Abstract
Free full text

miR-21 and miR-31 Converge on TIAM1 to Regulate Migration and Invasion of Colon Carcinoma Cells*
Abstract
TGF-β promotes cell migration and invasion, an attribute that is linked to the pro-metastasis function of this cytokine in late stage cancers. The LIM 1863 colon carcinoma organoid undergoes epithelial-mesenchymal transition (EMT) in response to TGF-β. This process is markedly accelerated by TNF-α, and we found that the levels of miR-21 and miR-31 were prominently elevated under the synergistic actions of TGF-β/TNF-α. Consistent with this, overexpression of either miR-21 or miR-31 significantly enhanced the effect of TGF-β alone on LIM 1863 morphological changes. More importantly, transwell assays demonstrated the positive effects of both miR-21 and miR-31 in TGF-β regulation of LIM 1863 motility and invasiveness. Elevated levels of miR-21 and miR-31 also enhanced motility and invasiveness of other colon carcinoma cell lines. We present compelling evidence that TIAM1, a guanidine exchange factor of the Rac GTPase, is a direct target of both miR-21 and miR-31. Indeed in LIM 1863 cells, suppression of TIAM1 is required for miR-21/miR-31 to enhance cell migration and invasion. Therefore, we have uncovered miR-21 and miR-31 as downstream effectors of TGF-β in facilitating invasion and metastasis of colon carcinoma cells.
Introduction
Transforming growth factor β (TGF-β)2 plays a complex role in cancer development (1). It is recognized as a tumor suppressor in that genetic lesions affecting TGF-β pathway components are found with high incidence particularly in pancreatic and colon cancers (2,–4). On the other hand, many clinical and basic studies point to elevated TGF-β signaling in late stage cancer, and suggest a pro-invasion/metastasis role for TGF-β (5, 6). Indeed, a high TGF-β signaling activity is often associated with a poor prognosis for breast and other cancer patients (7,–9). Therefore, it is important to delineate the downstream effectors mediating the different TGF-β responses at early versus late stages of cancer progression. Such information will be critical for designing strategies targeting specific aspects of TGF-β response to combat cancer.
The pro-metastasis function of TGF-β is directly linked to its ability to initiate epithelial- mesenchymal transition (EMT) in cell culture (10,–12). EMT is a complex transdifferentiation process in which epithelial cells lose junctional adhesion and adopt a mesenchymal phenotype and morphology (13). EMT takes place during embryonic development and wound healing, and is also believed to underlie the change into a more invasive behavior during cancer progression (14). TGF-β is a potent inducer of EMT. TGF-β directly activates the expression of transcription factors including SNAI1/2, Twist and ZEB1/2 (10,–12). These are master regulators of the EMT program, which suppress the levels of epithelial markers such as E-cadherin and ZO-1, and up-regulate mesenchymal markers including vimentin, fibronectin, and others (15).
MicroRNAs (miRNAs) are 20–22-nucleotide noncoding RNAs that regulate gene expression at post-transcriptional levels (16). Rapidly emerging evidence strongly suggest critical roles of miRNAs in the pathogenesis of cancer (17). Either oncogenic (e.g. miR-155, miR-17–5p) or tumor suppressor (e.g. let-7, miR-15a) functions have been assigned to various miRNAs (18, 19). Earlier profiling experiments have identified cohorts of miRNAs whose levels undergo significant changes upon TGF-β-induced EMT, suggesting possible involvements of miRNAs in this process (20). In particular, independent studies by several laboratories have found the miR-200 family as important suppressors of EMT in a number of different models (21,–24). MiR-200 inhibits EMT by directly recognizing complementary sites in the 3′-UTR of ZEB1/2 and repressing the translation of these positive regulators of EMT (21,–24). MiR-200 itself is repressed by TGF-β, through an unknown mechanism (24). MiRNAs such as miR-9 and miR-335 promote metastasis by directly suppressing the levels of E-cadherin (miR-9) or SOX4 (miR-335) (25, 26). MiR-10b has also been suggested to facilitate breast cancer metastasis, but this was contradicted by a more recent report in which high miR-10b appeared to suppress motility and invasiveness of breast cancer cells (27, 28). MiR-31 acts to repress breast cancer cell migration and invasion, and a low miR-31 level correlates with high metastatic potential (29, 30). The anti-metastasis function of miR-31 is attributable to down-regulation of three targets: integrin α5, radixin, and Rho A (29, 30). Therefore, miRNAs impose another layer of regulation on breast cancer metastasis.
In this study, we examined LIM 1863, a three-dimensional organoid culture derived from colon carcinoma and undergoes EMT in response to TGF-β (31, 32). We observed up-regulation of miR-21 and miR-31 during EMT of LIM 1863 organoid. Overexpression as well as inhibition experiments support the contributions of both miR-21 and miR-31 not only in the morphological changes of LIM 1863 in response to TGF-β, but also in cell motility and invasion. Furthermore, we show that TIAM1 (T lymphoma and metastasis gene 1) is a direct target of both miR-21 and miR-31, and that the suppression of TIAM1 is important for the pro-migration and -invasion activities of miR-21 and miR-31. Therefore, we have identified miR-21 and miR-31 as positive regulators of colon carcinoma migratory and invasive properties.
EXPERIMENTAL PROCEDURES
Cell Culture and Cytokine Treatment
LIM 1863 cells and DLD1 were maintained in RPMI 1640 supplemented with 5% fetal or 10% bovine serum (FBS), respectively. 293T, HaCaT, MDA-MB-231, and SW480 cells were maintained in Dulbecco's Modifed Eagle's (DME) media supplemented with 10% FBS. All cell culture media also contained penicillin (100 units/ml) and streptomycin (100 units/ml) (Invitrogen). For cytokine treatment, human TGF-β1 (R&D Systems) and human TNF-α (R&D Systems) were used at a final concentration of 2.5 ng/ml and 10 ng/ml, respectively. Cycloheximide (Sigma) was used at a final concentration of 15 μg/ml for 30 min.
miRNA Microarray
Total RNA was extracted using the mirVanaTM RNA Isolation kit (Ambion) according to the manufacturer's instructions from LIM 1863 cells that were treated with both TGF-β and TNF-α or media only for 24 h. Total RNA was submitted to Exiqon (Vebaek, Denmark) for miRNA microarray profiling services, in which RNA was labeled with Hy3 and Hy5 fluorophores and hybridized to a miRCURY LNA microRNA Array (version 8.1). All subsequent data analysis was performed by Exiqon.
Northern Blotting
Total RNA was extracted using the mirVanaTM RNA Isolation Kit according to manufacturer's instructions (Ambion). Twenty micrograms of total RNA was resolved on denaturing agarose gels and transferred to nylon membranes. Following UV-cross-linking, membranes were incubated with random-primed miR-21 or GAPDH cDNA probes in Church's hybridization buffer (0.5 m NaHPO4, pH 7.2; 1 mm EDTA; 7% SDS) at 42 °C for 18 h. For detecting the miR-21 primary transcript, the probe encompassed 250 bp on both sides of the mature miR-21 sequence. Membranes were washed three times in 2× SSC, 0.1% SDS at room temperature. To detect precursor and mature miRNA species, 20–40 μg of total RNA was resolved on 20% denaturing polyacrylamide gels, transferred to nylon membranes, UV-cross-linked and probed with 5′-end-labeled DNA oligonucleotide probes to detect miR-21 (5′-TCAACATCAGTCTGA TAAGCTA-3′), miR-31 (5′-CAGCTATGC CAGCATCTTGCC-3′), or 5 S rRNA (5′-TTAGCTTCCGAGATCA-3′) in Church's hybridization buffer at 37 °C for 18 h. Membranes were washed as above, except at 37 °C. All membranes were exposed to phosphor imaging screens, and scanned with a Storm 860 PhosphorImager (Molecular Dynamics).
Overexpression and Inhibition of miRNA Function
Human miR-21 and miR-31 precursors (AM17100) and a Cy3-labeled pre-miR negative control (AM17120) were purchased from Ambion. MiRNA activity was inhibited using chemically synthesized 2′O-methyl-modified RNA oligonucleotides (Dharmacon) that were antisense to miR-21 (5′-mUmCmAmAmCmAmUmCmAmG mUmCmUmGm-AmGmCmUmA-3′) or miR-31 (5′-mCmAmGmCmUmAmUmGmCmC mAmGmCmAmUmCmUmUmGmCmC-3′). A 5′-Cy3-labeled RNA oligonucleotide (Integrated DNA Technologies) targeting eGFP (5′-Cy3mA mAmGmGmCmAm-AmGmCmUmGmAmC mCmCmUmGmAmAmGmU-3′) was used as a negative control.
All transfections were performed utilizing Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. LIM 1863 organoids were resuspended in RPMI 1640 supplemented with 5% FBS and transfected with 100 nm of miRNA precursors in 24-well tissue culture plates. For migration and invasion assays, LIM 1863 organoids were trypsinized and dissociated before transfection. In 24-well tissue culture plates, 3 × 105 cells were transfected with 100 nm miRNA precursors or 250 nm miRNA inhibitors.
Real-time RT-PCR
Total RNA was isolated using the mirVanaTM RNA Isolation kit following manufacturer's instructions (Ambion). One microgram of RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). SYBR-green real-time quantitative PCR was performed using a Bio-Rad MyiQ PCR detection system with the following gene-specific primers: fibronectin-1 (FN1), forward 5′-GAGCCATGTGTCT-TACCATT-3′ and reverse 5′-AGTATTTCTGGTCCTGCTCA-3′; interleukin-8 (IL8), forward 5′-ATGACTTCCAAGCTGGCCGTGGCT-3′ and reverse 5′-TCTCAGCCCTCTTCAAAAAACTTCTC-3′; laminin-γ-2 (LAMC2), forward 5′-CTGCAGGTGGACAACA-GAAA-3′ and reverse 5′-TCTGCTGTCACATTGGCTTC-3′; matrix metalloproteinase-7 (MMP7), forward 5′-CATGAGTGAGCTACAGTGGG-A-3′ and reverse 5′-CTATGACGCGGGAGTTTAACAT-3′; T-lymphoma invasion and metastasis-1 (TIAM1), forward 5′-AAGACGTACTCAGGCCATGTCC-3′ and reverse 5′-GACCCAAATGTCGCAGTCAG-3′; and U6 snRNA, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
To measure mature miR-21 and miR-31 levels by quantitative real-time PCR, 10 ng of total RNA was reverse-transcribed using the TaqMan miRNA reverse transcription kit and RT primers for miR-21, miR-31, and U6 snRNA (Applied Biosystems). The cDNAs were then analyzed by real-time PCR using TaqMan probes for miR-21, miR-31, and U6 snRNA (Applied Biosystems).
Cell Migration and Invasion Assay
Uncoated or Matrigel-coated transwells containing 8 μm pores were used for the assays (BD Biosciences). Cells were seeded into the upper chamber in serum-free RPMI 1640 medium. Conditioned DME media from NIH 3T3 cells containing 10% FBS was added to the lower chamber. Cells were fixed in 100% methanol 72 h later and stained with a 1:5 dilution of Giemsa (Sigma) for 40 min at room temperature. Cells remaining on the upper side of the filter were removed with a cotton swab. The filters were then mounted onto slides and images were taken at 10X magnification. From these images, the number of migratory or invasive cells was counted.
Dual Luciferase Reporter Assay
The full-length TIAM1 3′-UTR (1970 nt) was cloned into the XbaI and NotI sites of a modified pRL-TK vector (Promega) immediately downstream of the Renilla luciferase stop codon and designated pRL-TIAM1. The putative miR-21 and miR-31 recognition elements (MRE) in the TIAM1 3′UTR were mutated by site-directed mutagenesis (Stratagene). For the dual luciferase assay, LIM 1863 cells were transfected with 100 nm of pre-miR-21, pre-miR-31, or a negative precursor control using Lipofectamine 2000 (Invitrogen). After 24 h, cells were co-transfected with 200 ng of pRL-TIAM and 100 ng of pGL3-luc as the internal control. Cell extracts were prepared 48 h later and the Dual-Glo luciferase reporter assay (Promega) was performed according to the manufacturer's protocol.
Lentiviral Transduction
To generate lentiviral particles, 293T cells in 10-cm plate were transfected with 12 μg of pLenti-CMV-puro (empty vector or containing TIAM1-FLAG), 8 μg of pCMV-dR8.74, and 4 μg of pMD2-VSVG. Forty-eight hours later, the medium was harvested, cleared by a 0.45 μm filter, mixed with polybrene, and applied to dissociated LIM 1863 cells. After overnight incubation, the virus-containing medium was replaced by fresh medium.
RESULTS
miR-21 and miR-31 Are Induced Synergistically by TGF-β and TNF-α in LIM 1863 Organoids
LIM 1863 is a colon carcinoma cell line that propagates as three-dimensional spheroids frequently referred to as organoids (31). The organoids grow in suspension and contain differentiated columnar and goblet cells around a central lumen. Upon TGF-β treatment, LIM 1863 undergoes profound morphological changes and becomes attached to the tissue culture plate (33). In 5–7 days after TGF-β stimulation, LIM 1863 cells assume a monolayer morphology (Fig. 1A). Co-treatment with TNF-α can accelerate the morphological changes to complete in 48 h, but by itself TNF-α has no effect (32). This morphological change is reversible upon removal of TGF-β and TNF-α. These cellular characteristics and other accompanying molecular changes led to the conclusion that this is a typical TGF-β-induced EMT (32). Therefore, the LIM 1863 cell line is a unique three-dimensional culture system to study TGF-β regulation of cancer cell migratory and invasive properties (34, 35).
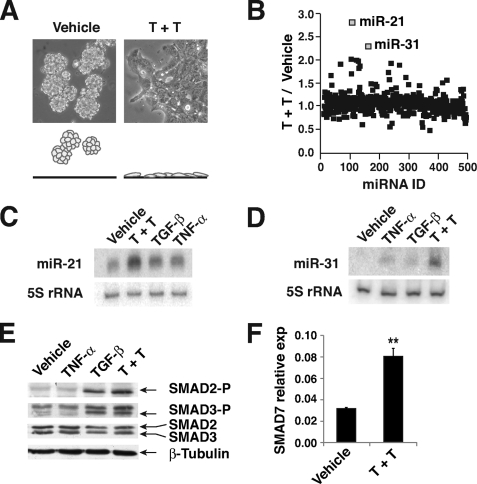
miR-21 and miR-31 are up-regulated during TGF-β/TNF-α-induced EMT in LIM 1863 organoids. A, phase contrast images of LIM 1863 organoids with or without TGF-β/TNF-α (T+T) treatment (24 h). A schematic depiction of the morphological and adhesive changes is shown (lower). B, LNA-based microarray was used to measure the relative ratio of each miRNA in LIM 1863 organoids with or without TGF-β/TNF-α (T+T) treatment. A total of 455 miRNAs were profiled. miR-21 and miR-31 were most significantly up-regulated by TGF-β/TNF-α. C and D, Northern blot analyses detecting miR-21 (C) or miR-31 (D) in LIM 1863 organoids with indicated cytokine treatment for 24 h. The ~21-nt mature miRNAs (miR-21 or miR-31) are shown. 5 S rRNA expression was used as an internal control. E, extracts from LIM 1863 cells after indicated cytokine treatments were analyzed by Western blotting using indicated antibodies. F, real-time quantitative RT-PCR analysis of SMAD7 mRNA levels in LIM 1863 cells after vehicle or TGF-β/TNF-α (T+T) stimulation (mean ± S.D., **, p < 0.01). U6 snRNA was used as the internal standard.
We became interested in whether miRNAs may play a role in regulating EMT of LIM 1863. Microarray profiling revealed miR-21 and miR-31 as the most elevated miRNAs after treatment of TGF-β and TNF-α (Fig. 1B). Northern blotting further validated the induction of the mature forms of these two miRNAs by TGF-β/TNF-α (Fig. 1, C and D). For both miR-21 and miR-31, co-treatment with TGF-β and TNF-α was more robust than each cytokine individually in elevating the levels of these two miRNAs (Fig. 1, C and D). Given the synergistic effect of TGF-β/TNF-α in both the up-regulation of miR-21/miR-31 and EMT, we reasoned that the increase in miR-21/miR-31 may have functional relevance to EMT of LIM 1863 organoid, and focused on these two miRNAs for further studies. The signal transduction of TGF-β in LIM 1863 cells appeared to be normal, as TGF-β induced a rapid C-terminal phosphorylation in both SMAD2 and SMAD3, and activated the transcription of a typical target gene SMAD7 (Fig. 1, E and F).
TGF-β/TNF-α Up-regulate miR-21/miR-31 at the Transcription and Processing Levels
In a time course analysis, we found that while the miR-21 precursor (a ~70-nt stem-loop processing intermediate) in LIM 1863 was rapidly increased and reached a plateau 2 h after TGF-β/TNF-α stimulation, the increase in the mature miR-21 was delayed and did not reach its peak until 24 h later (Fig. 2A). Whereas this observation suggests activation of miR-21 gene transcription by TGF-β/TNF-α, it is also clear that the processing from precursor to mature miR-21 is the more rate-limiting step in TGF-β/TNF-α induction of this miRNA. Interestingly, pretreating the LIM 1863 organoids with cycloheximide effectively abrogated the increase in mature miR-21 by TGF-β/TNF-α, and yet had no effect on the increase in miR-21 precursor (Fig. 2A). This suggests that TGF-β/TNF-α enhances the Argonaute-mediated processing of miR-21 precursor through an indirect mechanism, requiring the synthesis of unknown factors. Northern blotting of total RNA revealed three primary transcripts of the miR-21 gene, ranging in size from 1 to 4 kb (Fig. 2B). All three transcripts rapidly increased 2 h after TGF-β/TNF-α treatment, which was not affected by cycloheximide (Fig. 2B). Therefore, the level of miR-21 is regulated by TGF-β/TNF-α at the initial transcription and the processing steps, and only miR-21 gene transcription is a direct downstream target of TGF-β/TNF-α signaling.
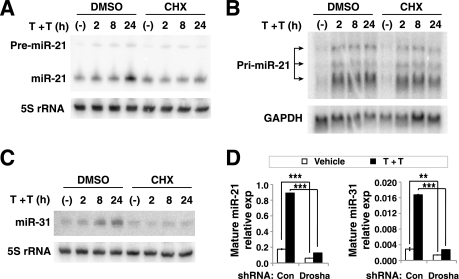
TGF-β/TNF-α regulates miR-21 and miR-31 abundance at the transcription and processing steps. A, TGF-β/TNF-α (T+T) induction of mature miR-21 was abrogated by cycloheximide (CHX). LIM 1863 organoids were incubated with DMSO or 15 μg/ml cycloheximide for 30 min followed by treatment with both TGF-β and TNF-α (T+T) for the indicated times. Shown are Northern blotting analysis of miR-21 precursor (pre-miR-21, ~70 nt) and mature miR-21 (~21 nt), with 5 S rRNA as the loading control. B, TGF-β/TNF-α (T+T) induced a rapid increase in the miR-21 primary transcript (pri-miR-21), which was not affected by cycloheximide. Northern blotting of total RNA shows three prominent miR-21 primary transcripts with ~1, ~2, and ~4 kb in size (arrows). GAPDH served as the loading control. C, similar experiment as in A showing the increase in mature miR-31 in response to TGF-β/TNF-α (T+T) was affected by cycloheximide. D, LIM 1863 cells expressing a control shRNA (con) or a Drosha shRNA vector were treated with TGF-β/TNF-α (T+T) for 36 h, and the levels of mature miR-21 and miR-31 were measured by the Taqman miRNA assay (mean ± S.D., **, p < 0.01; ***, p < 0.001). U6 snRNA was used as the internal standard.
The increase in mature miR-31 by TGF-β/TNF-α became noticeable 2 h after the cytokine treatment, and continued to accumulate until 24 h later (Fig. 2C). Cycloheximide also effectively inhibited such up-regulation of mature miR-31 by TGF-β/TNF-α (Fig. 2C). However, neither the primary nor the precursor forms of miR-31 were detectable by Northern blotting, so we could not determine whether miR-31 is also regulated at the primary transcription level like in the case of miR-21. Therefore, for both miR-21 and miR-31, the up-regulation by TGF-β/TNF-α appears to be indirect and requires new synthesis of unknown factors.
Furthermore, when we knocked down the essential miRNA-processing factor Drosha using a previously validated shRNA, TGF-β/TNF-α failed to elevate the level of mature miR-21 and miR-31 (Fig. 1D and supplemental Fig. S1) (36). This observation suggested that the increase in miR-21/miR-31 upon TGF-β/TNF-α stimulation is due to new biosynthesis and/or processing of miRNAs.
miR-21 and miR-31 Are Important Regulators in TGF-β-induced EMT of LIM 1863 Cells
TGF-β-induced EMT in LIM 1863 is a slow process. By the first 24 h, most organoids attached to the tissue culture plate, but only a small percentage of them began to spread out into a monolayer (Fig. 3A). By counting the organoids exhibiting either a “spreading” or “not spreading” morphology, we quantified TGF-β-induced EMT and evaluated the impact of miR-21 and miR-31 overexpression. Indeed, when examined 24 h after TGF-β addition, LIM 1863 organoids transfected with the precursors of either miR-21 or miR-31 had a significantly higher percentage adopting a “spreading” morphology, compared with organoids transfected with a non-targeting control miRNA precursor (Fig. 3B). The same was observed 48 h after TGF-β stimulation (Fig. 3B). These data suggest that miR-21 and miR-31 are able to accelerate the EMT process initiated by TGF-β.

TGF-β-induced LIM 1863 morphological change is potentiated by miR-21 and miR-31. A, representative LIM 1863 organoids exhibiting “spreading” or “not spreading” morphologies after 24 h TGF-β treatment (upper). A schematic drawing of “spreading” and “not spreading” morphologies is shown (lower). B, LIM 1863 organoids transfected with indicated miRNA precursors were stimulated with TGF-β, and the morphology was scored as “spreading” or “not spreading” at 24 h or 48 h post TGF-β addition. The results are plotted (mean ± S.D., > 200 organoids were counted in each experiment, data represent >3 experiments). *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, same experiment as in B (only the 24 h time point), and the mRNA levels of indicated markers were measured by quantitative real-time RT-PCR (mean ± S.D., n>3), using U6 snRNA as an internal reference. FN1: fibronectin 1; IL8: interleukin 8; LAMC2: laminin γ2; MMP7: matrix metalloproteinase 7. D, inhibition of miR-21 or miR-31 resulted in markedly diminished induction of EMT markers by TGF-β/TNF-α. LIM 1863 cells were transfected with 250 nm 2′O-methyl RNA inhibitors of miR-21, miR-31, or control (Con). Eighteen hours later, cells were stimulated with TGF-β/TNF-α (T+T), and the mRNA abundance of LAMC2 and MMP7 was measured by quantitative real-time RT-PCR at indicated time points (mean ± S.D.).
To better quantify the EMT process, we measured the mRNA levels of a number of EMT markers. In our LIM 1863 culture, TGF-β/TNF-α induced re-distribution of E-cadherin from cell surface to cytoplasm without much change in its protein level (data not shown), so we measured the expression of other known markers including fibronectin 1 (FN1), interleukin 8 (IL-8), laminin-γ-2 (LAMC2), and matrix metalloproteinase 7 (MMP7) (37, 38). The levels of these markers were all increased 24 h after TGF-β treatment (Fig. 3C). Transfection of either miR-21 or miR-31 precursors further potentiated TGF-β in increasing the mRNA levels of FN1, IL8, and LAMC2 (Fig. 3C). In the case of MMP7, only miR-31 had a significant effect (Fig. 3C). Even in cells without any exposure to TGF-β, either miR-21 or miR-31 elevated the expression of these EMT markers as well (Fig. 3C). These more quantitative analyses further substantiated our conclusion that a high level of miR-21 or miR-31 facilitates TGF-β-induced EMT of LIM 1863. Importantly, TGF-β-induced up-regulation of SMAD7 was not enhanced by either miR-21 or miR-31 overexpression, suggesting that these two miRNAs only facilitate a subset of the TGF-β responses, and do not enhance TGF-β signaling in general (supplemental Fig. S2).
We further carried out loss-of-function studies to evaluate the importance of miR-21 and miR-31 in TGF-β/TNF-α-induced EMT of LIM 1863 cells. Indeed, when miR-21 or miR-31 activity was neutralized by antisense 2′O-methyl RNA oligonucleotides, TGF-β/TNF-α induction of the mesenchymal markers LAMC2 and MMP7 was substantially decreased (Fig. 3D). This further supported the requirement of miR-21 and miR-31 in LIM 1863 cell EMT induced by TGF-β/TNF-α.
miR-21 and miR-31 Regulate the Migration and Invasion of LIM 1863 Cells
EMT is often linked to a gain in the migratory and invasive properties of the cell. Even though the LIM 1863 organoids hardly migrated in the standard transwell assay, we found that if cells were immediately plated after dissociation of the organoids by trypsin, there was migration in the transwell assay with 10% FBS and media from NIH 3T3 culture as the chemoattractant (Fig. 4A). The same was observed in the Matrigel invasion assay (Fig. 4B). When LIM 1863 cells transfected with miR-21 or miR-31 precursors were tested in these assays, they exhibited significantly enhanced migratory and invasive properties compared with cells transfected with a control miRNA precursor (Fig. 4, A and B). Interestingly, corroborating the results in Matrigel invasion assays, we noticed that when plated onto the Matrigel filter, LIM 1863 cells transfected with miR-21 or miR-31 readily adhered and spread out, whereas cells transfected with a control miRNA precursor did not show such characteristics (Fig. 4B, phase contrast images). These observations support the notion that an increase in miR-21 or miR-31 enhances the migration and invasion properties of LIM 1863 cells.
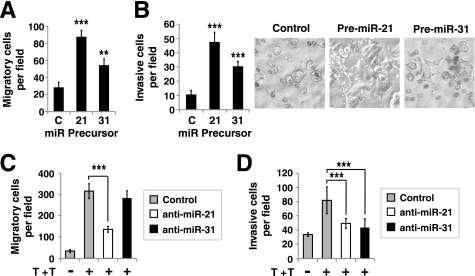
miR-21 and miR-31 regulate LIM 1863 migration and invasion. A and B, LIM 1863 organoids were transfected with 100 nm miR-21 or miR-31 precursors or a negative precursor control (C). Forty-eight hours later, the organoids were dissociated by trypsin and 1 × 105 cells were seeded into the upper wells of transwell chambers coated without (A) or with Matrigel (B). After 72 h, cells that migrated to the lower chambers were counted (mean ± S.D., 8 fields per filter were examined per experiment). LIM 1863 cells transfected with miR-21 and miR-31 precursors spontaneously attached and spread on Matrigel-coated filters (phase contrast images in B, right). C and D, Inhibition of miR-21 and miR-31 activities affects TGF-β//TNF-α-induced LIM 1863 cell migration and invasion. LIM 1863 cells were transfected with 250 nm 2′O-methyl RNA inhibitors of miR-21, miR-31, or control. Eight hours post-transfection, cells (5 × 105 for migration and 1 × 106 for invasion assay) were seeded into the upper wells of transwell chambers coated without (C) or with Matrigel (D) and treated with TGF-β//TNF-α (T+T) or without. After 72 h, the number of cells that migrated to the lower chamber was counted (mean ± S.D., 8 fields per filter were examined). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
If the dissociated LIM 1863 cells were treated with TGF-β/TNF-α upon plating into the transwells, the motility was markedly increased (Fig. 4C). Importantly, when miR-21 function was inhibited by the 2′O-methyl anti-miR-21 oligonucleotide, TGF-β/TNF-α-induction of LIM 1863 motility was significantly reduced (Fig. 4C). This suggests that miR-21 is required for TGF-β/TNF-α-regulation of LIM 1863 cell migration. In contrast, we observed little effect with anti-miR-31, suggesting miR-31 is not as rate-limiting as miR-21 is in regulating LIM 1863 motility. In Matrigel assays, TGF-β/TNF-α also stimulated invasion of the LIM 1863 cells (Fig. 4D). Interestingly, inhibition of either miR-21 or miR-31 significantly decreased LIM 1863 invasion in response to TGF-β/TNF-α, so both of these two miRNAs have non-overlapping targets that are important for LIM 1863 to invade through the extracellular matrix (Fig. 4D). From these gain- and loss- of function studies, we conclude that both miR-21 and miR-31 positively regulate migration and invasion properties of the LIM 1863 cancer cells.
Pro-migration and Pro-invasion Functions of miR-21 and miR-31 in Other Colon Cancer Cell Lines
To determine if the activities of miR-21/miR-31 we observed so far are unique to LIM 1863 cells, we overexpressed these two miRNAs in SW480 and DLD1 colon cancer cell lines. Because in our hands these two cell lines do not undergo EMT in response to TGF-β, we only evaluated their motility and invasion. Indeed in both cell lines, transfection of miR-21 and miR-31 precursors resulted in a marked increase in cell migration and invasion (Fig. 5, A and B). Given that SW480 cells do not express SMAD4 and experiments in Fig. 5 were all done without TGF-β stimulation, our observations also suggest that the pro-metastasis activities of miR-21 and miR-31 do not depend on a functional TGF-β/SMAD pathway.
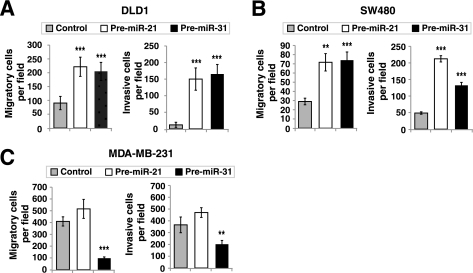
Context-dependent pro-migration and pro-invasion activities of miR-21 and miR-31 in colon and breast cancer cells. A–C, control, miR-21, or miR-31 precursors (100 nm) were transfected into DLD1 (A), SW480 (B), or MDA-MB-231 (C) cells. The motility and invasiveness of these cells were analyzed by transwell migration and invasion assays as in Fig. 4, A and B. Plotted is the number of cells that migrated to the lower chamber was counted (mean ± S.D., >3 fields per filter were examined, **, p < 0.01; ***, p < 0.001).
Interestingly, under exactly the same conditions, miR-31 overexpression substantially suppressed motility and invasion of a breast cancer cell line MDA-MB-231, consistent with previous reports of an anti-metastasis function of miR-31 in breast cancer (29, 30). Therefore, the impact of miR-31 on cancer cell migratory properties is dependent on cell contexts.
All of the above evidence points to pro-cancer invasion and metastasis functions of miR-21 and miR-31 in multiple colon cancer cell lines. These results also agree with previous studies of colon cancer tissues in which higher levels of miR-21 and miR-31 have been linked to colon cancer progression to a late stage and metastasis (39, 40).
TIAM1 Is a Target for Both miR-21 and miR-31
One critical question is the downstream targets of miR-21 and/or miR-31 that contribute to the cellular impact of these two miRNAs. Using TargetScan to search for 3′-UTR sequences with 7 nucleotide matches to the seed region of miR-21 or miR-31, TIAM1 (T lymphoma invasion and metastasis 1) emerged as a possible target for miR-21 (41). Upon visual inspection we found a weak recognition site for miR-31 (e.g. a 6 nucleotide match) in the TIAM1 3′-UTR as well. TIAM1 is a guanine nucleotide exchange factor (GEF) of Rac, and has been implicated in regulating cell migration, invasion and tumor progression (42,–44). Indeed, treating the LIM 1863 organoids with TGF-β/TNF-α substantially reduced the protein level of TIAM1, further prompting us to investigate TIAM1 as a possible target of miR-21 and/or miR-31 (Fig. 6A). Transfecting the LIM 1863 cells with precursors for either miR-21 or miR-31 resulted in markedly decreased abundance of TIAM1 protein when compared with the non-targeting miRNA precursor control (Fig. 6B). Quantitative real-time PCR revealed no change in the mRNA level of TIAM1 by either miR-21 or miR-31 (supplemental Fig. S3). Thus miR-21 and miR-31 appear to control TIAM1 expression mainly through repression of the protein translation, not degradation of the mRNA. Consistent with the functional data in Fig. 5, miR-21 and miR-31 also down-regulated TIAM1 in DLD1 and SW480 cells (Fig. 6C). Importantly, we found that transfection of miR-21 did not cause a significant change in mature miR-31 level, and vice versa (Fig. 6D). Therefore, miR-21 and miR-31 do not influence the expression levels of each other and likely impact the expression of TIAM1 independently.
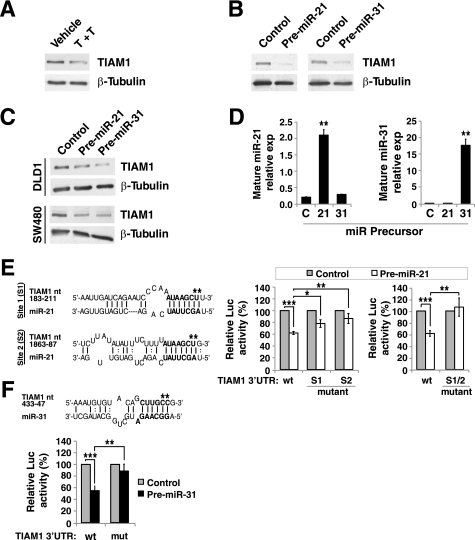
TIAM1 is an endogenous target of both miR-21 and miR-31. A, LIM 1863 organoids were treated with TGF-β/TNF-α (T+T) for 24 h and Western blotting was performed to determine TIAM1 levels (C16, Santa Cruz Biotechnology), with β-tubulin as a loading control. B and C, miR-21 and miR-31 down-regulate TIAM1 protein abundance in multiple colon cancer cell lines. LIM 1863 (B), DLD1 (C), or SW480 (C) cells were transfected with 100 nm miR-21 or miR-31 precursors, or a negative control precursor. After 72 h, TIAM1 protein level was measured by Western blotting. D, LIM 1863 cells were transfected with 100 nm indicated miRNA precursors and 48 h later, the levels of mature miR-21 and miR-31 were determined by the TaqMan miRNA assay (mean ± S.D., n>3). E, (left) Alignment of TIAM1 3′-UTR with the miR-21 sequence. The numbering starts from the first residue after the stop codon. Asterisks indicate the nucleotides that were mutated (CT->TG for both site 1 and site 2 mutations). (right), miR-21 directly targets the 3′-UTR of TIAM1. LIM 1863 cells were transfected first with 100 nm of miR-21 precursor or a control. Eighteen hours later the same cells were transfected with a Renilla luciferase reporter containing the wild type or mutant TIAM1 3′-UTR (e.g. S1: site 1 mutation; S2: site 2 mutation; S1/2: double mutation). A constitutively active firefly luciferase reporter was used as the internal control. The luciferase activities were measured and plotted (mean ± S.D., n>3). *, p < 0.05. F, Similar experiments as in E, analyzing the inhibitory impact of miR-31 on the 3′-UTR of TIAM1. The mutated TIAM1 3′-UTR residues are marked by * (CC->TT), the numbering is the same as in E. **, p < 0.05; ***, p < 0.001.
To further validate whether TIAM1 is a direct target of miR-21 or miR-31, we engineered a luciferase reporter construct containing the 1970 bp 3′-UTR of the TIAM1 gene. MiR-21 precursor indeed significantly reduced the reporter expression (Fig. 6E). We identified two potential miR-21 target sites (matching 7 nt of the miR-21 seed sequence) in the TIAM1 3′-UTR (Fig. 6E, left), and mutations in either one of them reduced, but did not fully abolish the suppression by miR-21 (Fig. 6E, left). Only when both sites were mutated did the miR-21 precursor completely fail to repress the reporter expression (Fig. 6F, right). These data strongly support the hypothesis that TIAM1 is a direct target of miR-21 in LIM 1863 cells. Transfection of miR-31 precursor also significantly reduced the TIAM1 3′-UTR reporter (Fig. 6F). However, we could only find one stretch of 6 nucleotides complementing the miR-31 seed sequence (Fig. 6F). Nevertheless, mutation of this sequence significantly alleviated the repression by miR-31 (Fig. 6F). This strongly suggests that miR-31 also directly represses TIAM1 translation. The remaining inhibitory effect of miR-31 may be due to additional cryptic sites within the TIAM1 3′-UTR or other indirect mechanisms.
Repression of TIAM1 Expression Is Important for Pro-metastasis Functions of miR-21 and miR-31 in LIM1863 Cells
To validate whether TIAM1 is a relevant factor in LIM 1863 morphological changes in response to TGF-β/TNF-α, we introduced exogenous TIAM1 through a lentiviral vector to override its suppression by miR-21 and miR-31. Ectopic TIAM1 expression did not cause any noticeable changes in the morphology of LIM 1863 organoids at basal state (Fig. 7A). However, upon TGF-β/TNF-α treatment, TIAM1-overexpressing LIM 1863 organoids completely failed to undergo morphological changes as the control cells did (Fig. 7B). Moreover, overexpression of TIAM1 also prevented TGF-β/TNF-α from enhancing motility and invasiveness of LIM 1863 cells (Fig. 7, C and D). Therefore, a low level of TIAM1 is important for TGF-β/TNF-α to promote LIM 1863 EMT, migration, and invasion.

Elevated TIAM1 level antagonizes promotion of LIM 1863 morphological changes, motility, and invasiveness by TGF-β/TNF-α and miR-21/miR-31. A, phase contrast images showing no obvious morphological changes in LIM 1863 organoids with or without TIAM1 overexpression. B, TIAM1 overexpression greatly reduced the ability of LIM 1863 cells to undergo EMT. Cells transduced with TIAM1 or empty vector (control) were stimulated with TGF-β/TNF-α (T+T). The morphology of the organoids (n >75 per group) was scored as “spreading” or “not spreading” as in Fig. 3B at 6 h and 24 h time points (mean ± S.D., >75 organoids were counted in each experiment, data represent >3 experiments). **, p < 0.01. C and D, Transwell migration (C) and Matrigel invasion (D) assays measuring LIM 1863 cell motility and invasiveness, respectively. LIM 1863 cells were transduced with lentiviral vectors encoding TIAM1 or control. Cells (5 × 105 for the migration assay and 1 × 106 for the invasion assay) were then seeded into the upper chambers and treated with vehicle or TGF-β/TNF-α (T+T) as indicated. After 72 h, the number of cells that migrated to the lower chambers was counted (mean ± S.D., 8 fields per filter were examined). ***, p < 0.001. E and F, LIM 1863 cells transduced with TIAM1-expressing or control vectors were further transfected with miR-21, miR-31 or control (C) precursors as indicated. Cells were then analyzed for motility (E) and invasion (F) (mean ± S.D., 8 fields per filter were examined). ***, p < 0.001.
Next we more directly tested whether the pro-migration and -invasion activities of miR-21 and miR-31 are also dependent on suppression of TIAM1. Indeed, miR-21/miR-31 precursors were no longer capable of enhancing motility and invasiveness of LIM 1863 cells overexpressing TIAM1 (Fig. 7, E and F). These data further substantiate our model that repression of TIAM1 is a critical component in miR-21/miR-31 regulation of migratory and invasive properties of LIM 1863 cells.
DISCUSSION
We conclude that miR-21 and miR-31 are downstream effectors of TGF-β and TNF-α signaling, and directly regulate motility and invasiveness of colon carcinoma cells. Although these two miRNAs are likely to have many different direct targets, they converge on TIAM1, a protein known to regulate migration and invasion of various cancer cells (42,–44). Therefore, we have uncovered a novel miRNA-mediated mechanism through which TGF-β, in conjunction with TNF-α, promotes invasion and metastasis of colon cancer. Our finding corroborates with previous clinical studies which associate elevated miR-21 and miR-31 levels to late stage colon cancer progression and metastasis (39, 40). With this new mechanistic understanding of miR-21 and miR-31 function in colon cancer cell biology, we suggest a possible utility of miR-21 and miR-31 as molecular markers or a therapeutic target of colon cancer.
Interestingly, although there have been several reports that down-regulation of the miR-200 family is a common element in EMT, we did not find evidence of a change in miR-200 expression during EMT in LIM 1863 organoid (21,–24). This could suggest that either the miR-200 level is already low in LIM 1863 or that ZEB1/2, the miR-200 target, is not rate-limiting in this system. Unlike miR-200, which suppresses an upstream master regulator of the EMT program such as ZEB1/2, miR-21 and miR-31 may impact on more downstream events such as TIAM1. Therefore, overexpression of miR-21 or miR-31 may not be sufficient to initiate the EMT program, but rather play a facilitating role.
One interesting observation is that the elevation of both miR-21 and miR-31 is not an immediate early signaling event downstream of TGF-β/TNF-α and requires de novo protein synthesis. Moreover, it is the processing from precursor to mature miR-21 that is more rate-limiting. This agrees with a report by Davis et al. (45) in which TGF-β up-regulation of miR-21 in vascular smooth muscle cells (VSMC) was attributed primarily to miRNA maturation, not transcription. However, in LIM 1863 cells at least, transcription of the miR-21 gene was robustly activated by TGF-β/TNF-α, with a time course typical of an immediate early target gene. Therefore, the transcriptional regulation of miR-21 by TGF-β/TNF-α is cell context-dependent. In VSMC, the processing of miR-21 was acutely activated by TGF-β, but in contrast the accumulation of mature miR-21 in LIM 1863 organoids was much delayed and dependent on new protein synthesis (45). Therefore, the underlying mechanism for TGF-β/TNF-α up-regulation of mature miR-21 in LIM 1863 remains to be determined and is likely to differ from that in VSMC.
In this study, we found both miR-21 and miR-31 target TIAM1 in LIM 1863 cells, and functional assays further confirmed the relevance of TIAM1 down-regulation in miR-21/miR-31 regulation of LIM 1863 motility and invasiveness. However, it is important to point out that miR-21 and miR-31 functions are not limited to suppressing TIAM1. These two miRNAs likely have other different targets, in addition to TIAM1, that may also be important for cell migration and invasion. Such a scenario could explain how in anti-miR-21-treated LIM 1863 cells, even though TIAM1 would still be repressed by miR-31 upon TGF-β/TNF-α treatment, the motility of LIM 1863 cells was inhibited (Fig. 4C). Furthermore, the anti-miR experiments also suggested that while miR-31 is not as critical as miR-21 in TGF-β/TNF-α-induced enhancement of migratioin, in terms of invasiveness, miR-21 and miR-31 play non-redundant roles and both are indispensable (Fig. 4D).
As a GEF for the Rac GTPase, TIAM1 has been implicated in many aspects of cellular regulation, and its roles in several types of cancer have been documented (44, 46). However, the contribution of TIAM1 to tumor growth, invasion and metastasis is rather complicated and context-dependent. Perhaps the more convincing evidence comes from studies with the Tiam−/− mice. Interestingly, while ablation of the TIAM1 gene significantly inhibited tumorigenesis in a Ras-induced skin cancer model, the tumor that did form progressed to malignance more efficiently (47). More relevant to our study, in an APC mutant Min mouse strain that develop intestinal tumors, TIAM1 deficiency reduced the incidence of polyps formation, but the tumors were more invasive (48). Consistent with this, knockdown of TIAM1 by siRNA suppressed the proliferation of DLD1 colon cancer cells and reduced cell adhesion (48). These all support our model that down-regulation of TIAM1 by miR-21/miR-31 facilitates colon cancer invasion and metastasis.
There is likely more complexity regarding the roles of miR-21 and miR-31 in cancer biology, considering that each miRNA impacts on many targets (16). From many miRNA profile studies of clinical samples, a high miR-21 level has emerged as a common molecular marker of several types of solid tumor including colon cancer (49, 50). Several targets for miR-21 have been demonstrated in many cell lines, and it is evident that miR-21 is involved in multiple aspects of cancer development from initiation to metastasis (51). Interestingly, opposite to our hypothesis in colon cancer cells, miR-31 appears to be a negative regulator of breast cancer metastasis (29, 30). In breast cancer cells, the anti-metastasis impact of miR-31 was attributed to suppression of integrin α5, radixin, and RhoA, and TIAM1 was not identified as a miR-31 target in that context (29, 30). Whether the difference in biological outcomes could be attributed to distinct cohorts of mRNAs regulated by miR-31 in colon versus breast cancer cells still awaits investigation. Equally possible is that TIAM1 function could also be cancer cell-type specific, and indeed previous studies have found that heightened TIAM1 serves to enhance motility and invasion of breast cancer cells, in contrast to our observation here in colon cancer cells (28, 52). How TIAM1 may function in a cell context-dependent manner is another interesting question in understanding the molecular underpinnings of cancer metastasis.
Acknowledgments
We thank Drs. A. Mercurio and Y. Kang for reagents, Dr. J. Straubhaar for statistical analysis of the microarray data, Ms. Aixa Navia for contribution, and Drs. A. Mercurio, B. Lewis, C. Mello, and A. Ross for critical discussions.
*This work was supported, in whole or in part, by National Institutes of Health Grant R01CA108509 (to L. X.) and a Ruth L. Kirschstein NRSA Individual Predoctoral Fellowship to Promote Diversity in Health-Related Research F31CA142216 (to C. L. C.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
2The abbreviations used are:
- TGF-β
- transforming growth factor β
- EMT
- epithelial-mesenchymal transition
- miRNA
- microRNA
- TIAM1
- T-lymphoma invasion and metastasis-1.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m110.160069
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/content/285/46/35293.full.pdf
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/content/full/285/46/35293
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/285/46/35293
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/285/46/35293.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Advances in microRNAs as Emerging Biomarkers for Colorectal Cancer Early Detection and Diagnosis.
Int J Mol Sci, 25(20):11060, 15 Oct 2024
Cited by: 0 articles | PMID: 39456841 | PMCID: PMC11507567
Review Free full text in Europe PMC
Cancer cell plasticity: from cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance.
Cancer Metastasis Rev, 43(1):197-228, 08 Feb 2024
Cited by: 9 articles | PMID: 38329598 | PMCID: PMC11016008
Review Free full text in Europe PMC
An Updated Review on Molecular Biomarkers in Diagnosis and Therapy of Colorectal Cancer.
Curr Cancer Drug Targets, 24(6):595-611, 01 Jan 2024
Cited by: 0 articles | PMID: 38031267
Review
Application of Nanoparticles in the Diagnosis and Treatment of Colorectal Cancer.
Anticancer Agents Med Chem, 24(18):1305-1326, 01 Jan 2024
Cited by: 0 articles | PMID: 39129164 | PMCID: PMC11497148
Review Free full text in Europe PMC
Exploring the interplay between Fusobacterium nucleatum with the expression of microRNA, and inflammatory mediators in colorectal cancer.
Front Microbiol, 14:1302719, 23 Nov 2023
Cited by: 4 articles | PMID: 38075864 | PMCID: PMC10701916
Go to all (178) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
miR-10b targets Tiam1: implications for Rac activation and carcinoma migration.
J Biol Chem, 285(27):20541-20546, 05 May 2010
Cited by: 70 articles | PMID: 20444703 | PMCID: PMC2898316
MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma.
Cancer Lett, 329(2):181-188, 08 Nov 2012
Cited by: 78 articles | PMID: 23142282
Thyroid hormone regulation of miR-21 enhances migration and invasion of hepatoma.
Cancer Res, 73(8):2505-2517, 26 Feb 2013
Cited by: 38 articles | PMID: 23442323
The CircRNA-ACAP2/Hsa-miR-21-5p/ Tiam1 Regulatory Feedback Circuit Affects the Proliferation, Migration, and Invasion of Colon Cancer SW480 Cells.
Cell Physiol Biochem, 49(4):1539-1550, 13 Sep 2018
Cited by: 80 articles | PMID: 30212824
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R01 CA108509
Grant ID: F31CA142216
Grant ID: F31 CA142216
Grant ID: R01CA108509





