Abstract
Free full text

Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein
Abstract
The cell cycle-regulated expression of core histone genes is required for DNA replication and proper cell cycle progression in eukaryotic cells. Although some factors involved in histone gene transcription are known, the molecular mechanisms that ensure proper induction of histone gene expression during S phase remain enigmatic. Here we demonstrate that S-phase transcription of the model histone gene HTA1 in yeast is regulated by a novel attach–release mechanism involving phosphorylation of the conserved chromatin boundary protein Yta7 by both cyclin-dependent kinase 1 (Cdk1) and casein kinase 2 (CK2). Outside S phase, integrity of the AAA-ATPase domain is required for Yta7 boundary function, as defined by correct positioning of the histone chaperone Rtt106 and the chromatin remodeling complex RSC. Conversely, in S phase, Yta7 is hyperphosphorylated, causing its release from HTA1 chromatin and productive transcription. Most importantly, abrogation of Yta7 phosphorylation results in constitutive attachment of Yta7 to HTA1 chromatin, preventing efficient transcription post-recruitment of RNA polymerase II (RNAPII). Our study identified the chromatin boundary protein Yta7 as a key regulator that links S-phase kinases with RNAPII function at cell cycle-regulated histone gene promoters.
The budding yeast has been a productive model for exploring the temporal control of transcription, which is likely a universal feature of cell cycles, with clear transcriptional programs in yeast, bacteria, and metazoans (Laub et al. 2000; RJ Cho et al. 2001; Rustici et al. 2004; Oliva et al. 2005; Lu et al. 2007). Bursts of gene expression tend to be associated with major cell cycle transitions, which are governed by cyclin-dependent kinases (Cdks), whose activation requires interaction with regulatory subunits called cyclins (Morgan 1997; Bloom and Cross 2007). In yeast, Cdk1 (Cdc28) is entirely devoted to cell cycle control (Mendenhall and Hodge 1998; Enserink and Kolodner 2010) and regulates a variety of cellular processes, including transcription (Wittenberg and Reed 2005). Although decades of research have produced an increasingly detailed view of how cell cycle-specific transcriptional programs are regulated, clear gaps remain in our mechanistic understanding of cell cycle biology.
One important set of cell cycle-regulated genes encodes the core histones—small, basic proteins that, together with DNA, form the nucleosome. In proliferating cells, the vast majority of histones are synthesized in S phase in parallel with DNA replication (Gunjan et al. 2005). Core histone overexpression outside of S phase is toxic, and eukaryotic cells have evolved a variety of mechanisms that likely work in concert to maintain the delicate equilibrium between DNA and histone synthesis. Recently, we used a functional genomic approach to discover several new regulators of core histone transcription in yeast (Fillingham et al. 2009), including the histone H3–H4 chaperone Rtt106 (Huang et al. 2005) and Yta7, a protein previously functionally connected to barrier activity on chromatin (Tackett et al. 2005). We showed that deletion of RTT106 or YTA7 has opposing effects, causing increased or decreased transcription of HTA1, respectively. Localization of both Rtt106 and Yta7 to HTA1 chromatin depends on the HIR histone H3–H4 chaperone protein complex (Fillingham et al. 2009), which represses core histone transcription outside of S phase through a specific DNA sequence—the negative regulatory element (NEG)—found in promoters of three of the four core histone gene pairs (Osley et al. 1986; Osley and Lycan 1987; Green et al. 2005; Prochasson et al. 2005). Rtt106 functions downstream from both HIR and another H3–H4 histone chaperone, Asf1, and appears to assemble repressive chromatin at the HTA1 regulatory region (Fillingham et al. 2009). Two Swi2/Snf2 family chromatin remodeling complexes, SWI/SNF and RSC, are recruited to histone gene promoters in an Rtt106- and HIR-dependent manner (Dimova et al. 1999; Ng et al. 2002), suggesting a role for these complexes ensuring an appropriate chromatin context at histone loci (Ferreira et al. 2011).
Yta7 is an evolutionarily conserved protein that contains both a bromodomain and an AAA-ATPase domain (Gradolatto et al. 2009) and was first described as a protein with chromatin barrier function that modulates chromatin states at the silent mating locus HMR (Tackett et al. 2005). The noncanonical bromodomain of Yta7 is involved in histone binding, but unlike other bromodomains, binding occurs independently of lysine acetylation (Gradolatto et al. 2009). Elimination of the bromodomain does not completely abolish histone binding; instead, the region of Yta7 with highest affinity for histones lies in its N terminus, which contains the AAA-ATPase domain (Gradolatto et al. 2009). To date, no link between AAA-ATPase function and histone binding has been established.
Here we present the first detailed mechanistic analysis of the regulation of cell cycle-dependent histone gene transcription by the cell cycle machinery. We demonstrate that the boundary function of Yta7 at HTA1 requires its AAA-ATPase domain and is established throughout the cell cycle, except during S phase when Yta7 dissociates from chromatin. This barrier function is required to correctly position Rtt106 and the RSC complex at the HTA1 locus. The region surrounding the AAA-ATPase domain of Yta7 is hyperphosphorylated by both the S-phase-specific forms of Cdk1 and casein kinase 2 (CK2). Our molecular analysis shows that phosphorylation causes dissociation of Yta7 from HTA1 chromatin during S phase, which in turn is essential for promoter escape and elongation of RNA polymerase II (RNAPII) to ensure productive transcription of HTA1 and other histone genes. Taken together, our data directly link regulation by S-phase Cdk1 and CK2 to histone gene transcription and provide a mechanism explaining the timing of S-phase-specific transcription.
Results
Yta7 binding at the HTA1 locus is cell cycle-regulated
To understand how Yta7 might influence the timing of histone gene expression, we first assessed RNAPII dynamics at the HTA1 locus through the cell cycle. We used synchronized yeast cells and chromatin immunoprecipitation (ChIP) to assess association of the RNAPII subunit Rpb3-TAP with the HTA1 locus. Figure 1A shows the primer sets used for our ChIP analysis of HTA1 (Ng et al. 2002). We observed minimal recruitment of Rpb3-TAP to the NEG region (Fig. 1B, Rpb3-TAP panel, primer set “NEG”) through the cell cycle. Instead, we saw specific recruitment of Rpb3-TAP to the HTA1 promoter (Fig. 1B, Rpb3-TAP panel, primer set “PRO”) and coding regions (Fig. 1B, Rpb3-TAP panel, primer set “ORF”) during the time period corresponding to productive HTA1 transcription (Fig. 1B, HTA1 mRNA panel, late G1/S and S). This result suggests that cell cycle regulation of HTA1 transcription occurs at the level of RNAPII recruitment.
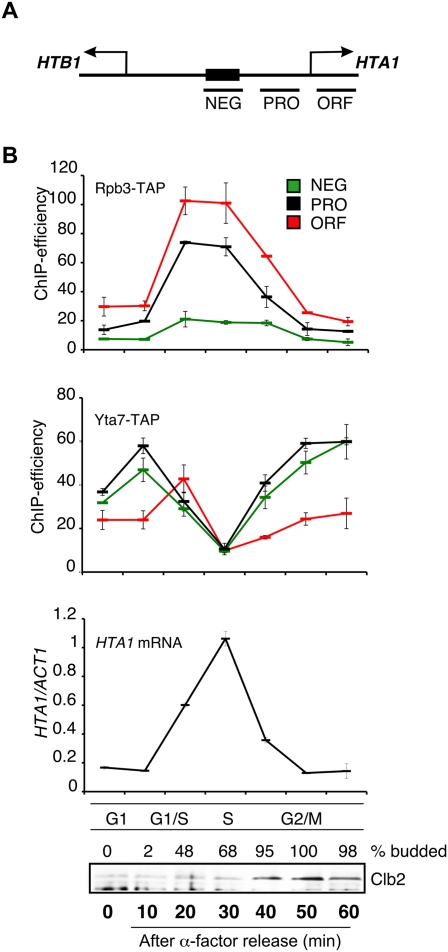
Yta7 and RNAPII binding to HTA1 chromatin is cell cycle-regulated. (A) Schematic representation of the PCR products (NEG, promoter [PRO], and ORF) used to cover the HTA1 locus in ChIP assays (Ng et al. 2002). (B) Rpb3 and Yta7 cross-link to HTA1 in a cell cycle-dependent manner. Rpb3-TAP and Yta7-TAP strains were arrested in G1 phase with 5 μM α-factor and released into fresh medium, and samples were taken at the indicated time points. IgG-sepharose ChIPs from the Rbp3-TAP or Yta7-TAP strains were analyzed for HTA1 (NEG, PRO, and ORF) by quantitative RT–PCR (qPCR), as indicated in the relevant panels. For each time point from the Yta7-TAP strain, cDNA was prepared and the ratio of HTA1 transcript to that of ACT1 was determined using qPCR (HTA1 mRNA panel). ChIP efficiency was calculated as described in the Materials and Methods. Cell cycle progression was monitored by assessing endogenous Clb2 cyclin levels (peak in G2/M phase) in the same samples by Western blotting with anti-Clb2 antibody and by quantification of budding (% budded). Similar results were seen for HTA1 transcript and Clb2 protein levels and budding indices in the Rpb3-TAP experiments (data not shown). Error bars in the experiments represent standard deviations from the mean for at least three replicate qPCR reactions.
It has been demonstrated previously that Yta7 localizes to the HTA1 promoter region in asynchronous cells (Gradolatto et al. 2008; Fillingham et al. 2009). Therefore, we next explored the relationship between Yta7, RNAPII binding, and cell cycle transcription. First, we used a ChIP assay with Yta7-TAP to discover that the proportion of Yta7 bound at HTA1 increased slightly in late G1, decreased significantly as cells progressed through S phase, and then increased again during G2/M (Fig. 1B, Yta7-TAP panel). Therefore, the presence of Yta7 at HTA1 correlates largely with gene repression, except during late G1 phase, when Yta7-TAP and Rbp3-TAP localization overlap, prior to the peak of histone gene expression (Fig. 1B).
Our ChIP experiments suggest a functional interaction between Yta7 and RNAPII, consistent with previous studies showing physical interactions on chromatin between Yta7 and RNAPII subunits (Tackett et al. 2005; Lambert et al. 2009). We confirmed these results by using either Western blotting (Fig. 2A) or mass spectrometry to identify proteins associated with Yta7 on chromatin (Fig. 2B; Lambert et al. 2009, 2010). Several of the most highly represented proteins associated with Yta7 on chromatin were known regulators of transcription: (1) Spt16 and Pob3, two subunits of the FACT complex, a histone chaperone protein complex that facilitates transcriptional elongation in a chromatin context (Krogan et al. 2002; Biswas et al. 2005) and is required for efficient HTB1 expression (Formosa et al. 2002); (2) Rtt106, a histone chaperone, and members of the RSC complex, a chromatin remodeler, both of which affect HTA1 transcription (Ng et al. 2002; Fillingham et al. 2009; Ferreira et al. 2011); (3) subunits of the CK2 complex, which has been linked to transcription elongation (Krogan et al. 2002); and (4) subunits of RNAPII as described previously.
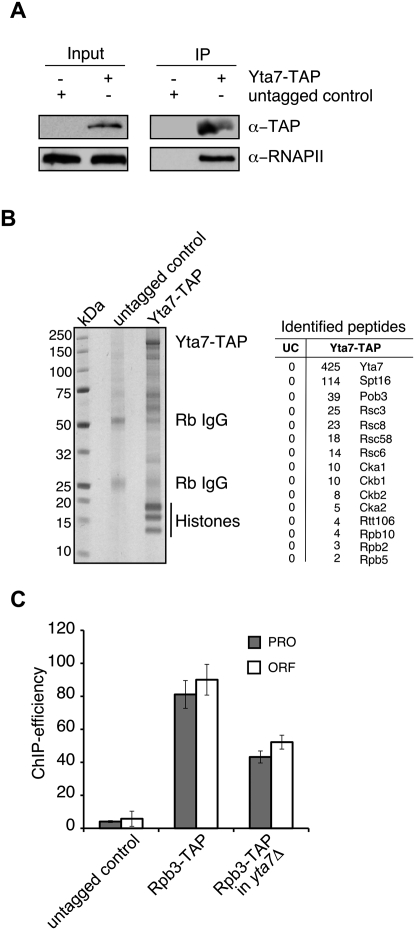
Yta7 functionally interacts with RNAPII. (A) Yta7 interacts with RNAPII. Western blotting was used to probe an mChIP (Lambert et al. 2009) from Yta7-TAP or untagged cells with an antibody that recognizes RNAPII. (B) Affinity purification and identification of Yta7-TAP-associated proteins. A Coomassie-stained SDS–polyacrylamide gel is shown with affinity-purified proteins from Yta7-TAP and an untagged wild-type control; (Rb IgG) rabbit IgG. Copurifying proteins were identified by LC-MS/MS as described previously (Lambert et al. 2009), and the numbers of the unique peptides for each interacting protein are listed; (UC) untagged control. (C) RNAPII association with HTA1 chromatin is partly dependent on Yta7. ChIP analyses from logarithmically growing cells were performed as described in Figure 1B using the indicated strains and primer sets (PRO and ORF). Error bars in the experiments represent standard deviations from the mean for at least three replicate qPCR reactions.
Finally, we tested RNAPII recruitment to HTA1 in the absence of YTA7. Association of Rbp3-TAP with both the PRO and ORF regions of HTA1 was reduced in a yta7Δ mutant (Fig. 2C), consistent with lower levels of HTA1 gene expression, as described previously (Fillingham et al. 2009). Cell cycle ChIP analyses revealed that RNAPII recruitment to both the PRO and ORF regions of the HTA1 promoter was reduced during G1 and early S phase in the yta7Δ mutant, whereas no RNAPII recruitment was observed at NEG in either the yta7Δ or wild-type strains (Supplemental Fig. 1). S-phase entry in yta7Δ was comparable with wild type, so the reduction of RNAPII recruitment is not due to a delay in cell cycle progression (data not shown). We conclude that Yta7 and RNAPII functionally interact and that Yta7 is important for efficient RNAPII recruitment during late G1 and early S phase to the HTA1 locus.
The AAA-ATPase domain is required for proper Yta7 function as a boundary protein
Our ChIP and protein interaction experiments reinforced our view that Yta7 functions as a critical element for efficient recruitment of RNAPII to HTA1 during S phase. Recognizable protein domains within Yta7 include a bromodomain and a canonical AAA-ATPase domain (Fig. 3A). We reasoned that the AAA-ATPase may be involved in HTA1 chromatin binding and/or boundary function. To test this idea, we mutated the conserved lysine residue in the Walker-A motif (GxxxxGKT) within the putative ATPase domain of Yta7 to alanine (yta7-K460A) and used several assays to examine how the boundary and chromatin-binding functions of Yta7were affected. First, we used our ChIP assay to examine specific localization of Yta7-K460A-TAP to HTA1 (Fig. 3B). Binding of the mutant version of Yta7 to the HTA1 locus was clearly detectable, although the pattern of chromatin association was altered such that more Yta7-K460A-TAP associated with the HTA1 ORF than was seen in wild-type cells (Fig. 3B). Importantly, dissociation of Yta7-K460A-TAP from the HTA1 locus during S phase was comparable with wild-type Yta7 (Fig. 3C). Next, we explored the effect of the AAA-ATPase domain mutation on the boundary function of Yta7 by assaying positioning of Rtt106 at the HTA1 locus. Similar to what we observed previously in yta7Δ (Fillingham et al. 2009), we found significantly increased association of Rtt106-TAP with all three regions of the HTA1 locus in a yta7-K460A background (Fig. 3D). Since Rtt106 is important for association of the RSC complex with HTA1 (Ferreira et al. 2011), and we detected subunit RSC in our Yta7 mChIP analysis (Fig. 2B), we also asked whether the ATPase domain of Yta7 influenced RSC localization to HTA1. Similar to Rtt106, Rsc8-TAP association with all three regions of the HTA1 locus was increased in the yta7-K460A mutant (Fig. 3E). Finally, we monitored HTA1 transcript levels in yta7-K460A mutants using quantitative RT–PCR (qPCR) and saw a reduction in HTA1 transcript levels comparable with that seen in yta7Δ mutants (Fig. 3F; Fillingham et al. 2009).
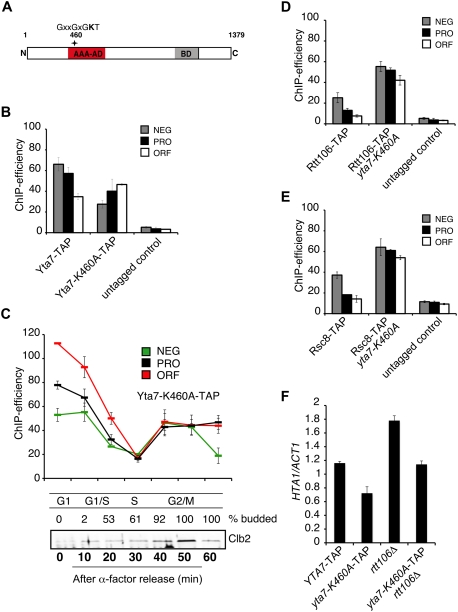
The AAA-ATPase domain is required for the boundary function of Yta7. (A) Schematic representation of full-length Yta7. The AAA-ATPase domain (AAA-AD) and bromodomain (BD) are depicted, and the putative catalytic active lysine residue (amino acid 460) is indicated by an asterisk. (B) Localization of Yta7-K460A at the HTA1 locus. ChIP analyses (primer set PRO, NEG, and ORF) with samples from log-phase Yta7-TAP and Yta7-K460A-TAP and an untagged wild-type strain as a negative control were performed as described in Figure 1B. (C) Localization of Yta7-K460A to the HTA1 locus throughout the cell cycle. Yta7-K460A-TAP mutants were arrested in G1 phase with 5 μM α-factor and released into fresh medium, and ChIP analyses were performed as in Figure 1B using primer sets NEG, PRO, and ORF. Cell cycle progression was monitored by quantification of budding (entry into S phase) and by assessing endogenous Clb2 cyclin levels (peak in G2/M phase). (D) Localization of Rtt106 to the HTA1 promoter is dependent on the AAA-ATPase site. ChIP analyses with Rtt106-TAP in wild-type and yta7-K460A backgrounds from log-phase cultures were performed as described in Figure 1B (primer sets NEG, PRO, and ORF). An untagged wild-type strain was used as a control. (E) Localization of RSC to the HTA1 promoter is dependent on the AAA-ATPase site in Yta7. ChIP analyses with Rsc8-TAP in wild-type and yta7-K460A strains from log-phase cultures were performed as described in Figure 1B (primer sets NEG, PRO, and ORF). An untagged wild-type strain was used as a control. (F) Mutation of Lys 460 results in reduced HTA1 transcription, which is suppressed by elimination of Rtt106. cDNA was prepared from log-phase YTA7-TAP wild type, yta7-K460A-TAP and rtt106Δ single mutants, and a yta7-K460A-TAP rtt106Δ double mutant, and HTA1 transcript levels were assessed as described in Figure 1B. Error bars in the experiments represent standard deviations from the mean for at least three replicate qPCR reactions.
We reasoned that reduced HTA1 transcript levels in the yta7-K460A mutant might reflect excessive Rtt106 (and RSC) attachment, which may generate repressive chromatin throughout the HTA1 locus and impede efficient transcription. To test this idea, we used qPCR to assess levels of HTA1 transcription in a yta7-K460A rtt106Δ double mutant. We found that the derepression of HTA1 transcription that occurs in an rtt106Δ strain (Fillingham et al. 2009) was suppressed in a yta7-K460A mutant background, with HTA1 transcription restored to wild-type levels (Fig. 3F). Consistent with this observation, we saw increased nucleosome occupancy at the promoter regions of HTA1–HTB1 and other core histone loci in an yta7-K460A mutant strain (Supplemental Fig. 6). Finally, cell cycle ChIP analyses revealed a decrease in RNAPII recruitment during G1 and early S phase in yta7-K460A (Supplemental Fig. 2), similar to what we observed in a yta7Δ mutant (Supplemental Fig. 1). S-phase entry in yta7-K460A was comparable with wild type, so the reduction of RNAPII recruitment is not due to a delay in cell cycle progression (data not shown). Together, these data suggest that the Yta7 AAA-ATPase domain, while not required for chromatin binding, is needed for correct positioning of Rtt106 and RSC, which is important for normal nucleosome positioning, RNAPII recruitment, and proper core histone gene expression.
Yta7 is phosphorylated by Cdk1 and CK2 during S phase
Our analysis of the yta7-K460A mutant implicated the AAA-ATPase domain in the function of Yta7 as a boundary protein. Importantly, like wild-type Yta7, Yta7-K460A was evicted from the HTA1 locus in S phase, suggesting an AAA-ATPase domain-independent mechanism for regulating Yta7 binding to chromatin during S phase. Prominent regulators of S-phase progression include S-phase-specific forms of Cdk1, and we therefore decided to explore possible regulation of Yta7 by Cdk1 and other kinases. Using mass spectrometry analyses, we and others discovered a number of phosphorylated residues concentrated within the N-terminal region of Yta7, extending into the AAA-ATPase domain (Fig. 4A; Chi et al. 2007; Li et al. 2007; Smolka et al. 2007; Albuquerque et al. 2008; Holt et al. 2009; Breitkreutz et al. 2010; data not shown). Seven of these phosphorylated residues occur within Cdk consensus sites (S/T-P-x-K/R or S/T-P), and six occur within CK2 consensus sites (S-xx-E/D) (Fig. 4A). Previous work has connected Cdk1 and Yta7: Yta7 is an in vitro substrate of Clb2–Cdk1 and Clb5–Cdk1 (Ubersax et al. 2003; Loog and Morgan 2005), and phosphorylation of Yta7 in vivo is dependent on Cdk1 (Holt et al. 2009). In addition, we also identified all four subunits of CK2 (Cka1, Cka2, Ckb1, and Ckb2) as physical interactors with Yta7 in mChIP analyses (Fig. 2B), consistent with a role for CK2 in Yta7 regulation.
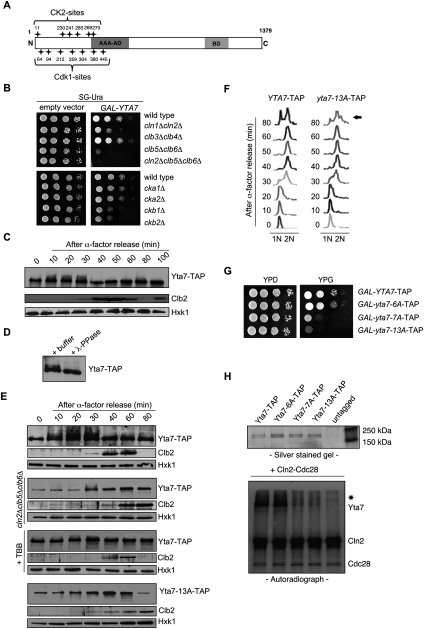
Yta7 is hyperphosphorylated by S-phase forms of Cdk1 and CK2. (A) Schematic representation of Cdk1 and CK2 phosphorylation sites in the N-terminal region of Yta7. (B) Growth defect caused by overexpression of YTA7 in the absence of Cdk1 G1–S-phase cyclins or CK2 subunits. Isogenic wild-type, cln1cln2, clb3clb4, clb5clb6, cln2clb5clb6, cka1, cka2, ckb1, or ckb2 deletion strains bearing either GAL-YTA7 (Hu et al. 2007) or empty vector were spotted in serial 10-fold dilutions on medium containing galactose and incubated for 2 d at 30°C. (C) Yta7 is phosphorylated in S phase. The phosphorylation of Yta7-TAP during a cell cycle was monitored by Western blotting using anti-TAP antibody. Cells were synchronized using 5 μM α-factor and released into fresh medium, and proteins extracts were prepared from samples taken at the indicated time points. Progression through the cell cycle was monitored by Western blotting for Clb2 (G2/M). Hxk1 was used as a gel loading control. (D) Mobility shift of Yta7 is due to phosphorylation. Yta7-TAP was isolated from wild-type cells in S phase and treated with λ-phosphatase (λ-PPase). (E) Phosphorylation of Yta7 is dependent on S-phase forms of Cdk1 and CK2 in vivo. (First panel) All experiments were performed under the same conditions and compared with wild type. (Second panel) Phosphoisoforms of Yta7-TAP were monitored during a cell cycle in a cln2Δclb5Δclb6Δ triple mutant exactly as described in C. (Third panel) Phosphorylation of Yta7-TAP after treatment with a CK2 inhibitor. Cells were synchronized as described in C and released into fresh medium supplemented with 100 mM CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) (Siepe and Jentsch 2009). Yta7-TAP, Clb2, and Hxt1 protein levels were assessed as described in C. (Fourth panel and F) Phenotypic assessment of a Yta7 phosphomutant. Phosphorylation of Yta7-13A-TAP was monitored during a cell cycle by Western blotting using anti-TAP antibody (Yta7-13A has all potential Cdk1 and CK2 sites [see A] converted to alanines). (Fourth panel) Samples were taken at the times indicated following α-factor block and release. (F) Corresponding FACS profiles indicate relative position in the cell cycle for the yta-13A-TAP strain and an isogenic wild-type control (YTA7-TAP). The arrow highlights the delayed mitosis in the yta7-13A mutant compared with wild type. (G) Growth defect caused by overexpression of yta7-13A. Wild-type, yta7-6A (lacking CK2 phospho-sites), yta7-7A (lacking Cdk1 phospho-sites) and yta7-13A (lacking both) strains in which the endogenous YTA7 promoter was replaced by the GAL1 promoter were spotted in serial 10-fold dilutions on glucose- or galactose-containing medium and incubated for 2 d at 30°C. (H) Phosphorylation of Yta7 by Cdk1 in vitro. (Top panel) Purification of Yta7-TAP, Yta7-6A-TAP, Yta7-7A-TAP, and Yta7-13A-TAP proteins from yeast was monitored by SDS-PAGE and silver staining. The purified proteins were incubated with Cln2–Cdk1 in kinase reactions along with [32P]-γ-ATP. (Bottom panel) Phosphorylation of proteins was analyzed by SDS-PAGE and autoradiography. The positions of migration of phosphorylated Yta7, Cln2, and Cdk1 are indicated. The asterisk marks the position of migration of a contaminant in the Cln2–Cdk1 preparation that is also phosphorylated in the reaction.
To determine whether Yta7 is regulated by Cdk1 and/or CK2 in vivo, we first performed a genetic test. We showed previously that overproduction of a kinase substrate in the absence of the kinase is often toxic, due to misregulation of the substrate, resulting in a so-called synthetic dosage lethal (SDL) interaction (Sopko et al. 2006; Huang et al. 2009). We therefore asked whether overexpressed YTA7 had an SDL interaction with mutation of either Cdk1 or CK2. Cdk1 is essential for cell viability and is activated sequentially by a series of cyclin subunits throughout the cell cycle (Bloom and Cross 2007). We overexpressed YTA7 in three double mutants lacking pairs of Cdk1 cyclins that function during all major cell cycle stages: cln1Δcln2Δ (deleted for late G1-phase cyclins), clb5Δclb6Δ (S-phase cyclins), and clb3Δclb4Δ (G2/M-phase cyclins). Overexpression of YTA7 in either wild-type cells or cells lacking the mitotic cyclins Clb3 and Clb4 caused a comparable mild slow growth phenotype (Fig. 4B) and resulted in moderate toxicity when the G1 cyclins Cln1 and Cln2 were absent. In contrast, overexpressed YTA7 was highly toxic in cells lacking the Cdk1 S-phase cyclins Clb5 and Clb6 and was lethal in a cln2Δclb5Δclb6Δ triple mutant (Fig. 4B). Overexpression of YTA7 in clb5Δclb6Δ cells was associated with an elongated bud phenotype typical of cells unable to transit G2/M (Supplemental Fig. 3A). The SDL phenotypes associated with YTA7 overexpression in cells lacking each of the four CK2 subunits were less dramatic, although a slow growth phenotype was clear when either of the two regulatory subunits of CK2 were deleted (Ckb1 or Ckb2) (Figure 4B). Our genetic tests are consistent with a significant overlapping function for G1–S-phase-specific forms of Cdk1 (Cln2, Clb5, and Clb6) in regulating Yta7 and suggest a role for CK2 as well.
We next examined Yta7 phosphorylation throughout the cell cycle (Fig. 4C) by analyzing Yta7 protein isolated from a synchronized strain expressing Yta7-TAP under control of its endogenous promoter. Slower-migrating forms of Yta7 appeared 10 min after release from an α-factor block, persisted until late S phase (30 min) (Figure 4C,F, left panel), and then decreased during G2 phase as levels of the mitotic cyclin Clb2 peaked (Fig. 4C,F). We note that Yta7 is a large protein, and only a subset of Yta7 is likely to be phosphorylated in vivo given the known association of Yta7 with many non-cell cycle-regulated promoters (Gradolatto et al. 2008). Although these properties make resolution of Yta7 isoforms difficult, we were able to consistently see collapse of Yta7 isoforms following treatment of extracts with phosphatase, indicating that the observed mobility shift was due to phosphorylation (Fig. 4D). We next asked whether the phospho-shifts of Yta7 were dependent on Cdk1 by assessing Yta7 isoforms in a synchronized cln2Δclb5Δclb6Δ mutant culture. We saw both reduced Yta7 isoforms and protein levels in the cln2Δclb5Δclb6Δ triple mutant compared with wild type (Fig. 4E, first and second panels), consistent with a role for Cdk1 in Yta7 phosphorylation. The reduced Yta7 protein levels may reflect a mechanism to prevent accumulation of unmodified Yta7, which our SDL experiments suggest is toxic (Fig. 4B). CK2 is an essential kinase, so we treated synchronized wild-type cells with the CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) to explore the effect on accumulation of Yta7 isoforms (Siepe and Jentsch 2009). Treatment of cells with CK2 inhibitor had a small effect on accumulation of Yta7 isoforms and no effect on protein levels (Fig. 4E, third panel). Taken together, these data are consistent with a prominent role for Cdk1 and a possible function for CK2 in Yta7 phosphorylation in vivo.
We sought to more directly test the in vivo consequences of a failure to phosphorylate Yta7. We used site-directed mutagenesis to generate versions of Yta7 with alanine substitutions in putative Cdk1 (yta7-7A), CK2 (yta7-6A), or both Cdk1 and CK2 phospho-sites (yta7-13A). We integrated all constructs at the YTA7 chromosomal locus and assessed Yta7 protein isoforms throughout the cell cycle. Although all mutant derivatives of Yta7 had comparable abundance, Yta7 phosphoforms were reduced in yta7-6A and yta7-7A mutants (data not shown) and virtually absent in extracts from the yta7-13A strain (Fig. 4E, fourth panel). When expressed at endogenous levels, the yta7-13A mutant had a minor growth defect (data not shown) and a prolonged G2/M phase (Fig. 4F), consistent with previous results showing that reduced histone levels produce a G2/M cell cycle delay (Han et al. 1987; Pinto and Winston 2000).
We predicted that expression of a hypophosphorylated form of Yta7 might mimic the effect we saw when we overexpressed wild-type YTA7 in strains compromised for G1/S Cdk1 activity. We therefore replaced the endogenous YTA7 promoter with the inducible GAL1 promoter in both a wild-type YTA7-TAP strain and the strains in which wild-type YTA7-TAP was replaced with the various YTA7 phosphomutants. Although galactose-induced overexpression of yta7-6A was of little phenotypic consequence, overexpression of yta7-7A led to a severe growth defect (Fig. 4G), and overexpression of the yta7-13A mutant was highly toxic. Morphological analysis and fluorescence-activated cell sorting (FACS) profiles showed that cells overexpressing yta7-13A accumulated in G2 phase, with a phenotype comparable with a clb5Δclb6Δ mutant (Supplemental Fig. 3A,B). Together, these data suggest that Cdk1- and CK2-dependent phosphorylation of Yta7 during late G1 and S phase is important for efficient progression through mitosis.
To corroborate our in vivo analyses, we next asked whether Yta7 is an in vitro substrate for Cdk1 and CK2 (Fig. 4H). Wild-type Yta7 and the Yta7-6A proteins were excellent substrates for Cln2–Cdk1, a G1-specific form of Cdk1 in vitro, while neither the Yta7-7A nor Yta7-13A proteins were phosphorylated above background levels (Fig. 4H). Since Yta7-7A and Yta7-13A both lack Cdk1 consensus sites, while Yta7-6A lacks only CK2 consensus sites, these data clearly demonstrate that Yta7 is an in vitro substrate for Cln2–Cdk1 and that the seven CDK sites are the major sites of phosphorylation. We also attempted comparable assays with recombinant CK2 as described previously (Siepe and Jentsch 2009) and detected phosphorylation of Yta7 in vitro, with a reduction in the mutants lacking CK2 sites (data not shown). Our results establish the importance of the N-terminal CDK sites for Cdk1-dependent phosphorylation in vitro and show that Yta7 can be also phosphorylated by CK2 in vitro, consistent with our in vivo analyses.
Phosphorylation-dependent dissociation of Yta7 from HTA1 chromatin during S phase ensures efficient transcription
Since Yta7 phosphorylation and dissociation from HTA1 chromatin occurred concurrently during S phase, we wondered whether these events might be interdependent. We first used ChIP to monitor Yta7-TAP association with the HTA1 regulatory region (NEG) throughout the cell cycle in strains compromised for Yta7 phosphorylation due to a lack of S-phase Cdk1 activity (clb5Δclb6Δ mutant) or through mutation of phosphorylation sites. Dissociation of Yta7-TAP from the HTA1 regulatory region (NEG) during S phase was reduced in the clb5Δclb6Δ mutant (Supplemental Fig. 4, top panel) and in strains harboring either the yta7-6A (lacking CK2 sites) or yta7-7A alleles (lacking Cdk1 sites), with an additive effect in a yta7-13A mutant (lacking both CK2 and Cdk1 sites) (Fig. 5A, top panel). A similar decrease in Yta7 association was seen at other regions of the HTA1 locus (PRO and ORF) (data not shown). Thus, phosphorylation of YTA7 appears to be required for S-phase dissociation of Yta7 from HTA1 chromatin.
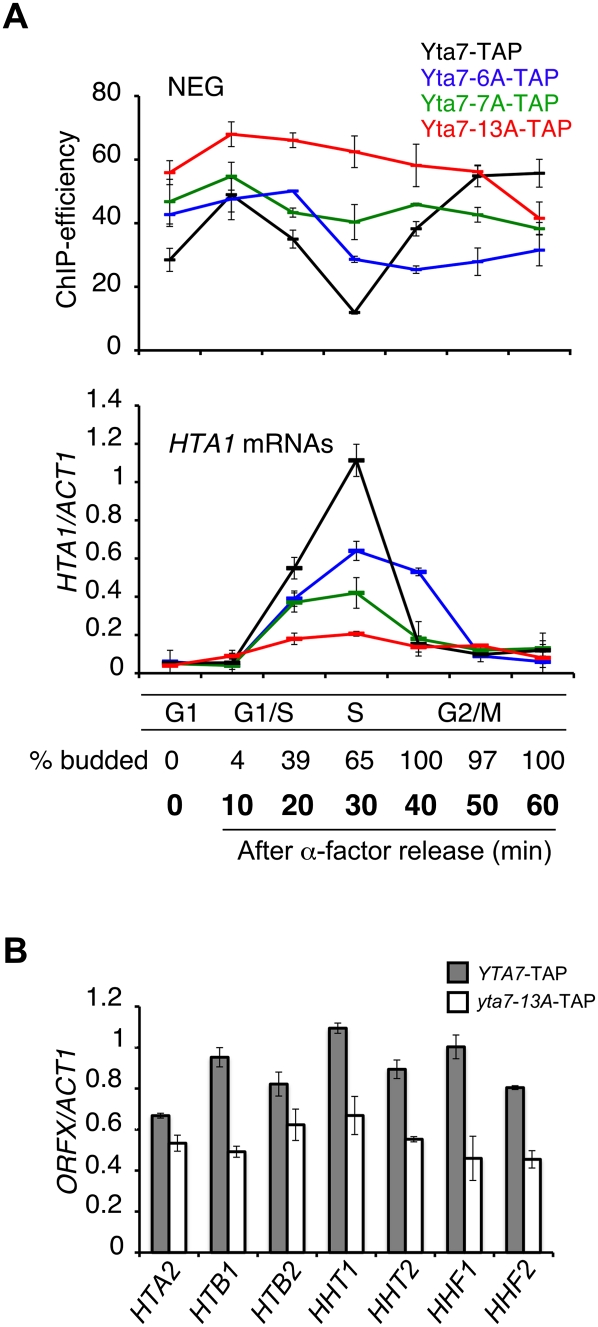
Dissociation of Yta7 from HTA1 chromatin ensures efficient transcription. (A) Phosphorylation of Yta7 regulates dissociation from HTA1 during S phase, which is essential for efficient HTA1 gene transcription. (Top panel) Yta7-TAP, Yta7-6A-TAP, Yta7-7A-TAP, and Yta7-13A-TAP strains were arrested with α-factor and released into fresh medium, and samples were taken at the time points indicated. ChIP analyses (primer set NEG) were performed as described in Figure 1B. (Bottom panel) S-phase induction of HTA1 transcription is markedly reduced in Yta7 phosphomutants. Samples of the same cultures used for the ChIP analyses were used for RNA isolation. cDNA was prepared from the samples, and HTA1 transcript levels were assessed as described in Figure 1B. Cell cycle progression was monitored by quantification of budding (entry into S phase). The budding indices for the Yta7-TAP strain are shown. S-phase entry was comparable for all strains (data not shown). (B) Analysis of transcript levels for other histone genes in a yta7 phosphomutant. YTA7-TAP and yta7-13A-TAP stains were arrested with α-factor and released into fresh medium, and samples were taken after 30 min (S phase). cDNA was prepared and analyzed as described in Figure 1B. Error bars in the experiments represent standard deviations from the mean for a least three replicate qPCR reactions.
In both the clb5Δclb6Δ mutant and the yta7-13A mutant, we also saw increased association of Yta7 with HTA1 chromatin (Fig. 5A, top panel; Supplemental Fig. 4, top panel). Consistent with increased binding of Yta7 to HTA1, particularly during S phase, we saw reduced HTA1 transcription in the clb5Δclb6Δ mutant (Supplemental Fig. 4, bottom panel) and the yta7-6A and yta7-7A mutants, and a near absence of S-phase induction of HTA1 transcription in the yta7-13A mutant (Fig. 5A, bottom panel). Transcription of other histone genes was also reduced in the yta7-13A mutant (Fig. 5B), with the smallest effect at the HTA2-HTB2 locus, consistent with previously demonstrated differences in Yta7 localization to HTA2–HTB2 (Gradolatto et al. 2008; Fillingham et al. 2009). Taken together, our data provide strong evidence that Yta7 phosphorylation by both Cdk1 and CK2 is required for S-phase dissociation from HTA1 chromatin, which is required for efficient transcription of HTA1 and other core histone genes.
Yta7 dissociation from chromatin may influence transcription elongation
We were interested in assessing the mechanistic significance of phosphorylation and timely dissociation of Yta7 from chromatin at HTA1. We reasoned that phosphorylation might be affecting several aspects of Yta7 function. First, a failure to phosphorylate Yta7 and ensure its appropriate dissociation from chromatin during S phase may reflect a defect in Rtt106 localization. For example, we showed that reduced expression of HTA1 in a yta7-K460A mutant was the consequence of excess Rtt106 (and RSC) throughout the HTA1 locus, including the ORF where they are normally not found (Fig. 3D,E). In this model (Fillingham et al. 2009), wild-type Yta7 effectively “activates” HTA1 by restricting the repressive function of Rtt106 to the regulatory region. We therefore assessed Rtt106-TAP localization in a yta7-13A strain and found a pattern of Rtt106-TAP at HTA1 that was comparable with wild type (Fig. 6A). Therefore, unlike yta7-K460A, the defect in HTA1 expression in the yta7-13A mutant (Fig. 5A, bottom panel) is not likely a consequence of altered Rtt106 localization.
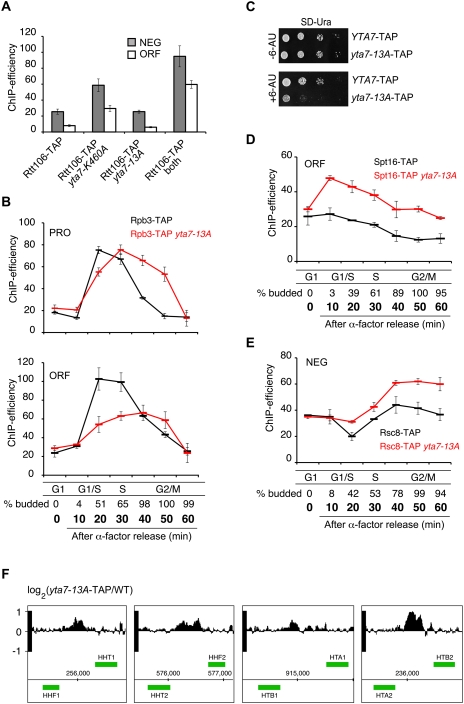
Phosphorylation of Yta7 is involved in transcript elongation by RNAPII. (A) Localization of Rtt106 in Yta7 mutants. Rtt106 localization to the HTA1 regulatory region (NEG) and ORF was assessed using ChIP in samples from asynchronous cultures of Rtt106-TAP strains harboring wild-type YTA7, yta7-K460A, yta7-13A, or both yta7-K460A and yta7-13A. ChIP analyses were performed as described in Figure 1B. (B) Analysis of RNAPII association with HTA1 during the cell cycle in a yta7-13A mutant. Rpb3-TAP localization to the promoter (primer set “PRO”) (top panel) and the ORF (primer set “ORF”) (bottom panel) of HTA1 was assayed in the indicated strains using qPCR as described (Fig. 1B). Cell cycle progression was monitored by quantification of budding (entry into S phase). Budding indices for the Rpb3-TAP strain are shown. S-phase entry was comparable for both strains (data not shown). (C) Sensitivity of yta7-13A to 6-azauracil. YTA7 and yta7-13A strains transformed with vector pRS-316 (for URA+ selection) were spotted in serial 10-fold dilutions on SD-URA plates with or without 6-azauracil (75 μg/mL) and incubated for 2 d at 30°C. (D) Increased localization of FACT to the HTA1 coding region in a yta7-13A strain during the cell cycle. Spt16-TAP and Spt16-TAP in a yta7-13A background were arrested in G1 phase with 5 μM α-factor and then released into fresh medium. ChIP samples were analyzed as described in Figure 1B (primer set ORF). Cell cycle progression was monitored by quantification of budding (entry into S phase). Budding indices for a Spt16-TAP strain are shown. S-phase entry was comparable for both strains (data not shown). (E) Increased localization of RSC to the HTA1 NEG region in a yta7-13A strain. Rsc8-TAP and Rsc8-TAP in yta7-13A were treated exactly as in D. ChIP samples were analyzed using primer set PRO. (F) Altered nucleosome occupancy in a yta7-13A mutant. Intergenic regions of all histone promoters are shown. A genome-wide nucleosome positioning assay was performed to identify regions of increased nucleosome occupancy in yta7-13A. Each track shows the change in nucleosome occupancy between mutant and wild type across the histone gene loci, expressed as the normalized log2 ratio of probe intensities.
Next, to determine whether there is any relationship between phosphorylation of the Yta7 N terminus and its AAA-ATPase function, we assessed Rtt106-TAP localization in an yta7-13A yta7K460A double mutant. We found significant Rtt106-TAP mislocalization throughout the HTA1 locus in the double mutant (Fig. 6A), even greater than in a yta7-K460A single mutant. Therefore, the function of Yta7 in Rtt106 localization appears unrelated to its phosphorylation and is instead dependent on its AAA-ATPase domain.
Because Yta7 functionally interacts with RNAPII, we next asked whether phosphorylation of Yta7 influenced recruitment of RNAPII to the HTA1 promoter. To test this possibility, we used ChIP to examine RNAPII recruitment to the HTA1 promoter in a yta7-13A mutant strain. The peak of Rpb3-TAP association with HTA1 during S phase was somewhat broader in the yta7-13A mutant than in wild type, but the pattern of Rpb3-TAP was not significantly different (Fig. 6B, primer set “PRO”). Therefore, the dramatic HTA1 expression defect in yta7-13A (Fig. 5A, bottom panel) cannot be explained by defective RNAPII recruitment, consistent with our demonstration that Yta7 is important for effective RNAPII localization to the HTA1 locus (Fig. 2C). Given our failure to discover a defect in RNAPII recruitment or Rtt106 spreading in the yta7-13A mutant, we next explored the possibility that Yta7 phosphorylation may be influencing transcriptional elongation by first assessing Rpb3-TAP recruitment to the coding region of HTA1. Significantly, we observed a clear decrease in RNAPII recruitment to the ORF of HTA1 during S phase (Fig. 6B, primer set “ORF”), indicative of a possible defect in transcriptional elongation. Consistent with a defect in transcriptional elongation, the yta7-13A mutant was highly sensitive to 6-azauracil (6-AU), which impairs transcriptional elongation by decreasing intracellular pools of UTP and GTP (Fig. 6C).
Our mass spectrometry experiments revealed a physical interaction between Yta7 and both subunits of the FACT complex (Spt16 and Pob3) (Fig. 2B), a histone chaperone that facilitates elongation by RNAPII by first destabilizing nucleosomes and evicting histones H2A and H2B from DNA, and then reforming chromatin after RNAPII has passed through a region (Orphanides et al. 1998, 1999; Belotserkovskaya et al. 2003; Mason and Struhl 2003; Schwabish and Struhl 2004). We previously localized FACT subunit Spt16 to the HTA1 coding region in asynchronous cells (Fillingham et al. 2009). Interestingly, we observed increased association of FACT with the HTA1 coding region in the yta7-13A mutant throughout the cell cycle (Fig. 6D), suggesting that, counter to its canonical role as a factor that stimulates transcriptional elongation, FACT interferes with efficient transcription of HTA1. Consistent with this hypothesis, reduction of FACT function using temperature-sensitive alleles of SPT16 (spt16-ts and spt16-197) resulted in elevated HTA1 transcript levels and nucleosome depletion over core histone promoters (Supplemental Fig. 6; data not shown). Thus, FACT may be antagonizing transcription throughout the HTA1 locus by contributing to a repressive chromatin structure.
Since Yta7 is known to influence RSC association with the HTA1 locus (Fig. 3E), and RSC dissociates from the HTA1 regulatory region in S phase (Ng et al. 2002), we next asked whether the presence of RSC at the HTA1 locus was influenced by Yta7 phosphorylation by comparing association of Rsc8-TAP with HTA1 in both wild type and yta7-13A. We saw increased Rsc8-TAP association with the HTA1 regulatory region throughout the cell cycle in the yta7-13A mutant strain (Fig. 6E), suggesting a role for RSC in generating repressive chromatin in the absence of proper Yta7 phosphorylation. Consistent with this idea, we observed a clear increase in nucleosome occupancy at all histone promoters in the yta7-13A mutant strain (Fig. 6F). Taken together, the results of our detailed phenotypic assessment of the yta7-13A mutant suggest that S-phase-specific phosphorylation of Yta7 is required for its removal from HTA1 chromatin in order to ensure efficient transcriptional elongation by RNAPII.
Discussion
Cell cycle-regulated expression of core histone genes is critical for maintaining the delicate balance between histone and DNA synthesis that is required for proper genome replication and cell division. Although a number of factors involved in histone transcription have been identified, molecular mechanisms linking histone gene expression to cell cycle regulators have remained obscure. Here we provide substantial evidence that the conserved AAA-ATPase domain-containing chromatin boundary protein Yta7 is a master regulator of core histone gene transcription and a conduit for signals from cell cycle kinases to the transcription machinery (Fig. 7): (1) Yta7 and subunits of RNAPII functionally interact, and both are recruited to the core histone HTA1 locus in a cell cycle-dependent manner. (2) Integrity of the AAA-ATPase domain in Yta7 is important for boundary function, which is necessary for appropriate repression of histone gene expression outside of S phase. (3) Phosphorylation of the N terminus of Yta7 by G1–S-phase-specific forms of Cdk1 and by CK2 inhibits chromatin binding of Yta7 at HTA1, causing its dissociation. (4) Dissociation of Yta7 from chromatin is necessary for RNAPII promoter escape and HTA1 transcript elongation, which in turn is essential for proper cell cycle progression. We propose that the Yta7 boundary protein temporally coordinates mutually exclusive domains of either repressed or accessible chromatin, ensuring appropriate cell cycle regulation of histone gene expression.
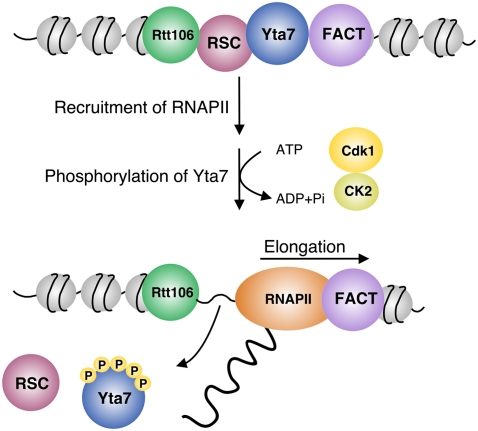
Summary of Yta7 action and regulation. Yta7 acts as a boundary element during early G1, G2, and M phases. In S phase, Yta7 is phosphorylated by Cdk1 and CK2, which in turn is important for effective elongation of RNAPII along the HTA1 gene. See the text for details.
How does Yta7 function as a boundary protein at HTA1? We showed that Yta7 boundary function, as defined by its regulation of Rtt106 localization, is dependent on its canonical AAA-ATPase domain. The AAA-ATPase family is composed of a large and functionally diverse group of proteins that hydrolyzes ATP to induce conformational changes in substrate proteins (Hanson and Whiteheart 2005). Mutation of the AAA-ATPase domain of Yta7 caused spreading of the Rtt106 histone chaperone and the RSC complex through the HTA1 coding region, a phenomenon that we also observed for Rtt106 in a yta7 deletion strain (Fillingham et al. 2009). These and other results suggest that the AAA-ATPase domain of Yta7 is required for its function as a boundary protein, preventing the ectopic presence of Rtt106, RSC, and associated repressive chromatin through the coding region of HTA1. Consistent with this idea, decreased HTA1 transcription in the yta7-K460A mutant was suppressed by eliminating Rtt106 (Fig. 3E).
As noted earlier, a recent study affirmed the important role for Rtt106 at the histone loci through all stages of the cell cycle via its recruitment of two chromatin remodeling complexes: SWI/SNF and RSC (Ferreira et al. 2011). Whereas SWI/SNF is required for histone gene activation during S phase (Dimova et al. 1999; Xu et al. 2005; Ferreira et al. 2011), RSC is implicated in histone gene repression outside of S phase. Rtt106 also functions upstream of SIR-based silencing at telomeric heterchromatin (Huang et al. 2007), but repression of HTA1 is unlikely to be SIR-dependent, since we observed no recruitment of Sir3-TAP to HTA1 chromatin (Supplemental Fig. 5). These data and our new observations illuminate a possible mechanism of Rtt106-mediated silencing of histone gene expression. We propose that Rtt106 recruits RSC outside of S phase to help position nucleosomes over histone gene promoters, blocking recruitment of RNAPII. This may occur indirectly via occlusion of TFIID binding to the TATA-box (recruitment of TBP to the HTA1 promoter happens in a cell cycle-dependent manner [J Fillingham and J Greenblatt, unpubl.]). Activation of histone gene expression in late G1/S then occurs via Rtt106-mediated recruitment of SWI/SNF, which removes nucleosomes over the promoter, permitting recruitment of RNAPII.
The presence of Yta7 at histone genes is correlated with periods of the cell cycle outside of S phase when histone gene expression is repressed. This result suggests that the boundary activity of Yta7 must be relieved during G1–S phase in order for histone gene expression to be induced. We made several observations that implicate G1–S-phase-specific forms of Cdk1 and CK2 in Yta7 regulation, allowing the timely activation of histone gene expression: (1) Yta7 genetically interacts with G1–S-phase-specific Cdk1 cyclins and with CK2 and physically interacts with all four subunits of CK2. (2) Yta7 is phosphorylated in a cell cycle-dependent manner, with peak phosphorylation during the G1–S-phase transition, and phosphorylation is reduced in mutants compromised for Cdk1 or CK2 activity. (3) Mutant versions of Yta7 lacking potential Cdk1 and CK2 phosphorylation (yta7-6A, yta7-7A, and yta7-13A) sites do not dissociate properly from the HTA1 promoter and prevent induction of histone gene expression during S phase. (4) Yta7 is an in vitro substrate for both Cdk1 and CK2. These and other observations lead us to propose that S-phase phosphorylation of Yta7 by Cdk1 and Ck2 is required to eject Yta7 from chromatin, relieving boundary activity and stimulating expression of HTA1.
Our results further suggest that the phosphorylation and eviction of Yta7 does not reflect a role for Yta7 in RNAPII recruitment to the HTA1 promoter; indeed, Yta7 appears to have a stimulatory role in RNAPII recruitment, and Yta7 and RNAPII can be copurified on chromatin (Fig. 2A,B). Instead, our data suggest that the sustained presence of Yta7 throughout S phase at histone promoters may interfere with elongation through improper recruitment of the FACT and RSC complexes to HTA1 chromatin. The FACT complex physically interacts with Yta7 on chromatin (Fig. 2B) and localizes to the HTA1 coding region (Fig. 6D; Fillingham et al. 2009). We observed increased FACT localization to the HTA1 ORF in a yta7-13A mutant (Fig. 6D), suggesting that phosphorylation of Yta7 may permit release of the Yta7–FACT complex, allowing efficient RNAPII elongation through HTA1 (Fig. 7). Like FACT, the RSC complex also physically interacts with Yta7 on chromatin (Fig. 2B), but, unlike FACT, it localizes to the HTA1 regulatory region. We saw increased RSC localization to the HTA1 regulatory region in a yta7-13A mutant during S phase when it is not normally present. We therefore propose that S-phase-specific transcription of histone genes reflects the presence of repressive chromatin at histone loci that is maintained by FACT and RSC outside of S phase, which interferes with efficient transcription by RNAPII.
Cdk1 is the major Cdk in yeast and regulates numerous cellular processes, including transcription. Although a connection between S-phase Cdk1 activity and histone gene transcription seems intuitive, to date, no protein involved in histone transcription has been identified as a Cdk1 substrate in yeast. In contrast, at least some activators of histone gene transcription are phosphorylated and activated by CDKs in mammals. In human cells, cyclin E–CDK2 colocalizes with the NPAT proteins to histone gene clusters in subnuclear Cajal bodies (Ma et al. 2000; Zhao et al. 2000). Notably, phosphorylation of NPAT by cyclin E–CDK2 activates histone gene transcription at the G1/S transition (Zhao et al. 1998, 2000; Ma et al. 2000; Ye et al. 2003). However, the underlying molecular mechanisms remain to be identified.
CK2 is a multifunctional kinase with known roles in transcription and RNA processing. In higher eukaryotes, one-third of CK2 substrates are involved in gene expression; half of those are transcription factors (Meggio and Pinna 2003). In yeast, CK2 physically interacts with the PAF and FACT complexes, both involved in RNAPII transcript elongation (Gavin et al. 2006; Krogan et al. 2006). Also, the RNAPII elongation factors Spt4 and Spt5, which also localize to HTA1 (Supplemental Fig. 5), are phosphorylated by CK2 in vivo (Krogan et al. 2002). However, the functional consequences of these phosphorylation events remain unclear. More recently it was shown that phosphorylation of the SR-like protein Npl3 by CK2 influences RNAPII-dependent elongation (Dermody et al. 2008). Although CK2 activity is not known to be cell cycle-regulated, our data suggest that phosphorylation of Yta7 by CK2 may be an important facet of regulation of transcript elongation by CK2, perhaps following a priming event involving phosphorylation of Yta7 by Cdk1.
Our study provides a detailed analysis of regulation of cell cycle-dependent histone gene transcription through phosphorylation of the chromatin boundary element Yta7 by S forms of Cdk1 and CK2. As overexpression of a yta7 phosphomutant is highly toxic and Yta7 localizes also to numerous other loci (Gradolatto et al. 2008), we propose that S-phase-specific regulation of Yta7 by Cdk1 and CK2 may be a mechanism that is broadly applied throughout the genome to regulate gene transcription. Interestingly, the human homolog of Yta7, ATAD2, is involved in chromatin dynamics and transcriptional activities and is up-regulated predominantly in G1/S-phase cells, consistent with a possible role in DNA replication and histone synthesis (Ciro et al. 2009; Caron et al. 2010). Like Yta7, ATAD2 is phosphorylated at CK and CDK consensus sites (Gnad et al. 2007), and overexpression of ATAD2 strongly correlates with poor prognosis and rapid mortality in lung and breast cancer patients (Caron et al. 2010; Kalashnikova et al. 2010). We suggest that further analysis of the Yta7 pathway in yeast will shed light on regulation by ATAD2 in mammalian cells and illuminate conserved pathways controlling regulatory chromatin domains throughout the eukaryotic genome.
Materials and methods
Yeast strains and plasmids
Yeast strains are listed in Supplemental Table 1. Strains were generated using standard yeast medium and genetic techniques. To generate yta7-K460A, yta7-6A, yta7-7A, yta7-13A, and yta7-13A-K460A mutants, the URA3-cassette was amplified from pFA6a-URA3 and integrated into the YTA7 locus to replace the N-terminal part of the gene. The N-terminal part of Yta7 was subcloned into a TOPO vector (Invitrogen), and site-directed mutagenesis was performed using QuikChange site-directed mutagenesis kit (Agilent). All primers are listed in Supplemental Table 2. Mutated versions of YTA7 were reintroduced into the chromosome, and positive clones were selected using replica plating on 5-FOA plates. Correct integration was verified by colony PCR and sequencing, and protein expression was tested by Western blotting.
ChIP
Soluble chromatin was prepared from cells treated with formaldehyde (1% final concentration) and immunoprecipitated using standard procedures (Kim et al. 2004). Chromatin from TAP-tagged strains was incubated overnight at 4°C with IgG-sepharose (Amersham Biosciences). Immunoprecipitated DNA was analyzed by qPCR, always including an internal control (a nontranscribed region of chromosome V). ChIP efficiency was calculated as the ratio of specific primer/control for immunoprecipitation divided by the same ratio for input (Kim et al. 2004). qPCR reactions were performed using the DyNAmo Flash SYBR Green qPCR kit (Thermo Scientific) on a 7500 Real-Time PCR block (Applied Biosystems). Primer annealing was 35 sec at 50°C and extension was 35 sec at 72°C.
Protein analysis and immunoblotting
Extracts for immunoblotting were prepared from pelleted cells that were incubated in 0.5 mL of breaking solution (0.2 M NaOH, 0.2% β-mercaptoethanol) for 10 min on ice. Proteins were precipitated with 5% trichloracetic acid (final concentration), and after an additional 10 min on ice, extracts were centrifuged in a microfuge at maximum speed for 5 min. Pellets were resuspended in 1× SDS loading buffer and incubated for 5 min at 95°C, and proteins were separated on 7.5% SDS–polyacryamide gels. After electrotransfer, nitrocellulose membranes (Bio-Rad) were blocked for 1 h in blocking solution containing 5% dry milk powder in TBST (10 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20 at pH 8.0). Antibodies used for immunoblotting were anti-Clb2 (Santa Cruz Biotechnology), anti-yeast hexokinase (Rockland), peroxidase anti-peroxidase (Sigma), and anti-RNAPII 8WG16 (EJ Cho et al. 2001).
Cell cycle synchronization by α-factor and FACS
Cells were grown to early log-phase (3 h) and arrested in G1 by α-factor treatment (5 μM) for 2.5 h at 30°C. Cells were washed twice with cold YP and released into prewarmed YPD without α-factor to resume cell cycle progression. At the indicated time points, 1-mL aliquots of cell culture were taken, fixed in 70% ethanol, and treated with 10 mg/mL RNaseA in 50 mM Tris-HCl (pH 8.0) for 3 h at 37°C. After resuspension in 50 mM Tris-HCl (pH 7.5), Proteinase K (2 mg/mL) was added and cells were further incubated for 60 min at 50°C. DNA was stained with 1 mM SYTOX Green (Invitrogen) in 50 mM Tris-HCl (pH 7.5), sonicated at low intensity, and scanned in a Guava Easycyte FACS from Guava Technologies using Flow Jo software (Flow Jo, LLC). λ-Phosphatase (λ-PPase) treatment (New England Biolabs) was performed as described previously (Kurat et al. 2009).
qPCR analysis of histone gene expression
RNA was prepared using RNeasy minikit (Qiagen). QuantiTect Reverse Transcription kit (Qiagen) was used to eliminate genomic DNA and synthesize cDNA from ~1 μg of RNA. qPCR reaction were performed as described above. To distinguish between similar copies of histone genes, reverse primers annealing to the 3′ untranslated region (UTR) of histone mRNA were used. All primers are listed in Supplemental Table 2.
Purification of TAP-tagged Yta7
The protocol can be found in the Supplemental Material.
In vitro kinase assays
Recombinant Cln2–Cdk1 kinase complex was expressed and purified from insect cells as described (Nash et al. 2001). Substrates including all versions of Yta7 (Yta7-TAP, Yta7-6A-TAP, Yta7-7A-TAP, and Yta7-13A-TAP) were expressed in yeast under control of the endogenous promoter and purified as described above. Kinase reactions were performed as described (Costanzo et al. 2004).
Acknowledgments
We thank Jack Greenblatt for support of J.F.'s work during the early stages of the project and for productive discussions, and Julia Petschnigg for helpful advice. S.K. was the recipient of an Ontario Graduate Scholarship, and C.F.K. holds an EMBO long-term fellowship. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to B.J.A. and Tim Hughes (BMB-210972), and a startup grant from Ryerson University and an NSERC Discovery grant to J.F.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.173427.111.
References
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7: 1389–1396 [Europe PMC free article] [Abstract] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [Abstract] [Google Scholar]
- Biswas D, Yu Y, Prall M, Formosa T, Stillman DJ 2005. The yeast FACT complex has a role in transcriptional initiation. Mol Cell Biol 25: 5812–5822 [Europe PMC free article] [Abstract] [Google Scholar]
- Bloom J, Cross FR 2007. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol 8: 149–160 [Abstract] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046 [Abstract] [Google Scholar]
- Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E, et al. 2010. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene 29: 5171–5181 [Abstract] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci 104: 2193–2198 [Europe PMC free article] [Abstract] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev 15: 3319–3329 [Europe PMC free article] [Abstract] [Google Scholar]
- Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ 2001. Transcriptional regulation and function during the human cell cycle. Nat Genet 27: 48–54 [Abstract] [Google Scholar]
- Ciro M, Prosperini E, Quarto M, Grazini U, Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M, Christensen J, et al. 2009. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res 69: 8491–8498 [Abstract] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913 [Abstract] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME 2008. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE 3: e3273 10.1371/journal.pone.0003273 [Europe PMC free article] [Abstract] [Google Scholar]
- Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell 4: 75–83 [Abstract] [Google Scholar]
- Enserink JM, Kolodner RD 2010. An overview of Cdk1-controlled targets and processes. Cell Div 5: 11 10.1186/1747-1028-5-11 [Europe PMC free article] [Abstract] [Google Scholar]
- Ferreira ME, Flaherty K, Prochasson P 2011. The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. PLoS ONE 6: e21113 10.1371/journal/pone.0021113 [Europe PMC free article] [Abstract] [Google Scholar]
- Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Pena-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, et al. 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol Cell 35: 340–351 [Abstract] [Google Scholar]
- Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: Polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571 [Europe PMC free article] [Abstract] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [Abstract] [Google Scholar]
- Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M 2007. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol 8: R250 10.1186/gb-2007-8-11-r250 [Europe PMC free article] [Abstract] [Google Scholar]
- Gradolatto A, Rogers RS, Lavender H, Taverna SD, Allis CD, Aitchison JD, Tackett AJ 2008. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179: 291–304 [Europe PMC free article] [Abstract] [Google Scholar]
- Gradolatto A, Smart SK, Byrum S, Blair LP, Rogers RS, Kolar EA, Lavender H, Larson SK, Aitchison JD, Taverna SD, et al. 2009. A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol Cell Biol 29: 4604–4611 [Europe PMC free article] [Abstract] [Google Scholar]
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR 3rd, Kaufman PD 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol 15: 2044–2049 [Europe PMC free article] [Abstract] [Google Scholar]
- Gunjan A, Paik J, Verreault A 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie 87: 625–635 [Abstract] [Google Scholar]
- Han M, Chang M, Kim UJ, Grunstein M 1987. Histone H2B repression causes cell-cycle-specific arrest in yeast: Effects on chromosomal segregation, replication, and transcription. Cell 48: 589–597 [Abstract] [Google Scholar]
- Hanson PI, Whiteheart SW 2005. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol 6: 519–529 [Abstract] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686 [Europe PMC free article] [Abstract] [Google Scholar]
- Hu Y, Rolfs A, Bhullar B, Murthy TV, Zhu C, Berger MF, Camargo AA, Kelley F, McCarron S, Jepson D, et al. 2007. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res 17: 536–543 [Europe PMC free article] [Abstract] [Google Scholar]
- Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z 2005. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc Natl Acad Sci 102: 13410–13415 [Europe PMC free article] [Abstract] [Google Scholar]
- Huang S, Zhou H, Tarara J, Zhang Z 2007. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J 26: 2274–2283 [Europe PMC free article] [Abstract] [Google Scholar]
- Huang D, Kaluarachchi S, van Dyk D, Friesen H, Sopko R, Ye W, Bastajian N, Moffat J, Sassi H, Costanzo M, et al. 2009. Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast. PLoS Biol 7: e1000188 10.1371/journal.pbio.1000188 [Europe PMC free article] [Abstract] [Google Scholar]
- Kalashnikova EV, Revenko AS, Gemo AT, Andrews NP, Tepper CG, Zou JX, Cardiff RD, Borowsky AD, Chen HW 2010. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res 70: 9402–9412 [Europe PMC free article] [Abstract] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [Europe PMC free article] [Abstract] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [Europe PMC free article] [Abstract] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [Abstract] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD 2009. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell 33: 53–63 [Abstract] [Google Scholar]
- Lambert JP, Mitchell L, Rudner A, Baetz K, Figeys D 2009. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol Cell Proteomics 8: 870–882 [Europe PMC free article] [Abstract] [Google Scholar]
- Lambert JP, Fillingham J, Siahbazi M, Greenblatt J, Baetz K, Figeys D 2010. Defining the budding yeast chromatin-associated interactome. Mol Syst Biol 6: 448 10,1038/msb.2010.104 [Europe PMC free article] [Abstract] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148 [Abstract] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP 2007. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J Proteome Res 6: 1190–1197 [Abstract] [Google Scholar]
- Loog M, Morgan DO 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434: 104–108 [Abstract] [Google Scholar]
- Lu Y, Mahony S, Benos PV, Rosenfeld R, Simon I, Breeden LL, Bar-Joseph Z 2007. Combined analysis reveals a core set of cycling genes. Genome Biol 8: R146 10.1186/gb-2007-8-7-r146 [Europe PMC free article] [Abstract] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev 14: 2298–2313 [Europe PMC free article] [Abstract] [Google Scholar]
- Mason PB, Struhl K 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 23: 8323–8333 [Europe PMC free article] [Abstract] [Google Scholar]
- Meggio F, Pinna LA 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J 17: 349–368 [Abstract] [Google Scholar]
- Mendenhall MD, Hodge AE 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1191–1243 [Europe PMC free article] [Abstract] [Google Scholar]
- Morgan DO 1997. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13: 261–291 [Abstract] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [Abstract] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16: 806–819 [Europe PMC free article] [Abstract] [Google Scholar]
- Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J 2005. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol 3: e225 10.1371/journal.pbio.0030225 [Europe PMC free article] [Abstract] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [Abstract] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288 [Abstract] [Google Scholar]
- Osley MA, Lycan D 1987. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol 7: 4204–4210 [Europe PMC free article] [Abstract] [Google Scholar]
- Osley MA, Gould J, Kim S, Kane MY, Hereford L 1986. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell 45: 537–544 [Abstract] [Google Scholar]
- Pinto I, Winston F 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J 19: 1598–1612 [Europe PMC free article] [Abstract] [Google Scholar]
- Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev 19: 2534–2539 [Europe PMC free article] [Abstract] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J 2004. Periodic gene expression program of the fission yeast cell cycle. Nat Genet 36: 809–817 [Abstract] [Google Scholar]
- Schwabish MA, Struhl K 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117 [Europe PMC free article] [Abstract] [Google Scholar]
- Siepe D, Jentsch S 2009. Prolyl isomerase Pin1 acts as a switch to control the degree of substrate ubiquitylation. Nat Cell Biol 11: 967–972 [Abstract] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci 104: 10364–10369 [Europe PMC free article] [Abstract] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21: 319–330 [Abstract] [Google Scholar]
- Tackett AJ, Dilworth DJ, Davey MJ, O'Donnell M, Aitchison JD, Rout MP, Chait BT 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol 169: 35–47 [Europe PMC free article] [Abstract] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [Abstract] [Google Scholar]
- Wittenberg C, Reed SI 2005. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene 24: 2746–2755 [Abstract] [Google Scholar]
- Xu F, Zhang K, Grunstein M 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385 [Abstract] [Google Scholar]
- Ye X, Wei Y, Nalepa G, Harper JW 2003. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol 23: 8586–8600 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao J, Dynlacht B, Imai T, Hori T, Harlow E 1998. Expression of NPAT, a novel substrate of cyclin E–CDK2, promotes S-phase entry. Genes Dev 12: 456–461 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E 2000. NPAT links cyclin E–Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev 14: 2283–2297 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.173427.111
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/25/23/2489.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6(1):993, 28 Sep 2023
Cited by: 0 articles | PMID: 37770645 | PMCID: PMC10539301
The Saccharomyces cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly.
J Biol Chem, 299(2):102852, 30 Dec 2022
Cited by: 2 articles | PMID: 36592926 | PMCID: PMC9898759
Tumor-Promoting ATAD2 and Its Preclinical Challenges.
Biomolecules, 12(8):1040, 28 Jul 2022
Cited by: 6 articles | PMID: 36008934 | PMCID: PMC9405547
Review Free full text in Europe PMC
SWI/SNF and the histone chaperone Rtt106 drive expression of the Pleiotropic Drug Resistance network genes.
Nat Commun, 13(1):1968, 12 Apr 2022
Cited by: 4 articles | PMID: 35413952 | PMCID: PMC9005695
Mitotic drive in asymmetric epigenetic inheritance.
Biochem Soc Trans, 50(2):675-688, 01 Apr 2022
Cited by: 4 articles | PMID: 35437581 | PMCID: PMC9162470
Review Free full text in Europe PMC
Go to all (32) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression.
Mol Cell Biol, 32(21):4337-4349, 20 Aug 2012
Cited by: 20 articles | PMID: 22907759 | PMCID: PMC3486138
A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity.
Mol Cell Biol, 29(17):4604-4611, 06 Jul 2009
Cited by: 28 articles | PMID: 19581291 | PMCID: PMC2725702
A CDK-regulated chromatin segregase promoting chromosome replication.
Nat Commun, 12(1):5224, 01 Sep 2021
Cited by: 4 articles | PMID: 34471130 | PMCID: PMC8410769
Lessons from yeast on emerging roles of the ATAD2 protein family in gene regulation and genome organization.
Mol Cells, 37(12):851-856, 05 Nov 2014
Cited by: 25 articles | PMID: 25377252 | PMCID: PMC4275701
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Canadian Institutes of Health Research (1)
Grant ID: BMB-210972




