Abstract
Free full text

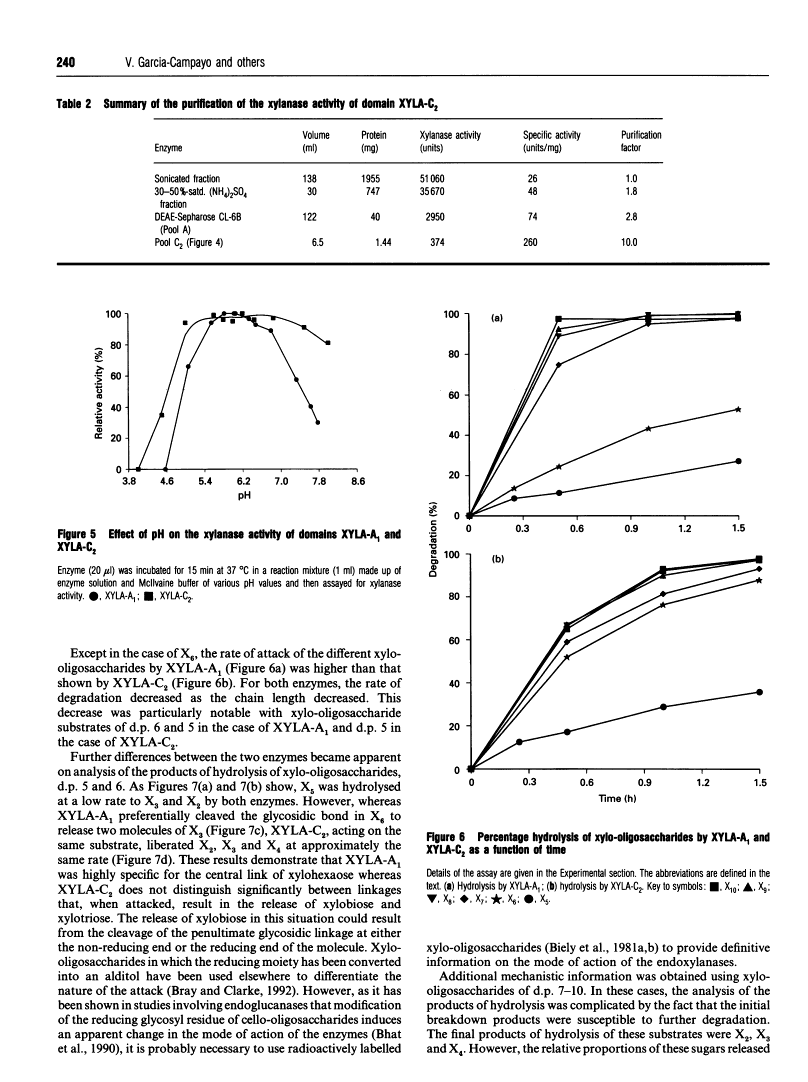
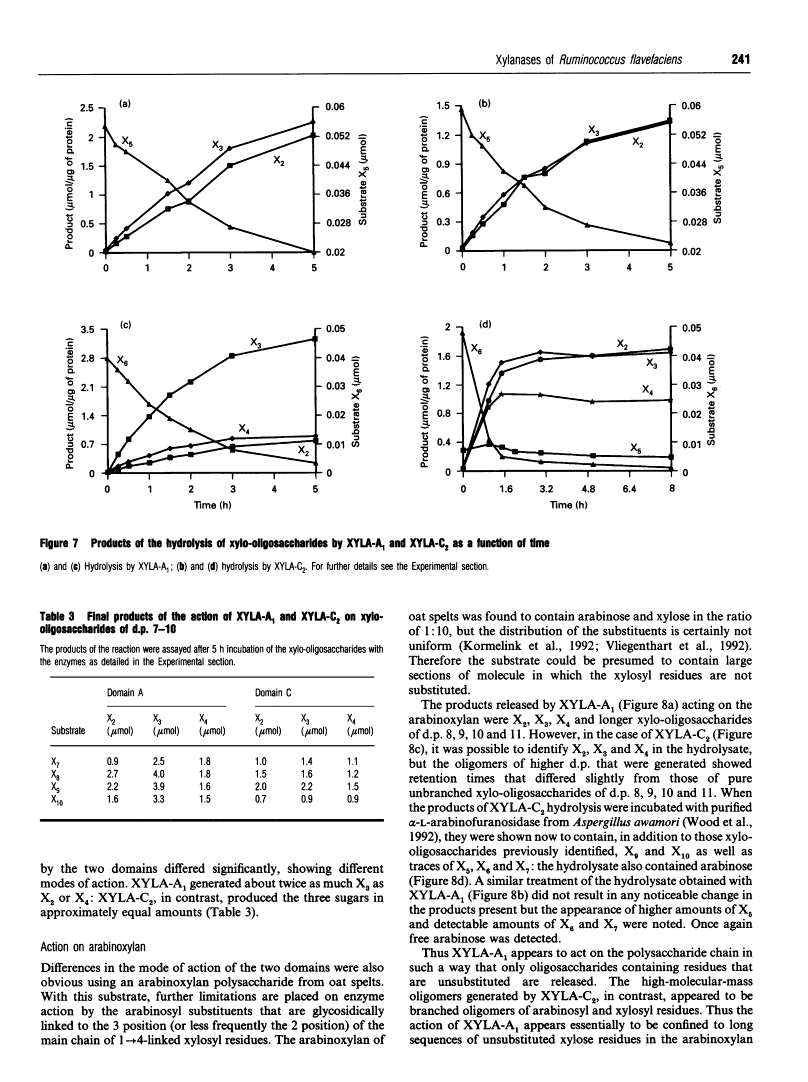
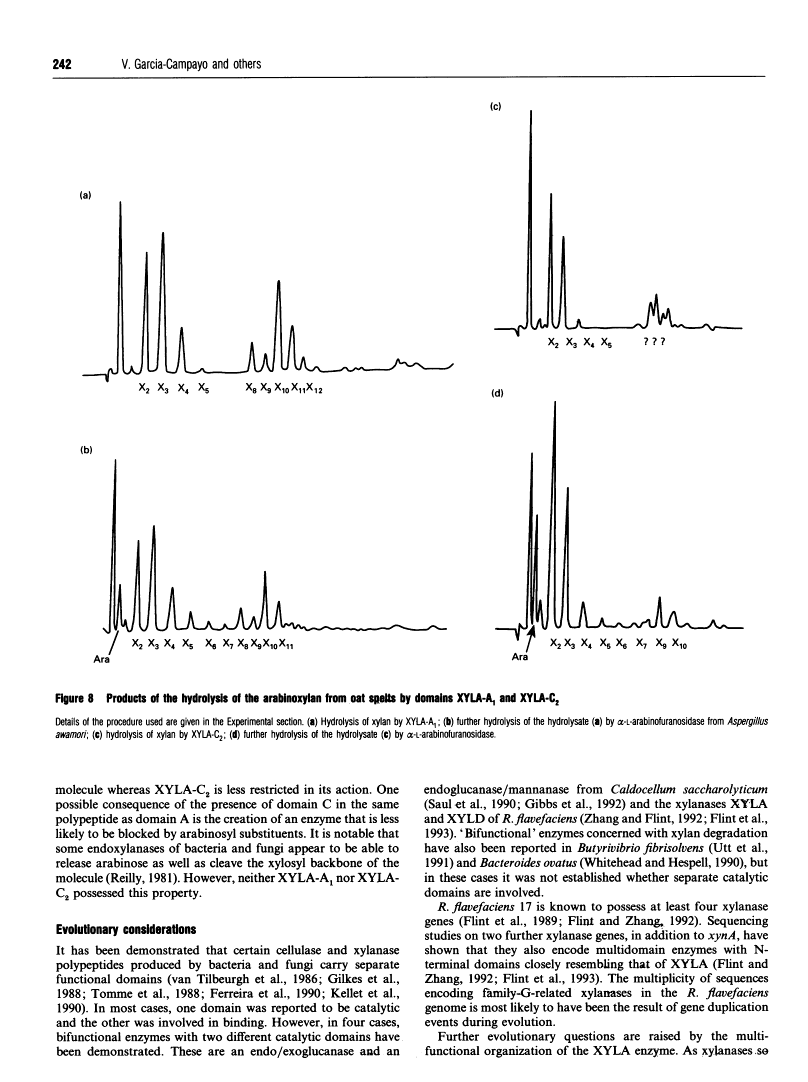
Mode of action, kinetic properties and physicochemical characterization of two different domains of a bifunctional (1-->4)-beta-D-xylanase from Ruminococcus flavefaciens expressed separately in Escherichia coli.
Abstract
Two catalytic domains, A and C, of xylanase A (XYLA) from Ruminococcus flavefaciens were expressed separately as truncated gene products from lacZ fusions in Escherichia coli. The fusion products, referred to respectively as XYLA-A1 and XYLA-C2, were purified to homogeneity by anion-exchange chromatography and chromatofocusing. XYLA-A1 was isoelectric at pH 5.0 and had a molecular mass of 30 kDa, whereas XYLA-C2 had a pI of 5.4 and a molecular mass of 44 kDa. The catalytic activity shown by both domains was optimal at 50 degrees C, but XYLA-A1 was more sensitive than XYLA-C2 to temperatures higher than the optimum. XYLA-A1 showed a higher sensitivity to pH than XYLA-C2. The enzyme activity of both domains was completely inactivated in the presence of copper or silver ions and partially inactivated by iron or zinc ions. Neither domain was active on xylo-oligosaccharides shorter than xylopentaose: the rate of degradation of longer xylo-oligosaccharides (degree of polymerization 5-10) increased as the chain length increased. Analysis of the products of hydrolysis of xylo-oligosaccharides and xylan (arabinoxylan) polysaccharide showed that the two domains differed in their modes of action: xylobiose was the shortest product of the hydrolysis. With oat spelt xylan as substrate, XYLA-A1 activity was apparently restricted to regions where xylopyranosyl residues did not carry arabinofuranosyl substituents, whereas XYLA-C2 was able to release hetero-oligosaccharides carrying arabinofuranosyl residues. Neither domain was able to release arabinose from oat spelt xylan.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.4M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhat KM, Hay AJ, Claeyssens M, Wood TM. Study of the mode of action and site-specificity of the endo-(1----4)-beta-D-glucanases of the fungus Penicillium pinophilum with normal, 1-3H-labelled, reduced and chromogenic cello-oligosaccharides. Biochem J. 1990 Mar 1;266(2):371–378. [Europe PMC free article] [Abstract] [Google Scholar]
- Biely P, Krátký Z, Vrsanská M. Substrate-binding site of endo-1,4-beta-xylanase of the yeast Cryptococcus albidus. Eur J Biochem. 1981 Oct;119(3):559–564. [Abstract] [Google Scholar]
- Biely P, Vrsanská M, Krátký Z. Mechanisms of substrate digestion by endo-1,4-beta-xylanase of Cryptococcus albidus. Lysozyme-type pattern of action. Eur J Biochem. 1981 Oct;119(3):565–571. [Abstract] [Google Scholar]
- Bray MR, Clarke AJ. Action pattern of xylo-oligosaccharide hydrolysis by Schizophyllum commune xylanase A. Eur J Biochem. 1992 Feb 15;204(1):191–196. [Abstract] [Google Scholar]
- Crawford IP. Gene rearrangements in the evolution of the tryptophan synthetic pathway. Bacteriol Rev. 1975 Jun;39(2):87–120. [Europe PMC free article] [Abstract] [Google Scholar]
- Ferreira LM, Durrant AJ, Hall J, Hazlewood GP, Gilbert HJ. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990 Jul 1;269(1):261–264. [Europe PMC free article] [Abstract] [Google Scholar]
- Flint HJ, McPherson CA, Bisset J. Molecular cloning of genes from Ruminococcus flavefaciens encoding xylanase and beta(1-3,1-4)glucanase activities. Appl Environ Microbiol. 1989 May;55(5):1230–1233. [Europe PMC free article] [Abstract] [Google Scholar]
- Flint HJ, Martin J, McPherson CA, Daniel AS, Zhang JX. A bifunctional enzyme, with separate xylanase and beta(1,3-1,4)-glucanase domains, encoded by the xynD gene of Ruminococcus flavefaciens. J Bacteriol. 1993 May;175(10):2943–2951. [Europe PMC free article] [Abstract] [Google Scholar]
- Gibbs MD, Saul DJ, Lüthi E, Bergquist PL. The beta-mannanase from "Caldocellum saccharolyticum" is part of a multidomain enzyme. Appl Environ Microbiol. 1992 Dec;58(12):3864–3867. [Europe PMC free article] [Abstract] [Google Scholar]
- Gilkes NR, Warren RA, Miller RC, Jr, Kilburn DG. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988 Jul 25;263(21):10401–10407. [Abstract] [Google Scholar]
- Gilkes NR, Henrissat B, Kilburn DG, Miller RC, Jr, Warren RA. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. [Europe PMC free article] [Abstract] [Google Scholar]
- Kellett LE, Poole DM, Ferreira LM, Durrant AJ, Hazlewood GP, Gilbert HJ. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. [Europe PMC free article] [Abstract] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. [Abstract] [Google Scholar]
- Meagher MM, Tao BY, Chow JM, Reilly PJ. Kinetics and subsite mapping of a D-xylobiose- and D-xylose-producing Aspergillus niger endo-(1----4)-beta-D-xylanase. Carbohydr Res. 1988 Mar 1;173(2):273–283. [Abstract] [Google Scholar]
- Redinbaugh MG, Turley RB. Adaptation of the bicinchoninic acid protein assay for use with microtiter plates and sucrose gradient fractions. Anal Biochem. 1986 Mar;153(2):267–271. [Abstract] [Google Scholar]
- Saul DJ, Williams LC, Grayling RA, Chamley LW, Love DR, Bergquist PL. celB, a gene coding for a bifunctional cellulase from the extreme thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Oct;56(10):3117–3124. [Europe PMC free article] [Abstract] [Google Scholar]
- Shareck F, Roy C, Yaguchi M, Morosoli R, Kluepfel D. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene. 1991 Oct 30;107(1):75–82. [Abstract] [Google Scholar]
- Tomme P, Van Tilbeurgh H, Pettersson G, Van Damme J, Vandekerckhove J, Knowles J, Teeri T, Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988 Jan 4;170(3):575–581. [Abstract] [Google Scholar]
- Utt EA, Eddy CK, Keshav KF, Ingram LO. Sequencing and expression of the Butyrivibrio fibrisolvens xylB gene encoding a novel bifunctional protein with beta-D-xylosidase and alpha-L-arabinofuranosidase activities. Appl Environ Microbiol. 1991 Apr;57(4):1227–1234. [Europe PMC free article] [Abstract] [Google Scholar]
- Whitehead TR, Hespell RB. The genes for three xylan-degrading activities from Bacteroides ovatus are clustered in a 3.8-kilobase region. J Bacteriol. 1990 May;172(5):2408–2412. [Europe PMC free article] [Abstract] [Google Scholar]
- Wong KK, Tan LU, Saddler JN. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988 Sep;52(3):305–317. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang JX, Flint HJ. A bifunctional xylanase encoded by the xynA gene of the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 comprises two dissimilar domains linked by an asparagine/glutamine-rich sequence. Mol Microbiol. 1992 Apr;6(8):1013–1023. [Abstract] [Google Scholar]
Associated Data
Articles from Biochemical Journal are provided here courtesy of The Biochemical Society
Full text links
Read article at publisher's site: https://doi.org/10.1042/bj2960235
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1137679?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Bacterial Branched-Chain Amino Acid Biosynthesis: Structures, Mechanisms, and Drugability.
Biochemistry, 56(44):5849-5865, 01 Nov 2017
Cited by: 77 articles | PMID: 28977745 | PMCID: PMC5839172
Review Free full text in Europe PMC
Identification of GH10 xylanases in strains 2 and Mz5 of Pseudobutyrivibrio xylanivorans.
Folia Microbiol (Praha), 59(6):507-514, 20 Jun 2014
Cited by: 0 articles | PMID: 24942109
Paradigmatic status of an endo- and exoglucanase and its effect on crystalline cellulose degradation.
Biotechnol Biofuels, 5(1):78, 24 Oct 2012
Cited by: 11 articles | PMID: 23095278 | PMCID: PMC3502487
The effects of adding xylanase, vitamin C and copper sulphate to wheat-based diets on broiler performance.
Br Poult Sci, 42(4):493-500, 01 Sep 2001
Cited by: 8 articles | PMID: 11572625
The endoxylanases from family 11: computer analysis of protein sequences reveals important structural and phylogenetic relationships.
J Biotechnol, 95(2):109-131, 01 May 2002
Cited by: 63 articles | PMID: 11911922
Go to all (7) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A bifunctional xylanase encoded by the xynA gene of the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 comprises two dissimilar domains linked by an asparagine/glutamine-rich sequence.
Mol Microbiol, 6(8):1013-1023, 01 Apr 1992
Cited by: 43 articles | PMID: 1584021
A bifunctional enzyme, with separate xylanase and beta(1,3-1,4)-glucanase domains, encoded by the xynD gene of Ruminococcus flavefaciens.
J Bacteriol, 175(10):2943-2951, 01 May 1993
Cited by: 73 articles | PMID: 8491715 | PMCID: PMC204612
Expression of two xylanase genes from the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 cloned in pUC13.
J Gen Microbiol, 137(1):123-129, 01 Jan 1991
Cited by: 20 articles | PMID: 2045775
Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin.
Mol Microbiol, 6(15):2065-2072, 01 Aug 1992
Cited by: 74 articles | PMID: 1406248