Abstract
Free full text

Rational HIV immunogen design to target specific germline B cell receptors
Abstract
Vaccine development to induce broadly neutralizing antibodies (bNAbs) against HIV-1 is a global health priority. Potent VRC01-class bNAbs against the CD4 binding site of HIV gp120 have been isolated from HIV-1-infected individuals; however, such bNAbs have not been induced by vaccination. Wild-type gp120 proteins lack detectable affinity for predicted germline precursors of VRC01-class bNAbs, making them poor immunogens to prime a VRC01-class response. We employed computation-guided, in vitro screening to engineer a germline-targeting gp120 outer domain immunogen that binds to multiple VRC01-class bNAbs and their germline precursors. When multimerized on nanoparticles, this immunogen (eOD-GT6) activates both germline and mature VRC01-class B cells. Thus, eOD-GT6 nanoparticles have promise as a vaccine prime candidate. In principle, similar germline-targeting strategies can be applied to other epitopes and pathogens.
Protection against disease by nearly all licensed vaccines is associated with the induction of antibodies (1). Viruses with high antigenic diversity, such as HIV, Influenza, and hepatitis C virus, pose major challenges for vaccine development (2). Most exposed surfaces on the Envelope glycoproteins (Env) of these viruses are hypervariable or shielded by glycans (3), and traditional vaccine approaches tend to induce neutralizing antibodies against only a small subset of viral strains (4–6). However, discoveries of bNAbs against each of these viruses have identified conserved epitopes as leads for vaccine design (2), and structural analysis has provided atomic definition for many of these epitopes (7, 8). Structure-based approaches are, therefore, needed to reverse-engineer vaccines capable of inducing bNAbs against these conserved epitopes (9).
High potency VRC01-class bNAbs against the HIV gp120 CD4 binding site (CD4bs) have been isolated from several rare individuals infected with different strains of HIV-1 (10–12). VRC01-class bNAbs all derive from the human VH1-2*02 variable heavy gene, but differ significantly in amino acid sequence and complementarity-determining region H3 (CDRH3) length and use a few different variable light chain genes (figs. S1–2). Structural studies have revealed that VRC01-class bNAbs employ a common mode of gp120 binding in which the VH1-2 framework mimics CD4 and provides additional electrostatic and hydrophobic contacts (Fig. 1A) (12–15). A short CDRL3 loop is also required for interaction with gp120 V5 and Loop D, and a CDRL1 deletion in many VRC01-class bNAbs avoids clashes with a glycan linked to gp120 Asn276 (N276). Vaccine design to induce VRC01-class bNAbs is attractive because VH1-2 genes are estimated to be present in ~2% of the human Ab repertoire (16) and, even considering restrictions on light chain usage, suitable precursors should be present in the naïve B cell repertoire of most individuals. However, predicted germline (GL) precursors for VRC01-class bNAbs exhibit no detectable affinity for wild-type Env (11, 13) (Table 1 and table S1), a potential explanation for the rarity of VRC01-class bNAbs in HIV-1 infection (13). More importantly, wild-type Env constructs lacking GL affinity are poor vaccine candidates to prime VRC01-class responses, as they are unlikely to reliably stimulate GL precursors to initiate antibody maturation.
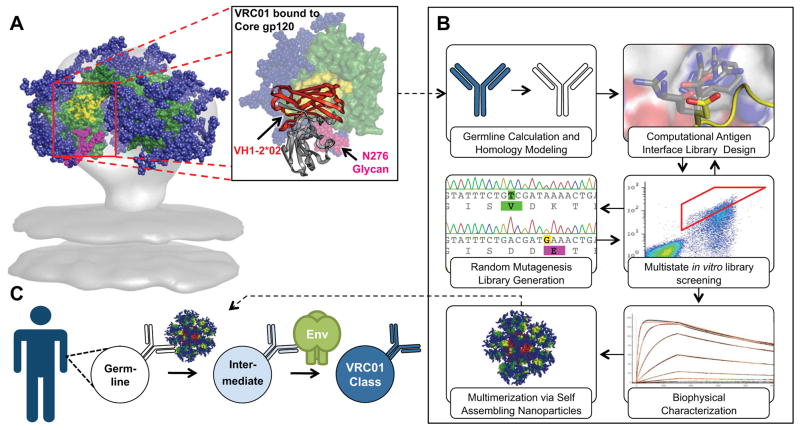
Development of a germline (GL)-targeted HIV immunogen.
(A) VRC01-class bNAbs bind to gp120 primarily through paratope residues encoded by VH 1-2*02. Residues on gp120 that interact with CD4 are colored yellow on an Env trimer model and on the surface of core gp120 (green) in complex with VRC01 (PDBID: 3GNB). The region encoded by VH 1-2*02 on VRC01 is colored red and shown as secondary structure rendering. Glycans are shown as blue spheres, except for the critical N276 glycan that is shown in magenta. (B) Engineering of a modified gp120-based nanoparticle capable of activating GL VRC01-class B-cells. (C) This nanoparticle can be used in an HIV-1 vaccine GL-prime-boost strategy to bridge this initial recognition gap and initiate clonal expansion and start somatic hypermutation of VRC01-class bNAbs precursors.
Table 1
Binding of GL and mature (Mat) Abs to gp120 and eOD variants. Values represent KDs in nM measured by SPR. Binding to GL antibodies highlighted in red.
| Antigen | Antibody | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| VRC01 | 12A12 | 3BNC60 | NIH45-46 | PGV04 | PGV19 | PGV20 | VRC-CH31 | ||||||||||
| GL | Mat | GL | Mat | GL | Mat | GL | Mat | GL | Mat | GL | Mat | GL | Mat | GL | Mat | ||
|
| |||||||||||||||||
| WT CD4Binding Site | Core.HXB2 | >105 | 5 | >105 | 6 | >105 | 36 | >105 | 35 | >105 | 48 | >105 | 20 | >105 | 19 | >105 | 47 |
| eOD Base N276D | >105 | 5 | >105 | 380 | >105 | 4,100 | >105 | 14 | >105 | 110 | >105 | 3,100 | 17,000 | 16 | >105 | 30,000 | |
|
| |||||||||||||||||
| Modified CD4 Binding Site | Core.BaL-GT1 | 1,800 | 0.5 | 3,200 | 1 | 16,400 | 4 | >105 | 0.6 | >105 | 170 | 25 | 14 | 5,500 | 4 | 35,000 | 1,800 |
| eOD-GT1 | 44,000 | 1 | >105 | 2,300 | >105 | 83 | >105 | 3 | >105 | 4 | 7,800 | 1,000 | 1,100 | 10 | >105 | >105 | |
| eOD-GT6 | 44 | 2 | 2,000 | 400 | 14,000 | 200 | 410 | 4 | 52,000 | 10 | 19 | 88 | 3 | 6 | 28,000 | 29,000 | |
Immunogen Design Strategy
To address the problem described above, we modified the CD4bs on a minimal, “engineered outer domain (eOD)” (17) to produce a “germline-targeting” vaccine prime (Fig. 1) with two important binding properties: (i) moderate affinity for diverse predicted VH1-2*02 GL-Abs to enhance the ability to activate VH1-2 GL B cells with appropriate light chains; (ii) high affinity for VRC01-class bNAbs to provide an affinity gradient to guide early somatic mutation toward the mature bNAbs. Furthermore, we developed self-assembling nanoparticles presenting 60 copies of the germline-targeting eOD, to enhance B cell activation and to improve trafficking to lymph nodes (Fig. 1).
Engineering and Biophysical Analysis of Germline-Targeting Antigens
Modifications to the VRC01 epitope included removing clashes and building new contacts between the CD4bs and the GL-Abs, as well as rigidifying the CD4bs in a conformation that is favorable for binding. Initially, we constructed a homology model of GL-VRC01 bound to gp120 and identified a likely clash between CDRL1 and the N276 glycan. Therefore, we evaluated the GL-VRC01 binding of an eOD (eOD-Base) that lacks glycans at 276 and nearby on the V5 loop owing to N276D and N463D mutations. The eOD-Base barely interacted with GL-VRC01 (KD ~ 1 mM) and had low affinity for only 2 of 8 other GL VH1-2*02 Abs tested (Table 1). We then used Rosetta computational protein interface design (18) to identify other mutations in and around the CD4bs that were predicted to improve GL-VRC01 binding, and created directed libraries that included all possible combinations of the computationally identified mutations. These libraries were screened for core gp120 and eOD variants that showed GL-VRC01 binding using yeast cell surface display (19). This strategy generated germline-targeting (GT) variants of core gp120 (CoreBal-GT1) and eOD (eOD-GT1) that bound GL-VRC01 with KDs of 1.8 μM and 44 μM, respectively (table S2). We focused further development on the smaller eOD because it lacks potentially distracting epitopes on core gp120. A second round of computational design and directed-library screening produced eOD-GT2 with a three-fold improvement in KD for GL-VRC01 (table S2). Subsequent screening of mixed computational/random mutagenesis libraries led to larger improvements and resulted in eOD-GT3 and eOD-GT4, which had KDs for GL-VRC01 of 220 and 34 nM, respectively (table S2). Interestingly, screening for GL-VRC01 binding also improved binding to other GL VH1-2 Abs, as eOD-GT4 bound to GL-NIH45-46 and GL-PGV19 with KDs of 1.0 μM and 28 nM, respectively (table S2). To achieve these improvements, eOD-GT4 had accumulated 17 mutations relative to eOD-Base.
To retain as native a CD4bs as possible (by reducing the number of mutations) while also maintaining or improving binding to GL VH1-2 Abs and mature bNAbs, we employed multi-target optimization. Here, libraries with either the wild-type HIV-1 strain HxB2 residue or the mutation in eOD-GT4 were sorted in parallel against multiple Abs, and mutations were retained only if they were beneficial to at least one GL Ab and tolerated by all Abs in the sort. eOD-GT6 was generated by sorting against GL Abs for VRC01, NIH45-46, PGV19, PGV04 and VRC-CH31, as well as mature VRC01 and PGV04. eOD-GT6 had only 8 mutations relative to eOD-base (10 mutations relative to HxB2 gp120) and retained excellent binding to diverse GL VH1-2*02 Abs, with a KD of 44 nM for GL-VRC01 and KDs < 500 nM for 5 of 9 GL Abs tested (Table 1, table S2, fig. S3). eOD-GT6 also had high affinity for several mature bNAbs, with KDs of 2, 4, and 88 nM for VRC01, NIH45-46, and PGV19, respectively. eOD-GT6 also maintained the desired affinity gradient for 6 of 8 Abs tested (Table 1). Further, eOD-GT6 bound with high affinity to GL VRC01-class Abs derived from VH1-2*03 and *04 alleles (table S3, fig. S4). eOD-GT6 had no affinity for VH1-2*01, probably due to the absence of TrpH50, however, recent data from sequencing of 1092 human genomes(20)shows that the W50R mutation in VH1-2*01occurs at a frequency of 0.21, indicating that only ~4% of the population are VH1-2*01 homozygotes not amenable to eOD-GT6 priming.
Crystallographic Analysis
To understand the atomic interaction of eOD-GT6 with GL-VRC01, we solved three crystal structures: unliganded GL-VRC01, unliganded eOD-GT6, and the complex of GL-VRC01 bound to eOD-GT6, to resolutions of 2.1 Å, 2.5 Å, and 2.4 Å, respectively (Fig 2 and table S4). The structure of unliganded GL-VRC01 revealed that the gp120 contacting loops closely resemble those of VRC01 despite extensive affinity maturation of the latter (Fig. 2A, and figs. S5–7). Unliganded eOD-GT6 showed a similar structure to the outer domains of unliganded and VRC01-bound gp120 core (1.2 Å Cα root-mean-square-deviation (RMSD) in both instances) (Figs. 2B and fig. S8), suggesting good mimicry of the CD4bs. The eOD-GT6 unliganded structure was also similar to the structure of the eOD-GT6 bound to GL-VRC01 Fab (0.9 Å RMSD), with the largest differences occurring in the flexible loops and in the eOD exit loop (fig. S9). In addition, the crystal structure of the eOD-GT6+GL-VRC01 complex indicated that the VH1-2-encoded domain of GL-VRC01 has an angle of approach to eOD-GT6 nearly identical to that of VRC01 to gp120 (Fig. 2C) (4.2° angular difference when the complexes are superposed on the CD4b loop). Overall, the buried surface area (BSA) of GL-VRC01 (1075.6 Å2) on eOD-GT6 (1102.0 Å2) is nearly identical to VRC01 (1125.7 Å2) on core gp120 (1158.2 Å2), further demonstrating the high degree of similarity between the two structures (tables S5–6). Key hydrogen-bonding networks are preserved in the GL-VRC01+eOD-GT6 interaction, particularly in the CD4b loop (fig. S10). On the other hand, key differences in hydrogen bonds, BSA and Cα positions are observed for interactions in loop D, V5 and the OD exit loop, which we hypothesize confer GL-VRC01 reactivity to eOD-GT6 (Fig. 2D and fig. S11).
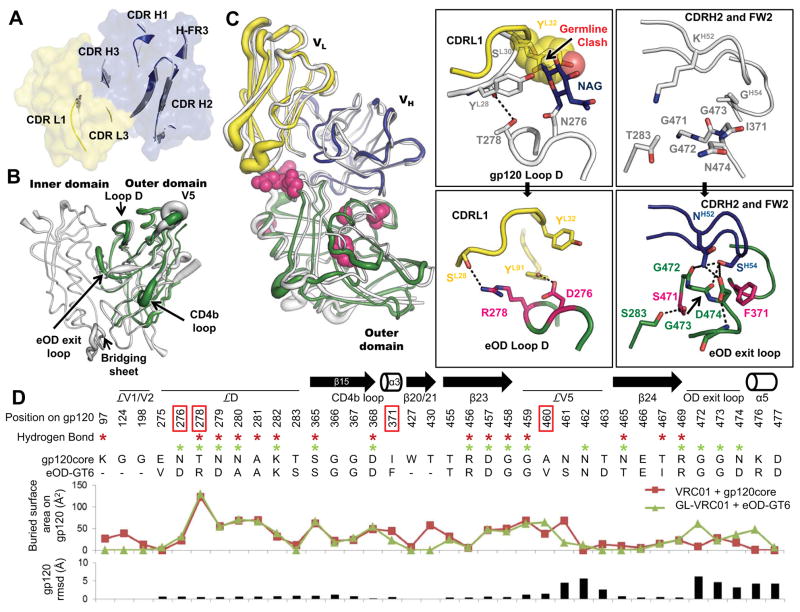
(A) Comparison between the crystal structure of an unliganded predicted GL-VRC01 antibody (heavy and light chains colored blue and yellow, respectively) and VRC01 bound to gp120 (gray). Variable regions of GL-VRC01 are rendered as a surface. Areas of GL-VRC01 and VRC01 that contact gp120, shown as secondary structure cartoons, are similar in conformation. (B) Comparison between the crystal structures of unliganded eOD-GT6 (green) and unliganded gp120 core from HIV-1 strain 93TH057 (PDBID: 3TGT, gray). Structures are rendered according to B-values, with thin and thick lines representing areas possessing low and high flexibility, respectively. (C) Comparison between the crystal structures of GL-VRC01+eOD-GT6 and VRC01+gp120 core. Components are colored as in (A) and (B) and shown as tubes. The positions of the recovered mutations in eOD-GT6 that enable binding of GL-VRC-class Abs are shown as space-fill magenta spheres. The angle of approach of GL-VRC01 and VRC01 to the CD4bs is nearly identical. Key areas where interactions are different between VRC01 on gp120 (upper panels) and GL-VRC01 on eOD-GT6 (bottom panels) are shown in insets. eOD-GT6 confers germline reactivity by removing a potential clash with the N276 glycan, as well as by creating additional contacts with loop D (inset I), the OD exit loop (inset II) and V5 (fig. S12). (D) gp120 residues involved in the VRC01+gp120 and GL-VRC01+eOD-GT6 interfaces are compared in sequence, H-bond (stars), surface buried area and RMSD. Interfaces were calculated using PDBePISA (29) and Cα rmsd using Chimera (30).
Reversion Analysis
To understand the affinity contributions of individual mutations, we measured GL-VRC01 binding affinities for point reversions of each mutation (Table 2). Six mutations on eOD-GT6 conferred improved affinity for GL-Abs relative to the starting construct (eOD-Base) that lacked glycans at 276 and 463. The eOD-GT6+GL-VRC01 complex crystal structure revealed that these mutations are either directly involved in the binding interface (T278R, I371F and N460V) or stabilize loops involved in the interface (L260F, K357R and G471S) (Fig. 2C). The two mutations with the largest effect on GL-VRC01 binding were G471S and I371F; reversion at these positions reduced GL-VRC01 affinity by factors of 39 and 10, respectively (Table 2). Ser471, together with Phe371 and Phe260 appears to play a role in altering the conformation of the OD exit loop to allow the GL-VRC01 CDRH2 to make H-bonds with three additional gp120 residues (G472, G473 and D474) and bury an additional 119.6 Å2 on gp120, resulting in improved binding (Fig. 2C–D, inset 2, tables S5–6, fig. S9). Also at the interface, the N460V mutation located in V5 improves packing with the antibody and appears to contribute to an altered V5 conformation and pattern of V5 H-bonding with VRC01, as compared to Clade A/E 93TH057 gp120 recognition of VRC01 (Fig. 2D and fig. S12). Reversion of the N460V mutation reduced GL-VRC01 binding by a factor of 2.5 (Table 2).
Table 2
Binding of GL VRC01 to eOD-GT6 point reversions. Values represent KDs in nM measured by SPR. Amino acid frequencies determined from 2867 HIV-1 sequences from http://www.hiv.lanl.gov/.
| eOD-GT6 | Single Reversions | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Reversion from eOD-GT6 | - | F260L | R278T | R357K | F371I | V460N | S471G |
| Frequency of WT AA | - | L (97.3%) | T (78.4%) | K (68.9%) | I (85.8%) | N (36.2%) | G (78,7%) |
| Frequency of Mutation | - | F (0.0%) | R (0.1%) | R (0.5%) | F (0.0%) | V (3.4%) | S (0.5%) |
| Method of Discovery | - | epPCR | Rosetta | epPCR | Rosetta | Rosetta | epPCR |
| GL-VRC01 Affinity (nM) | 44 | 310 | 160 | 64 | 450 | 120 | 1,700 |
| Change from eOD-GT6 | - | 7.0 | 3.6 | 1.5 | 10.3 | 2.7 | 39 |
Removal of key glycosylation sites was necessary for GL affinity. Reintroduction of the N276 glycosylation site in eOD-GT6 (by a double mutation, D276N/R278T) reduced binding by a factor of 140, and the remaining binding was likely due to a small fraction of the eOD-GT6-D276N/R278T that underutilized the N276 glycosylation position (fig. S13). Reversion of R278T alone reduced affinity by a factor of only 3.6 (Table 2). Thus, removal of the 276 glycan appears to release a block on GL-VRC01 binding but does not confer appreciable eOD affinity; further interface modification was required to achieve high affinity. Indeed, the eOD-GT6+GL-VRC01 complex crystal structure revealed that, in addition to removing a clash between the N276 glycan on gp120 and the GL-VRC01 CDRL1 (Fig. 2C, inset 1), eOD-GT6 residues D276 and R278 make two additional H-bonds with GL-VRC01. eOD-GT6 also lacks glycans at positions 386 (β12) and 463 (V5). Restoration of these glycosylation sites reduced affinity for GL-VRC01 by a factor of 3 (table S7).
eOD-GT6 Nanoparticle Generation
To enable eOD-GT6 to activate GL B cells via cross-linking of B cell receptors, and to develop a multivalent platform for eOD-GT6 that mimics the size, shape, and symmetric surface geometry of many viruses for improved immunogenicity (21), we sought to fuse eOD-GT6 to a self-assembling virus-like nanoparticle. From a search of large homomeric particles in the Protein Data Bank (PDB) we prioritized 60mers of Lumazine Synthase from the hyperthermophile Aquifex Aeolicus for experimental testing, due to their thermal stability and because modeling suggested that, with a suitable linker length, 60 copies of glycosylated eOD-GT6 could be sterically accommodated in an orientation that would expose the VRC01 epitope (Fig 3A). Though expression of the wild-type particle had been reported in E. coli (22), we found that such nanoparticles presenting glycosylated eOD-GT6 could be secreted from mammalian (293) cells and purified by lectin chromatography with good yield (~10 mg/L) and structural homogeneity (Fig 3B and figs S14–15).
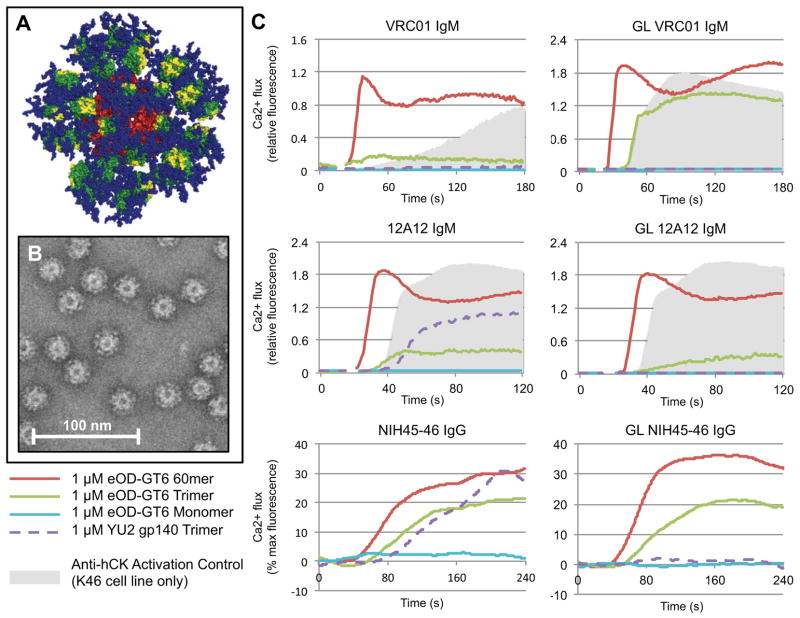
A 60-mer eOD-GT6 nanoparticle activates GL and mature VRC01-class B cells. (A) Model representation of the 60-mer eOD-GT6 nanoparticle. eOD-GT6 is colored in green, with residues that interact with CD4 colored yellow. Glycans are shown as blue spheres and the self-assembling 60mer Lumazine Synthase to which eOD-GT6 is fused is colored red. (B) Raw negative stain electron microscopy images of the 60-mer eOD-GT6 nanoparticle. (C) Calcium flux experiments with various gp120 constructs show that the 60-mer eOD-GT6 nanoparticle activates B cell lines engineered to express either GL or mature VRC01 IgM, 12A12 IgM or NIH45-46 IgG, while a recombinant soluble gp140 trimer activates the B cells expressing mature but not GL VRC01-class Abs. Data for each antibody are representative of at least two separate experiments.
In Vitro B Cell Activation
The ability of eOD-GT6 nanoparticles to activate B-cells expressing GL and mature VRC01 (IgM) (23), 12A12 (IgM) and NIH45-46 (IgG) (24), was tested in Ca2+-dependent activation assays. The 60-mers potently activated both GL and mature B cells with 1 μM outer domain (16 nM particle) and modestly activated all three cell lines at 1000-fold lower concentrations (Fig. 3C and fig. S16). In contrast, monomeric eOD-GT6 was non-stimulatory, probably due to an inability to cross-link B cell receptors (23). Trimeric eOD-GT6 activated both GL and mature B cells but less potently and rapidly than the 60mers, and a soluble gp140 trimer from HIV-1 strain YU2 (25) showed no activation of GL B cells but did activate the mature counterparts (Fig. 3C). Both IgM and IgG B cell lines were generated for GL 12A12 and we observed no significant difference between activation behaviors of the two antibody isotypes (fig. S16).
Animal Models for Human VH1-2 Germline-Targeting
We then assessed whether eOD-GT6 might interact with related GL-Abs in animal models. Analysis of VH genes from rabbit (fig. S17) (26), mouse (figs. S18–19) (27) and macaque (fig. S20) revealed that none of these commonly used model organisms have a known VH gene containing all of the critical residues for GL binding (15). To measure binding experimentally, chimeric GL-Abs were produced in which the human VH1-2*02 gene from GL-VRC01 was replaced with GL VH genes from mice or macaques containing the essential ArgH71 and as many other critical residues as possible. Chimeric GL-Abs with mouse VH genes had no detectable binding to eOD-GT6. Abs derived from two of the three rhesus VH genes bound only weakly to eOD-GT6, with KDs of ~30 μM and ~40 μM (table S8). Despite being >94% identical in sequence to human GL-VRC01, the rhesus chimeric GL-Abs had three orders of magnitude lower affinity. Annotation of the rhesus macaque antibody repertoire and analysis of gene usage frequencies will be useful to construct bona fide macaque GL VH1-2 Abs. These analyses illustrate the potential difficulty for using animal models to produce VRC01-class bNAbs, and suggest that immunization of humans or mice engineered to produce human Abs may be essential for testing and iteratively optimizing such immunogens.
The events that led to GL VH1-2*02 B cell activation in the HIV-infected individuals from which VRC01-class bNAbs were isolated remain unclear. Our finding that a small number of rare or previously undocumented Env mutations confers high affinity GL binding suggests that Env variants might have acquired one or more such mutations stochastically during infection and thereby gained the ability to prime GL VRC01-class B cells. Vaccines to induce VRC01-class responses will need to activate such B cells dependably and drive appropriate somatic mutation to produce high affinity bNAbs (28). We propose the eOD-GT6 nanoparticle as a promising candidate for a vaccine prime based on its ability to bind diverse VH1-2*02 GL Abs, activate VRC01, 12A12 and NIH45-46 B cells in vitro, and provide an affinity gradient for early somatic mutation. We hypothesize that ultimate elicitation of mature VRC01-class bNAbs will require, at minimum, boosting with different immunogens that present a less engineered, more native CD4bs including the glycans around the CD4bs.
Acknowledgments
We thank J. Mascola and X. Wu for providing sequences of mature PGV19 and PGV20, R. Wyatt for providing the YU2 gp140 trimer, C. Sundling and G. Karlsson Hedestam for the sequence of Rhe1 VH gene, and A. Bradley, R.L. Stanfield, and D.C. Diwanji for technical assistance and insightful discussions. The data presented in this manuscript are tabulated in the main paper and the supplementary materials. Coordinates and structure factors for GL-VRC01 Fab, eOD-GT6 and GL-VRC01+eODGT6 structures will be deposited with the Protein Data Bank and released upon publication. The International AIDS Vaccine Initiative (IAVI) has filed a patent relating to immunogens in this manuscript: PCT Application No. PCT/US12/60062, titled “Engineered Outer Domain (EOD) Of HIV GP120 And Mutants Thereof”, with inventors WRS, JJ, SM, PH. Materials and information will be provided under MTA. This work was supported by the International AIDS Vaccine Initiative Neutralizing Antibody Center, CHAVI-ID (UM1 AI100663), NIH grant AI84817 (I.A.W.), National Institute of Allergy and Infectious Diseases grants P01AI081625 (L.S.) and AI33292 (D.R.B.), NIH NRSA Training Grant fellowship T32CA080416 (J.J.), Canadian Institutes of Health Research fellowship (J.-P.J.), CNIHR grant P30 AI027767-24 (T.O.), NIH Interdisciplinary Training Program in Immunology 5T32AI007606-10 (D.S.) and the Ragon Institute. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a Directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research; NIH’s National Center for Research Resources, Biomedical Technology Program (P41RR001209); and the National Institute of General Medical Sciences (NIGMS). Use of the Advanced Photon Source was supported by the DOE, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. GM/CA CAT has been funded in whole or in part with federal funds from NCI (grant Y1-CO-1020) and NIGMS (grant Y1-GM-1104). This is manuscript 23050 from The Scripps Research Institute.
References
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.1234150
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3689846?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/science.1234150
Article citations
Advanced technologies for the development of infectious disease vaccines.
Nat Rev Drug Discov, 21 Oct 2024
Cited by: 0 articles | PMID: 39433939
Review
Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies.
Nat Commun, 15(1):9503, 03 Nov 2024
Cited by: 0 articles | PMID: 39489734 | PMCID: PMC11532496
An Integrated Signaling Threshold Initiates IgG Response toward Virus-like Immunogens.
J Immunol, 213(8):1061-1075, 01 Oct 2024
Cited by: 0 articles | PMID: 39212443
Interventions during Early Infection: Opening a Window for an HIV Cure?
Viruses, 16(10):1588, 09 Oct 2024
Cited by: 0 articles | PMID: 39459922 | PMCID: PMC11512236
Review Free full text in Europe PMC
A mathematical model simulating the adaptive immune response in various vaccines and vaccination strategies.
Sci Rep, 14(1):23995, 14 Oct 2024
Cited by: 0 articles | PMID: 39402093 | PMCID: PMC11473516
Go to all (521) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 3GNBView structure
-
(1 citation)
PDBe - 3TGTView structure
Protocols & materials
Related Immune Epitope Information - Immune Epitope Database and Analysis Resource
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Short CDRL1 in intermediate VRC01-like mAbs is not sufficient to overcome key glycan barriers on HIV-1 Env.
J Virol, 98(10):e0074424, 06 Sep 2024
Cited by: 0 articles | PMID: 39240111 | PMCID: PMC11495006
HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen.
Science, 349(6244):156-161, 18 Jun 2015
Cited by: 292 articles | PMID: 26089355 | PMCID: PMC4669217
Bacterially expressed HIV-1 gp120 outer-domain fragment immunogens with improved stability and affinity for CD4-binding site neutralizing antibodies.
J Biol Chem, 293(39):15002-15020, 09 Aug 2018
Cited by: 8 articles | PMID: 30093409 | PMCID: PMC6166733
Targeting broadly neutralizing antibody precursors: a naïve approach to vaccine design.
Curr Opin HIV AIDS, 14(4):294-301, 01 Jul 2019
Cited by: 11 articles | PMID: 30946041
Review
Funding
Funders who supported this work.
CIHR
NCI NIH HHS (3)
Grant ID: T32 CA080416
Grant ID: Y1-CO-1020
Grant ID: T32CA080416
NCRR NIH HHS (2)
Grant ID: P41 RR001209
Grant ID: P41RR001209
NIAID NIH HHS (15)
Grant ID: AI081625
Grant ID: R01 AI073148
Grant ID: R56 AI084817
Grant ID: UM1 AI100663
Grant ID: P30 AI027767
Grant ID: P30 AI027767-24
Grant ID: R01 AI084817
Grant ID: 5T32AI007606-10
Grant ID: AI84817
Grant ID: T32 AI007606
Grant ID: P01 AI094419
Grant ID: R01 AI081625
Grant ID: R37 AI033292
Grant ID: AI33292
Grant ID: R01 AI033292
NIGMS NIH HHS (1)
Grant ID: Y1-GM-1104





