Abstract
Free full text

Quasispecies Development of Helicobacter pylori Observed in Paired Isolates Obtained Years Apart from the Same Host
Abstract
Helicobacter pylori isolates show greater genetic diversity than other bacterial species studied, but the basis for this phenomenon is unknown. Whether detectable genomic mutation appears within an H. pylori population during persistent colonization was investigated. Paired H. pylori populations obtained across 7- to 10-year intervals from 13 patients were characterized by use of methods including polymerase chain reaction (PCR) genotyping for cagA, vacA, iceA, recA, and IS605; random arbitrarily primed DNA (RAPD)–PCR and amplified fragment length polymorphism (AFLP) analysis; and ELISA, to determine Lewis phenotypes. Genotyping, including recA sequence analysis, revealed that initial and follow-up populations represented the same population in 11 patients (85%). Nevertheless, distinct dissimilarities were shown within each of these 11 pairs by both RAPD-PCR and AFLP analyses. During follow-up, Lewis-y levels, but not Lewis-x levels, decreased significantly. The changes detected by RAPD-PCR and AFLP indicate that genetic drift occurs within H. pylori populations over the course of years of colonization of a single host.
Helicobacter pylori isolates obtained from different individuals show substantial genomic diversity [1, 2]. Differences among strains include point mutations in conserved genes [3]; the presence of nonconserved genes, such as in the cag region [4, 5]; the presence of insertion elements [4, 6], plasmids [7], or gene mosaicisms [8]; gene size heterogeneity [9–11]; and chromosomal rearrangements [12]. For example, 80 clinical H. pylori isolates studied in the same laboratory each had a unique nucleotide sequence within a small region of the conserved ureC gene [3]. This genomic diversity can be used for epidemiologic analysis of the mechanisms of H. pylori transmission [13] and has been shown to have clinical relevance. In the United States and Europe, patients carrying strains containing the cag pathogenicity island, or having the iceA1 and s1/m1 vacA alleles, are at increased risk for peptic ulcer disease, compared with those carrying strains that lack these genetic characteristics [9, 11, 14]. In addition, cagA-positive strains have been associated with severe gastritis and the development of atrophic gastritis [15, 16] and noncardia gastric cancer [17], whereas recent data suggest that they may protect against cancer of the cardia and distal esophagus [18].
A number of explanations for this diversity have been advanced, including a long evolutionary history, a high level of mutation, and frequent horizontal gene transfer between strains [19]. Diversity also might reflect variation and changes in host niches for H. pylori. For example, many persons who carry H. pylori eventually develop atrophic changes of the gastric body mucosa that are associated with a reduction in acid production [20].
We hypothesize that mutations frequently develop over the course of prolonged colonization of a host by H. pylori and can be detected during long-term assessment. An earlier study reported distinct differences among H. pylori isolates obtained within a single family [21]. By means of random arbitrarily primed DNA (RAPD)–polymerase chain reaction (PCR), van der Ende et al. [21] observed highly similar H. pylori strains among members of the same family, with variation between and within individuals. Another study reported different susceptibilities to metronidazole for genotypically identical single colonies of clinical H. pylori isolates [22]. By use of pulsed-field electrophoresis, others did not observe development of genomic diversity in repeated H. pylori isolates from 20 patients followed for up to 2 years [23]. We therefore sought to detect genomic diversity among H. pylori populations that were sampled between 7 and 10 years apart, by use of several sensitive genotyping methods.
Methods
Patients, bacteria, and growth media
Thirteen patients who presented with dyspeptic symptoms and were colonized with H. pylori were studied by initial and follow-up upper gastrointestinal endoscopy, with a mean (±SD) interval of 8.3 ± 1.1 years (range, 7–10; table 1). There were 8 men and 5 women, and their mean age (±SD) at the first visit was 52 ± 15 years (range, 29–84). For each patient, H. pylori was isolated from antral biopsy specimens at both the initial and follow-up endoscopies. After isolation, cultures of sweeps of the biopsy plates were stored at –70°C. They were regrown under microaerobic conditions (5% CO2) at 37°C on trypticase soy agar (TSA) plates with 5% sheep blood (BBL Microbiology Systems, Cockeysville, MD). The entire population of bacterial cells was harvested after 48 h of growth on TSA plates and was subjected to genotypic and phenotypic analysis.
Table 1
Characteristics of patients sampled by endoscopy at 7- to 10-year intervals and genotypes and phenotypes of Helicobacter pylori pools cultured from each sampling.
| Patient | Genotype | Phenotype | ||||||
|---|---|---|---|---|---|---|---|---|
| Pool | Sex/age (years) at first visit | Interval between samplings (years) | cagA | vacA | iceA | IS605 | CagA | Antibiotic resistance |
| 1A | M/65 | 8.8 | + | s1a/m1 | 2 | – | – | |
| 1B | + | s1a/m1 | 1 | + | Mtz | |||
| 2A | F/41 | 7.2 | – | s2/m2 | 2 | – | – | |
| 2B | + | s1a/m1 | 2 | – | – | |||
| 3A | M/74 | 7.9 | – | s2/m2 | 2 | + | – | – |
| 3B | ||||||||
| 4A | F/57 | 8.7 | + | s1a/m1 | 1 | + | + | Mtz |
| 4B | Mtz | |||||||
| 5A | M/29 | 7.0 | – | s2/m2 | 2 | – | – | – |
| 5B | ||||||||
| 6A | F/35 | 8.1 | + | s1a/m1 | 1 | + | + | – |
| 6B | ||||||||
| 7A | F/49 | 7.0 | + | s1a/m1 | 2 | – | + | – |
| 7B | ||||||||
| 8A | M/61 | 9.8 | + | s1a/m1 | 1 | + | + | – |
| 8B | ||||||||
| 9A | M/64 | 8.8 | – | s2/m2 | 1 | – | – | – |
| 9B | ||||||||
| 10A | M/84 | 10.2 | + | s1b/m1 | 2 | – | + | – |
| 10B | ||||||||
| 11A | M/41 | 7.8 | + | s1a/m1 | 2 | – | + | Mtz |
| 11B | – | |||||||
| 12A | F/47 | 9.0 | + | s1a/m1 | 1 | – | + | – |
| 12B | ||||||||
| 13A | M/46 | 7.4 | + | s1a/m1 | 1 | + | + | – |
| 13B | Mtz | |||||||
NOTE. If no data are shown for 2d pool in pair, it is identical to 1st. cagA [11], vacA [8], and iceA [9] genotypes were determined by polymerase chain reaction (PCR), as described elsewhere. IS605 status was determined by PCR [6] and hybridization by use of 323-bp probe to IS200 homologue. CagA phenotype was determined by Western blot for pairs 3–13. All isolates were susceptible to amoxicillin, tetracycline, and clarithromycin. F, female; M, male; plus sign (+), present; minus sign (–), absent; Mtz, metronidazole resistant (MIC, >8 μg/mL).
Genotyping for cagA, vacA, iceA, and IS605
From each pool, chromosomal DNA was prepared as described previously [24]. DNA concentrations were measured by fluorometry (Dynaquant 200; Hoefer Pharmacia Biotech, Piscataway, NJ). The cagA [11], vacA [8], iceA [9], and IS605 [6] status for each population was determined by PCR and analyzed by gel electrophoresis, as described previously. In addition, the results of cagA, vacA, and iceA genotyping were confirmed by PCR followed by line probe assay [9]. For IS605, primers were complementary to orfA, homologous to the transposase of IS200, as described elsewhere [6]. Ten nanograms of DNA template was used for each reaction. Results were analyzed by electrophoresis in 1% agarose gels (Eastman Kodak, Rochester, NY).
RAPD-PCR
Forty nanograms of DNA template was used for RAPD-PCR with primers 1254, 1281, 8635, 9355, and d11344, as described elsewhere [25]. Results were analyzed by electrophoresis on 1% agarose gels. RAPD photographs obtained with primers 1254, 9355, and d11344 were scanned with a densitoscanner (Scanjet 4C; Hewlett-Packard, Bristol, UK), and the images were stored as TIFF (Tagged Image File Format) files with Deskscan II version 2.3 software (Hewlett-Packard).
Amplified fragment length polymorphism (AFLP)
AFLP analysis was done as described elsewhere [26, 27], with the following modifications. In a final volume of 20 μL, 20 ng of purified chromosomal DNA was digested with 1 U of EcoRI (Pharmacia, Uppsala, Sweden) and 1 U of MseI (New England Biolabs, Beverly, MA). The final volume then was increased to 30 μL by addition of 50 pmol each of the EcoRI adaptor and MseI adaptor, 1.2 U of T4 DNA ligase (Pharmacia), 1 mM ATP, and ligase buffer. The adaptors were allowed to ligate to the restriction fragments for 16 h at 16°C, after which the sample was diluted with distilled water in a final volume of 500 μL. A Texas Red fluorescent-labeled EcoO primer (5′-AGACTGCGTACCAATTC-3′; Isogen Bioscience, Maarssen, The Netherlands) was used for DNA amplification in 10 μL of a reaction mixture containing 0.3 ng of template DNA, 20 ng of labeled EcoO primer, 1 μL of 2 mM dNTP, 60 ng of unlabeled MseO primer (5′-GACGATGAGTCCTGAG-3′), 1 U of Taq polymerase (Perkin-Elmer, Norwalk, CT) in 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 1.5 mM MgCl2. Amplification was done in a GeneAmp PCR system 9700 thermal cycler (Perkin-Elmer) with 35 cycles: denaturation (30 s at 94°C), annealing (30 s at 65–56°C), and DNA molecule extension (60 s at 72°C). In the first 12 cycles, the annealing temperature was lowered 0.7°C per cycle. After completion of the cycle program, 3 μL of loading buffer (Amersham Life Science, Cleveland) was added to the reaction mixtures, after which 2.5 μL of each sample was analyzed according to the manufacturer's instructions in a Vistra 725 automated DNA sequencer (Amersham Life Science). Fluorescent AFLP images were stored as TIFF files with the Vistra 2 TIFF software (Amersham).
Densitometric scanning and data processing
Computer-assisted analysis of the digitized RAPD images and the AFLP images was done by means of the GelCompar 4.0 software (Applied Maths, Kortrijk, Belgium). The obtained images were normalized for lane-to-lane differences in mobility by the alignment of patterns obtained on multiple specimens of standard DNA size markers (Amersham) included at regular intervals in each gel, and subsequently the background was subtracted by use of a curve-fitting algorithm. The levels of similarity of these digitized patterns then were calculated with the Pearson product moment correlation coefficient (r) of the GelCompar program, and the UPGMA method (unweighted-pair group method using arithmetic averages) was used to cluster the obtained patterns [28].
Sequencing of recA and IS605 PCR products
From each of the 26 pools, a 246-bp segment of H. pylori recA was amplified, as described elsewhere [29]. From each of the pools possessing IS605, a 371-bp segment from the IS200 homologous region also was PCR-amplified, as described elsewhere [6]. From each PCR run, 10 μL of product was analyzed by gel electrophoresis to exclude the presence of additional bands. The PCR product was purified by use of a Qiaquick PCR purification kit (Qiagen, Chatsworth, CA), and the DNA sequence was determined on both strands by use of an automated sequencer (ABI 377; Applied Biosystems, Foster City, CA). For the recA products from the strains from patient 3, AciI (New England Biolabs) digestion was done. Interstrain comparisons of 206-bp recA and 323-bp IS200 regions were made by use of the GCG Pileup program (Genetics Computer Group, Madison, WI) [30]. Statistical confidence in the trees was tested with 100 bootstrap replicates as implemented by PAUP (Smithsonian Institute, Washington, DC).
Southern hybridization
Two micrograms of DNA derived from IS605-positive isolates was digested with HindIII and electrophoresed on a 1.2% Tris-acetate/EDTA agarose gel (0.04 M Tris-acetate, 0.001 M EDTA). DNA was transferred to a nylon membrane by capillary transfer [31]. The probe for IS200 was made by PCR, as described elsewhere [6], and labeled by nick translation and hybridization by means of the Renaissance chemiluminescent kit (DuPont NEN Research Products, Boston), following the manufacturer's instructions.
Lewis phenotype analysis
Bacteria were harvested from 2 TSA plates after 48 h of growth for ELISA analysis of Lewis-x and -y expression, as described elsewhere [32]. In brief, microtiter plates were coated with whole H. pylori cells and incubated with mouse monoclonal antibodies to Lewis-x or -y, goat anti-mouse antibodies coupled to alkaline phosphatase, and p-nitrophenylphosphate as substrate. The results were expressed as optical density (OD) at 410 nm × 1000 U for H. pylori coating concentrations corresponding to 1 μg of protein per well. In addition to controls without primary or secondary antibodies, OD values of wells coated with equal concentrations of Escherichia coli HB101 cells were subtracted from the values of the wells coated with H. pylori. As controls, aliquots of the same preparation of cell suspensions of reference H. pylori strains previously determined to strongly express Lewis-x or Lewis-y, or to express neither (C1, MO19, and 88-22), were included; their OD values varied within a range of ± 10% between different assays [33].
Antibiotic susceptibility assays
Bacteria were harvested from two TSA plates after 48 h of growth and were assayed for susceptibility to amoxicillin, clarithromycin, tetracycline, and metronidazole by inoculation onto fresh TSA plates containing E-strips (AB Biodisk, Solna, Sweden) with single antibiotics in increasing concentrations. After 72 h of growth under microaerobic conditions, the MIC was defined as the boundary of the contact zone between bacterial growth and the strip.
CagA Western blot
Bacteria were harvested from TSA plates after 48 h of growth, and cellular proteins were extracted by solubilizing pelleted bacteria in 100 μL of SDS sample buffer. After SDS-PAGE and transfer onto a nitrocellulose membrane (Schleicher & Schuell, Keene, NH), blotting was done as described elsewhere [34], by use of rabbit polyclonal antiserum against purified CagA protein of H. pylori strain 88-23.
Results
Genotyping for cagA, vacA, iceA, and IS605
We first sought to characterize the 26 paired bacterial pools by means of PCR genotyping for cagA, vacA, iceA, and IS605. The initial and follow-up pools of 2 of the 13 pairs differed in genotype (table 1). The results for pair 1 differed with respect to iceA and IS605 and those for pair 2 with respect to cagA and vacA. For the remaining 11 pairs, the initial and follow-up pools from each patient had concordant cagA, vacA, iceA, and IS605 genotypes. These findings indicate that the pools of pairs 1 and 2 represent different strains, whereas the initial and follow-up pools of the other 11 pairs are likely to represent the same population within each patient.
Southern hybridization
To further differentiate the IS605-positive pairs among the 5 patients possessing them and to determine the degree of similarity within the pairs, we performed Southern hybridization by use of a probe within orfA, the region homologous to IS200. Each patient's initial IS605-positive strain showed its own unique hybridization pattern, with the number of hybridizing fragments within the chromosomal DNA varying from 1 to 5 (results not shown). In each case, the patterns for the initial and follow-up pools were identical, indicating IS605 stability within the H. pylori genome for a total of 503 person-months.
Sequence analysis of recA and IS605 PCR products
To provide an estimate of the rate and nature of point mutations, we determined both the nucleotide and deduced amino acid sequences for a 206-bp recA PCR product from each of the 26 pools and for a 323-bp PCR product of the IS605 region from the pools possessing this insertion element. For patients 3–13, for whom the above-described genotyping studies did not reveal any differences between the paired pools, the 206-bp recA products from the initial and follow-up samples were 100% identical. In contrast, there was 96.7% and 98.0% identity, respectively, within pairs 1 and 2 (figure 1), for which the above-described studies revealed differences with respect to cagA, vacA, iceA, or IS605 genotypes. As such, the probability of identity of the baseline and follow-up pools of pairs 3–13 was significantly higher than that of the isolates from pairs 1 and 2 (P = .0009; t test) and significantly higher than any out-of-pair comparison (mean ± SE identity, 96.8% ± 0.06%; P = .0002; t test). For pair 3, repeated direct sequencing of the PCR products from both pools showed either a C or a T at position 39. We then isolated DNA from multiple single colonies of both the initial and follow-up pools of this strain, performed recA PCR, and did AciI restriction digestion on each product. By agarose gel electrophoresis, AciI digestion yields 3 bands if C is at position 39 or 2 bands in case of a T. Of 16 colonies from the initial pool, 15 (94%) had a T and 1 (6%) a C at this position. Of 19 colonies of the follow-up pool, 17 (89%) had a T and 2 (11%) a C. Thus, the C/T polymorphism at position 39, which was synonymous, existed within the H. pylori population at both baseline and follow-up in patient 3 and did not represent a sequencing artifact. For the 5 patient pairs (3, 4, 6, 8, and 13) that contained IS605 (table 1), the intrapair identity of a 323-bp PCR sequence was 100%, whereas the out-of-pair identity was 95.9% ± 0.03% (P < .001; t test; figure 2). These studies confirm the high degree of relatedness of the paired pools from each person. The clustering of in-pair sequences in the dendrograms for patients 3–13 was supported in a majority of bootstrap replicates and therefore was statistically significant. No out-of-pair sequences were grouped in a majority of bootstraps.
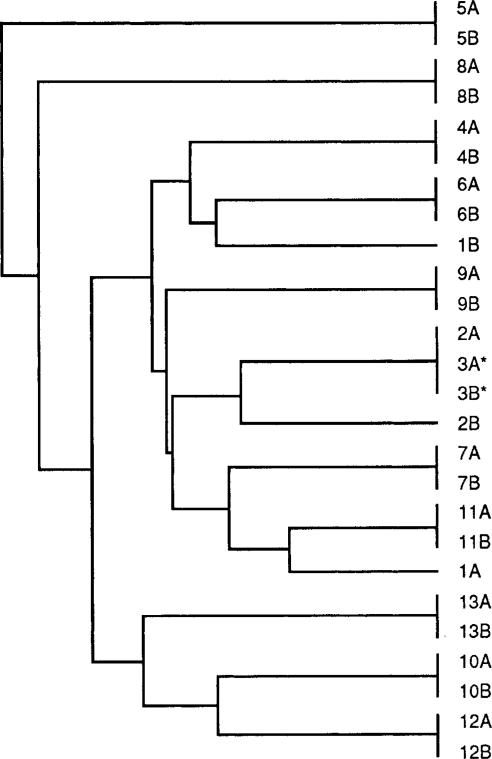
Sequences of 206-bp polymerase chain reaction product derived from Helicobacter pylori recA for 26 paired H. pylori pools. For each pair shown in the dendrogram, original and follow-up pools are designated A and B, respectively. Sequences were identical for initial and follow-up pools of pairs 3–13, each pair having its unique sequence. In contrast, there was 96.7% and 98.0% identity within pairs 1 and 2, respectively. Pools 3A and 3B (*) consisted of mixed populations with silent C/T polymorphism at position 39. In-pair clusters each were supported in majority of bootstrap replicates.
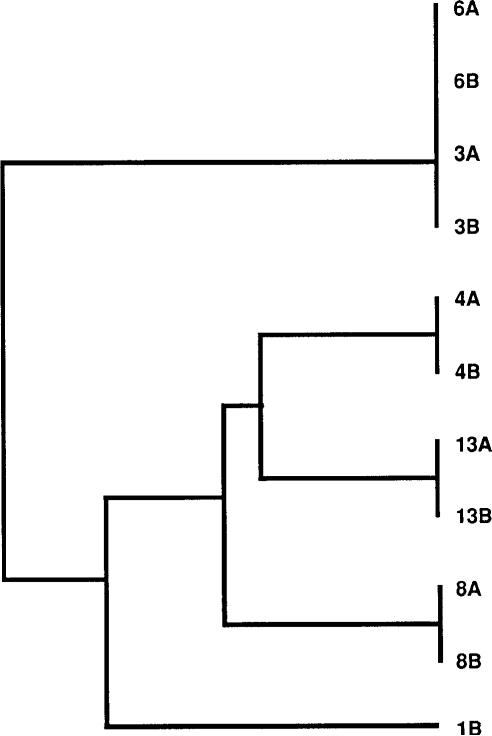
Sequences of 323-bp polymerase chain reaction product derived from Helicobacter pylori IS605 insertion element, for those isolates containing this element (pairs 3, 4, 6, 8, and 13 and second pool of pair 1). Pool designations in this dendrogram are identical to those in figure 1. Intrapair identity was 100%, whereas out-of-pair identity was 95.9% ± 0.03%. In-pair clusters each were supported in majority of bootstrap replicates.
Lewis antigen expression
To determine whether prolonged colonization was associated with phenotypic differences, we examined expression of Lewis antigens, which represent the terminal structures of the O antigens of the H. pylori lipopolysaccharide molecules [32, 35–37]. For the 11 related pairs (3–13), no significant changes were noted in Lewis-x expression over time (mean ± SE, baseline vs. follow-up, 870 ± 240 vs. 737 ± 248; P = .70; paired t test). However, for these same pairs, the expression of Lewis-y decreased over time from 737 ± 196 to 452 ± 197 (P = .05; paired t test). These results suggest that there is in vivo selection for particular phenotypes (e.g., Lewis-y expression).
Other phenotypes
All pools were susceptible to amoxicillin, tetracycline, and clarithromycin (MIC, <0.016, <1.5, and <2 μg/mL, respectively). Pool 1B was resistant to metronidazole (MIC, >8 μg/mL), as were pools 4A, 4B, 11A, and 13B (table 1). Because cagA has a variable number of 3′ repeats that affect the size of the cagA protein, we examined immunoblots of cells from pairs 3–13. For 8 of the 11 pairs, as expected, cagA products were present (table 1). No size shifts were noted for any of these pairs.
RAPD-PCR
The sequence and Southern analyses examine only a small part of the H. pylori genome. We next used more comprehensive methods for assessing genetic drift without focus on specific genomic loci. We performed RAPD-PCR, because this technique yields strain-specific profiles based on the presence of particular sequences, which vary with different primers, within the entire bacterial genome [25]. As expected, and confirming the sensitivity of this method, the pools in pairs 1 and 2 showed clearly dissimilar RAPD patterns with each of the primers used (figure 3). For the remaining 11 pairs, the RAPD profiles of the initial and follow-up pools were similar. The profiles for each pair differed from those of other pairs. However, within each pair, differences were noted in the number and locations of amplimers generated by 1–4 primers. In an analysis of the profiles obtained with primers 1254, 9355, and d11344 and pairs 3–13, the range of interpair similarity was 27%–69% (mean ± SD, 44% ± 14%), whereas, as expected, the intrapair similarity, with a range of 65%–95% (mean ± SD, 82% ± 9%), was substantially higher (figure 3). The RAPD profiles of each pool and intrapair difference were reproducible on repetition of RAPD testing with the same or newly isolated DNA. In addition, for 10 pools (5 pairs), these profiles also were examined after prolonged culture on TSA plates with passage of strains every 48 h for 20 times. In each case, the initial and follow-up isolates had identical profiles. The differences also were reproducible when the process of culture, DNA isolation, and RAPD testing was repeated in another laboratory by another operator. These data suggest that intrapair differences in RAPD patterns were not due to laboratory- or operator-related conditions, nor to in vitro handling of the strains, but represent genetic changes that occurred in vivo within the individual pools over time.
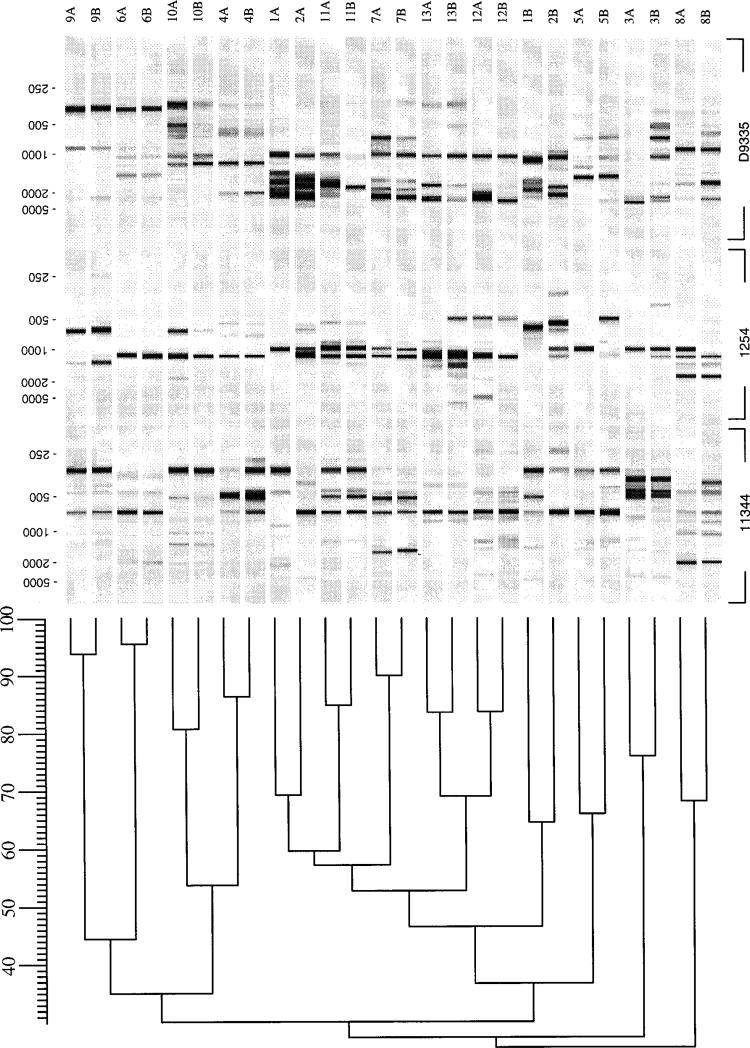
Analysis of random arbitrarily primed DNA (RAPD) polymerase chain reaction profiles of 26 paired Helicobacter pylori pools obtained with primers 1254, 9355, and d11344. Pool designations are identical to those in figure 1. RAPD photographs were digitized, and levels of similarity between patterns were calculated (GelCompar 4.0 software; Applied Maths, Kortrijk, Belgium). The range of intrapair similarity for pairs 3–13 was 65%–95%, but for pairs 1 and 2, similarities were only 46% each.
AFLP analysis
To confirm this diversity by means of an independent method that also is based on the complete bacterial genome, AFLP-PCR was performed for the same 26 pools. With the use of restriction digestion followed by PCR amplification of resulting products, this method allows the production of genomic patterns with even higher detail. As expected, the pools of pairs 1 and 2 showed highly dissimilar AFLP patterns. For the remaining 11 pairs, the AFLP profiles of the initial and follow-up pools were highly similar to one another (figure 4). The profiles of each pair clearly differed from those of other pairs; the range of interpair similarity was 54%–77% (mean ± SD, 70% ± 7%) for pairs 3–13. However, within each pair, distinct differences again were noted. The intrapair similarity was 73% and 74% for pairs 1 and 2, respectively, but ranged from 83% to 96% (mean ± SD, 90% ± 4%) for pairs 3, 4, and 6–13. For pair 5, the intrapair similarity was only 21%, a result that was reproducible with repeated AFLP with the same and newly isolated DNA with two different sets of primers.
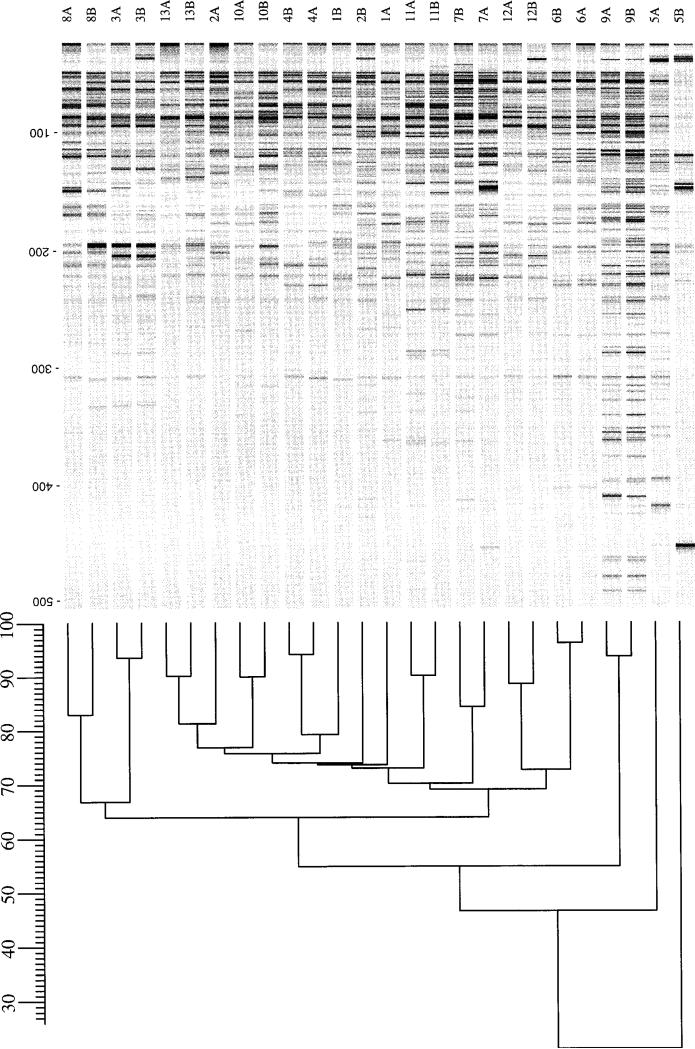
Analysis of amplified fragment length polymorphism (AFLP) profiles of 26 paired Helicobacter pylori pools. Isolate designations are identical to those in figure 1. Fluorescent-labeled AFLP profiles were scanned and digitized, and levels of similarity between profiles were calculated. The range of intrapair similarity was 83%–96% for pairs 3–13, but similarities for pairs 1 and 2 were only 73% and 74%, respectively.
Discussion
H. pylori has high genomic diversity [1], but the origins of this variation are unknown. It may reflect a long evolution for H. pylori and its ancestors, which are believed to have been in the stomachs of vertebrates for much of their history (≤400 million years) [38]. The diversity also could reflect a process of persistent accumulation of mutations within individual strains as a result of either spontaneous mutations or horizontal uptake and incorporation of homologous (H. pylori) or heterologous DNA. H. pylori strains are naturally competent [39] and also possess a conjugation-like mechanism for DNA uptake [40], properties that allow efficient incorporation of foreign DNA. Persistent accumulation of mutations could provide a pool of H. pylori variants that can be selected for optimal colonization of particular gastric niches (leading to maximum resource utilization) that develop because of host changes during the long duration of colonization. For example, over decades of H. pylori colonization, many hosts develop atrophic gastritis [20], a process accompanied by a decline in acid production [41] and changes in mucosal expression of Lewis antigens [42].
Our hypothesis was that mutations frequently develop over the course of prolonged colonization of an individual host by H. pylori. These mutations then would be detected during long-term assessment. The lack of difference in one previous study [23] could indicate that this hypothesis is incorrect or, alternatively, that the follow-up period (<2 years) in that study was too brief or that the RFLP analysis was not sufficiently sensitive. We therefore tested our hypothesis by use of paired pools of H. pylori isolates obtained ≥7 years apart, making use of a broad panel of genotyping and phenotyping methods. We applied these methods to the entire population of bacterial cells cultured from a biopsy sample instead of to single colonies. This approach has the potential disadvantage of failure to recognize mixed colonization with different strains and any changes in these individual strains over time. In theory, a repeated analysis of such a strain pool also may give the impression of a changing genotype if the ratio of the individual strains in the pool varies over time. Results obtained by examination of single colonies could be influenced by sampling errors. Had we used such an approach, observed differences as well as similarities might have been based on colony selection and would have been less representative of the entire bacterial population. The utility of this approach was supported by the demonstration of 100% identity in the specific genotyping results within the 11 pairs in which the baseline and follow-up samples represented the same population. Further support came from the demonstration of ≥99% similarity in AFLP patterns obtained with DNA from single colonies of each of these patients (results not shown) and from our ability to detect a 1-bp difference in the recA sequences of 6% of the bacteria. We believe that this sensitivity was adequate for the purpose of our study. With this approach, we first showed that the paired H. pylori populations from 2 (15%) of our 13 patients represented different strains, a frequency similar to that observed from simultaneous examination of colonies from different biopsy samples in a single host, as reported elsewhere [43–45]. The populations sampled from these two patients differed with respect to specific genotypes, recA sequences, and RAPD-PCR and AFLP profiles and thus represented clearly different bacterial strains. Thus, these patients had become colonized with a new strain during follow-up, or they had been colonized with the second strain throughout the study period, in which case, by chance, one strain was isolated at the first endoscopy and the other at follow-up. Frequent cocolonization can explain the high rates of recombination observed for H. pylori genes [1, 46]. Within each of the remaining 11 pairs, the initial and follow-up pools were fully concordant with respect to their cagA, vacA, iceA, and IS605 genotypes and recA sequence, and RAPD-PCR and AFLP analysis yielded similar patterns for each pair, which differed from those of other pairs. We therefore concluded that each pair of pools from these 11 patients encompassed the same dominant genotype.
Nevertheless, the RAPD-PCR and AFLP analyses showed distinct differences between initial and follow-up isolates of each of these 11 pairs. To examine whether the RAPD differences were artifactual, we repeated the RAPD-PCRs with multiple primers, making use of the same DNA sample, newly extracted DNA samples obtained after renewed culture of pools stored at –70°C, and DNA samples obtained from cells after 20 further passages in vitro. In addition, new DNA was prepared and PCR testing repeated in a second laboratory by another investigator. With each of these repetitions, the distinct intrapair differences in RAPD-PCR profiles that we initially noted remained the same, indicating that they truly reflect genomic diversity within a population over time. We cannot exclude that this genomic diversity already existed within each host at baseline, instead of occurring during follow-up. However, on the basis of identical overall genotypes for each pair of pools and the dendrograms matching any pool with its own partner obtained from the same host, we conclude that patients 3–13 had been colonized at one point by a single strain and that the changes observed represent in vivo genetic drift. The intrapair differences with both RAPD and AFLP were most pronounced for the pools obtained from patient 5. These results suggest that the evolution of pool 5B might be more substantial; further studies are in progress to test this hypothesis.
In addition to these observed differences in patterns for pairs 3–13, we also observed heterogeneity at the same bp position within recA in both the initial and follow-up pools of pair 3. This observation suggests that the two clonal variants exist in a long-term equilibrium. As these different recA sequences are synonymous, they do not represent differences between the strains due to selective pressure. In addition to these genomic differences, we also observed a change in expression of the Lewis-y antigen. Although these phenotypic changes may result from selection due to altered Lewis expression of the host gastric mucosa, we could not directly test that hypothesis because we lacked adequate histologic material. However, with the progressive development of atrophic gastritis during the course of aging, Lewis-y expression is lost out of proportion to Lewis-x changes [42, 47]. If H. pylori strains are under selective pressure to express Lewis antigens matching those of the host, for which preliminary evidence exists [33, 48], then the phenotypic variation we observed is consistent with the presence of adaptive rather than directionless mutation. For the other variation detected, we cannot now discern whether it is directionless or adaptive.
Finally, the concept of quasispecies development of H. pylori also is supported by the distribution of pairs in the recA and IS200 dendrograms (figures 1 and and2).2). Pools that are closely related on the basis of one sequence may be only distantly related on the basis of the other sequence. Such data are consistent with interstrain recombination, suggesting a continuum of strain differences instead of division into a limited number of strain families. This phenomenon is well-known for RNA viruses, including human immunodeficiency viruses, which can exist within their hosts as pools of related genetic variants, referred to as quasispecies [49–53]. Such a population of viruses share a common origin but have distinct genomic sequences as a result of mutation, drift, and the impact of selection [52]. For human immunodeficiency virus type 1, genomic evolution within an individual host over time is continuous [53, 54]. The level of genetic diversity among related human immunodeficiency virus strains in a geographic locus reflects the length of time since introduction of the virus to that locus [49].
In conclusion, paired H. pylori pools obtained 7–10 years apart from the same host show consistent genotypic differences in RAPD and AFLP analyses and phenotypic differences in the level of Lewis-y detected by ELISA. These findings indicate that genetic drift occurs within an H. pylori population during long-term host colonization. Our observations thus demonstrate quasispecies in H. pylori, which may be considered the development over time of a pool of genetic variants from a single strain within a host, providing a bacterial population that can be selected by the changing gastric environment of the host. This phenomenon may have several implications. First, the development of variation and subsequent selection may allow adaptation to new hosts, enabling successful colonization. Second, such variation also may allow adaptation to different compartments of the human stomach that have different selective pressures, such as the acid-producing corpus and the non–acid-producing antrum. Conditions within these environments can change, for example, because of the development of atrophic gastritis with declining acid production. Genetic drift and host selection would enable H. pylori populations to adapt to such environmental changes, including the development of antibiotic resistance. In the ancient past, such drift and selection may have provided one mechanism for an ancestral Helicobacter species to adapt to different mammals that now are known to be colonized with related organisms.
Acknowledgments
Financial support: Stichting van Helten of the Royal Dutch Academy of Sciences; NIH (DK-53707); the Medical Research Service of the Department of Veterans Affairs; and the Iris and Homer Akers Fellowship in Infectious Disease.
References
Full text links
Read article at publisher's site: https://doi.org/10.1086/315173
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jid/article-pdf/181/1/273/17995304/181-1-273.pdf
Citations & impact
Impact metrics
Article citations
Subinhibitory concentrations of antibiotics affect development and parameters of <i>Helicobacter pylori</i> biofilm.
Front Pharmacol, 15:1477317, 14 Oct 2024
Cited by: 0 articles | PMID: 39469629 | PMCID: PMC11513322
Molecular insights into the fine-tuning of pH-dependent ArsR-mediated regulation of the SabA adhesin in Helicobacter pylori.
Nucleic Acids Res, 52(10):5572-5595, 01 Jun 2024
Cited by: 0 articles | PMID: 38499492
Fatty acids of Helicobacter pylori lipoproteins CagT and Lpp20.
Microbiol Spectr, 12(5):e0047024, 19 Mar 2024
Cited by: 0 articles | PMID: 38501821 | PMCID: PMC11064636
Essential role of Helicobacter pylori apolipoprotein N-acyltransferase (Lnt) in stomach colonization.
Infect Immun, 91(12):e0036923, 08 Nov 2023
Cited by: 1 article | PMID: 37937999 | PMCID: PMC10715074
Helicobacter pylori genomes reveal Paleolithic human migration to the east end of Asia.
iScience, 25(7):104477, 30 May 2022
Cited by: 4 articles | PMID: 35720267 | PMCID: PMC9204748
Go to all (117) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genotype Diversity and Quasispecies Development of Helicobacter pylori in a Single Host.
Jpn J Infect Dis, 68(3):176-180, 01 Jan 2015
Cited by: 11 articles | PMID: 25672355
Genetic characterisation of Helicobacter pylori isolates from an Argentinean adult population based on cag pathogenicity island right-end motifs, lspA-glmM polymorphism and iceA and vacA genotypes.
Clin Microbiol Infect, 10(9):811-819, 01 Sep 2004
Cited by: 14 articles | PMID: 15355412
High prevalence of multiple strain colonization of Helicobacter pylori in Korean patients: DNA diversity among clinical isolates from the gastric corpus, antrum and duodenum.
Korean J Intern Med, 19(1):1-9, 01 Mar 2004
Cited by: 12 articles | PMID: 15053036 | PMCID: PMC4531544
Microevolution between paired antral and paired antrum and corpus Helicobacter pylori isolates recovered from individual patients.
J Med Microbiol, 53(pt 7):669-677, 01 Jul 2004
Cited by: 27 articles | PMID: 15184540
Funding
Funders who supported this work.
NIAID NIH HHS (1)
Grant ID: R29 AI043548-02
NIDDK NIH HHS (1)
Grant ID: DK-53707





