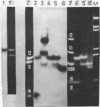
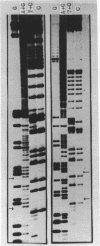
Abstract
Free full text

The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA.
Abstract
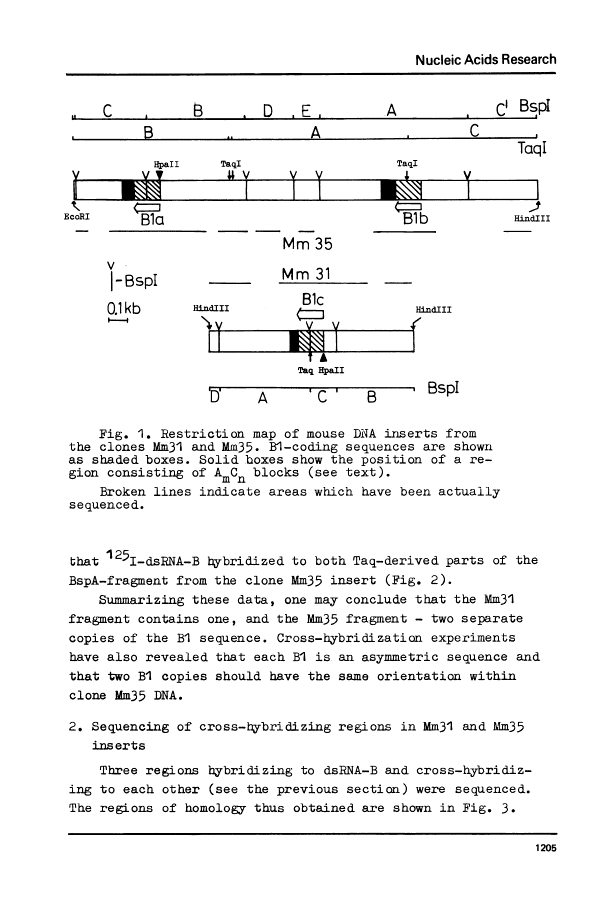
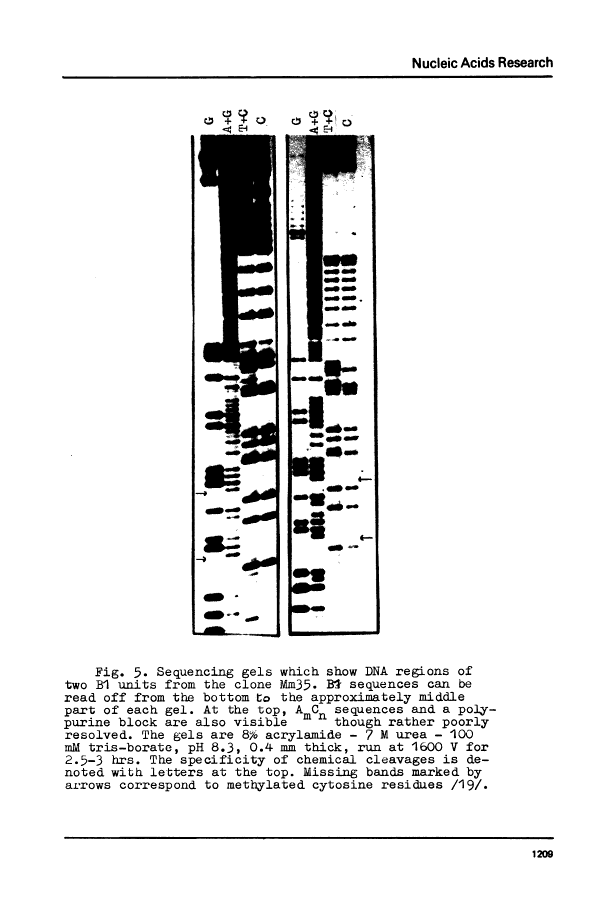
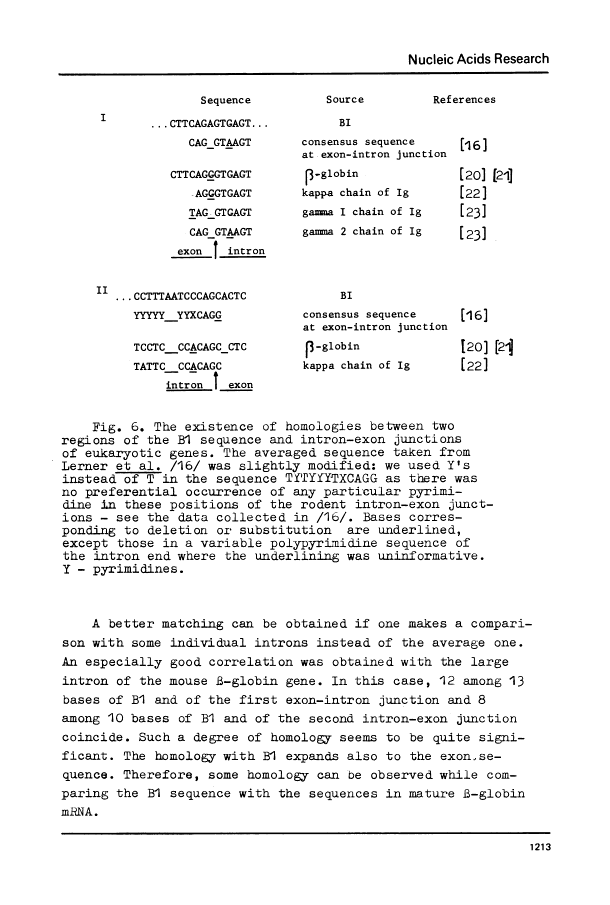
Three copies of a highly repetitive DNA sequence B1 which is complementary to the most abundant class of mouse fold-back RNA have been cloned in pBR322 plasmid and sequenced by the method of Maxam and Gilbert. All the three have a length of about 130 base pairs and are very similar in their base sequence. The deviation from the average sequence is equal to 4% and the overall mismatch between each two is not higher than 8%. One of the recombinant clones used contained two copies of B1 oriented in the same direction. All of the B1 copies are flanked with sequences which possess nonidentical but very similar structure. They consist of a number of AmCn blocks (where m varies from 2 to 8 and n equals 1-2). These peculiar sequences in all cases are separated from B1 by non-homologous DNA stretches of 2-8 residues. In one case, a long polypurine stretch is located next to such a block. It consists of 74 residues most of which represent a reiteration of the basic sequence AAAAG. We have found two regions within the B1 sequence which are homologous to the intron-exon junctions, especially to those present in the large intron of the mouse beta-globin gene. It may indicate the involvement of the B1 sequence in pre-mRNA splicing.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.4M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ryskov AP, Farashyan VR, Georgiev GP. Ribonuclease-stable base sequences specific exclusively for giant dRNA. Biochim Biophys Acta. 1972 Apr 12;262(4):568–572. [Abstract] [Google Scholar]
- Jelinek W, Darnell JE. Double-stranded regions in heterogeneous nuclear RNA from Hela cells. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2537–2541. [Europe PMC free article] [Abstract] [Google Scholar]
- Ryskov AP, Saunders GF, Farashyan VR, Georgiev GP. Double-helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim Biophys Acta. 1973 Jun 8;312(1):152–164. [Abstract] [Google Scholar]
- Kramerov DA, Ryskov AP, Georgiev GP. The structural organization of nuclear pre-mRNA. II. Very long double-stranded structures in nuclear pre-mRNA. Biochim Biophys Acta. 1977 Apr 4;475(3):461–475. [Abstract] [Google Scholar]
- Ryskov AP, Kramerov DA, Georgiev GP. The structural organization of nuclear messenger RNA precursor. I. Reassociation and hybridization properties of double-stranded hairpin-like loops in messenger RNA precursor. Biochim Biophys Acta. 1976 Oct 4;447(2):214–229. [Abstract] [Google Scholar]
- Kramerov DA, Grigoryan AA, Ryskov AP, Georgiev GP. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. [Europe PMC free article] [Abstract] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [Abstract] [Google Scholar]
- Bickle TA, Pirrotta V, Imber R. A simple, general procedure for purifying restriction endonucleases. Nucleic Acids Res. 1977 Aug;4(8):2561–2572. [Europe PMC free article] [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. [Europe PMC free article] [Abstract] [Google Scholar]
- Szalay AA, Grohmann K, Sinsheimer RL. Separation of the complementary strands of DNA fragments on polyacrylamide gels. Nucleic Acids Res. 1977;4(5):1569–1578. [Europe PMC free article] [Abstract] [Google Scholar]
- Laskey RA, Mills AD. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. [Abstract] [Google Scholar]
- Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. [Europe PMC free article] [Abstract] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. [Abstract] [Google Scholar]
- Ryskov AP, Tokarskaya OV, Georgiev GP, Coutelle C, Thiele B. Globin mRNA contains a sequence complementary to double-stranded region of nuclear pre-mRNA. Nucleic Acids Res. 1976 Jun;3(6):1487–1498. [Europe PMC free article] [Abstract] [Google Scholar]
- Jelinek WR. Specific nucleotide sequences in HeLa cell inverted repeated DNA: enrichment for sequences found in double-stranded regions of heterogeneous nuclear RNA. J Mol Biol. 1977 Oct 5;115(4):591–601. [Abstract] [Google Scholar]
- Ohmori H, Tomizawa JI, Maxam AM. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. [Europe PMC free article] [Abstract] [Google Scholar]
- van den Berg J, van Ooyen A, Mantei N, Schamböck A, Grosveld G, Flavell RA, Weissmann C. Comparison of cloned rabbit and mouse beta-globin genes showing strong evolutionary divergence of two homologous pairs of introns. Nature. 1978 Nov 2;276(5683):37–44. [Abstract] [Google Scholar]
- Konkel DA, Maizel JV, Jr, Leder P. The evolution and sequence comparison of two recently diverged mouse chromosomal beta--globin genes. Cell. 1979 Nov;18(3):865–873. [Abstract] [Google Scholar]
- Max EE, Seidman JG, Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3450–3454. [Europe PMC free article] [Abstract] [Google Scholar]
- Sakano H, Rogers JH, Hüppi K, Brack C, Traunecker A, Maki R, Wall R, Tonegawa S. Domains and the hinge region of an immunoglobulin heavy chain are encoded in separate DNA segments. Nature. 1979 Feb 22;277(5698):627–633. [Abstract] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. [Abstract] [Google Scholar]
Associated Data
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/8.6.1201
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc323986?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Analysis of SINE Families B2, Dip, and Ves with Special Reference to Polyadenylation Signals and Transcription Terminators.
Int J Mol Sci, 22(18):9897, 13 Sep 2021
Cited by: 6 articles | PMID: 34576060 | PMCID: PMC8466645
TFs for TEs: the transcription factor repertoire of mammalian transposable elements.
Genes Dev, 35(1-2):22-39, 01 Jan 2021
Cited by: 38 articles | PMID: 33397727 | PMCID: PMC7778262
Review Free full text in Europe PMC
Linkage disequilibrium and functional analysis of PRE1 insertion together with SNPs in the promoter region of IGFBP7 gene in different pig breeds.
J Appl Genet, 59(2):231-241, 25 Mar 2018
Cited by: 3 articles | PMID: 29574509
Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes.
Viruses, 9(12):E386, 18 Dec 2017
Cited by: 6 articles | PMID: 29258254 | PMCID: PMC5744160
Review Free full text in Europe PMC
Host Noncoding Retrotransposons Induced by DNA Viruses: a SINE of Infection?
J Virol, 91(23):e00982-17, 14 Nov 2017
Cited by: 10 articles | PMID: 28931686 | PMCID: PMC5686761
Review Free full text in Europe PMC
Go to all (233) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[Mapping of 2 cloned DNA fragments containing repetitive mouse genome regions, actively transcribing into pre-mRNA].
Genetika, 18(5):713-723, 01 May 1982
Cited by: 0 articles | PMID: 6284588
[Certain properties of transcribed fragments of the mouse genome cloned in plasmid pBR322].
Genetika, 16(10):1741-1752, 01 Jan 1980
Cited by: 0 articles | PMID: 6257585
[Nucleotide sequence of the repetitive B2 region of the mouse genome].
Mol Biol (Mosk), 17(6):1272-1279, 01 Nov 1983
Cited by: 1 article | PMID: 6656756
Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments.
Nucleic Acids Res, 6(2):697-713, 01 Feb 1979
Cited by: 70 articles | PMID: 370792 | PMCID: PMC327722