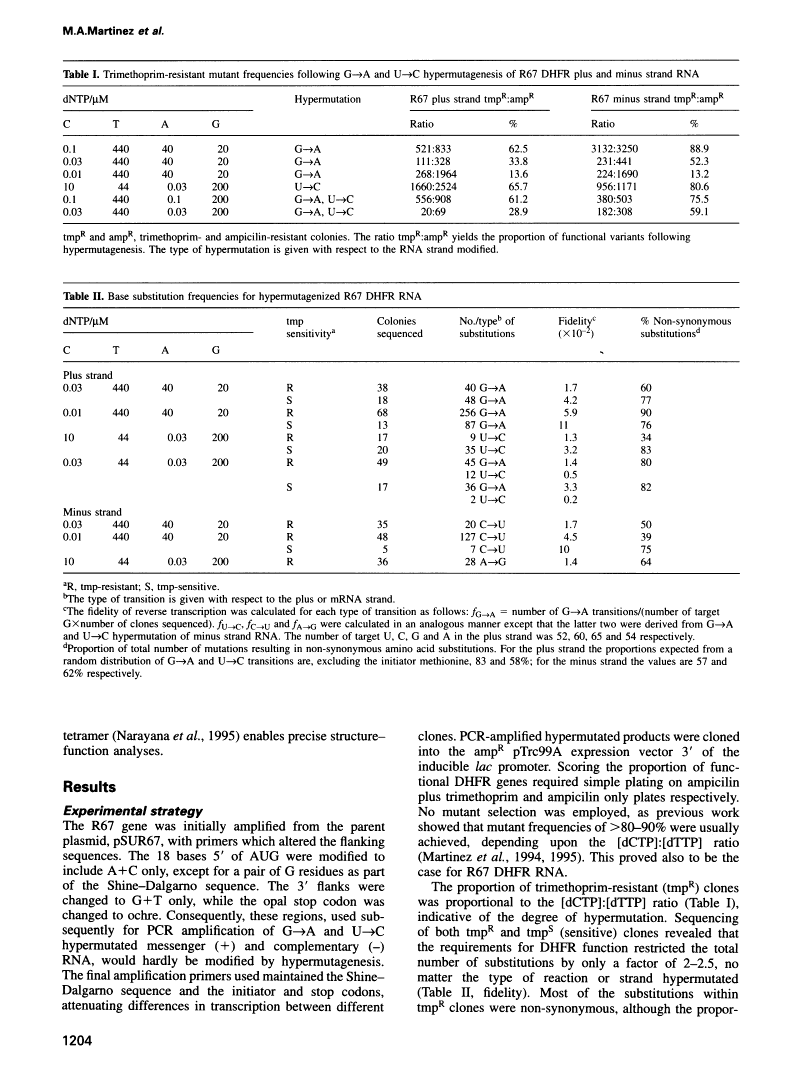
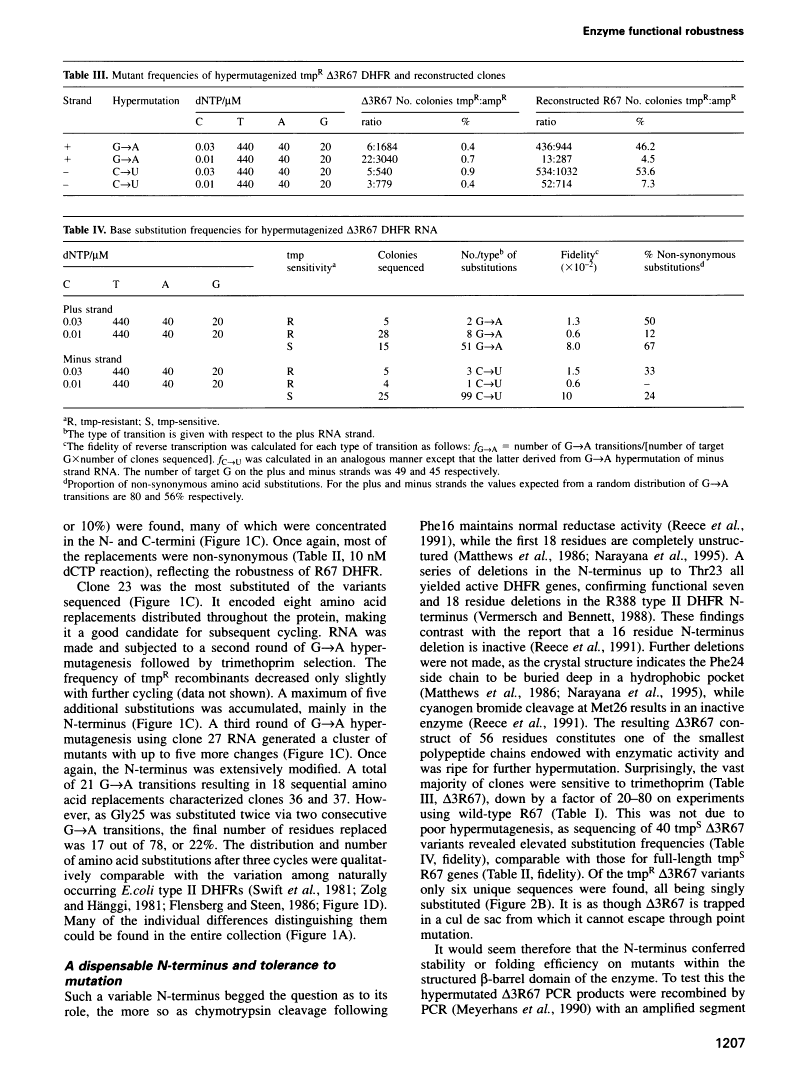
Abstract
Free full text

Exploring the functional robustness of an enzyme by in vitro evolution.
Abstract
The evolution of natural proteins is thought to have occurred by successive fixation of individual mutations. In vitro protein evolution seeks to accelerate this process. RNA hypermutagenesis, cDNA synthesis in the presence of biased dNTP concentrations, delivers elevated mutant and mutation frequencies. Here lineages of active enzymes descended from the homotetrameric 78 residue dihydrofolate reductase (DHFR) encoded by the Escherichia coli R67 plasmid were generated by iterative RNA hypermutagenesis, resulting in >20% amino acid replacement. The 22 residue N-terminus could be deleted yielding a minimum functional entity refractory to further changes, designating it as a determinant of R67 robustness. Complete substitution of the segment still allowed fixation of mutations. By the facile introduction of multiple mutations, RNA hypermutagenesis allows the generation of active proteins derived from extant genes through a mode unexplored by natural selection.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.4M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbas CF, 3rd, Bain JD, Hoekstra DM, Lerner RA. Semisynthetic combinatorial antibody libraries: a chemical solution to the diversity problem. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4457–4461. [Europe PMC free article] [Abstract] [Google Scholar]
- Barbas CF, 3rd, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry RM, Nara PL, Burton DR. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3809–3813. [Europe PMC free article] [Abstract] [Google Scholar]
- Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. [Abstract] [Google Scholar]
- Brisson N, Hohn T. Nucleotide sequence of the dihydrofolate-reductase gene borne by the plasmid R67 and conferring methotrexate resistance. Gene. 1984 May;28(2):271–274. [Abstract] [Google Scholar]
- Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992 Aug;2(1):28–33. [Abstract] [Google Scholar]
- Dao-pin S, Söderlind E, Baase WA, Wozniak JA, Sauer U, Matthews BW. Cumulative site-directed charge-change replacements in bacteriophage T4 lysozyme suggest that long-range electrostatic interactions contribute little to protein stability. J Mol Biol. 1991 Oct 5;221(3):873–887. [Abstract] [Google Scholar]
- DeGrado WF, Wasserman ZR, Lear JD. Protein design, a minimalist approach. Science. 1989 Feb 3;243(4891):622–628. [Abstract] [Google Scholar]
- Doolittle RF, Feng DF, Johnson MS, McClure MA. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. [Abstract] [Google Scholar]
- Flensburg J, Steen R. Nucleotide sequence analysis of the trimethoprim resistant dihydrofolate reductase encoded by R plasmid R751. Nucleic Acids Res. 1986 Jul 25;14(14):5933–5933. [Europe PMC free article] [Abstract] [Google Scholar]
- French DL, Laskov R, Scharff MD. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989 Jun 9;244(4909):1152–1157. [Abstract] [Google Scholar]
- Gram H, Marconi LA, Barbas CF, 3rd, Collet TA, Lerner RA, Kang AS. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3576–3580. [Europe PMC free article] [Abstract] [Google Scholar]
- Lehtovaara PM, Koivula AK, Bamford J, Knowles JK. A new method for random mutagenesis of complete genes: enzymatic generation of mutant libraries in vitro. Protein Eng. 1988 Apr;2(1):63–68. [Abstract] [Google Scholar]
- Loeb DD, Swanstrom R, Everitt L, Manchester M, Stamper SE, Hutchison CA., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989 Aug 3;340(6232):397–400. [Abstract] [Google Scholar]
- Martinez MA, Vartanian JP, Wain-Hobson S. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11787–11791. [Europe PMC free article] [Abstract] [Google Scholar]
- Martínez MA, Sala M, Vartanian JP, Wain-Hobson S. Reverse transcriptase and substrate dependence of the RNA hypermutagenesis reaction. Nucleic Acids Res. 1995 Jul 25;23(14):2573–2578. [Europe PMC free article] [Abstract] [Google Scholar]
- Matthews DA, Smith SL, Baccanari DP, Burchall JJ, Oatley SJ, Kraut J. Crystal structure of a novel trimethoprim-resistant dihydrofolate reductase specified in Escherichia coli by R-plasmid R67. Biochemistry. 1986 Jul 29;25(15):4194–4204. [Abstract] [Google Scholar]
- Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990 Apr 11;18(7):1687–1691. [Europe PMC free article] [Abstract] [Google Scholar]
- Narayana N, Matthews DA, Howell EE, Nguyen-huu X. A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site. Nat Struct Biol. 1995 Nov;2(11):1018–1025. [Abstract] [Google Scholar]
- Pakula AA, Young VB, Sauer RT. Bacteriophage lambda cro mutations: effects on activity and intracellular degradation. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8829–8833. [Europe PMC free article] [Abstract] [Google Scholar]
- Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6019–6023. [Europe PMC free article] [Abstract] [Google Scholar]
- Pattishall KH, Acar J, Burchall JJ, Goldstein FW, Harvey RJ. Two distinct types of trimethoprim-resistant dihydrofolate reductase specified by R-plasmids of different compatibility groups. J Biol Chem. 1977 Apr 10;252(7):2319–2323. [Abstract] [Google Scholar]
- Pjura P, Matsumura M, Baase WA, Matthews BW. Development of an in vivo method to identify mutants of phage T4 lysozyme of enhanced thermostability. Protein Sci. 1993 Dec;2(12):2217–2225. [Europe PMC free article] [Abstract] [Google Scholar]
- Reece LJ, Nichols R, Ogden RC, Howell EE. Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of the N-terminus in the protein. Biochemistry. 1991 Nov 12;30(45):10895–10904. [Abstract] [Google Scholar]
- Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991 Nov 5;222(1):67–88. [Abstract] [Google Scholar]
- Shortle D. Mutational studies of protein structures and their stabilities. Q Rev Biophys. 1992 May;25(2):205–250. [Abstract] [Google Scholar]
- Shortle D, Lin B. Genetic analysis of staphylococcal nuclease: identification of three intragenic "global" suppressors of nuclease-minus mutations. Genetics. 1985 Aug;110(4):539–555. [Europe PMC free article] [Abstract] [Google Scholar]
- Siderovski DP, Matsuyama T, Frigerio E, Chui S, Min X, Erfle H, Sumner-Smith M, Barnett RW, Mak TW. Random mutagenesis of the human immunodeficiency virus type-1 trans-activator of transcription (HIV-1 Tat). Nucleic Acids Res. 1992 Oct 25;20(20):5311–5320. [Europe PMC free article] [Abstract] [Google Scholar]
- Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10747–10751. [Europe PMC free article] [Abstract] [Google Scholar]
- Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994 Aug 4;370(6488):389–391. [Abstract] [Google Scholar]
- Swift G, McCarthy BJ, Heffron F. DNA sequence of a plasmid-encoded dihydrofolate reductase. Mol Gen Genet. 1981;181(4):441–447. [Abstract] [Google Scholar]
- Vartanian JP, Meyerhans A, Asjö B, Wain-Hobson S. Selection, recombination, and G----A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991 Apr;65(4):1779–1788. [Europe PMC free article] [Abstract] [Google Scholar]
- Vartanian JP, Meyerhans A, Sala M, Wain-Hobson S. G-->A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3092–3096. [Europe PMC free article] [Abstract] [Google Scholar]
- Vermersch PS, Bennett GN. Synthesis and expression of a gene for a mini type II dihydrofolate reductase. DNA. 1988 May;7(4):243–251. [Abstract] [Google Scholar]
- Zhou YH, Zhang XP, Ebright RH. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991 Nov 11;19(21):6052–6052. [Europe PMC free article] [Abstract] [Google Scholar]
- Zolg JW, Hänggi UJ. Characterization of a R plasmid-associated, trimethoprim-resistant dihydrofolate reductase and determination of the nucleotide sequence of the reductase gene. Nucleic Acids Res. 1981 Feb 11;9(3):697–710. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1002/j.1460-2075.1996.tb00461.x
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc450021?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1002/j.1460-2075.1996.tb00461.x
Article citations
Computational Development of Inhibitors of Plasmid-Borne Bacterial Dihydrofolate Reductase.
Antibiotics (Basel), 11(6):779, 07 Jun 2022
Cited by: 1 article | PMID: 35740185 | PMCID: PMC9220120
The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health.
Antibiotics (Basel), 10(4):433, 13 Apr 2021
Cited by: 7 articles | PMID: 33924456 | PMCID: PMC8103504
Small Angle Neutron Scattering Studies of R67 Dihydrofolate Reductase, a Tetrameric Protein with Intrinsically Disordered N-Termini.
Biochemistry, 56(44):5886-5899, 01 Nov 2017
Cited by: 4 articles | PMID: 29020453 | PMCID: PMC5678894
Base-Modified Nucleic Acids as a Powerful Tool for Synthetic Biology and Biotechnology.
Chemistry, 23(40):9560-9576, 27 Jun 2017
Cited by: 13 articles | PMID: 28513881
Quality control test for sequence-phenotype assignments.
PLoS One, 10(2):e0118288, 20 Feb 2015
Cited by: 2 articles | PMID: 25700273 | PMCID: PMC4336291
Go to all (38) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Directed evolution of trimethoprim resistance in Escherichia coli.
FEBS J, 274(10):2661-2671, 19 Apr 2007
Cited by: 29 articles | PMID: 17451440
Complementation between dimeric mutants as a probe of dimer-dimer interactions in tetrameric dihydrofolate reductase encoded by R67 plasmid of E. coli.
J Mol Biol, 302(1):235-250, 01 Sep 2000
Cited by: 9 articles | PMID: 10964572
Combinatorial exploration of the catalytic site of a drug-resistant dihydrofolate reductase: creating alternative functional configurations.
Protein Eng Des Sel, 17(11):809-819, 01 Nov 2004
Cited by: 30 articles | PMID: 15576381
Searching sequence space: two different approaches to dihydrofolate reductase catalysis.
Chembiochem, 6(4):590-600, 01 Apr 2005
Cited by: 30 articles | PMID: 15812782
Review