastic population (mCherry+) was transplanted into sublethally irradiated (2 ×
× 475
475 cGy) secondary recipients. A week after secondary transplant, CXCR4 deletion was induced in some of the recipient mice. The untreated mice were used as controls. Cxcr4 deletion occurrence increases the latency of the neoplasms derived from Gprasp‐deficient progenitors. Data in (A–C) from three independent biological replicates. Data in (A, B) represented as mean
cGy) secondary recipients. A week after secondary transplant, CXCR4 deletion was induced in some of the recipient mice. The untreated mice were used as controls. Cxcr4 deletion occurrence increases the latency of the neoplasms derived from Gprasp‐deficient progenitors. Data in (A–C) from three independent biological replicates. Data in (A, B) represented as mean ±
± SEM and individual values. Data in (D, F) from two independent transplants with N
SEM and individual values. Data in (D, F) from two independent transplants with N =
= 5 recipients/condition/transplant. */#
p
5 recipients/condition/transplant. */#
p <
< 0.05; **/##
p
0.05; **/##
p <
< 0.005, ***/###
p
0.005, ***/###
p <
< 0.001 relative to control (and relative to tamoxifen‐treated mice in (F). * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
0.001 relative to control (and relative to tamoxifen‐treated mice in (F). * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
To test if CXCR4 is required for tumor development following knockdown of Gprasp1 or Gprasp2, we isolated HSPCs from Cxcr4
fl/fl
Rosa26
+/+ or Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ mice and transduced them with control, Gprasp1‐ or Gprasp2‐shRNAs. 48 h posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+). Three months later, Cxcr4 deletion was induced via treatment with tamoxifen for five consecutive days (Figure 5D). Notably, Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ recipients experienced a lower incidence of B‐cell tumors than controls following Gprasp1 or Gprasp2 knockdown (Figure 5D). Only one Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ recipient developed a B‐cell tumor, which was found to have retained the unrecombined Cxcr4
fl allele (Supporting Information S1: Figure 5G). Typically, Cxcr4 deletion efficiency in the Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ mice is around 90%, which could have allowed some HSPCs to escape in this transplant recipient. Predictably, 40‐50% of Gprasp1‐ or Gprasp2‐deficient Cxcr4
fl/fl
Rosa26
+/+ HSPC recipients developed B‐cell tumors, consistent with prior observations (Figure 1C). These results confirmed that CXCR4 is essential for the emergence of a malignant phenotype in recipients of Gprasp1‐ or Gprasp2‐deficient HSPCs.
h posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+). Three months later, Cxcr4 deletion was induced via treatment with tamoxifen for five consecutive days (Figure 5D). Notably, Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ recipients experienced a lower incidence of B‐cell tumors than controls following Gprasp1 or Gprasp2 knockdown (Figure 5D). Only one Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ recipient developed a B‐cell tumor, which was found to have retained the unrecombined Cxcr4
fl allele (Supporting Information S1: Figure 5G). Typically, Cxcr4 deletion efficiency in the Cxcr4
fl/fl
Rosa26
ERT2‐Cre/+ mice is around 90%, which could have allowed some HSPCs to escape in this transplant recipient. Predictably, 40‐50% of Gprasp1‐ or Gprasp2‐deficient Cxcr4
fl/fl
Rosa26
+/+ HSPC recipients developed B‐cell tumors, consistent with prior observations (Figure 1C). These results confirmed that CXCR4 is essential for the emergence of a malignant phenotype in recipients of Gprasp1‐ or Gprasp2‐deficient HSPCs.
We next tested if CXCR4 is required for tumor progression in this context. Toward this, we isolated HSPCs from Cxcr4 fl/fl Rosa26 ERT2‐Cre/+ mice and transduced them with control, Gprasp1‐ or Gprasp2‐shRNAs. Forty‐eight hours posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+). We waited for the appearance of B‐cell tumors in the recipient mice (confirmed by pathology analysis as described in Figure 1E) and then we isolated and transplanted 200,000 mCherry+ WBM cells into sublethally irradiated secondary recipients (CD45.1+/CD45.2+). One week later, Cxcr4 deletion was induced in 50% of secondary recipients via 5 days of tamoxifen treatment (Figure 5E). Fewer tumors and extended tumor latency were observed in tamoxifen‐treated secondary recipients relative to untreated controls (Figure 5E). These data reveal that CXCR4 also contributes to tumor maintenance.
Gprasp1 or Gprasp2 deficiency leads to increased somatic hypermutation which contributes to malignant transformation
Given CXCR4's known role in B‐cell trafficking within the GC,
35
we reasoned that Gprasp‐deficient B‐cells might linger in the GC and be subject to excessive SHM. Indeed, pathology and immunophenotype of premalignant Gprasp1‐ or Gprasp2‐deficient spleen suggests a GC origin for this B‐cell malignancy (Figures 1, ,2,2, ,3).3). In the GC, activated B‐cells are subjected to SHM of the rearranged immunoglobulin locus.
2
If Gprasp1‐ or Gprasp2‐deficient B‐cells linger in the GC, they might be subject to prolonged SHM, as has been seen in other models.
38
In this case, we would expect to observe high mutational burden, diverse mutation profiles, and stereotypic SHM‐rearrangements and mutation in immunoglobulin genes.
2
To investigate this, we examined the mutation profiles of Gprasp1 and Gprasp2‐deficient tumor samples by whole exome sequencing (WES) and observed a high number of somatic mutations and large sample‐to‐sample variance in mutation numbers (Supporting Information S1: Figure 6A,B and Supporting Information S1: Table 3). B‐cell immunoglobulin genes rearrange during the development of B‐cell. Class switch recombination occurs in the GC, which produces antibodies with altered constant regions of the heavy chain. Gprasp‐deficient lymphoma samples were oligoclonal and displayed reduced clonal complexity than control samples (Supporting Information S1: Figure 6C). Variant allele frequency (VAF) analysis supports this reduction in clonal complexity, as most samples have at least one mutation with VAF >
> 0.5 (Supporting Information S1: Figure 6D). Comparison of mutation VAFs in primary and secondary recipients shows a general increase, suggesting clonal selection upon serial transplantation (Supporting Information S1: Figure 6E).
0.5 (Supporting Information S1: Figure 6D). Comparison of mutation VAFs in primary and secondary recipients shows a general increase, suggesting clonal selection upon serial transplantation (Supporting Information S1: Figure 6E).
A defining feature of human mature B‐cell lymphomas is their heterogeneous mutation landscape.
2
Despite a high mutation burden, human mature B‐cell lymphoma are classified based on presumed driver mutations.
2
Many driver mutations associated with human mature B‐cell lymphoma subtypes, such as GC B‐cell‐like DLBCLs, were detected in Gprasp1‐ and Gprasp2‐deficient tumors, such as Ebf1, Kmt2d, and Bcl2 (Figure 6A,B and Supporting Information S1: Table 3). AID, the enzyme responsible for SHM in the GC encoded by Aicda,
3
leaves behind a specific mutation signature typified by DGYW/WRCH base changes (G:C is the mutable position; D =
= A/G/T, H
A/G/T, H =
= T/C/A).
39
Mutations detected in Gprasp1‐ and Gprasp2‐malignant samples were enriched for this signature (Supporting Information S1: Figure 6F). Similarly, mutations found in Gprasp1‐ and Gprasp2‐malignant samples are enriched in superenhancer regions when compared to random genomic sequences (Supporting Information S1: Figure 6G), which has been proposed as a feature of AID‐derived mutations.
9
,
40
Additionally, using the Catalogue of Somatic Mutations in Cancer (COSMIC) single base substitution (SBS) signatures revealed high similarity values for Gprasp1‐ and Gprasp2‐malignant samples with AID mutational signatures (SBS84 and SBS85) relative to other cytidine deaminase signatures, such as APOBEC (SBS2 and SBS13) (Supporting Information S1: Figure 6H).
T/C/A).
39
Mutations detected in Gprasp1‐ and Gprasp2‐malignant samples were enriched for this signature (Supporting Information S1: Figure 6F). Similarly, mutations found in Gprasp1‐ and Gprasp2‐malignant samples are enriched in superenhancer regions when compared to random genomic sequences (Supporting Information S1: Figure 6G), which has been proposed as a feature of AID‐derived mutations.
9
,
40
Additionally, using the Catalogue of Somatic Mutations in Cancer (COSMIC) single base substitution (SBS) signatures revealed high similarity values for Gprasp1‐ and Gprasp2‐malignant samples with AID mutational signatures (SBS84 and SBS85) relative to other cytidine deaminase signatures, such as APOBEC (SBS2 and SBS13) (Supporting Information S1: Figure 6H).
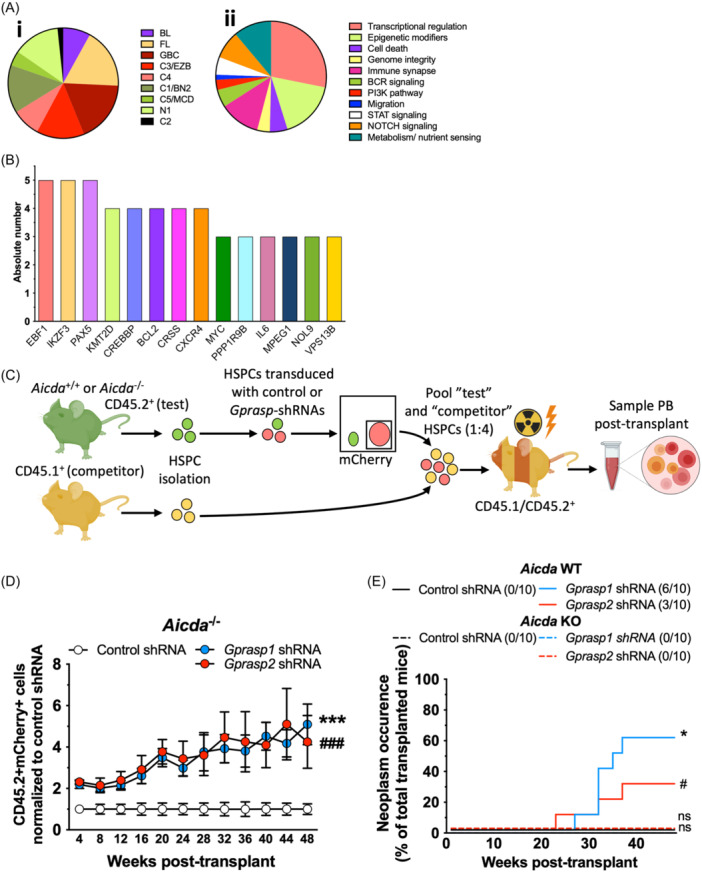
The effects of
Gprasp1
and
Gprasp2
in lymphomagenesis depends on AID. (A) Distribution of the mutations in the malignant samples representing commonly perturbed genes and pathways in B cell lymphoma subtypes. Classified by (i) tumor type and (ii) pathway type.
2
BL, Burkitt lymphoma; C1/BN2, Activated B cell‐like lymphoma (ABC) cluster 1 lymphoma (BCL6 fusions and NOTCH2 mutations); C2, ABC/GCB‐independent cluster 2 lymphoma; C3/EZB, GCB cluster 3 lymphoma (EZH2 mutations and BCL2 translocations); C4, GCB cluster 4 lymphoma; C5/MCD, ABC cluster 5 lymphoma (co‐occurrence of MYD88L265P and CD79B mutations); FL, follicular lymphoma; GCB, germinal center B cell‐like lymphoma; N1, NOTCH1 mutations. (B) Number of malignant samples with somatic mutations in genes commonly mutated in human B cell lymphoma. (C) Schematic for transplantation assay. CD45.2+ “test” LSK cells from Aicda
+/+ or Aicda
−/− mice were transduced with control or Gprasp‐shRNAs and then transplanted along with CD45.1+ “competitor” LSK cells into lethally irradiated (2 ×
× 550
550 cGy) recipients. Recipient PB was analyzed for CD45.2+ mCherry+ cells. (D)
Gprasp‐deficient HSPCs display enhanced PB repopulating activity relative to HPSCs treated with control‐shRNA, both in the presence and the absence of AID. (E) Recipients of Gprasp‐deficient HSPCs do not show increased frequency of neoplasm in the absence of AID. Data in (A, B) are from 28 independent neoplasms. Data in (D, E) from two independent transplants with N
cGy) recipients. Recipient PB was analyzed for CD45.2+ mCherry+ cells. (D)
Gprasp‐deficient HSPCs display enhanced PB repopulating activity relative to HPSCs treated with control‐shRNA, both in the presence and the absence of AID. (E) Recipients of Gprasp‐deficient HSPCs do not show increased frequency of neoplasm in the absence of AID. Data in (A, B) are from 28 independent neoplasms. Data in (D, E) from two independent transplants with N =
= 5 recipients/condition/transplant. Data in (D) represented as mean
5 recipients/condition/transplant. Data in (D) represented as mean ±
± SEM. */#
p
SEM. */#
p <
< 0.05, ***/###
p
0.05, ***/###
p <
< 0.001 relative to control. * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
0.001 relative to control. * refers to Gprasp1‐shRNA, # refers to Gprasp2‐shRNA.
These data suggest that elevated levels of SHM may contribute to the malignant transformation of Gprasp1 and Gprasp2‐deficient B‐cells. To test this, CD45.2+ HSPCs from Aicda
+/+ and Aicda
−/− mice were transduced with control, Gprasp1‐ or Gprasp2‐shRNAs. Forty‐eight hours posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+) (Figure 6C). Gprasp1 and Gprasp2‐deficient Aicda
+/+ and Aicda
−/− HSPCs displayed enhanced PB repopulating activity relative to Aicda
+/+ and Aicda
−/− HSPCs treated with control‐shRNAs, consistent with previous results (Figure 6D). 30%–60% recipients of Gprasp1 and Gprasp2‐deficient Aicda
+/+ HSPCs developed B‐cell tumors by 40 weeks posttransplant. No mice transplanted with Gprasp1‐ or Gprasp2‐deficient Aicda
−/− HSPCs developed B‐cell tumors (Figure 6E). These data strongly support our model that prolonged exposure to SHM mechanistically contributes to lymphomagenesis of transplanted Gprasp1‐ and Gprasp2‐deficient HSPCs.
hours posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+) (Figure 6C). Gprasp1 and Gprasp2‐deficient Aicda
+/+ and Aicda
−/− HSPCs displayed enhanced PB repopulating activity relative to Aicda
+/+ and Aicda
−/− HSPCs treated with control‐shRNAs, consistent with previous results (Figure 6D). 30%–60% recipients of Gprasp1 and Gprasp2‐deficient Aicda
+/+ HSPCs developed B‐cell tumors by 40 weeks posttransplant. No mice transplanted with Gprasp1‐ or Gprasp2‐deficient Aicda
−/− HSPCs developed B‐cell tumors (Figure 6E). These data strongly support our model that prolonged exposure to SHM mechanistically contributes to lymphomagenesis of transplanted Gprasp1‐ and Gprasp2‐deficient HSPCs.
GPRASP1 and GPRASP2 present genomic and transcriptional perturbations in human DLBCL
The pathology analysis of the Gprasp‐deficient neoplasms indicates that their immunohistological features resembles those of human DLBCL (Figure 1E and Supporting Information S1: Figure 1H,I). We analyzed the mutational profile of human DLCBL samples and found mutations in GPRASP1 or GPRASP2 in about 1% (Figure 7A). 41 , 42 , 43 , 44 Then, we compared GPRASP1 and GPRASP2 expression in the different DLBCL subtypes (e.g., Activated B‐cell [ABC] DLBCL, germinal center B‐cell [GCB] DLBCL and unclassified DLBCL) with healthy B‐cell subpopulations (e.g., naïve, centroblast and memory B‐cells) (Figure 7B). Notably, GPRASP1 and GPRASP2 expression positively correlates in all DLBCL subtypes. Additionally, there is a subset of samples with low GPRASP1 or GPRASP2 expression relative to healthy B cells that represents ~70%–90% of all lymphoma samples for GPRASP1 and ~20–60% GPRASP2 (Figure 7B). Further, we found that AICDA expression negatively correlates with both GPRASP1 and GPRASP2 expression in unclassified DLBCL samples, indicating an increase of AICDA as GPRASP genes decrease (Figure 7CI and Supporting Information S1: Figure 7A). We also found CXCR4 expression to positively correlates with GPRASP1 and GPRASP2 (Figure 7CII and Supporting Information S1: Figure 7B). Although further efforts would be necessary to clarify the potential role of GPRASPs in human B‐cell lymphoma biology, this analysis suggests a link between GPRASPs expression and DLBCL.
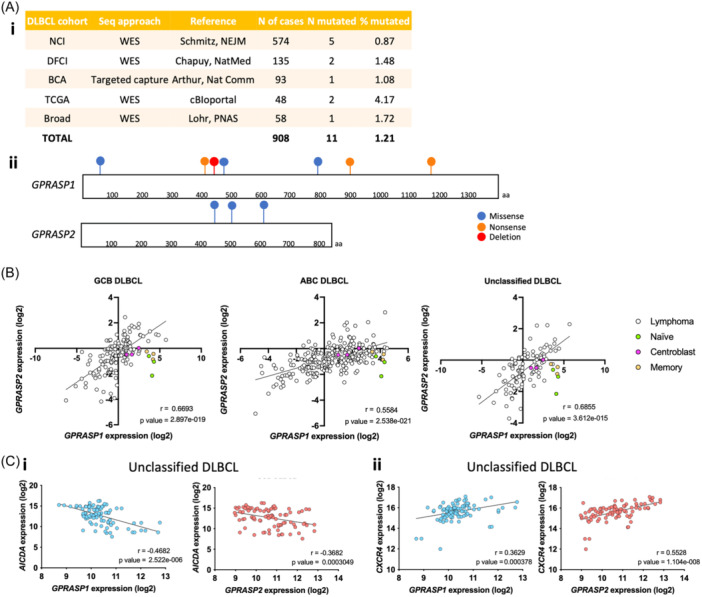
GPRASP1 and GPRASP2 are mutated and show low expression in a subset of human DLBCL samples. (A) Frequency of GPRASP1 and GPRASP2 mutations found in DLBCL patients (i) and location in their protein sequence (ii). (B) GPRASP1 and GPRASP2 gene expression in healthy B‐cell subpopulations and DLBCL subtypes. (C) Correlation between GPRASP1 or GPRASP2 gene expression and AICDA (i) or CXCR4 (ii) in DLBCL subtypes. r, Pearson correlation coefficient.
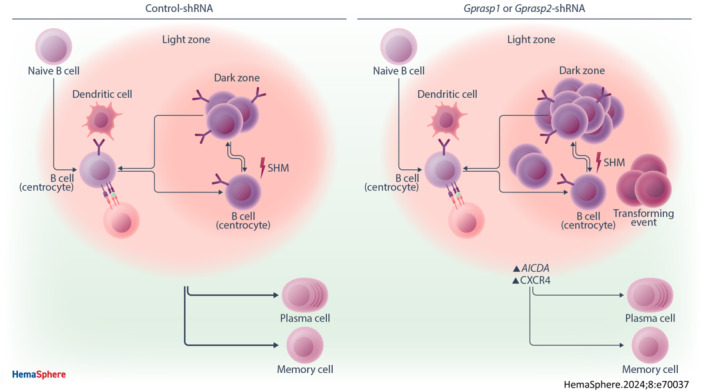
In summary, we have shown that silencing Gprasp1 or Gprasp2 in HSPCs perturbs B‐cell differentiation at the GC, causing an accumulation of centrocytes and centroblasts. Additionally, the transcriptional identity of B‐cells transitioning through the GC is perturbed in the absence of Gprasp1 or Gprasp2, revealing dysregulation of components of the SHM mechanism (e.g., Aicda) and its regulators (e.g., Id2 and Id3). This pool of cells appears to express an active SHM program for an elongated time, increasing the chances of aberrant mutations and consequent malignant transformation/lymphoma development, for which both Aicda and CXCR4 are critical (Figure 8).
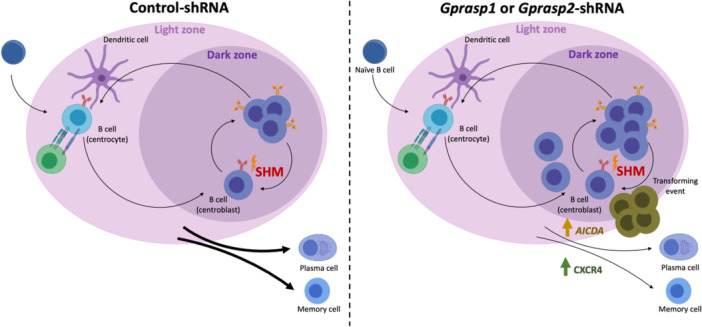
Gprasp1 and Gprasp2 deficiency affect CXCR4 and SHM dynamics increasing lymphomagenesis. Gprasp1 and Gprasp2‐deficient B‐cells accumulate at the GC and present transcriptional differences affecting critical components of the SHM (i.e., Aicda) and regulators of the GC migration processes (i.e., CXCR4), increasing the risk of lymphomagenesis.
DISCUSSION
We recently described GPRASP1 and GPRASP2 as novel regulators of CXCR4 stability in HSCs. 10 We now report that downregulation of GPRASP1 and GPRASP2 in maturing B‐cells results in pathology that is also dependent on CXCR4. CXCR4 is a critical regulator of B‐cell migration in and around the GC. 36 We find that Gprasp1‐ and Gprasp2‐knockdown causes mature B‐cells to sustain high levels of CXCR4, accumulate in the GC, and increase AID gene expression. This likely subjects Gprasp1‐ and Gprasp2‐deficient cells to excess somatic hypermutation, which increases the odds of malignant transformation. Consistently, we observe Cxcr4‐ and Aicda‐dependent B‐cell lymphomagenesis in mice transplanted with Gprasp1‐ or Gprasp2‐deficient cells. Progression of these lymphomas also depends on CXCR4. Thus, Gprasp1 and Gprasp2 expression dynamics are necessary for B‐cell normal maturation and their disruption has critical influence in B‐cell lymphomagenesis at the level of the GC. There are more than 30 different categories of human B‐cell lymphomas. 45 This complex classification, which groups lymphomas according to their putative cell of origin, as well as their clinical, pathological, and genetic features, reflects our improved understanding of the immune system, where lymphocytes undergo complex multi‐step maturation and each step has the potential for malignant transformation. 46 , 47 Immunohistochemical subtyping of human GC‐origin DLBCLs is defined using key markers including BCL6, IRF4/MUM1, and CD10, although staining patterns are not entirely correlative with defining gene rearrangements and molecular subgroupings. 45 Similar, but species‐specific IHC and molecular findings, were observed for the high‐grade GC murine B‐cell tumors that developed following Gprasp1 or Gprasp2 deficiency. All tumors expressed LMO2, a marker of normal germinal center centroblasts and centrocytes and some postgerminal center lymphomas and in GC‐derived non‐Hodgkins lymphomas. 48 LMO2 is also a putative marker for long‐term survival and responsiveness to chemotherapy in positive cases of human DLBCL. 49 Tumors were immunopositive for IRF4, which may be expressed in lineage‐committed germinal center B cells having either plasmacytic or memory cell differentiation phenotypes. 50 Staining for CD10 by either flow cytometry or IHC was not performed since there are species‐specific differences in expression in human hematolymphoid cells expressing the antigen and murine hematolymphoid cells, which are negative. 51 Immunohistochemistry for BCL2 was not performed since this marker is over expressed in many murine B‐cell lymphoma subtypes, unlike in human lymphoma, and is not correlative with gene rearrangements for BCL2. 52 The histomorphology and immunophenotype of the B‐cell tumors correlated with other data showing that neoplastic B cells had undergone a GC reaction. GC B‐cells remodel their immunoglobulin genes by SHM and class switching. 3 Most human B‐cell lymphomas originate from B‐cells that have passed through the GC and thus contain both chromosomal translocations and many point mutations. 2 , 3 AID initiates class switching and SHM by generating U:G mismatches on immunoglobulin DNA that can then be processed by Uracyl‐N‐glycosylase (UNG). 3 AID promotes collateral damage in the form of chromosome translocations and off‐target SHM. 53 However, the exact contribution of AID activity to lymphoma generation and progression is not completely understood. Additionally, AID‐induced SHM may contribute to lymphoma progression by generating a diverse array of oncogenic variants that drive tumor evolution and aggressiveness. 54 This results in inter‐ and intra‐tumoral molecular heterogeneity that hinders identifying the key genetic drives of transformation. For this reason, a deeper knowledge of the molecular regulators of normal B‐cell development and signaling factors that influence lymphocyte migration and may also increase the likelihood of transformation are needed.
Thus, our work not only reveals the importance of Gprasp genes for murine B‐cell development but also suggests that perturbed Gprasp1 and Gprasp2 expression dynamics may be a risk factor for B‐cell lymphoma development. As an alternative to genetically introducing known lymphogenic mutations, manipulation of Gprasp1 and Gprasp2 provides a powerful model to study the dynamics and initial transforming events that result in a variety of GC‐associated B‐cell lymphomas, rather than the genetic transformation of a particular lymphoma type. Indeed, the transcriptional and mutational profile of the Gprasp1‐ or Gprasp2‐deficient derived malignant samples include a fair representation of the genes, and the pathways under their control, that are most commonly disrupted in many high‐grade B‐cell lymphoma sub‐types. In addition, this molecular profiling exposed an extraordinary heterogeneity, characteristic of this disease, which is not surprising, since aberrant SHM contributes to malignant transformation and molecular heterogeneity in most B‐cell lymphomas. 2 In short, the introduction of random mutations across the genome creates mutational (and, consequently, transcriptional) profiles that differ from case to case.
Although more studies are necessary to investigate a role for Gprasp1 and Gprasp2 in human lymphomagenesis, their expression level in HSPCs or B‐cells may be indicative of B‐cell lymphoma predisposition. Similarly, given CXCR4's known role in many different malignancies, 55 , 56 , 57 , 58 , 59 , 60 , 61 our data suggest that disruption or downregulation of GPRASPs in tumors may also contribute to the progression of the malignancy. In this context, it is noteworthy that improved HSPC transplantation efficiency following Gprasp1 or Gprasp2 downregulation could be compromised by the possibility of a long‐term lymphoproliferative disease, 10 when considering a potential clinical application. Thus, transient disruption of Gprasp expression should be considered to promote an HSPC repopulating advantage without increasing the risk of malignant transformation.
Altogether, our study reveals GPRASP1 and GPRASP2 as novel regulators of B‐cell maturation and lymphomagenesis. Further, Gprasp1 or Gprasp2 knockdown provide an alternative and simple means to model B‐cell lymphoma genetic and transcriptional heterogeneity for high‐grade B‐cell lymphomas of germinal center origin.
AUTHOR CONTRIBUTIONS
Antonio Morales‐Hernández, Heather Sheppard, Gabriela Gheorghe, and Shannon McKinney‐Freeman wrote, reviewed and revised the manuscript and designed the experiments. Antonio Morales‐Hernández, Emilia Kooienga, Claire Caprio, and Ashley Chabot performed the experiments. Heather Sheppard and Gabriela Gheorghe performed the pathology analyses.
FUNDING
This work was supported by the American Lebanese Syrian Associated Charities (ALSAC) (S.M.F.) and the National Institute for Diabetes and Digestive and Kidney Diseases at the National Institute of Health (R01 DK104028, S.M.F.; R01 DK116835, S.M.F.; DK123395, A.M.H.). S.M.F. is a Scholar of the Leukemia and Lymphoma Society. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
We thank the McKinney‐Freeman laboratory, Clements laboratory, the Obeng laboratory, and Dr. Laura Pasqualucci for critical discussions and reading of the manuscript; D. Ashmun, L. He, S. Schwemberger and S. Wollard, as well as D. Cullins at the Flow Cytometry Core in the Hematology Department at St. Jude Children's Hospital for FACS support; J. McCommon and K. Millican for help with mouse injections; and the Center for Applied Bioinformatics at St. Jude Children's Hospital for bioinformatic support. Aicda −/− were generously provided by Dr. Ari Melnick (Weill Cornell Medicine).
Contributor Information
Antonio Morales‐Hernández, Email: ude.hcimu@romtna.
Shannon McKinney‐Freeman, Email: [email protected].
DATA AVAILABILITY STATEMENT
All RNAseq and whole exome sequencing (WES) files are submitted to the Sequence Read Archive (SRA) of the NIH. Bioproject numbers: PRJNA1029555, PRJNA1047147 and PRJNA1092532.
REFERENCES
Articles from HemaSphere are provided here courtesy of Wiley
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/169881582
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
GPRASP proteins are critical negative regulators of hematopoietic stem cell transplantation.
Blood, 135(14):1111-1123, 01 Apr 2020
Cited by: 2 articles | PMID: 32027737 | PMCID: PMC7118811
Cohesin Core Complex Gene Dosage Contributes to Germinal Center Derived Lymphoma Phenotypes and Outcomes.
Front Immunol, 12:688493, 21 Sep 2021
Cited by: 5 articles | PMID: 34621263 | PMCID: PMC8490713
Determining the Origin of Human Germinal Center B Cell-Derived Malignancies.
Methods Mol Biol, 1623:253-279, 01 Jan 2017
Cited by: 2 articles | PMID: 28589362
GPRASP/ARMCX Protein Family: Potential Involvement in Health and Diseases Revealed by their Novel Interacting Partners.
Curr Top Med Chem, 21(3):227-254, 01 Jan 2021
Cited by: 2 articles | PMID: 33267763
Review
Funding
Funders who supported this work.
American Lebanese Syrian Associated Charities
Leukemia and Lymphoma Society
National Institutes of Health (3)
Grant ID: DK104028
Grant ID: DK123395
Grant ID: DK116835

 1
1