Abstract
Free full text

Comparison of Phenotypic and Genotypic Techniques for Identification of Unusual Aerobic Pathogenic Gram-Negative Bacilli
Abstract
Rapid and accurate identification of bacterial pathogens is a fundamental goal of clinical microbiology, but one that is difficult or impossible for many slow-growing and fastidious organisms. We used identification systems based on cellular fatty acid profiles (Sherlock; MIDI, Inc., Newark, Del.), carbon source utilization (Microlog; Biolog, Inc., Hayward, Calif.), and 16S rRNA gene sequence (MicroSeq; Perkin-Elmer Applied Biosystems Division, Foster City, Calif.) to evaluate 72 unusual aerobic gram-negative bacilli isolated from clinical specimens at the Mayo Clinic. Compared to lengthy conventional methods, Sherlock, Microlog, and MicroSeq were able to identify 56 of 72 (77.8%), 63 of 72 (87.5%), and 70 of 72 (97.2%) isolates to the genus level (P = 0.002) and 44 to 65 (67.7%), 55 of 65 (84.6%), and 58 of 65 (89.2%) isolates to the species level (P = 0.005), respectively. Four Acinetobacter and three Bordetella isolates which could not be identified to the species level by conventional methods were identified by MicroSeq. In comparison to the full 16S rDNA sequences, the first 527 bp provided identical genus information for all 72 isolates and identical species information for 67 (93.1%) isolates. These data show that MicroSeq provides rapid, unambiguous identification of clinical bacterial isolates. The improved turnaround time provided by genotypic identification systems may translate into improved clinical outcomes.
Accurate identification of bacterial isolates is an essential task of the clinical microbiology laboratory. For many slow-growing and fastidious organisms, traditional phenotypic identification is difficult and time-consuming. In addition, when phenotypic methods are used to identify bacteria, interpretation of test results involves substantial subjective judgement (34). Several commercial systems offer computer-assisted identification of a wide variety of bacterial organisms. Such systems include the carbon source utilization system developed by Biolog, Inc., based on panels of biochemical reactions (18, 24), and the gas-liquid chromatography system developed by MIDI, Inc. (Newark, Del.), based on the cellular fatty acid profile (27, 37). Such systems may reduce subjectivity and turnaround time, but they still rely on phenotypic identification.
Genotypic identification is emerging as an alternative or complement to established phenotypic methods. Typically, genotypic identification of bacteria involves the use of conserved sequences within phylogenetically informative genetic targets, such as the small-subunit (16S) rRNA gene (20, 23, 30, 41, 43). Broad-range PCR primers recognize conserved sequences in a variety of bacteria, while amplifying highly variable regions between the primer binding sites (3, 6, 14, 31, 39, 41). The amplified segment is sequenced and compared with known databases to identify a close relative (8, 20, 22, 25, 29, 30).
The MicroSeq 16S rRNA gene kit (Perkin-Elmer Applied Biosystems Division [PE-ABD], Foster City, Calif.) allows identification of bacteria based on the sequences of their 16S rRNA gene (21, 32). Genomic DNA extracted from bacteria is amplified and then sequenced. Sequence analysis software compares genes analyzed from unknown bacteria to a proprietary 16S rDNA sequence library. Although the MicroSeq system was developed specifically for the identification of environmental microorganisms, we have been interested in the use of this system for identifying clinical isolates, some of which may be synonymous with environmental isolates.
We report a comparative evaluation of three bacterial identification systems which are based on cellular fatty acid profiles (Sherlock), biochemical reactions (Microlog), and 16S rRNA gene sequence analysis (MicroSeq). These three methods were tested for their ability to differentiate 72 clinical isolates of unusual aerobic gram-negative bacilli. All these commercial systems were also compared with conventional phenotypic evaluation standards.
(This study was presented in part at the 98th General Meeting of the American Society for Microbiology, Atlanta, Ga., 17 to 21 May 1998.)
MATERIALS AND METHODS
Clinical isolates.
With the exception of two isolates, the gram-negative bacilli evaluated for this study were clinical consecutive isolates received by the Mayo Referral Bacteriology Laboratory during February and March 1997. Each isolate was screened by the computer-assisted replica plating (CARP) system developed at the Mayo Clinic (36, 38). CARP identifies gram-negative bacilli based on citrate, lysine and ornithine decarboxylase, urease, DNase, colistin, cephalosporinase, hydrogen sulfide, esculin hydrolysis, and arginine dihydrolase and on the fermentation of d-glucose, lactose, d-mannitol, sucrose, inositol, and l-arabinose. Isolates unidentifiable by CARP (approximately 10 to 15% of all isolates) were considered unusual and evaluated in our study.
Conventional methods—evaluation standard.
Unusual gram-negative bacilli were tested with one of two biochemical panels, one for glucose fermenters and one for nonfermenters. Classification was based on criteria used in the Enteric Bacteriology Laboratory and the Special Bacteriology Laboratory at the Centers for Disease Control and Prevention, Atlanta, Ga. (11, 26, 35, 36, 40). Taxonomy was based on newly published nomenclature (5).
Cellular fatty acid profile.
After 24 to 48 h growth at 28°C on Trypticase soy broth agar plates or at 35°C on 5% sheep blood agar plates, bacteria were saponified, and the liberated fatty acids were methylated and analyzed by capillary gas-liquid chromatography (1, 9) by the Sherlock system with the trypticase soy broth agar or CLIN aerobe database 3.9 (MIDI, Inc.) precisely according to the manufacturer’s instructions. The results for each isolate were computed as a similarity index. Similarity indices from 0.5 to 0.9 were considered reliable for identifying individual species (27, 37).
Biolog microstation system.
Single colonies were chosen, subcultured on 5% sheep blood agar plates, and incubated overnight at 35°C. A homogenous suspension of inoculum was made in 0.85% saline and diluted to a transmittance of 55 to 60% at 590 nm. One hundred fifty microliters of the suspension was dispensed into each well of the GN Microlog microplate, which was incubated for 24 h at 35°C. Microplates were read at 590 nm at 4 and 24 h with a computer-controlled MicroPlate reader (18, 24). Each metabolic profile was compared automatically with the GN Microlog database (release 3.50).
Sequence analysis of 16S rRNA gene.
The MicroSeq system contains a PCR and cycle sequencing module, bacterial identification and analysis software, and a 16S rDNA sequence database library (Fig. (Fig.1).1). For DNA preparation (step A, Fig. Fig.1),1), a loopful of bacterial cells was washed with distilled water and incubated with 200 μl of 5% Chelex solution (PE-ABD) for 15 min at 56°C. The suspension was vortexed and heated for 8 min at 100°C and centrifuged at 8,000 × g for 2 min. Two microliters of a 10-fold dilution of the supernatant was used for PCR amplification.
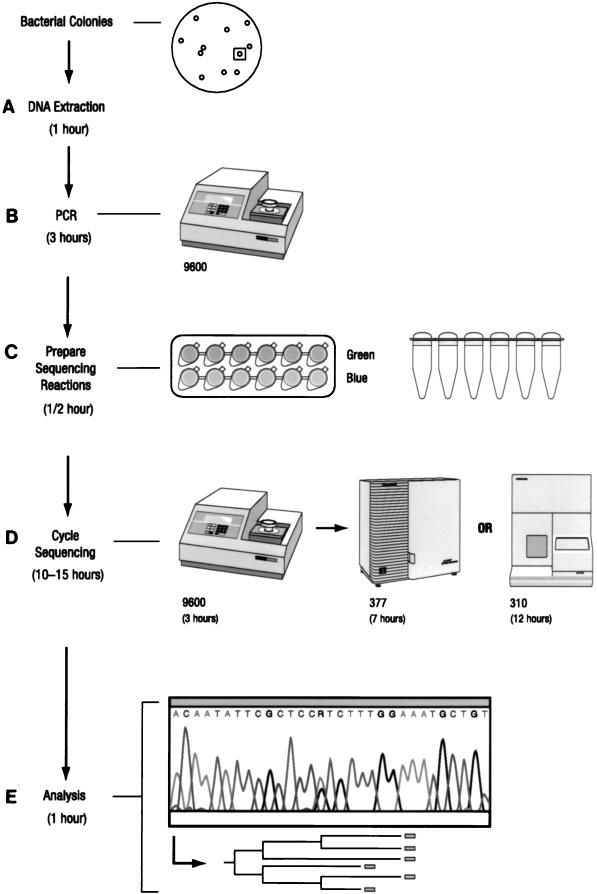
Flowchart of the MicroSeq process from culture to sequence. The total elapsed time was 15.5 to 18.5 h, comprising bacterial DNA extraction (A), PCR (B), sequencing reaction preparation (C), cycle sequencing (D), and analysis (E). The time required for each step is indicated.
PCR amplification of the full 16S rRNA gene (4) was accomplished by adding 2 μl of genomic DNA extract plus 48 μl of water to 50 μl of MicroSeq 16S rRNA gene PCR master mix (including primers 0005F and 1540R) in 0.2-ml MicroAmp PCR tubes (PE-ABD). A negative control (sterile deionized water) and a positive control (Escherichia coli genomic DNA; Sigma Corporation) were included. Tubes were capped and placed in the GeneAmp PCR System 9600 (PE-ABD). Thermal cycling parameters included an initial 10 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 60°C, and 45 s at 72°C; and a final extension at 72°C for 10 min (step B, Fig. Fig.1).1). The PCR products were purified prior to sequencing by Microcon-100 microconcentrator columns (Amicon).
To prepare cycle sequencing reactions, 45 μl of PCR product was diluted with 60 μl of water, and 7 μl was aliquoted into each of 12 tubes containing dye terminator sequencing mix which included 1 of 12 16S sequencing primers (0005F, 0338F, 0357R, 0515F, 0531R, 0776F, 0810R, 1087F, 1104R, 1174F, 1193R, or 1540R) (step C, Fig. Fig.1).1). Cycle sequencing was performed by the GeneAmp PCR 9600 system. The cycle sequencing profile consisted of a 96°C denaturation step for 10 s followed by an anneal/extension step starting at 65°C and decreasing 1°C every six cycles until a touchdown temperature of 55°C was reached, for a total of 66 cycles. Sequences were determined by electrophoresis with the ABI PRISM 377 DNA sequencer (step D, Fig. Fig.11).
Three subregions of the 16S rRNA gene were defined as follows. Region A was defined as the 527-bp sequences between primers 0005F and 0531R, region B was defined as the 418 bp between primers 0776F and 1193R, and region AB was defined as the 1,189 bp between primers 0005F and 1193R.
With the MicroSeq microbial identification and analysis software, sequence sample files were assembled and the final consensus sequence was compared with over 1,100 validated full 16S rDNA sequences in the MicroSeq database library. Polymorphic positions present in those organisms containing multiple copies of the gene were included to ensure the highest degree of accuracy (step E, Fig. Fig.11).
Statistics.
Comparisons of agreement rate with results of conventional methods (based on rate ratios) were performed with Epiinfo, version 6, Centers for Disease Control and Prevention.
RESULTS
All 72 isolates in our study were identified to genus level, and 65 isolates were identified to species level by conventional Mayo methods, which served as our evaluation standard. The isolates identified to the species level included 25 fermenters (1 Citrobacter braakii species, 1 Citrobacter farmeri species, 2 Citrobacter koseri species, 2 Enterobacter aerogenes species, 1 Enterobacter agglomerans species, 6 Enterobacter cloacae species, 3 E. coli species, 1 Klebsiella pneumoniae species, 1 Klebsiella oxytoca species, 1 Kluyvera cryocrescens species, 1 Leclercia adecarboxylata species, 2 Pasteurella multocida species, 1 Providencia rettgeri species, 1 Providencia stuartii species, and 1 Serratia odorifera species) and 40 nonfermenters (2 Acinetobacter lwoffii species, 1 Alcaligenes faecalis species, 3 Alcaligenes xylosoxidans species, 1 Brevundimonas dimunuta species, 1 Brevundimonas vesicularis species, 3 Burkholderia cepacia species, 1 Moraxella osloensis species, 2 Oligella urethralis species, 7 Pseudomonas aeruginosa species, 6 Pseudomonas fluorescens species or Pseudomonas putida species, 1 Pseudomonas pictorum species, 1 Pseudomonas stutzeri species, and 11 Stenotrophomonas maltophilia species).
Seven isolates could not be identified to the species level by phenotypic identification systems including Sherlock, Microlog, and the conventional Mayo methods (Table (Table1).1). These were all identified at species level by the MicroSeq system. These organisms were identified as two Acinetobacter baumannii species, one Acinetobacter calcoaceticus species, one Acinetobacter junii species, and three Bordetella holmesii species (35).
TABLE 1
Rapid techniques for identification of unusual pathogenic aerobic gram-negative bacilli compared with conventional phenotypic methods
methods
| Organisma and level | No. tested | No. matched (%) by indicated test
| ||
|---|---|---|---|---|
| Sherlock | Biolog | MicroSeq | ||
| Fermentersb | ||||
 Genus Genus | 25 | 17 (68.0) (68.0) | 21 (84.0) (84.0) | 23 (92.0) (92.0) |
 Species Species | 25 | 13 (52.0) (52.0) | 20 (80.0) (80.0) | 18 (72.0) (72.0) |
| Nonfermentersc | ||||
 Genus Genus | 47 | 39 (83.0) (83.0) | 42 (89.4) (89.4) | 47 (100.0) (100.0) |
 Species Species | 40 | 31 (77.5) (77.5) | 35 (87.5) (87.5) | 40 (100.0) (100.0) |
| Total | ||||
 Genus Genus | 72 | 56 (77.8) (77.8) | 63 (87.5) (87.5) | 70 (97.2) (97.2) |
 Species Species | 65 | 44 (67.7) (67.7) | 55 (84.6) (84.6) | 58 (89.2) (89.2) |
Among the three identification systems evaluated, genotypic identification by MicroSeq had the highest concordance with conventional methods (Table (Table1).1). At the genus level, agreement with conventional methods was 97.2% for MicroSeq compared to 87.5% for Microlog (χ2 = 4.82, P = 0.028) and 77.8% for Sherlock (χ2 = 12.44, P < 0.001). MicroSeq demonstrated better performance with nonfermentative bacilli than with fermenters (principally Enterobacteriaceae).
We tested the efficacy of identification based on subregion analysis in comparison to the full 16S rDNA sequence. Table Table22 showed that region A, which covered first 527 bp, gave the same genus information for all 72 isolates as the full 16S rRNA gene. Region A provided same species identification as full sequence data for 46 (97.9%) of 47 nonfermenters and 21 (84.0%) of 25 fermenters.
TABLE 2
Identification of unusual gram-negative bacilli based on sequence analysis of different regions of the 16S rRNA genea
genea
| Region | No. agreed to full sequences (%)
| |||||
|---|---|---|---|---|---|---|
| Fermenters (n = 25)
| Nonfermenters (n = 47)
| Total (n = 72)
| ||||
| Genus level | Species level | Genus level | Species level | Genus level | Species level | |
| A | 25 (100.0) (100.0) | 21 (84.0) (84.0) | 47 (100.0) (100.0) | 46 (97.9) (97.9) | 72 (100.0) (100.0) | 67 (93.1) (93.1) |
| B | 19 (76.0) (76.0) | 17 (68.0) (68.0) | 46 (97.9) (97.9) | 38 (80.9) (80.9) | 65 (90.3) (90.3) | 55 (76.4) (76.4) |
| AB | 25 (100.0) (100.0) | 24 (96.0) (96.0) | 47 (100.0) (100.0) | 46 (97.9) (97.9) | 72 (100.0) (100.0) | 70 (97.2) (97.2) |
The identification of seven isolates by the MicroSeq system differed from that provided by the conventional Mayo method, according to which six of these isolates were identified as E. cloacae species. We analyzed the 16S rDNA sequence derived from these 6 isolates, 11 Enterobacteriaceae stored in the MicroSeq database, and 8 Enterobacter species available in GenBank (15, 16). The six mismatched isolates fell into three clusters. Genotypically related isolates within a defined cluster had different biochemical profiles, including those generated by both the conventional Mayo method and the Microlog methods (data not shown). The sequences of these six E. cloacae isolates had a difference rate of 0.98 to 2.42% (mean ± standard deviation, 1.90% ± 0.47%) from the reference E. cloacae strain. The difference rate was 1.39 to 1.62% (mean ± standard deviation, 1.52% ± 0.10%) among three clusters and <0.5% within the clusters (Fig. (Fig.2).2).
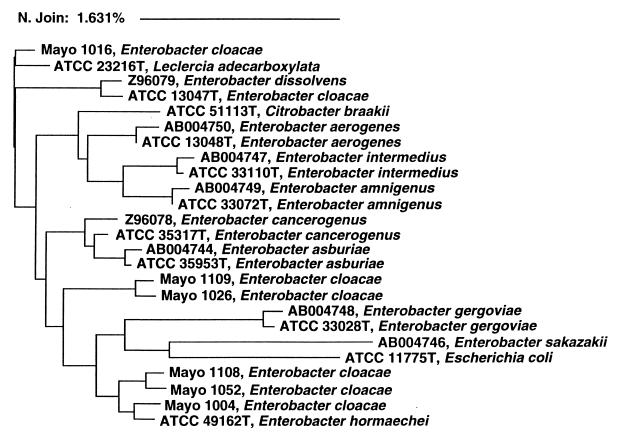
Neighbor-joining analysis of DNA sequences from several Enterobacter spp. Phylogenetic analysis was based on full 16S rRNA gene sequences, and the scale reflects relative phylogenetic distance. Isolates with names beginning with Mayo were evaluated in this study. Isolates with names beginning with accession numbers were retrieved from GenBank (15, 16). The remaining isolates, whose names begin with ATCC numbers, were type strains stored in the MicroSeq database.
DISCUSSION
Clinical and public health interventions depend on rapid, accurate identification of pathogenic bacteria. We have shown that genotypic methods based on 16S rDNA sequence analysis improved the identification of aerobic gram-negative pathogens compared to conventional phenotypic methods, cellular fatty acid profiles, and a carbon source utilization system. Four Acinetobacter and three Bordetella isolates which could not be identified to the species level by conventional methods were identified by the MicroSeq system by using phylogenetic analysis.
For decades, microbiologists have sought improved pathogen identification through the use of phenotypic methods. Abel and colleagues first suggested that microorganisms could be classified by gas chromatographic analysis (1). These concepts found application in the form of the Sherlock system. The Sherlock system bases organism identification solely on computer comparison of the unknown organism’s fatty acid methyl ester profile with the profiles of a predetermined library of known isolates with pattern recognition software. Osterhout and associates found that the system correctly identified 478 of 532 (89.8%) clinical isolates and reference strains of gram-negative nonfermentative bacteria. The majority of the strains for which gas-liquid chromatography identification did not agree with biochemical criteria belonged to the genera Acinetobacter, Moraxella, Alcaligenes, and Pseudomonas, all of which were among our isolates (27). In our study, Sherlock identified only 67.7% of isolates to the species level, presumably because the easily identified species had already been screened out by CARP (see Materials and Methods).
Traditional phenotypic identification relies upon biochemical pathways and carbon source utilization. Investigators over the years have tried a variety of improvements through automation. One such effort is the Microlog system, comprising a microtiter plate that tests for the ability of a microorganism to utilize different carbon sources. The test yields a pattern of colored wells that constitutes a metabolic fingerprint of the inoculated organism. Preliminary evaluation revealed that the system performed well with many genera, but problems were encountered with some strains of Klebsiella, Enterobacter, and Serratia (24). Holmes and colleagues have evaluated the system with 214 strains of nonfermenters representing 15 species (18). The authors noted that no other commercial identification system has as many organisms in a single database as that supplied by Microlog, but even the large number of tests available may not be adequate for discriminating all clinically relevant pairs of taxa (18). The Microlog system possesses the limitations of all biochemical identification schemes, since (i) individual tests may not be highly reproducible and (ii) the species metabolic phenotype is not an absolute property but may exhibit variability.
We found that 16S rRNA gene sequences frequently provide phylogenetically useful information. Signature nucleotides allow classification even if a particular sequence has no match in the database, since otherwise-unrecognizable isolates can be assigned to a phylogenetic branch at the class, family, genus, or subgenus level. We also note that identification of slow-growing or biochemically inert gram-negative bacilli to the species level is difficult and time-consuming by conventional methods (13, 17, 19, 35). In the present study, seven species (four Acinetobacter spp. and three Bordetella spp.) that could not be identified by exhaustive phenotypic methods were all assigned a definite species identification by the MicroSeq system. Direct sequence determination of 16S rRNA gene fragments represents a highly accurate and versatile method for identification of bacteria to the species level, even when the species in question is notoriously difficult to identify by biochemical means.
Some of the potential difficulties of the interpretation 16S sequence analysis were also noted in this study. Reliable genotypic identification requires accurate and complete genetic databases. In our study, six isolates identified as E. cloacae species by the conventional Mayo methods were assigned to other taxa by the MicroSeq system. Phylogenetic analysis indicated that these six isolates fell into three clusters, with a 1.52% difference rate among them (Fig. (Fig.2).2). Since a difference rate of >0.5% is generally considered a new species within a given genus (28, 33), the 16S sequence divergence among these strains suggests that they may in fact be new species. E. cloacae is one of the most-difficult Enterobacteriaceae to identify due to its variable biochemical and antibiogram profiles (2, 7, 42). We conclude from the evaluation of these isolates that more E. cloacae strains need to be included in genetic databases and that subdivision of E. cloacae into several new species may be indicated.
Another potential limitation of the 16S rDNA sequencing application in general is the inability to assign a species for recently diverged species (10, 12, 44). Since our study was limited to gram-negative bacilli and to a relatively small number of analyses, we did not encounter this problem. However, if this problem does become significant, 16S sequence analysis could be envisioned as a screening platform on which to assign additional analytical procedures such as sequencing protein coding genes or the intergenic spacer region of the ribosomal gene complex for increased variability.
The speed of microbial identification results can have a major impact on clinical management. Identification of bacteria by conventional methods usually requires ≥48 h after a discrete colony has been isolated. Two weeks are required for the identification of many slow-growing and fastidious organisms by conventional methods. In some circumstances, no identification can be made after weeks of analysis, even by an experienced technologist. Although commercially available identification systems such as Biolog and Sherlock sometimes shorten turnaround time, these systems often require prolonged growth before a fastidious organism can be assayed and a percentage of unusual organisms is always unidentifiable. In contrast, the MicroSeq system can be completed within 48 h with a minimal amount of starting material.
Cost is a critical issue in the evaluation of 16S rDNA sequence analysis as a diagnostic tool. Driven in part by the technology underlying the human and microbial genome projects, sequencing costs will probably continue their rapid trend downward, bringing this technology within the reach of many microbiology laboratories (32). Our results indicate that the first 527 bp gave the same genus identification as full sequence data from all 72 clinical isolates evaluated. Sequence analysis of this region needs only one amplification step and two sequence reactions; therefore, the price of reagents approaches the costs of the reagents and labor for many phenotypic methods. Since genus-level information is usually sufficient for clinical diagnostic purposes, sequence analysis of the first 527 bp of the 16S rRNA gene may be the first-tier test to be considered in laboratories with automated sequencing equipment. A MicroSeq 500 16S bacterial sequencing kit designed to sequence the first 527 bp of the 16S rRNA gene is currently being developed.
ACKNOWLEDGMENTS
We thank Scott Anderson, Michael Waddington, John Bartell, Maggie Riehman, Patrick Collins, Dan Chapman, Stacy Montogomery, Kelly Doerr, Sherri Wohlfiel, Jean Gutschenritter, Robert Segner, Janice Gillard, Patricia Schams, Jeanne Licari, Dianne McGrath, Sandra Borsheim, Carol McKibbon, Shirley Pokorski, and Jonathan Hibbs for their assistance.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.36.12.3674-3679.1998
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/36/12/3674.full.pdf
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/36/12/3674.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/36/12/3674
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/full/36/12/3674
Citations & impact
Impact metrics
Article citations
Occurrence of Multidrug-Resistant Strains of Acinetobacter spp.: An Emerging Threat for Nosocomial-Borne Infection in Najran Region, KSA.
Trop Med Infect Dis, 8(2):108, 09 Feb 2023
Cited by: 4 articles | PMID: 36828524 | PMCID: PMC9963463
A Comprehensive Overview of Antibiotic Selection and the Factors Affecting It.
Cureus, 13(3):e13925, 16 Mar 2021
Cited by: 4 articles | PMID: 33868859 | PMCID: PMC8049037
Review Free full text in Europe PMC
Molecular Phylogenetic Analysis of 16S rRNA Sequences Identified Two Lineages of Helicobacter pylori Strains Detected from Different Regions in Sudan Suggestive of Differential Evolution.
Int J Microbiol, 2020:8825718, 27 Oct 2020
Cited by: 6 articles | PMID: 33178282 | PMCID: PMC7609147
Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory.
Clin Microbiol Rev, 33(4):e00053-19, 09 Sep 2020
Cited by: 90 articles | PMID: 32907806 | PMCID: PMC7484979
Review Free full text in Europe PMC
Diversity and enzymatic potential of thermophilic bacteria associated with terrestrial hot springs in Algeria.
Braz J Microbiol, 51(4):1987-2007, 21 Sep 2020
Cited by: 3 articles | PMID: 32959204 | PMCID: PMC7688810
Go to all (133) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis.
J Clin Microbiol, 38(4):1676-1678, 01 Apr 2000
Cited by: 51 articles | PMID: 10747168 | PMCID: PMC86524
Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles.
J Clin Microbiol, 41(5):1996-2001, 01 May 2003
Cited by: 102 articles | PMID: 12734240 | PMCID: PMC154750
Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system.
J Clin Microbiol, 38(1):246-251, 01 Jan 2000
Cited by: 99 articles | PMID: 10618095 | PMCID: PMC88703
Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories.
Clin Microbiol Infect, 14(10):908-934, 01 Oct 2008
Cited by: 345 articles | PMID: 18828852
Review





