Abstract
Free full text

miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis
Associated Data
Abstract
MicroRNAs (miRNAs) are increasingly implicated in regulating the malignant progression of cancer. Here we show that miR-9, the level of which is upregulated in breast cancer cells, directly targets CDH1, the E-cadherin-encoding mRNA, leading to increased cell motility and invasiveness. miR-9-mediated E-cadherin downregulation results in the activation of β-catenin signaling, which contributes to upregulated expression of the gene encoding vascular endothelial growth factor (VEGF); this leads, in turn, to increased tumor angiogenesis. Overexpression of miR-9 in otherwise-non-metastatic breast tumor cells enables these cells to form pulmonary micrometastases in mice. Conversely, inhibiting miR-9 using a ‘miRNA sponge’ in highly malignant cells inhibits metastasis formation. Expression of miR-9 is activated by MYC and MYCN, both of which directly bind to the mir-9-3 locus. Significantly, in human cancers, miR-9 levels correlate with MYCN amplification, tumor grade, and metastatic status. These findings uncover a regulatory and signaling pathway involving a metastasis-promoting miRNA that is predicted to directly target expression of the key metastasis-suppressing protein E-cadherin.
Metastases are responsible for >90% of cancer-related mortality. These secondary growths arise as products of a multi-step process that begins when caner cells within primary tumors break away from neighboring cells and invade through the basement membrane1. This initial step of local invasion may frequently be triggered by contextual signals that carcinoma cells receive from the nearby stroma, causing them to undergo an epithelial-mesenchymal transition (EMT), a multi-faceted transdifferentiation program that enables tumor cells to acquire malignancy-associated phenotypes2. Subsequently, metastasizing cells can enter the circulation, doing so either directly or via lymphatics. Size constraints in the microvasculature cause many of these cells to be arrested at distant tissue sites, where they may extravasate and enter the foreign tissue parenchyma. At this point, they may remain dormant or, with low efficiency, proliferate from occult micrometastases to form angiogenic, clinically detectable metastases. The absence of EMT-inducing signals in the microenvironment of distant tissues may cause such disseminated cells to revert to an epithelial phenotype via a mesenchymal-epithelial transition (MET). Much research has been focused on identifying the critical regulators of the metastatic process; these regulatory molecules include both proteins and microRNAs (miRNAs)3,4.
MiRNAs are small non-coding RNA molecules that suppress gene expression by interacting with the 3′ untranslated regions (UTRs) of target mRNAs. These interactions may result in either inhibition of translation of the targeted mRNAs or their degradation5. In an initial real-time RT-PCR-based screen for differentially expressed miRNAs, we identified three miRNAs that are most significantly upregulated in human breast cancer cell lines – miR-155, miR-9, and miR-10b6. The subsequent functional studies of miR-10b validated its candidacy as a mechanistically important miRNA in cancer progression, as demonstrated by experiments showing that overexpression of miR-10b in otherwise-non-metastatic breast tumors initiated tumor invasion and distant metastasis in xenograft models6. Subsequently, several other miRNAs, including miR-373, miR-520c, miR-335, miR-206, miR-126, miR-21, and miR-31, have also been identified as either promoters or suppressors of metastasis7–11. In addition, the miR-200 family, whose role in regulating metastasis remains unclear, has emerged as a silencer of ZEB1 and ZEB2, two established EMT-inducing and metastasis-promoting transcription factors12,13, thereby representing yet another set of regulators of the EMT program.
A second miRNA that stood out in our initial screen is miR-96, a miRNA that is selectively expressed in neural tissues under normal conditions14 and regulates their development15. Expression of this miRNA is higher in brain tumors than in tumors of other histological types, further demonstrating a tissue-specific expression pattern16. In the context of clinical breast cancer, miR-9 has been found to be upregulated in primary tumors relative to its expression in normal mammary tissues17. Interestingly, miR-9 was recently shown to be upregulated by 1,000-fold in c-myc-induced mouse mammary tumors18.
In a preliminary survey, we used several computational algorithms, including the two most widely tested programs, TargetScan19 and PicTar20, to search for miRNAs that target evolutionarily conserved sequences present in the CDH1 mRNA; this survey revealed that miR-9 was the only known miRNA that was predicted to target the CDH1 mRNA (Fig. 1a). CDH1 encodes the epithelial cell adhesion molecule E-cadherin, a trans-membrane glycoprotein that forms the core of the adherens junctions between adjacent epithelial cells21. The cytoplasmic tail of E-cadherin associates with a number of intracellular proteins that link E-cadherin to the actin cytoskeleton21. Given its well-established function in maintaining adherens junctions, E-cadherin inactivation presumably promotes metastasis by enabling the first step of the metastatic cascade – the dissociation of carcinoma cells from one another. In addition, its loss liberates β–catenin molecules that may move into the nucleus and activate pro-metastatic genes22. The significance of E-cadherin inactivation for metastasis has been demonstrated in a variety of in vitro and in vivo models22–27. Recently, we have found that E-cadherin loss in certain cell types can also trigger an EMT and a wide range of transcriptional and signaling changes that contribute to metastatic dissemination27. Thus, miR-9’s potential role as a suppressor of E-cadherin expression made this miRNA a strong candidate for promoting the acquisition of malignant phenotypes by carcinoma cells.
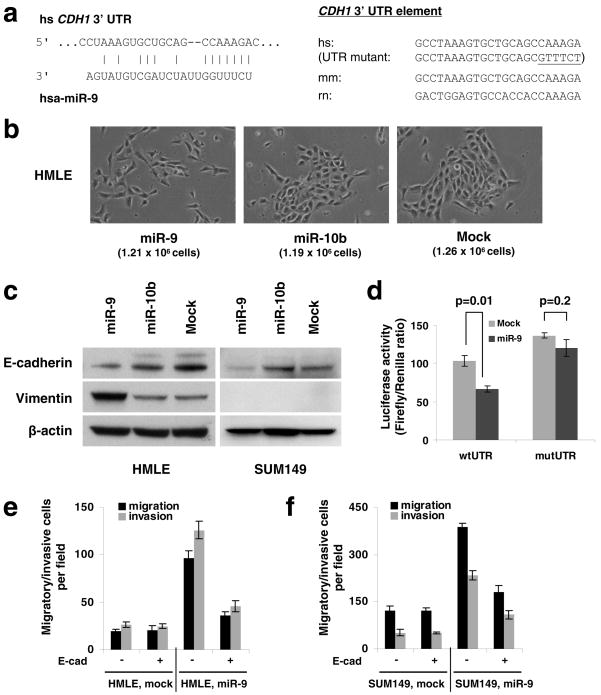
(a) Left panel: predicted duplex formation between human CDH1 3′UTR and miR-9. Right panel: sequence of the miR-9 binding site within the CDH1 3′UTR of human (hs), mouse (mm), and rat (rn).
(b) Phase contrast images of HMLE cells infected with the miR-9-expressing, miR-10b-expressing, or empty vector. Cells were plated on 10 cm dishes at the same density (1 × 106 cells in 10 ml medium). Two days after plating, images were taken and cells were then counted (cell numbers are shown in parentheses). Magnification: x200.
(c) Immunoblotting of E-cadherin and vimentin in HMLE and SUM149 cells infected with the miR-9-expressing, miR-10b-expressing, or empty vector.
(d) Luciferase activity of the wild-type or mutant CDH1 3′UTR reporter gene in SUM149 cells infected with the miR-9-expressing or empty vector.
(e, f) Transwell migration assay and Matrigel invasion assay of miR-9-transduced or mock-infected HMLE (e) and SUM149 (f) cells with or without ectopic expression of E-cadherin. A representative experiment is shown in triplicate along with s.e.m. in d–f.
RESULTS
Effect of miR-9 on the expression of E-cadherin
To determine whether miR-9 can indeed downregulate E-cadherin expression, we stably expressed miR-9 and, as a control, miR-10b, in two epithelial cell lines (Supplementary Information, Fig. S1). Ectopic expression of miR-9, but not miR-10b, led to an EMT-like conversion in HMLE non-transformed, immortalized human mammary epithelial cells28: these cells became scattered and assumed a spindle-like or star-like morphology (Fig. 1b) and displayed a 70% reduction in E-cadherin and a 5-fold increase in the mesenchymal marker vimentin (Fig. 1c). In contrast, in SUM149 human breast carcinoma cells29, miR-9 downregulated E-cadherin expression by ~50% but failed to induce vimentin, other mesenchymal markers, and a fibroblastic cell morphology (Fig. 1c, and data not shown). These differences in response were intrinsic to the two cell lines rather than a consequence of different degrees of E-cadherin suppression, because knockdown of E-cadherin by >90% using small-interfering RNA (siRNA) in both cell lines caused an EMT in HMLE cells27 but not in SUM149 cells (T.T.O. and R.A.W., unpublished observations). Hence, while miR-9 succeeds in suppressing E-cadherin expression in two epithelial cell lines, it only induces an EMT in one of them.
To determine whether miR-9 directly targets the CDH1 mRNA, we performed reporter assays and found that miR-9 reduced the activity of a luciferase reporter that was fused to the wild-type 3′ UTR of the CDH1 mRNA but not to a mutant 3′UTR (Fig. 1d); the latter carried altered nucleotides that were introduced in the miR-9 ‘seed-pairing’19 recognition site (Fig. 1a). Hence, the observed downregulation of E-cadherin by miR-9 depends directly on a single cognate recognition site in the CDH1 3′UTR.
Inactivation of E-cadherin has been shown to promote cell migration and invasion in the presence or absence of an EMT27. Consistent with this, we found that ectopic expression of miR-9 led to a 3- to 5-fold increase in the motility and invasiveness of both HMLE and SUM149 cells in vitro (Fig. 1e, f). In order to determine whether these effects depend specifically on E-cadherin suppression, we employed an expression construct that encodes the entire E-cadherin coding sequence but lacks the 3′ UTR, yielding an mRNA that is resistant to miRNA-mediated suppression. Ectopic expression of E-cadherin with this construct reduced migration and invasion in the miR-9-overexpressing cells, but not in the control cells, which have a low basal level of miR-9 (Fig. 1e, f). This suggests that following miR-9 overexpression, a resulting reduction in E-cadherin is required in order for cells to exhibit increased motility and invasiveness. However, we cannot exclude the possibility that miR-9-mediated suppression of other targets is also required for the observed effects of this miRNA on cell phenotypes.
Regulation of β-catenin signaling and VEGF expression by miR-9
Having validated E-cadherin as a miR-9 target, we next sought to determine whether miR-9-mediated E-cadherin suppression would affect intracellular signaling. Binding of Wnt ligands to their receptors results in the stabilization of β-catenin, allowing it to enter the cell nucleus, interact with the TCF/LEF family of transcription factors, and promote the transcription of genes 30. Independent of this, E-cadherin binds and sequesters a large pool of β-catenin at the cytoplasmic membrane, thereby preventing its nuclear translocation and its function as a component of the TCF/LEF transcription factor complex22,31,32. Our previous work demonstrated that knockdown of E-cadherin in the experimentally transformed human mammary epithelial cells caused relocalization of β-catenin from adherens junctions to the cytoplasm and nucleus. Of note, this suppression of E-cadherin expression also led to a reduction of β-catenin phosphorylation by GSK-3β through unknown regulatory mechanisms, thereby enabling the liberated β-catenin to escape proteasome-mediated degradation27.
In the present work, we found that miR-9-expressing SUM149 cells exhibited both cytoplasmic and nuclear localization of β-catenin, whereas in the control SUM149 cells this protein was predominantly associated with cell-cell junctions, presumably with the cytoplasmic tails of E-cadherin molecules (Fig. 2a). Moreover, the β-catenin in miR-9-expressing cells showed a reduced level of GSK-3β-dependent inhibitory phosphorylation (Fig. 2b) and was therefore present in a more active functional state27,30.
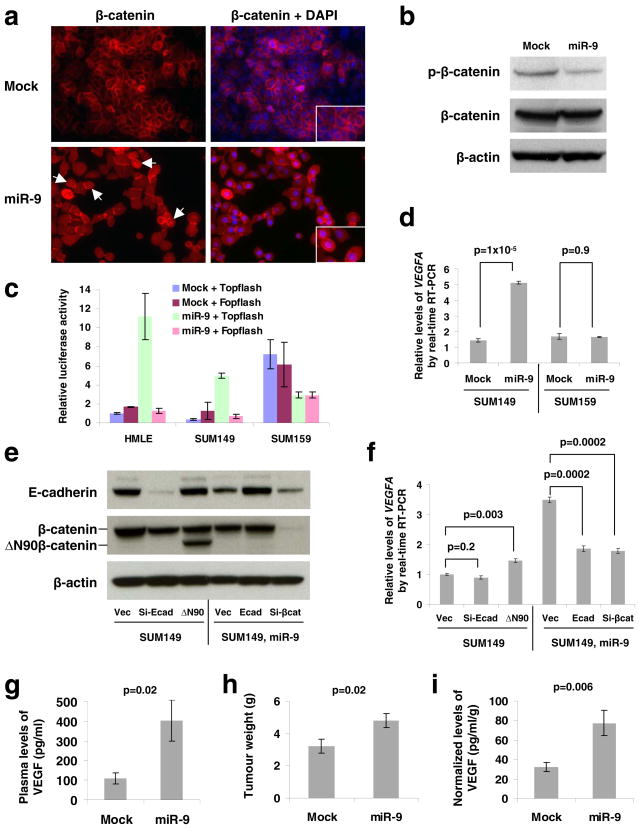
(a) Immunofluorescence staining of β-catenin (red) in mock-infected or miR-9-expressing SUM149 cells demonstrates differential localization. Right panels are the overlay of β-catenin and nuclear 4′,6-diamidino-2-phenylindole (DAPI; blue) staining of the same field. White arrows indicate cells positive for nuclear β-catenin. Insets: blow-up images of particular cells. Magnification: x200.
(b) Immunoblotting of phospho-β-catenin (Ser33/37/Thr41, GSK-3β phosphorylation sites) and β-catenin in SUM149 cells infected with the miR-9-expressing or empty vector.
(c) Topflash reporter assay in HMLE, SUM149, and SUM159 cells infected with the miR-9-expressing or empty vector.
(d) Real-time RT-PCR of total VEGFA mRNA in SUM149 and SUM159 cells infected with the miR-9-expressing or empty vector.
(e) Immunoblotting of E-cadherin and β-catenin in SUM149 cells infected with E-cadherin siRNA (si-Ecad) or ΔN90β-catenin (ΔN90), and in SUM149-miR-9 cells infected with E-cadherin (Ecad) or β-catenin siRNA (si-βcat). Vec: the pLKO-puro vector with a scrambled sequence that does not target any mRNA.
(f) Real-time RT-PCR of total VEGFA mRNA in the same cells described in e. A representative experiment is shown in triplicate along with s.e.m. in c, d and f.
(g–i) Plasma levels of VEGF (g), primary tumor weight (h), and normalized VEGF levels (i) in mice that received orthotopic injection of miR-9-transduced or mock-infected SUM149 cells, at week 12 after transplantation. Data are presented as mean ± s.e.m. (n = 8 mice per group).
To provide the definitive proof of β-catenin functional activation, we performed β-catenin reporter assays using both the Topflash construct with multiple TCF/LEF-binding sites in the promoter of a firefly luciferase reporter gene and the derived Fopflash construct with mutated TCF/LEF binding sites33. These assays demonstrated that miR-9 increased β-catenin activity by >10-fold in E-cadherin-positive cells (HMLE and SUM149) but not in E-cadherin-negative human breast cancer cells (SUM159) (Fig. 2c). Of note, the basal β-catenin activity was far higher in SUM159 cells than in HMLE and SUM149 cells (Fig. 2c), which provided further indication of the inverse correlation between E-cadherin expression and β-catenin activity.
Because E-cadherin inactivation has recently been shown to promote tumor angiogenesis in genetically engineered mouse models22,26, and because the pro-angiogenic factor VEGFA has been described as a transcriptional target gene of β-catenin22,34, we used the SUM149 and SUM159 breast cancer cell lines, described above and in further detail in Supplementary Information, Fig. S2, to determine whether miR-9 would increase the levels of VEGFA. We found that upon miR-9 overexpression, the VEGFA mRNA levels increased by 3- to 4-fold in the SUM149 cells but not in the SUM159 cells (Fig. 2d). In addition, miR-9 also suppressed E-cadherin expression and induced VEGFA expression by 3-fold in MCF7-RAS breast carcinoma cells (Supplementary Information, Fig. S3). Thus, the ability of miR-9 to upregulate VEGFA is correlated with its ability to modulate E-cadherin expression.
We undertook to determine whether the ability of miR-9 to upregulate VEGFA mRNA depended on its ability to downregulate E-cadherin expression and activate β-catenin-mediated transcription. We discovered that expression of either an E-cadherin siRNA or a constitutively active β-catenin (the ΔN90 non-degradable mutant27, Fig. 2e) in parental SUM149 cells was not sufficient to phenocopy the observed induction of VEGFA mRNA expression by miR-9; however, ectopic expression of either an E-cadherin mRNA (without 3′UTR) or a β-catenin siRNA (Fig. 2e) in miR-9-expressing SUM149 cells was indeed able to reverse the ability of miR-9 to induce VEGFA mRNA (Fig. 2f). These data indicate that E-cadherin downregulation and β-catenin activation are necessary but not sufficient for mediating miR-9-dependent VEGF upregulation. Stated differently, E-cadherin is a critical target of miR-9, but other functionally important miR-9 targets remain to be identified.
We next implanted miR-9-expressing or mock-infected SUM149 cells into orthotopic sites – the mammary fat pads of non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice. At 12 weeks post implantation, we found an approximately 4-fold increase in the plasma level of human-specific VEGF in mice bearing miR-9-expressing SUM149 tumors compared to mice implanted with mock-infected SUM149 cells (Fig. 2g), which was accompanied by a 1.5-fold increase in primary tumor weight (Fig. 2h). We then normalized the VEGF level to the primary tumor weight for individual recipients and found that the normalized VEGF levels were still significantly higher in the miR-9 group (Fig. 2i), indicating that miR-9-expressing tumor cells do indeed secrete more VEGF in vivo. Finally, we found in a control experiment that miR-10b, which did not reduce E-cadherin expression in SUM149 cells (Fig. 1c), could not upregulate VEGF levels in these cells (Supplementary Information, Fig. S4).
Effects of miR-9 on tumor angiogenesis, mesenchymal traits, and the formation of micrometastases
SUM149 cells forced to express miR-9 grew more slowly in vitro than the corresponding control cells (Supplementary Information, Fig. S5); in contrast, in vivo the miR-9-expressing SUM149 tumors displayed an approximately 2-fold increase in the levels of the Ki-67 cell proliferation marker compared with those having basal levels of miR-9 expression (Fig. 3a). Hence, the observed increase in primary tumor size appeared to reflect a cell-non-autonomous effect of this miRNA, such as its ability to enhance tumor-associated angiogenesis. To address this possibility, we performed immunohistochemistry to detect expression of the MECA-32 mouse endothelial cell antigen, which should serve as a specific marker of angiogenesis. Consistent with previous observations6, the control SUM149 tumors displayed only low levels of staining. In stark contrast, miR-9-expressing SUM149 tumors exhibited a >10-fold increase in the density of intratumoral microvessels (Fig. 3b), which is in consonance with the observed higher levels of VEGF secreted by these tumor cells.
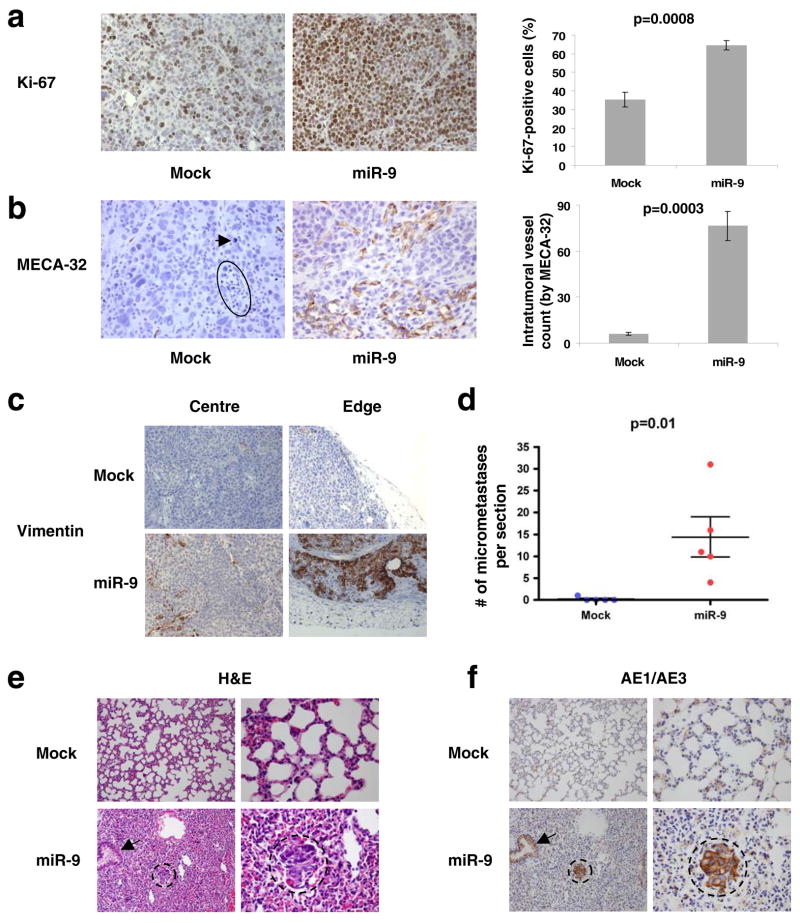
(a, b) Left panels: Ki-67- (a) and MECA-32-stained sections (b) of primary mammary tumors formed by mock-infected or miR-9-transduced SUM149 cells, at week 12 after orthotopic transplantation. The circle and arrow indicate pyknotic nuclei. Magnification: x200 for Ki-67; x400 for MECA-32. Right panels: counting of Ki-67-positive cells (a) and intratumoral vessels (the number of microvessels per field, b). Data are presented as mean ± s.e.m. (we counted three fields per section and analyzed four mice per group).
(c) Human-specific vimentin staining of primary mammary tumors formed by mock-infected or miR-9-transduced SUM149 cells, at week 12 after orthotopic transplantation. Both intratumoral regions (center) and tumor-stroma interfaces (edge) are shown. Magnification: x200. n = 3 mice per group were analyzed for vimentin.
(d) Numbers of lung micrometastases per section in individual mice that received orthotopic injection of miR-9-transduced or mock-infected SUM149 cells, at week 12 after transplantation. Data are presented as mean ± s.e.m. (each data point represents a different mouse; n = 5 mice per group).
(e, f) H&E- (e) and AE1/AE3 (a cocktail of two distinct anti-cytokeratin monoclonal antibodies, f)-stained sections of lungs isolated from mice that received orthotopic injection of miR-9-transduced or mock-infected SUM149 cells, at week 12 after transplantation. Circles indicate clusters of micrometastatic cells. Arrows indicate normal bronchial epithelium. Magnification: x200 in left columns; x600 in right columns.
In fact, an even more robust vessel formation was observed at the outer edges of these miR-9-expressing tumors than in the intratumoral regions (Supplementary Information, Fig. S6a). Of additional interest, some of the cells within the lumina of vessels at the tumor edge were nucleated and were positive for cytokeratin expression, providing evidence for miR-9-induced intravasation of carcinoma cells (Supplementary Information, Fig. S6a, b). Although the great majority of carcinoma cells within the miR-9-expressing and control tumors exhibited similar morphology (Fig. 3a, b), the carcinoma cells located near vessels at the tumor edge (Supplementary Information, Fig. S6a) were cytologically different from the bulk populations of carcinoma cells (Fig. 3a, b) in the miR-9-overexpressing tumors. We reasoned that these cytologically distinctive cells were either stromal cells of host origin or, alternatively, carcinoma cells undergoing an EMT in vivo.
To distinguish between these possibilities, we performed immunostaining with antibodies reactive with E-cadherin and human vimentin. As expected, the miR-9-expressing human tumor xenografts had weaker E-cadherin staining than control tumors (data not shown). More strikingly, a significant number of carcinoma cells at the edges of the miR-9-expressing tumors exhibited strong human vimentin staining, while intratumoral regions only displayed occasional human vimentin-positive cells (Fig. 3c). The control tumors, which expressed lower levels of miR-9, were essentially vimentin-negative (Fig. 3c). Moreover, as anticipated, mouse fibroblasts in the tumor stroma were completely negative for human vimentin (Fig. 3c), demonstrating that the observed vimentin-positive cells did indeed derive from implanted human carcinoma cells. Because miR-9-induced vimentin expression in cultured HMLE cells but not in SUM149 cells (Fig. 1c), we propose that this miRNA can induce an EMT in a cell type- and context-dependent manner, and that in certain cell types, such as SUM149 carcinoma cells, miR-9 sensitizes cells to EMT-inducing signals emanating from the tumor microenvironment rather than eliciting an EMT on its own.
SUM149 cells have been reported to exhibit little, if any, metastatic ability6,35. Indeed, we observed that mouse hosts bearing control SUM149 primary tumors were essentially free of lung micrometastases except for one mouse, which showed a single micrometastatic cluster that was immunostained by the AE1/AE3 anti-cytokeratin antibodies at 12 weeks after cancer cell implantation (Fig. 3d). In stark contrast, the lungs of mice implanted in the mammary fat pads with miR-9-overexpressing SUM149 cells exhibited clusters of dense hyperchromatic cells that were positive for cytokeratins, as demonstrated by both histological examination (Fig. 3e) and AE1/AE3 immunostaining (Fig. 3f). On average, we found ~14 micrometastases per 5-μm section (Fig. 3d). This provided direct evidence that miR-9 operates as a pro-metastatic miRNA.
Effects of miR-9 silencing on metastasis
We also wished to determine whether miR-9 expression is required for metastasis formation by cancer cells that are naturally highly malignant. To address this, we used ‘miRNA sponges’11,36 to stably knock down miR-9 in the highly metastatic 4T1 mouse mammary tumor cells that naturally express a very high level of miR-9 (data not shown). A ‘miRNA sponge’ is a construct encoding an mRNA (e.g., the gfp mRNA) that contains in its 3′UTR multiple tandem binding sites for a miRNA of interest36. We assessed the level of functional knockdown of miR-9 by a reporter assay, in which the predicted miR-9 binding site was introduced into the 3′UTR of a luciferase reporter gene6, and found that infection of 4T1 cells with the miR-9 sponge caused a >2-fold increase in luciferase activity compared with the control sponge (Fig. 4a); this suggested that a greater than 50% inhibition of the actions of miR-9 had been achieved by the miR-9 sponge. We then implanted the infected 4T1 cells into the mammary fat fads of syngeneic immunocompetent Balb/c mice. Four weeks later, mice were moribund due to primary tumor burdens in both groups, and no difference in primary tumor size was observed between the 4T1 tumors expressing a control sponge and those expressing the miR-9 sponge (Fig. 4b). Examination of lungs revealed an average of 35 visible metastases in mice implanted with 4T1 cells expressing the control sponge (Fig. 4c, d); in contrast, an approximately 50% reduction in lung metastases (average: 17 visible metastases per mouse) was observed in mice bearing 4T1 tumors expressing the miR-9 sponge (Fig. 4c–e). Thus, inhibition of miR-9 can suppress metastasis formation by otherwise-highly malignant cancer cells.
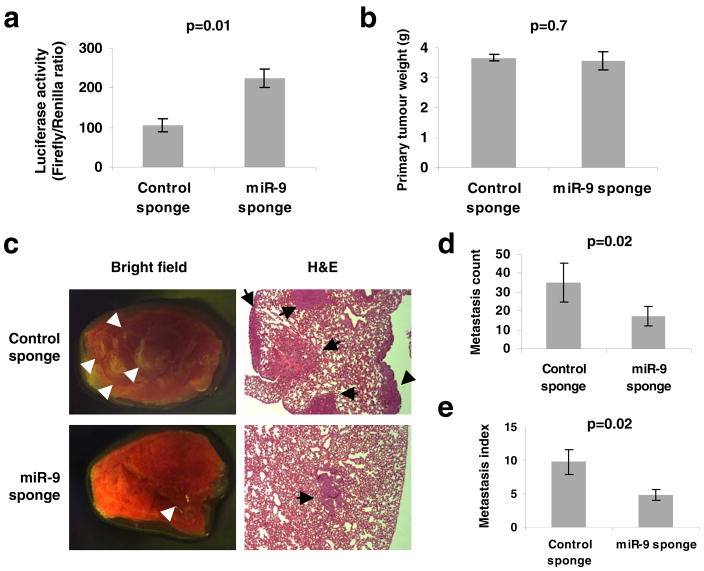
(a) Enhanced activity of miR-9-regulated reporter by infection of 4T1 cells with the miR-9 sponge. A representative experiment is shown in triplicate along with s.e.m.
(b) Primary tumor weight in Balb/c mice that received orthotopic injection of 4T1 cells infected with the miR-9 sponge or control sponge, at 4 weeks after transplantation.
(c) Bright field imaging and H&E staining of lungs isolated from mice that received orthotopic injection of 4T1 cells infected with the miR-9 sponge or control sponge, at 4 weeks after transplantation. Arrows indicate metastatic nodules. Magnification: x8 for bright field imaging; x40 for H&E staining.
(d, e) Numbers of visible lung metastases (d) and metastasis index (= number of metastases/primary tumor weight, e) in mice that received orthotopic injection of 4T1 cells infected with the miR-9 or control sponge, at 4 weeks after transplantation. Data in b, d and e are presented as mean ± s.e.m. (n = 8 mice per group).
Activation of miR-9 expression by MYC/MYCN
We undertook to determine how miR-9 expression is regulated. Consistent with our previous findings6, immortalized, non-transformed HMLE cells naturally expressed a low level of miR-9, whereas the breast cancer cell lines examined expressed higher levels of this miRNA (Fig. 5a). Interestingly, a correlation between RAS status and miR-9 levels was observed in various cell lines; for instance, both the MCF7-RAS human breast cancer cells, which express an introduced, activated RAS oncoprotein (H-RASV12)37, and the MDA-MB-231 human breast cancer cells, which contain an activating K-RAS, codon 12 mutation38, showed significantly higher miR-9 levels than did the HMLE and MCF7 cells, both of which lack an activated RAS oncogene38 (Fig. 5a). Of note, relative to miRNA expression in normal mammary tissues, miR-9 stood out as the most significantly upregulated miRNA in mammary tumors arising in the MMTV-c-myc transgenic mice18. We found this to be particularly interesting, because RAS can potentiate MYC activity to promote tumor angiogenesis39. Moreover, oncogenic RAS is capable of inducing VEGF expression40.
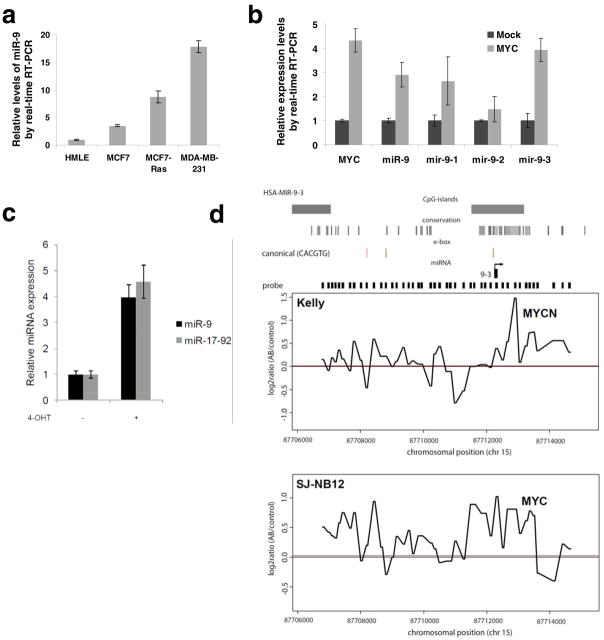
(a) Real-time RT-PCR of miR-9 in HMLE, MCF7, MCF7-RAS, and MDA-MB-231 cells.
(b) Real-time RT-PCR of MYC, mature miR-9, mir-9-1, mir-9-2, and mir-9-3 in mock-infected or MYC-transduced HMLE cells.
(c) Mature miRNA expression in SH-EP-MYCN-ER cells upon MYCN induction with 4-OHT, 48 h after treatment. MiRNA expression values were rescaled relative to the control (no 4-OHT). The miR-17-92 expression value represents the expression of miR-20a, a miRNA residing within the miR-17-92 cluster representative for miR-17-92 expression. Data in a–c are presented as mean ± s.e.m. of triplate samples.
(d) ChIP-on-chip data showing occupancy of the mir-9-3 genomic sequence by MYCN and MYC in Kelly (MYCN-amplified) and SJ-NB-12 (MYC-amplified) neuroblastoma cells, respectively. The genomic positions for probes and their enrichment ratios are displayed on the X and Y axes, respectively. Smoothed local enrichment ratios are given for either MYCN or MYC at the mir-9-3 locus in Kelly and SJ-NB-12 cells. The red line indicates median enrichment ratio for MYCN or MYC versus input as calculated of all probes for chromosome 15.
Responding to these various observations, we were particularly interested in the actions of MYC, because this transcription factor has been shown to directly regulate expression of a diverse set of miRNAs (e.g., the oncogenic miR-17-92 miRNA cluster) that contribute in one way or another to tumorigenesis41–43. For this reason, we ectopically expressed MYC in HMLE cells and found that a 4-fold increase in MYC expression level resulted in an approximately 3-fold increase in the level of mature miR-9 (Fig. 5b). miR-9 can be generated by the processing of any of three primary transcripts encoded by three distinct genes. Among the three encoding genes, the transcription of mir-9-3 is upregulated most strongly (3.9-fold, Fig. 5b) by the MYC transcription factor.
Another MYC gene family member, N-MYC (MYCN), is frequently amplified in neuroblastomas. Operating as transcription factors, MYC and MYCN share a common set of target genes and are, from this perspective, functionally interchangeable. To evaluate whether MYCN can also upregulate miR-9, we measured its expression in a MYCN-inducible model system, SHEP-MYCN-ER42. Upon 4-hydroxy-tamoxifen (4-OHT)-mediated activation of the MYCN-ER fusion protein, miR-9 expression level was increased by 4-fold, which was comparable to the increase in the expression of the miR-17-92 positive control, whose expression is known to be activated by MYCN through direct promoter binding44 (Fig. 5c).
To determine whether MYC and MYCN regulate the expression of miR-9 by binding directly to the corresponding genomic sequences, we performed genome-wide ChIP-on-chip experiments using the Kelly and SJ-NB-12 neuroblastoma cells, which express high levels of endogenous MYCN and MYC, respectively, due in both cases to gene amplification. We observed that the endogenously expressed MYCN and MYC proteins bound directly to a conserved CpG island region encompassing canonical MYC binding sites (CACGTG) in the mir-9-3 locus; this was observed in both the Kelly and SJ-NB-12 neuroblastoma cells (Fig. 5d).
The association of chromatin-bound MYC with H3K4Me3 (a mark of activated chromatin) at MYC-regulated genes has been described previously45. Indeed, we found that MYC binding at the mir-9-3 locus was associated with elevated H3K4me3 binding (Supplementary Information, Fig. S7). Although we also observed a peak for MYC binding at mir-9-1 and mir-9-2 loci, respectively, these two regions exhibited low occupancy by H3K4me3 and relatively high occupancy by H3K27me3 (a mark of repressed chromatin, data not shown). Hence, among the three miR-9-encoding genes, the transcription of mir-9-3 is the most responsive to transcriptional activation by MYC/MYCN.
Recently, others have identified candidate transcriptional start sites (TSSs) and promoters of the great majority of human and mouse miRNA genes46. They exploited yet other findings that histone H3 is trimethylated at lysine 4 (H3K4me3) at the TSSs and promoters of most genes (>90%) in the genome. This study identified position 87712233 of chromosome 15 as the putative human mir-9-3 TSS46. In the present study, we observed co-occupancy by H3K4me3 and MYC of this putative mir-9-3 TSS region (close to position 87712000; Supplementary Information, Fig. S7), as well as of a region upstream of this putative mir-9-3 TSS (near position 87708500, where a canonical E-box is located; Supplementary Information, Fig. S7). Hence, the regions in which we observed MYC binding appear to encompass the mir-9-3 TSS and promoter, providing further evidence of transcriptional regulation of the mir-9-3 gene by MYC.
Correlation of miR-9 with MYCN amplification, tumor grade, and metastatic status in human cancers
To extend our analysis to clinical cancers, we measured the expression of miR-9 in a cohort of 45 neuroblastoma tumor samples. Twenty-two tumors exhibited normal MYCN copy number and were classified as Stage 1 (n = 10), Stage 2 (n = 6) or Stage 3 (n = 6) neuroblastomas. Twenty-three tumors were classified as Stage 4 neuroblastomas and these showed MYCN gene amplification. Compared to tumors with normal MYCN copy number, tumors that exhibited an amplification of the MYCN gene showed a 2.5-fold upregulation of miR-9 expression (p = 9 × 10−4, Fig. 6a). The observed increase of miR-9 expression was similar to that of the miRNAs belonging to the miR-17-92 cluster (average miR-17-92 fold increase = 2.6, Supplementary Information, Fig. S8), which was previously shown to be upregulated in MYC/MYCN-overexpressing tumors42,47.
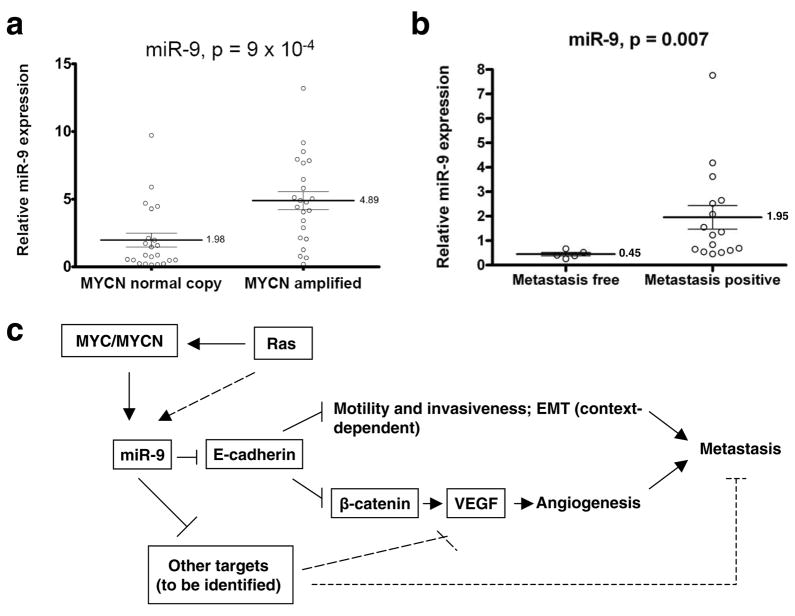
(a) Expression of mature miR-9 in MYCN-normal copy (n = 22) and MYCN-amplified (n = 23) neuroblastoma tumor samples.
(b) Expression of mature miR-9 in primary breast tumor samples from metastasis-free and metastasis-positive patients. Data in a and b are presented as mean ± s.e.m.
(c) Model for miR-9-mediated pathway in cancer metastasis.
We also examined miR-9 levels in clinical breast cancers. Consistent with our expression analyses of cultured cell lines, miR-9 levels were significantly elevated in primary breast tumors from patients with diagnosed metastases compared to those from metastasis-free patients (fold increase = 4.33, p = 0.007, Fig. 6b).
DISCUSSION
Model for miR-9-mediated pathway in cancer metastasis
Our findings identify miR-9 as a pro-metastatic miRNA and a negative regulator of the key metastasis suppressor, E-cadherin. On the basis of our results, we propose that the MYC and MYCN oncoproteins, acting on the mir-9-3 locus, cause activation of miR-9 expression in tumor cells. The resulting miR-9 miRNA can suppress the expression of E-cadherin, resulting in the promotion of carcinoma cell motility and invasiveness and, in addition, causing the activation of β-catenin signaling; the latter, in turn, contributes to elevated expression of VEGFA, leading to induction of tumor-associated angiogenesis. Ultimately, both increased cell motility and invasiveness, as well as enhanced angiogenesis, can contribute to metastasis formation (Fig. 6c). Since E-cadherin downregulation and β-catenin activation appear to be necessary but not sufficient for mediating miR-9-dependent VEGF upregulation (Fig. 2f), yet other miR-9 targets must also operate to enable VEGF induction by this miRNA (Fig. 6c). Indeed, the E-cadherin-independent roles of miR-9 in tumor cells warrant further investigation.
Paradoxically, a recent study proposed that miR-9 might be a candidate metastasis-suppressing miRNA, based on the observation that miR-9-encoding gene promoters, as well as the promoters of genes encoding two other miRNAs (miR-34b/c and miR-148a), were methylated in a relatively large fraction of primary tumors present in patients with lymph node metastases; in contrast, only a small fraction of primary tumors from patients without lymph node metastases exhibited this promoter methylation, while the status of distant metastases of these tumors was not determined48. Importantly, because the miR-9 expression levels in these tumors were not determined, it remains unclear whether there was a correlation between miR-9 gene methylation status and miR-9 miRNA levels. Furthermore, when ectopically expressed in several highly malignant cells, miR-34b/c and miR-148a suppressed metastasis formation, while miR-9 did not48.
Yet other lines of evidence indicate that miR-9 is positively associated with malignancy of human cancers. Some have demonstrated that miR-9 expression is significantly upregulated in both clinical breast cancers17 and in c-myc-induced mouse mammary tumors18. We have found that miR-9 is upregulated in various breast cancer cell lines compared with non-transformed mammary epithelial cell lines, and that metastatic breast cancer cells express even higher miR-9 levels than non-metastatic tumor cells. miR-9 is also upregulated in metastatic breast tumors compared to non-metastatic breast tumors from patients (Fig. 6b). These results suggest that methylation of the promoters of miR-9 genes reported in the cited study48 does not indicate a wider role of this miRNA in inhibiting metastasis.
miR-9 expression and EMT
In contrast to miR-10b, which is specifically upregulated in metastatic cancer cells6, miR-9 expression is upregulated in both metastatic and non-metastatic tumor cells relative to non-transformed cells. These observations, together with the fact that miR-9 expression can be increased by the actions of MYC and MYCN oncoproteins, cause us to propose that miR-9 expression is often induced at earlier stages of multi-step tumor progression when elevated MYC and MYCN expression is already apparent49 but before tumors become actively invasive and metastatic.
Can miR-9, acting on its own, initiate the EMT program? In the SUM149 carcinoma cells, the partial reduction of E-cadherin levels mediated by this miRNA does not directly induce an EMT in vitro (Fig. 1c and data not shown); instead, it seems to sensitize these tumor cells to EMT-inducing signals arising from the tumor microenvironment. This leads to the acquisition by carcinoma cells of mesenchymal traits late in tumor progression (Fig. 3c), which could in turn contribute to metastatic dissemination.
A contrasting response was observed in HMLE cells, in which we did indeed observe an EMT-like conversion resulting from overexpression of miR-9 (Fig. 1b, c). One possibility is that miR-9 might induce an EMT in a cell type-dependent and context-dependent manner; for example, HMLE cells have undergone functional inactivation of both p53 and Rb, which might prime these cells for induction of an EMT. Alternatively, the apparent ability of miR-9 to induce an EMT in HMLE cells might be explained by selection for a pre-existing rare mesenchymal subpopulation present within this particular cell line50. Unlike the HMLE cell line, the SUM149 cell line used in this study does not contain a detectable mesenchymal subpopulation, based on the absence of detectable levels of the mesenchymal marker vimentin (Figure 1c) and on FACS profiles (data not shown). For this reason, the selection mechanism, if it ever occurs, applies to HMLE cells but not to SUM149 cells, and the conclusions drawn from SUM149 experiments are unaffected.
Inactivation of the key metastasis suppressor E-cadherin can occur via a variety of distinct mechanisms, such as promoter hypermethylation and the actions of various EMT-inducing transcription factors. Our findings identify an additional mechanism for downregulating E-cadherin and indicate that elevated expression of the miR-9 miRNA contributes to EMT and metastasis in some and possibly many human tumors.
METHODS
Methods and any associated references are available in the online version of the paper.
Supplementary Material
1
2
3
Acknowledgments
We thank Johannes Schulte, Martin Eilers, Rosa Noguera, Margaret Ebert, and Phillip Sharp for providing tumor samples and reagents; the Histology Core Lab at MIT and MSKCC for assistance with sectioning and IHC; and members of the Weinberg Lab for useful discussions. L.M. is a recipient of a Life Sciences Research Foundation Fellowship, a Margaret and Herman Sokol Award, and an NIH Pathway to Independence Award (K99/R00). J.Y. and E.P. are supported by a Howard Hughes Medical Institute Undergraduate Fellowship. J.T-F. is supported by the MSKCC Cancer Core Grant. T.T.O. and S.V. are recipients of a U.S. Department of Defense Breast Cancer Research Program Predoctoral Fellowship. R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Cancer Research Professor. This research is supported by an NIH grant to R.A.W. and the Ludwig Center for Molecular Oncology at MIT.
Footnotes
AUTHOR CONTRIBUTIONS
L.M. conceived the project. R.A.W. supervised research. L.M. and H.P. designed experiments. L.M., J.Y., H.P., E. P., J.T-F., and F.R. performed most of the experiments and analyzed data. P.M., D.M., F.W., F.P., and J.V. contributed MYCN and ChIP-on-chip data. T.T.O. contributed some of the constructs and shared unpublished observations. S.V. modified the miRNA sponge design for stable expression. L.M. and R.A.W. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ncb2024
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2845545?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101801680
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/ncb2024
Article citations
The highly metastatic 4T1 breast carcinoma model possesses features of a hybrid epithelial/mesenchymal phenotype.
Dis Model Mech, 17(9):dmm050771, 04 Sep 2024
Cited by: 0 articles | PMID: 39104192 | PMCID: PMC11391819
MicroRNA as Key Players in Hepatocellular Carcinoma: Insights into Their Role in Metastasis.
Biochem Genet, 05 Aug 2024
Cited by: 0 articles | PMID: 39103713
Review
Up-regulation of LPCAT1 is correlated with poor prognosis and promotes tumor progression in glioblastoma.
Cytotechnology, 76(6):735-748, 10 Aug 2024
Cited by: 0 articles | PMID: 39435418
Tumor-associated genetic amplifications impact extracellular vesicle miRNA cargo and their recruitment of nerves in head and neck cancer.
FASEB J, 38(13):e23803, 01 Jul 2024
Cited by: 0 articles | PMID: 38963404
MicroRNA biomarkers as next-generation diagnostic tools for neurodegenerative diseases: a comprehensive review.
Front Mol Neurosci, 17:1386735, 31 May 2024
Cited by: 0 articles | PMID: 38883980 | PMCID: PMC11177777
Review Free full text in Europe PMC
Go to all (879) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Myc-modulated miR-9 makes more metastases.
Nat Cell Biol, 12(3):209-211, 21 Feb 2010
Cited by: 81 articles | PMID: 20173743
MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas.
FASEB J, 25(12):4138-4149, 19 Aug 2011
Cited by: 54 articles | PMID: 21856782 | PMCID: PMC3236625
MicroRNA-506 inhibits tumor growth and metastasis in nasopharyngeal carcinoma through the inactivation of the Wnt/β-catenin signaling pathway by down-regulating LHX2.
J Exp Clin Cancer Res, 38(1):97, 21 Feb 2019
Cited by: 44 articles | PMID: 30791932 | PMCID: PMC6385449
MYCN, neuroblastoma and focal adhesion kinase (FAK).
Front Biosci (Elite Ed), 3(2):421-433, 01 Jan 2011
Cited by: 4 articles | PMID: 21196322 | PMCID: PMC3171213
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: K99 CA138572-01
Grant ID: K99 CA138572




