Abstract
Free full text

Collaborative evaluation of the Radiometer Sensititre AP80 for identification of gram-negative bacilli.
Abstract
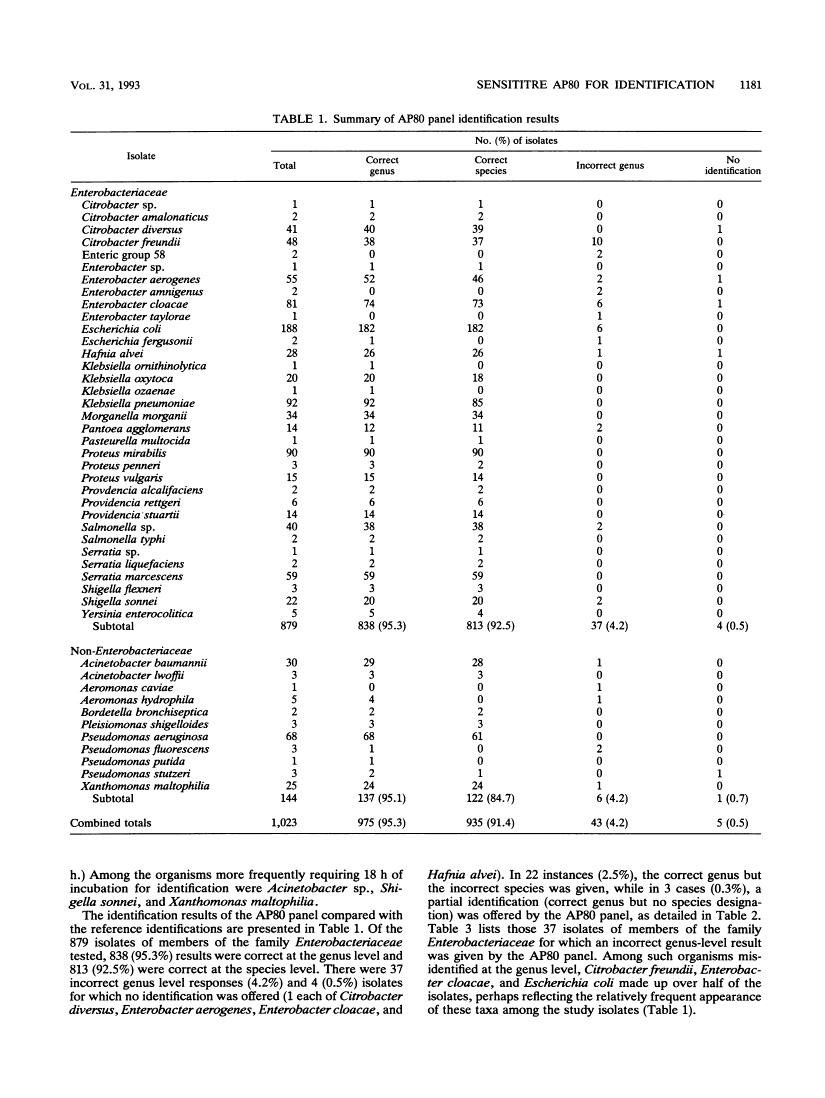
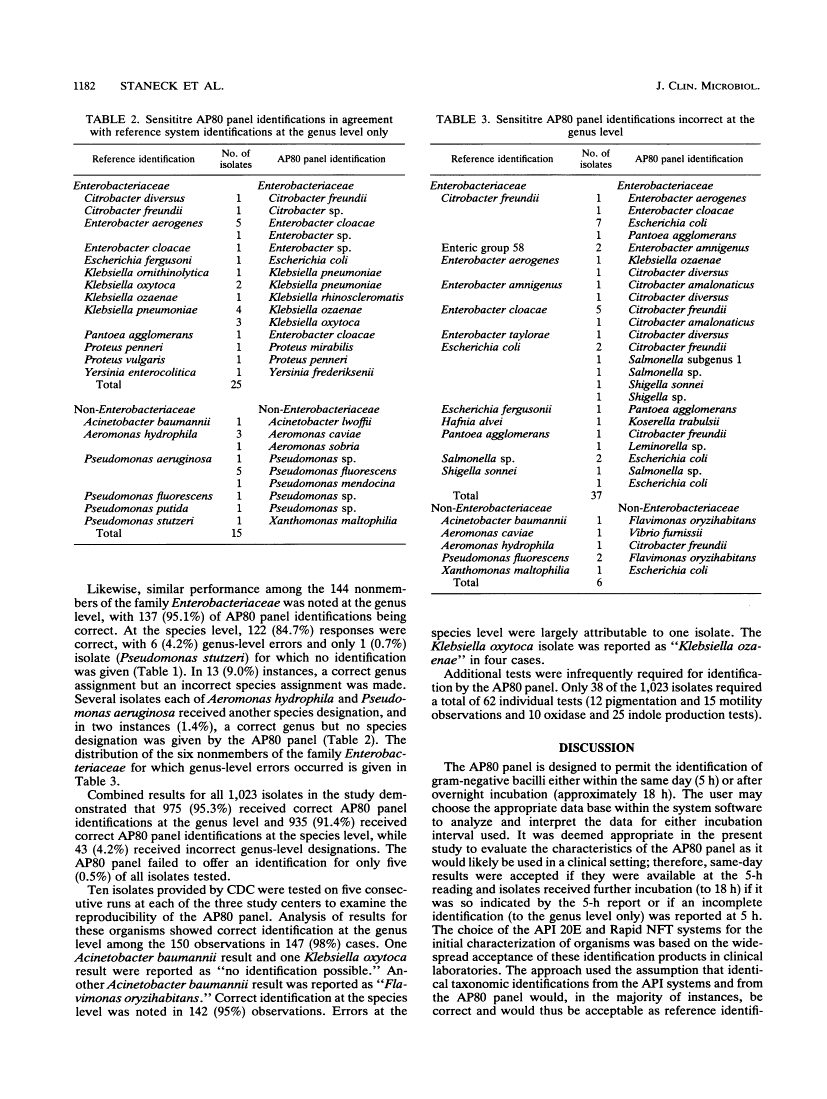
A multicenter trial of the Sensititre AP80 panel read on the Sensititre AutoReader (Radiometer America, Westlake, Ohio) for the automated identification of gram-negative bacilli was conducted with 1,023 clinical isolates (879 members of the family Enterobacteriaceae plus 144 nonenteric organisms). Assignment of taxa was based on the computer-assisted interpretation of the results of a series of reactions with fluorogenic enzyme substrates after 5 h of incubation, with an incubation interval of approximately 18 h used when indicated. Accuracy was determined initially by comparison with the results obtained with the API 20E or Rapid NFT system (Analytab Products, Plainview, N.Y.). Isolates showing discrepancies were identified by using conventional biochemical profiles. Identifications were available after 5 h of incubation for 918 isolates (90%). Agreements with reference results for members of the family Enterobacteriaceae were 95.3 and 92.5% at the genus and species levels, respectively, and for the nonmembers of the family Enterobacteriaceae, the agreements with reference results were 95.1 and 84.7%, respectively. The Sensititre AP80 panel was found to be simple and convenient to use, allowed for the testing of three isolates per panel, required minimal supplementary testing for completion of identification, performed in a reproducible fashion, and demonstrated an accuracy of same-day identification comparable to that reported for other automated systems. The AP80 panel appears well suited for routine use in the clinical microbiology laboratory as an automated means of identifying both members of the family Enterobacteriaceae and nonenteric gram-negative bacilli.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- DiPersio JR, Dyke JW, Vannest RD. Evaluation of the updated MS-2 Bacterial Identification system in comparison with the API 20E system. J Clin Microbiol. 1983 Jul;18(1):128–135. [Europe PMC free article] [Abstract] [Google Scholar]
- Doern GV, Staneck JL, Needham C, Tubert T. Sensititre autoreader for same-day breakpoint broth microdilution susceptibility testing of members of the family Enterobacteriaceae. J Clin Microbiol. 1987 Aug;25(8):1481–1485. [Europe PMC free article] [Abstract] [Google Scholar]
- Nolte FS, Krisher KK, Beltran LA, Christianson NP, Sheridan GE. Rapid and overnight microdilution antibiotic susceptibility testing with the Sensititre Breakpoint Autoreader system. J Clin Microbiol. 1988 Jun;26(6):1079–1084. [Europe PMC free article] [Abstract] [Google Scholar]
- Pfaller MA, Sahm D, O'Hara C, Ciaglia C, Yu M, Yamane N, Scharnweber G, Rhoden D. Comparison of the autoSCAN-W/A rapid bacterial identification system and the Vitek AutoMicrobic system for identification of gram-negative bacilli. J Clin Microbiol. 1991 Jul;29(7):1422–1428. [Europe PMC free article] [Abstract] [Google Scholar]
- Staneck JL, Allen SD, Harris EE, Tilton RC. Automated reading of MIC microdilution trays containing fluorogenic enzyme substrates with the Sensititre Autoreader. J Clin Microbiol. 1985 Aug;22(2):187–191. [Europe PMC free article] [Abstract] [Google Scholar]
- Staneck JL, Allen SD, Harris EE, Tilton RC. Rapid MIC testing with the sensititre autoreader. J Clin Microbiol. 1988 Jan;26(1):1–7. [Europe PMC free article] [Abstract] [Google Scholar]
- York MK, Brooks GF, Fiss EH. Evaluation of the autoSCAN-W/A rapid system for identification and susceptibility testing of gram-negative fermentative bacilli. J Clin Microbiol. 1992 Nov;30(11):2903–2910. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.31.5.1179-1184.1993
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/31/5/1179.full.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/31/5/1179
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/31/5/1179.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Adding a selective enrichment step to the iQ-Check real-time PCR improves the detection of Salmonella in naturally contaminated retail turkey meat products.
Lett Appl Microbiol, 43(1):78-83, 01 Jul 2006
Cited by: 9 articles | PMID: 16834725
An evaluation of sampling methods for the detection of Escherichia coli and Salmonella on Turkey carcasses.
J Food Prot, 68(1):34-39, 01 Jan 2005
Cited by: 10 articles | PMID: 15690801
Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia.
Clin Microbiol Rev, 11(1):57-80, 01 Jan 1998
Cited by: 446 articles | PMID: 9457429 | PMCID: PMC121376
Review Free full text in Europe PMC
Comparison of Crystal Enteric/Nonfermenter system, API 20E system, and Vitek AutoMicrobic system for identification of gram-negative bacilli.
J Clin Microbiol, 33(2):364-370, 01 Feb 1995
Cited by: 26 articles | PMID: 7714193 | PMCID: PMC227949
Recovery of uncommon bacteria from blood: association with neoplastic disease.
Clin Microbiol Rev, 8(3):336-356, 01 Jul 1995
Cited by: 23 articles | PMID: 7553569 | PMCID: PMC174628
Review Free full text in Europe PMC
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evaluation of the Phoenix 100 ID/AST system and NID panel for identification of Enterobacteriaceae, Vibrionaceae, and commonly isolated nonenteric gram-negative bacilli.
J Clin Microbiol, 44(3):928-933, 01 Mar 2006
Cited by: 20 articles | PMID: 16517878 | PMCID: PMC1393076
Evaluation of the Vitek 2 ID-GNB assay for identification of members of the family Enterobacteriaceae and other nonenteric gram-negative bacilli and comparison with the Vitek GNI+ card.
J Clin Microbiol, 41(5):2096-2101, 01 May 2003
Cited by: 18 articles | PMID: 12734254 | PMCID: PMC154679
Comparison of Crystal Enteric/Nonfermenter system, API 20E system, and Vitek AutoMicrobic system for identification of gram-negative bacilli.
J Clin Microbiol, 33(2):364-370, 01 Feb 1995
Cited by: 26 articles | PMID: 7714193 | PMCID: PMC227949
Comparison of the autoSCAN-W/A rapid bacterial identification system and the Vitek AutoMicrobic system for identification of gram-negative bacilli.
J Clin Microbiol, 29(7):1422-1428, 01 Jul 1991
Cited by: 23 articles | PMID: 1885737 | PMCID: PMC270128