Abstract
Free full text

MODOMICS: a database of RNA modification pathways—2013 update
Abstract
MODOMICS is a database of RNA modifications that provides comprehensive information concerning the chemical structures of modified ribonucleosides, their biosynthetic pathways, RNA-modifying enzymes and location of modified residues in RNA sequences. In the current database version, accessible at http://modomics.genesilico.pl, we included new features: a census of human and yeast snoRNAs involved in RNA-guided RNA modification, a new section covering the 5′-end capping process, and a catalogue of ‘building blocks’ for chemical synthesis of a large variety of modified nucleosides. The MODOMICS collections of RNA modifications, RNA-modifying enzymes and modified RNAs have been also updated. A number of newly identified modified ribonucleosides and more than one hundred functionally and structurally characterized proteins from various organisms have been added. In the RNA sequences section, snRNAs and snoRNAs with experimentally mapped modified nucleosides have been added and the current collection of rRNA and tRNA sequences has been substantially enlarged. To facilitate literature searches, each record in MODOMICS has been cross-referenced to other databases and to selected key publications. New options for database searching and querying have been implemented, including a BLAST search of protein sequences and a PARALIGN search of the collected nucleic acid sequences.
INTRODUCTION
During the course of RNA maturation, various enzymes are able to introduce chemical modifications into ribonucleotide residues. Chemical alteration may occur in the base, at the 2′-hydroxyl of the ribose, or both. Indeed, many modified residues in fact correspond to intermediates in the sequential, multistep formation of hypermodified nucleotides (1). There are also modifications whose biosynthesis starts outside the RNA. For example, queuosine derivatives arise from azaguanine, in which extra chemical groups are attached to C7 (rather than N7 as in guanine). These compounds are introduced into RNA by transglycosylation involving the replacement of an original unmodified guanine by a modified pre-Q base (2). Likewise, in some positive-sense RNA viruses, such as those of the family Alphaviridae, the mRNA cap is formed by nucleotidyltransfer involving a GTP nucleotide pre-modified to N7-methyl-GTP (m7GTP) (3). The variety of chemical groups and their positions in natively modified nucleosides in RNA is illustrated in Figure 1.
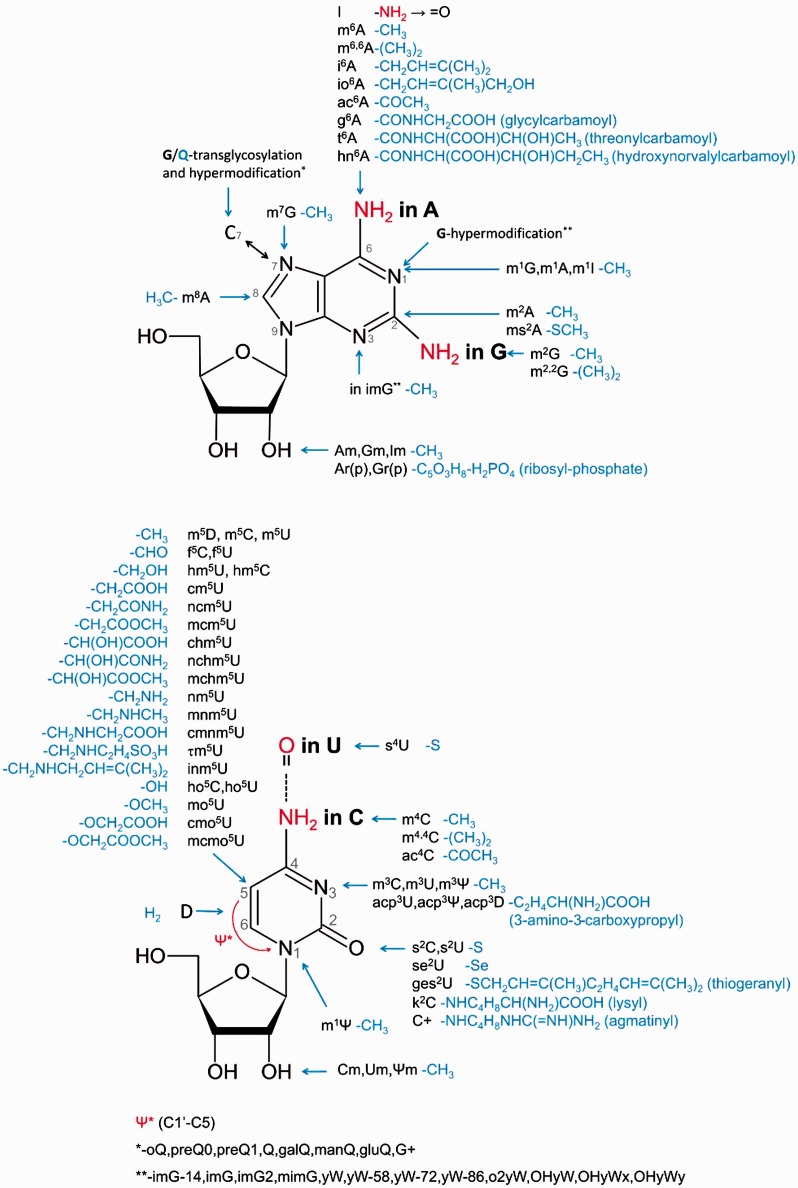
The diversity of chemical groups introduced enzymatically during the process of RNA maturation at various locations of the four common ribonucleosides A, G, C and U linked via 5′- to 3′-phosphodiester bonds. Only conventional acronyms/symbols are shown. The complete scientific names of all modified nucleosides presented in this figure are provided in the MODOMICS database. Nucleosides in RNA can also be doubly or triply modified by the introduction of several of these chemical groups.
The location, abundance and distribution of various types of modification vary greatly between different RNA molecules, organisms and organelles. Physiological environment and growth conditions of the cell also affect the pattern of RNA modification and/or the degree of individual modifications among different RNA molecules of the same type (4). While the majority of modified nucleosides are present in transfer and ribosomal RNAs of all types of cells, they have also been found to occur in small non-coding RNAs, such as spliceosomal (sn)RNAs, small nucleolar (sno)RNAs and more recently in regulatory RNAs, such as siRNAs, miRNAs and piRNAs (5,6). The presence of modified bases in mRNAs and viral RNAs and their potential role in the regulation of gene expression has been recently intensively studied (7–9). New types of modifications have been found (10–16), and biochemical and physiological roles have been revealed for many known modified ribonucleosides. Examples include the immune response to rRNA and tRNA methylation (17,18), the linkage between tRNA modification and host resistance to viral infection (19) and stress-induced cleavage of small RNAs (20). Many of these advances were driven by the use of synthetic RNA containing naturally occurring modified nucleotides. Moreover, numerous new RNA-modifying enzymes have been identified and characterized (21–26). To adequately represent this rapid accumulation of knowledge, we have added both to the variety and volume of data in the MODOMICS database. The most significant additions are: (i) snoRNAs linked to the corresponding modification sites in human and yeast RNAs; (ii) modifications in snRNAs and snoRNAs; (iii) update of the recently identified enzymes and pathways; (iv) a catalogue of ‘building blocks’ for the chemical synthesis of naturally occurring modified nucleosides. The implementation of BLAST (27) and PARALIGN (28) sequence search engines facilitates access to MODOMICS data on the level of protein and nucleic acid sequences. Finally, greater functionality has been added to the user interface.
DATABASE CONTENT
The MODOMICS database (http://modomics.genesilico.pl) has been developed to house and distribute collections of RNA modification pathways, chemical structures of modified nucleosides, sequences of modified RNAs, enzymes responsible for individual reactions and a catalogue of ‘building blocks’ for chemical synthesis of modified RNA. MODOMICS was created as a single resource to organize and present all these data in a convenient and straightforward way. Information about modified residues is also available in the RNMDB database (29). General-purpose pathway databases, such as REACTOME (30) also present some aspects of RNA modification pathways. However, MODOMICS is currently the most comprehensive source of information among all existing RNA modification databases.
MODIFICATIONS
At present, MODOMICS contains 144 different modifications that have been identified in RNA molecules [34 were added since the previous database release (31)]. A typical entry for a modified ribonucleoside contains information about its basic chemical properties, localization in known RNA molecule types, the phylogenetic distribution with respect to Domains of Life and known enzymes responsible for its biosynthesis. The list of modified nucleosides can be browsed by the modification names, the standard bases (A, G, C and U) from which they originate and the chemical groups they contain. The available details contain full and short names, the sum formula, PubChem ID and—to facilitate MS analyses of modified RNAs—the monoisotopic, HRMS and average masses. The chemical structures of the modified nucleosides are represented by 1D SMILE codes, 2D structure plots and 3D structures in the mol format displayed interactively on the website by a Jmol applet. Reactions linking a modified nucleoside to its precursor(s) and to hypermodifications are listed. Many of the products of modification reactions are substrates of further reactions, and the formation of hypermodified residues occurs in complex pathways, which are displayed as graphs. All modified nucleosides found in RNA structures deposited in the RCSB Protein Data Bank are also indicated and appropriate hyperlinks are provided.
PATHWAYS
MODOMICS comprises a collection of RNA modification pathways divided into six different categories according to their starting point: four categories correspond to the standard bases (A, G, C and U), another presents the incorporation and hypermodification pathway of queuosine and the other the modifications of the RNA 5′-cap. The pathway display in MODOMICS has undergone a radical overhaul. The new display, which is based on the Cytoscape Web network visualization tool (32), allows users to zoom and change the graph layout, and to save the obtained result as an image (png or svg), pdf or xml file. Pathway graphs are now easier to navigate and present information about the structures of modified nucleosides and the type of chemical reactions involved. Information about enzymatic transformations that have been verified experimentally (plain arrows) are distinguished from ‘putative’ reactions that are predicted but not yet experimentally confirmed (dashed arrows). Users can access detailed information about each reaction (including the type of transformation and enzymes responsible for its execution) by simply clicking the chosen arrow of the graph. The display of pathway graphs is currently supported for Google Chrome, Mozilla Firefox and for Microsoft Internet Explorer 9 (with Document Mode set to Internet Explorer 9 standards).
RNA SEQUENCES
MODOMICS provides a collection of modified RNA sequences of different types, such as tRNAs, rRNAs, snRNAs and snoRNAs. For families of homologous RNAs, multiple sequence alignments are available. Sequences are visualized with all modifications highlighted and linked to the corresponding modification record. The alignments can be displayed directly on the webpage or in a Jalview applet, or downloaded in plain text format. In comparison to the previous release of MODOMICS, the set of tRNA sequences has been updated and greatly expanded. In particular, in addition to the previously collected manually curated sequences, MODOMICS contains now all tRNAs imported from the Transfer RNA database (33), which were found to possess modified nucleosides (over 500 tRNA sequences in total). The secondary structure of tRNAs is also indicated now. rRNA sequence alignments and secondary structure indications have been updated as well, based on the data from the Comparative RNA Website (CRW) (34). MODOMICS includes an arbitrarily selected subset of rRNA sequences representing different phylogenetic taxa, based on those from the CRW database. For these sequences, both the positions of modified residues and the identities of rRNA-modifying enzymes are known. The current release (as of September 2012) contains 10 SSU rRNA sequences (5 from Bacteria, 2 from Archaea and 3 from Eukaryota) and 9 LSU rRNA sequences (4 from Bacteria, 2 from Archaea and 3 from Eukaryota). For the cytoplasmic LSU in Eukaryota, 5.8S and 28S rRNAs are presented as a single-fused molecule, as in the CRW database. Currently, only one representative rRNA SSU and one LSU sequence per species is included. In future, we intend to expand this data set to include rRNAs from additional species, as well as to cover all rRNA variants encoded by a given genome. To map modifications onto rRNA sequences, we used data from the ‘The Small Subunit rRNA Modification Database’ (35), the 3D rRNA modification maps database (36) and the recently published data concerning modifications in both ribosomal subunits. Finally, we have improved MODOMICS by adding modified snRNA and snoRNA sequences. Unmodified snRNA and snoRNA sequences and alignments were obtained from the Rfam database (37). Positions of modifications in snRNA and snoRNA sequences were included based on the published data.
A new utility that allows mapping the modified positions on secondary structure diagrams of RNA molecules has been implemented. All modified positions from sequences collected in MODOMICS can be mapped onto reference diagrams based on the sequence alignments. For rRNAs, we used the structure of Escherichia coli SSU and LSU rRNAs obtained from the CRW as a reference. For tRNAs, we generated a consensus secondary structure diagram using VARNA (38), based on the data obtained from the Transfer RNA database. Graphics are presented using the JavaScript InfoVis Toolkit library (http://thejit.org/: Nicolas Garcia Belmonte). It is possible to map information from a user-selected set of sequences onto the diagram. In such a case, the percentage of modified ribonucleosides of any type in each alignment position is calculated and displayed. The resulting diagrams can be downloaded as image files.
PROTEINS
The MODOMICS database currently contains information about 274 proteins involved in RNA modification, both functional enzymes and protein co-factors necessary for multi-protein enzymatic activities. More than one hundred functionally and structurally characterized proteins have been added since the previous release (31) and the collection of protein sequences has been updated accordingly. We expanded the collection to include not only the functionally characterized RNA-modifying enzymes from E. coli and Saccharomyces cerevisiae, but also from other organisms, in particular if their crystal structures were available. ‘Predicted’ enzymes, whose activity has not been experimentally validated by genetic or biochemical methods, are currently excluded from MODOMICS. Enzymes that have been characterized biochemically in vitro or in vivo, but for which the corresponding genes have not been identified, are also excluded (although the corresponding reactions are collated).
The MODOMICS catalogue of proteins can be browsed by the source organism and/or type of the enzyme activity (methyltransferase, pseudouridine synthase, etc.). A list of matches can be further edited by the user, based on the features, such as name, position of modification, GI, COGs, PDB ID of structure, etc. At the level of individual protein entries, the database provides information about protein name(s), synonyms, amino acid sequence, corresponding ORF, modified RNA(s) and the position of the residues modified (if available). For proteins that are parts of enzymatic complexes, the name of the complex is provided. Reactions known to be catalysed by each protein with enzymatic activity are listed. Accession numbers from the Swiss-Prot (39) and GenPept (40) databases are provided and proteins with experimentally determined structures are linked to appropriate entries in the Protein Data Bank (41).
snoRNAs
As a new feature, we have included a census of human and yeast snoRNAs, involved in RNA-guided RNA modification by the C/D box and H/ACA box ribonucleoproteins, and we linked these snoRNAs to the corresponding modification sites in human and yeast RNAs. For this section of MODOMICS, we used data from the Saccharomyces Genome Database (42), the Yeast snoRNA Database (43) and snoRNA-LBME-db (44). The list of snoRNAs can be browsed by organism and/or type of modification found in the target position. Links to the HGNC database (45) and the Yeast snoRNA Database are provided for human and yeast snoRNAs, respectively.
BUILDING BLOCKS
In this MODOMICS database release, we have added a catalogue of ‘building blocks’ for the chemical synthesis of naturally occurring modified nucleosides. Each modification is uniquely characterized by its IUPAC name and CAS number, but more than one building block may be available for a given modification. The compilation is intended to facilitate solid phase synthesis of modified RNA, and thus to foster biophysical and biochemical studies. It provides a rapid overview of which modifications may be chemically incorporated with little or moderate synthetic effort, and inversely, which modifications remain attractive targets for synthetic bioorganic chemistry endeavours. The listing was compiled from the CAS database. As not all CAS entries are contained in PubMed, the relevant literature pertaining to synthesis and incorporation of building blocks is given. Its contents reflect the overall dominance of ‘classical’ phosphoramidite chemistry, featuring an acid labile 5′O-protecting group and a fluoride labile 2′O-protecting group. Protective groups that have proven useful in published syntheses are given, including conditions for chemical deprotection after RNA synthesis. The protective groups themselves have separate entries, and further information is available on the reagent used for their introduction, and cross references to other building blocks containing the same protecting group.
SEARCH
New options for database searching and querying have been implemented, including a BLAST (27) search of protein sequences and a PARALIGN (28) search of nucleic acid sequences collected in MODOMICS, as well as a utility that sends a protein sequence from a MODOMICS entry to BLAST on the NCBI webserver. Hits and query-hit alignments resulting from MODOMICS searches can be downloaded in fasta format.
FUTURE PROSPECTS
The total number of confirmed modifications and RNA-modifying enzymes is growing continuously. Though there is a considerable amount of experimentally derived information available, there are still many modified positions in well-characterized RNA molecules for which the responsible enzymes are not known. New modified nucleosides are also being discovered, especially in RNA originating from more recently adopted model systems, such as extremophilic prokaryotes. Projects aimed at systematically studying genomes of related organisms holds promise of many more surprises [e.g. the recent initiative to sequence 5000 insects genomes (46)]. Thus, characterization of RNA modification pathways appears to be a moving target, and we appreciate feedback from users who bring newly discovered modifications and enzymes to our attention, and make suggestions of new methods of data presentation. In the future, we plan to further develop the graphical presentation of RNA modification data to allow comparative analysis of modification pathways and modification positions in sequences with respect to chosen taxa. An ultimate goal is to integrate MODOMICS with databases on other aspects of RNA metabolism. We also intend to link the information available in MODOMICS with ‘RNAcentral’, the planned database of RNA sequences (47).
AVAILABILITY
The data are freely accessible for research purposes at http://modomics.genesilico.pl. Most of the data are available for download in plain text formats. Modified nucleosides and building blocks are also available as structure files and images. Images of pathways are available for download from the web page in several formats. The pathway graphs can be also downloaded as an xml file (graphml format). Program code for parsing the plain text formats is available on request.
FUNDING
Foundation for Polish Science (FNP) [grant TEAM/2009-4/2 to J.M.B.]; Polish Ministry of Science and Higher Education [POIG.02.03.00-00-003/09 to J.M.B.]; Deutsche Forschungsgemeinschaft [FOR1082; HE3397/6 to M.H.]; E.U. Framework Programme 7 [HEALTH-PROT, contract number 229676 for collaboration of the J.M.B. group with H.G. and with M.H.]; National Science Centre [N N301 072 640 to K.M.]; statutory funds of the Adam Mickiewicz University (to K.M.). Funding for open access charge: FNP [TEAM/2009-4/2].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Murray Coles for critical reading of the article and Juliusz Stasiewicz for help in the curation of Modifications section of the database. We would like to thank all previous contributors to the MODOMICS database for their work that provided a solid basis for the present updates. We are indebted to the authors of the primary databases and services, whose content could be reused or linked to by MODOMICS. Last, but not least, we thank all the users of MODOMICS who provided feedback and made suggestions, and who cited MODOMICS in their publications.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gks1007
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/41/D1/D262/3583955/gks1007.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/gks1007
Article citations
Regulation of m<sup>6</sup>A (N<sup>6</sup>-Methyladenosine) methylation modifiers in solid cancers.
Funct Integr Genomics, 24(6):193, 23 Oct 2024
Cited by: 0 articles | PMID: 39438339
Review
Domain-knowledge enabled ensemble learning of 5-formylcytosine (f5C) modification sites.
Comput Struct Biotechnol J, 23:3175-3185, 08 Aug 2024
Cited by: 0 articles | PMID: 39253057 | PMCID: PMC11381828
RNA-modifying enzyme Alkbh8 is involved in mouse embryonic development.
iScience, 27(9):110777, 22 Aug 2024
Cited by: 0 articles | PMID: 39280612 | PMCID: PMC11402254
Progression of m6A in the tumor microenvironment: hypoxia, immune and metabolic reprogramming.
Cell Death Discov, 10(1):331, 20 Jul 2024
Cited by: 1 article | PMID: 39033180 | PMCID: PMC11271487
Review Free full text in Europe PMC
Liquid-liquid phase separation in diseases.
MedComm (2020), 5(7):e640, 13 Jul 2024
Cited by: 2 articles | PMID: 39006762 | PMCID: PMC11245632
Review Free full text in Europe PMC
Go to all (647) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
MODOMICS: a database of RNA modification pathways. 2017 update.
Nucleic Acids Res, 46(d1):D303-D307, 01 Jan 2018
Cited by: 1009 articles | PMID: 29106616 | PMCID: PMC5753262
MODOMICS: An Operational Guide to the Use of the RNA Modification Pathways Database.
Methods Mol Biol, 2284:481-505, 01 Jan 2021
Cited by: 28 articles | PMID: 33835459
MODOMICS: a database of RNA modification pathways. 2008 update.
Nucleic Acids Res, 37(database issue):D118-21, 14 Oct 2008
Cited by: 130 articles | PMID: 18854352 | PMCID: PMC2686465
Brain-specific small nucleolar RNAs.
J Mol Neurosci, 28(2):103-109, 01 Jan 2006
Cited by: 23 articles | PMID: 16679551
Review





