Abstract
Free full text

Multiple interactions between regulatory regions are required to stabilize an active chromatin hub
Abstract
The human β-globin locus control region (LCR) is required for the maintenance of an open chromatin configuration of the locus. It interacts with the genes and the hypersensitive regions flanking the locus to form an active chromatin hub (ACH) transcribing the genes. Proper developmental control of globin genes is largely determined by gene proximal regulatory sequences. Here, we provide the first functional evidence of the role of the most active sites of the LCR and the promoter of the β-globin gene in the maintenance of the ACH. When the human β-globin gene promoter is deleted in the context of a full LCR, the ACH is maintained with the β-globin gene remaining in proximity. Additional deletion of hypersensitive site HS3 or HS2 of the LCR shows that HS3, but not HS2, in combination with the β-globin promoter is crucial for the maintenance of the ACH at the definitive stage. We conclude that multiple interactions between the LCR and the β-globin gene are required to maintain the appropriate spatial configuration in vivo.
Gene transcription and the underlying modification of the chromatin structure are dynamic interrelated nuclear processes (Urnov and Wolffe 2001 and references therein). The human β-globin locus is an excellent model system to study this interplay. The locus consists of five functional genes, arranged in the order that they expressed during human development (Fig. 1). The most important control element is the locus control region (LCR), which consists of five hypersensitive sites (HS1–5; for review, see Stamatoyannopoulos and Grosveld 2001). These HSs interact with each other and three additional 5′ and 3′ distant HSs to form an active chromatin hub (ACH) through looping (de Laat and Grosveld 2003), which interacts with the different genes (Carter et al. 2002; Tolhuis et al. 2002) to form an active transcription “factory” throughout development (Fig. 1B; Palstra et al. 2003). Two developmental switches occur during human ontogeny, controlled exclusively at the transcriptional level. The embryonic ε- and fetal γ-globin genes are expressed during primitive hematopoiesis. Expression switches to the fetal γ- and adult δ- and β-globin genes during early definitive hematopoiesis, with the balance gradually shifting to δ- and β-genes. The transition from ε- to γ-globin genes and γ- to δ- and β-globin genes transcription is largely controlled by gene cis-regulatory sequences flanking the genes (for review, see Stamatoyannopoulos and Grosveld 2001). The LCR is required for the expression of all of the genes and provides integration site independent expression in transgenic mice (Grosveld et al. 1987). Also, patients lacking the LCR (but retaining the 5′ and 3′ distant HS and at least one of the genes) fail to express the genes and have an inactive chromatin configuration (Kioussis et al. 1983; Forrester et al. 1990). The role of the individual HS is as yet not clear, although HS2 and HS3 appear to be the most important sites for the efficiency of transcription both in the human and murine locus, each leading to an ~30% loss of transcription when deleted (Philipsen et al. 1990; Talbot et al. 1990; Ellis et al. 1993; Fraser et al. 1993; Bungert et al. 1995; Fiering et al. 1995; Hug et al. 1996; Peterson et al. 1996), whereas developmental regulation appears largely unaffected. The deletion of HS2 and HS3 from the human locus in transgenic mice also leads to position effects when the locus is integrated in heterochromatic regions (Milot et al. 1996), suggesting they are important for the stability of the ACH. The globin genes are expressed in a competitive manner (Hanscombe et al. 1991; Dillon et al. 1997) by an alternating transcription process (Wijgerde et al. 1995); that is, only a single gene in the locus is transcribed at any given moment, suggesting that only a single gene can interact with the hypersensitive sites to form an ACH.
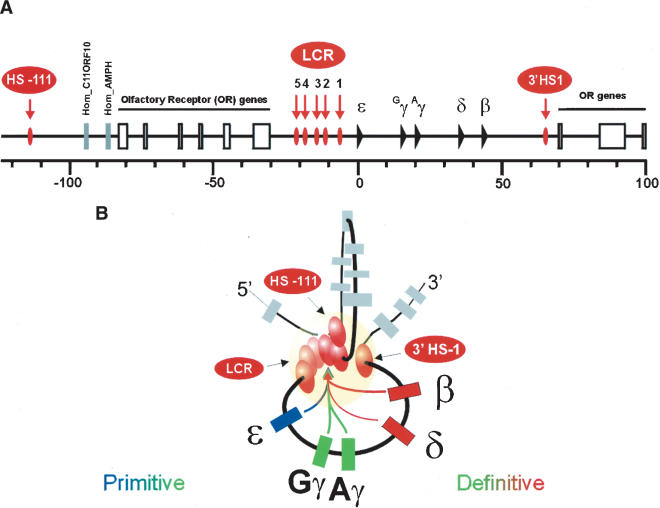
(A) Schematic representation of the human β-globin locus. (B) Two-dimensional representation of the 3-D interactions between the HSs and the globin genes in erythroid cells. The LCR, together with most 5′ and 3′ HSs are forming the ACH (indicated as a yellow sphere), with which the globin genes stably interact for high-level transcription. The genes are depicted in different colors, indicative of their interaction with the LCR within the ACH during development (see also Palstra et al. 2003).
Although much of the molecular basis of globin gene transcriptional activation has been elucidated in recent years, the dynamic conformation of the LCR and the precise role of the individual HS of the LCR remain unclear. In particular, we have previously proposed that the LCR acts as a holo-complex (Ellis et al. 1996). However, to date there has been little insight about whether this “holo-complex” is formed by the interaction between individual hypersensitive sites or whether it only acts as such a complex because of simultaneous interactions between the different HSs of the LCR and any one of the genes, or a combination of both of these possibilities.
Here we report results obtained from transgenic mice carrying the human β-globin locus with a β-globin promoter deletion in the context of a full or an incomplete LCR. We show that the β-globin gene remains in proximity to the ACH when its promoter has been deleted, suggesting that the association of the gene with the ACH is not the result of transcription. However, when HS3 (but not HS2) and the β-globin gene promoter are deleted in cis, the histone acetylation is lost during definitive hematopoiesis. Importantly, this double deletion leads to a loss of the ACH, yielding a structure of the locus that is very similar to that observed in other nonexpressing tissues.
Results
Generation of transgenic mice with the mutant β-globin loci
The basal promoter and 5′ untranslated regions of the human β-globin gene, spanning from positions -139 to +49 relative to the Cap site of the gene, were deleted (Fig. 2A), as this fragment includes all of the known promoter mutations leading to β-thalassemia (Hardison et al. 2002). A 288-bp HincII/SmaI fragment, including the β-globin promoter deletion, was subcloned into the pDF-25 recombination vector and used to delete the promoter in a PAC containing the human β-globin locus (Imam et al. 2000). DNA from individual Escherichia coli clones was analyzed by Southern blot hybridization. Electrophoresis of EcoRI digests using cosLCRε and cosγγδβ cosmid probes confirmed that the resulting locus (Δβp) was identical to the wild type, except for the β-globin promoter deletion (Fig. 2B). The same result was obtained using BamHI, HindIII, SacI, PstI, and BlnI digests (data not shown). After the β-globin promoter deletion was successfully generated, homologous recombination was used to generate the same HS2 and HS3 deletions, as published previously (Milot et al. 1996), in both the wildtype and Δβp PACs. Following modification, DNA from the resulting PAC clones, containing the HS2 or HS3 deletion in cis to the normal or deleted β-promoter (Δ2, Δ2Δβp, Δ3, and Δ3Δβp, respectively, Fig. 2A), were analyzed by Southern blot hybridization, which confirmed the presence of the targeted modifications in an otherwise unchanged PAC (Fig. 2B). The normal PAC (wild type) and each of the constructs were digested with NotI and subjected to salt gradient ultracentrifugation to isolate the inserts. These were checked for integrity by pulse field gel electrophoresis and microinjected into fertilized mouse oocytes, resulting in several wild-type and modified PAC transgenic founder animals. Integrity of the transgene was assessed as described previously for the isolated PAC DNA (Wai et al. 2003; data not shown). Copy numbers varied between one and three, as determined by the ratio of human β-globin intron II signal versus an endogenous mouse carbonic anhydrase II signal on Southern blots. DNA in situ hybridization, using a probe against the complete human β-globin LCR, was used to determine the transgene integration sites. Subsequently, the founder animals were bred to establish two to four lines for each of the constructs.
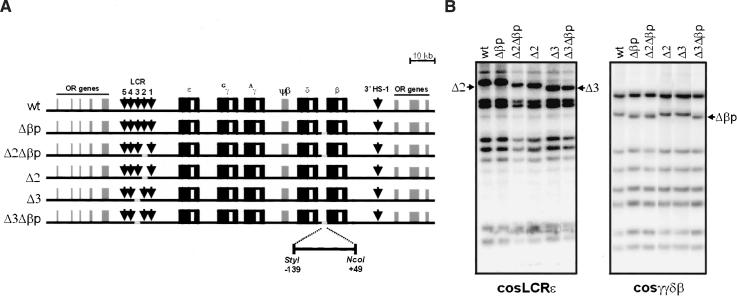
(A) Construction of the human β-globin locus deletion mutants, within the 185-kb PAC insert. Vertical arrows correspond to the individual LCR HS sites, and globin exonic and intronic sequences are designated with black and white boxes, respectively (not to scale). Individual HS and β-globin gene promoter deletions (depicted in gray) were generated by homologous recombination in E. coli DH10B strain (Imam et al. 2000). The 188-bp StyI/NcoI β-globin promoter deletion is indicated below. (B) EcoRI restriction analysis of the different β-globin PAC deletion mutants. The presence of the desired modification is highlighted, whereas the remaining sequence is unaltered. The probes used for Southern blot analysis of the deletion constructs are cosmids cosLCRε and cosγγδβ (Strouboulis et al. 1992).
Effects of the deletion of the human β-globin promoter on the locus
In our previous studies on the mouse and human β-globin locus, we have shown that the β-globin gene only interacts with the ACH when it is transcribed in definitive cells (Palstra et al. 2003). In differentiated primitive cells, the early genes (ε- and γ-) interact with the ACH, whereas in precursor cells, the LCR and the genes do not interact with the ACH. This poses the question of whether the genes need to associate with the ACH to be transcribed or whether the association with the ACH is the result of transcription. This question was addressed by deletion of the β-globin promoter without affecting any other parts of the locus. Two independent single copy transgenic lines were analyzed for expression of the ε-, γ-, δ-, and β-genes (Fig. 3A). The results show that early gene expression in the primitive erythroid cells (embryonic day 10.5 [E10.5] yolk sac) is not affected by the deletion of the β-globin promoter when compared with mice containing the wild-type locus. However, the level of γ-globin gene expression is higher in the E12.5 fetal liver. Similarly, δ-globin gene expression, when measured in adult blood cells, although still very low (note the scale in Fig. 3A), is higher compared with the wild-type locus. This result is in agreement with the notion that a single gene is expressed at any given time (Wijgerde et al. 1995) and that there is transcriptional competition between the genes (Dillon et al. 1997). When the chromatin structure of the Δβp locus was compared with the wild type locus both by DNaseI sensitivity and histone H3 acetylation (see Figs. Figs.5,5, ,6,6, respectively [below]), the chromatin structure of the locus was essentially the same. A similar result was obtained for histone H4 acetylation (data not shown). We next asked the question of whether the β-globin gene still interacts with the ACH in the definitive differentiated erythroid cells when compared with control fetal brain cells. For this purpose, Chromosome Conformation Capture (3C) technology (Dekker et al. 2002; Tolhuis et al. 2002; Palstra et al. 2003) was used, which gives a measure of proximity of two DNA restriction fragments in the nuclear space using quantitative PCR of the cross-linking frequencies between the fragments in the nucleus. The assay is not biased for particular fragments, because of preferential restriction enzyme digestion of one site over the other as a result of different chromatin structure (Tolhuis et al. 2002; Palstra et al. 2003; Splinter et al. 2004). In the wild-type locus, the HS1–5 of the LCR, the transcribed gene, and the HS in the 3′ flanking region (3′HS1) interact with each other to form the ACH of the human β-globin locus in transgenic mice (Palstra et al. 2003). The result of the 3C analysis (Fig. 3B,C) shows that the spatial structure of the locus is essentially unchanged in the Δβp mutant locus. The ACH, consisting of the LCR and 3′HS1 (Palstra et al. 2003), is maintained. However, the relative frequency of association of the β-globin gene with the fragment containing the HS4, HS3, and HS2 of the LCR, but not the other regions, is reduced by 30% in the Δβp line when compared with the wildtype control. This reduction is presumably due to the absence of the factor-binding sites normally present in the promoter. In nonexpressing brain, both transgenic lines show an essentially linear conformation of the locus (Fig. 3B,C; data not shown; Palstra et al. 2003). Whether the remaining association of the β-promoterless gene is (in part) the result of γ- and δ-gene expression is presently not clear. In any case, we suggest that the association of the β-globin gene with the ACH is not the result of transcription and that it would normally precede transcription.
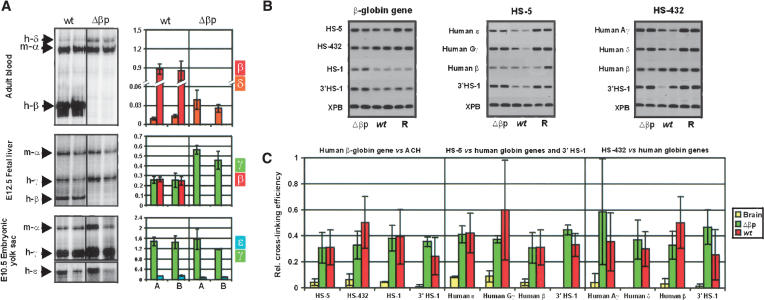
The effect of the human β-globin promoter deletion on the locus. (A) S1 nuclease protection assays showing the up-regulation of the δ- and γ-globin genes in the absence of the human β-globin promoter in the adult and fetal developmental stages, respectively. S1 assays were quantitated by PhosphorImager analysis and corrected for copy number and probe specific activity. The levels of expression shown in the histogram on the right are per copy of the human globin genes, normalized to the level seen for the endogenous mouse-α globin gene. (B) Combined agarose gels of the PCR fragments resulting from the 3C ligation experiments. (R) Random control. (C) Histogram of the relative cross-linking efficiencies after quantitation and normalization of the gels shown in B. The histogram is the average of at least three separate experiments, each in duplicate.
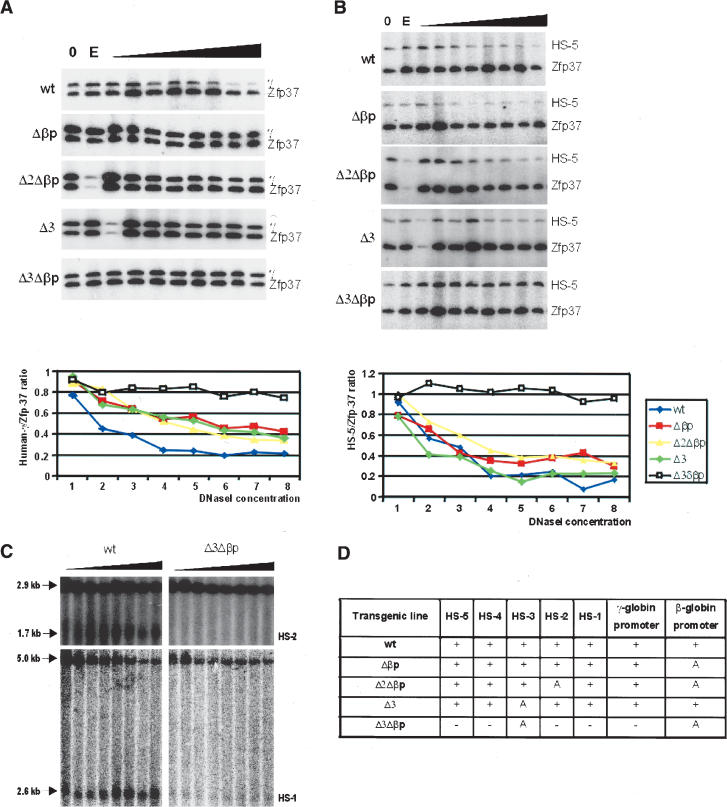
DNaseI accessibility assessed by DNaseI–PCR analysis. PCR product series of the human γ-globin promoter (A) and HS5 (B), in the wild-type, Δβp, Δ2Δβp, Δ3, and Δ3Δβp PAC transgenic mouse lines. An increase in the relative amount of the PCR product, normalized to the signal of mouse Zfp37 PCR products, reflects a decreased DNaseI sensitivity. Quantitation of DNaseI sensitivity in the human γ-globin gene promoter or HS5, relative to the mouse Zfp37 promoter, is shown below each panel. (C) Southern blot analysis to show the loss of hypersensitivity in the LCR of the Δ3Δβp. Sizes of the PstI restriction fragments and the fragments resulting from DNaseI digestion of HS1 and HS2 are indicated. Probes were PCR fragments from the 5′ end of the HS1 fragment and the 3′ end of the HS2 fragment. (D) Summary of the data obtained during chromatin hypersensitivity mapping in various sites in the human β-globin locus and the LCR, as determined by DNaseI PCR assays in the transgenic lines analyzed. (A) Absence of HS due to the targeted deletion.
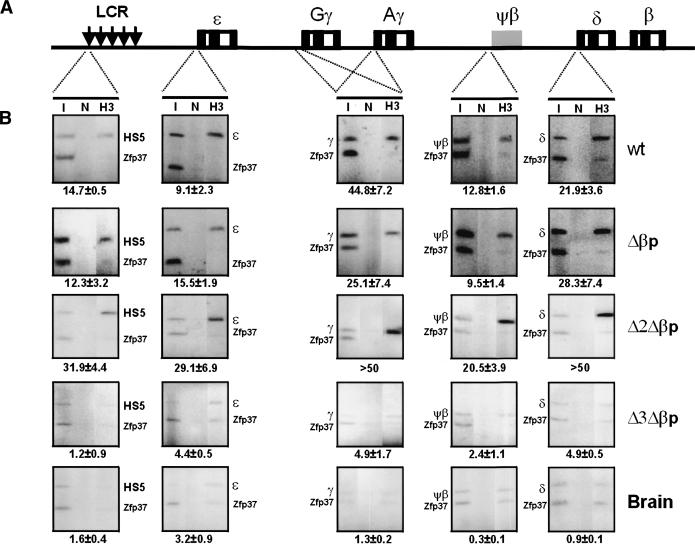
ChIP assays. (A) The various regions within the human β-globin locus that have been analyzed for the acetylation status of histones H3 and H4. (B) Acetylation status of histone H3 throughout the human β-globin locus in the wild-type, Δβp, Δ2Δβp, and Δ3Δβp E12.5 transgenic fetal livers and brain. ChIP assays were carried out as described in Materials and Methods. The PCR products corresponding to the human β-globin locus were normalized against mouse Zpf37 promoter PCR products and inputs. The enrichment over Zfp37 is given below the panel. (I) Input; (N) no antibody control; (H3) acetylated histone H3 antibody.
Expression of the mutant Δ2Δβp and Δ3Δβp β-globin loci
The second aim of this work was to determine which of the most active sites of the LCR (HS2 and HS3) interact with the promoter of the β-globin gene, using double mutants. In addition, we wanted to know what effect these mutations would have on the 3-D structure of the locus. To this end, the HS2 or HS3 were deleted from the PAC, leading to the Δ2 and Δ3 lines (see earlier), and these deletions were combined with the deletion of the β-globin promoter, resulting in the Δ2Δβp and Δ3Δβp lines, respectively (see earlier). Like the Δ2 and Δ3 lines that have been previously analyzed by several laboratories as part of cosmid or YAC-based human β-globin loci (Bungert et al. 1995; Milot et al. 1996; Peterson et al. 1996), or in the endogenous mouse locus (Fiering et al. 1995; Hug et al. 1996), the deletions in our PACs result in a transcriptional loss of the β-globin gene of ~30% at the definitive stage (Fig. 4A), unless the constructs were integrated in heterochromatin, resulting in position effect variegation (data not shown; Milot et al. 1996). The double mutants Δ2Δβp and Δ3Δβp were first analyzed at the adult stage for δ- and β-globin gene expression (Fig. 4A). This shows that the level of δ-globin gene has increased in all four Δ2Δβp lines (single or multicopy) when compared with the levels observed in the wildtype locus and is very similar to what is observed with the single Δβp mutant locus. Thus, the additional ΔHS2 mutation has no effect, suggesting that HS2 normally interacts with the promoter of the β-globin gene at the adult stage. This would be in agreement with its role as a classical transcriptional enhancer. Surprisingly, the expression of the δ-globin gene in the Δ3Δβp double-mutant transgenic lines is completely extinguished. To ensure that the double Δ3Δβp mutant is not completely defective, we carried out timed mating for two lines of each construct.
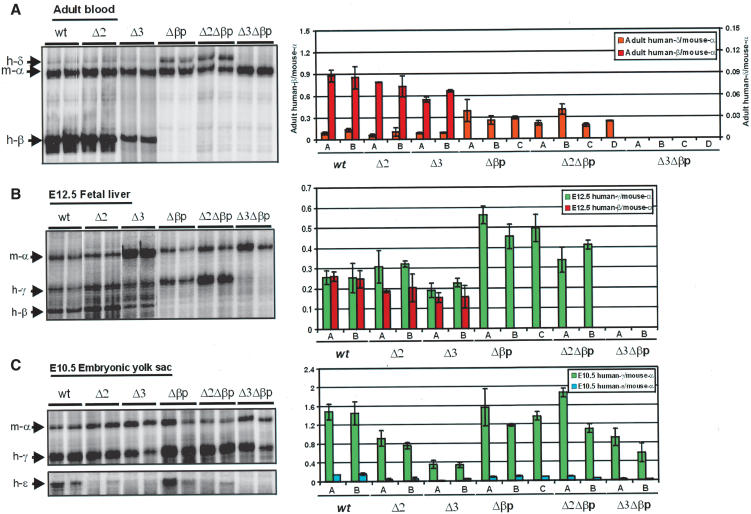
S1 nuclease protection assays. S1 nuclease protection assays are shown for one transgenic line for the Δβp deletion constructs in the context of a full or incomplete LCR at the adult (A), fetal (E12.5, B), and embryonic (E10.5, C) developmental stages (see also Materials and Methods). Two littermates for each developmental stage were analyzed, using 32P-labeled probes for mouse-α and human ε-, γ-, δ-, and β-globin genes. Each of the protected fragment positions is indicated. S1 assays were quantitated by PhosphorImager analysis and corrected for copy number and probe specific activity. The values are per copy levels of expression of human globin genes, as a ratio of the endogenous mouse-α globin gene and shown as a histogram on the right. The histogram shows the analysis of all transgenic lines for each construct. Each experiment has been done in triplicate.
The results (Fig. 4B,C) show that ε- and γ-globin gene expression at the embryonic stage and the early fetal liver stage for the Δ2Δβp transgenic line is very similar to that observed for the Δβp line. At the early fetal liver stage, γ-globin gene expression has approximately doubled when compared with a wild-type locus because of the absence of competition of the β-globin gene. This was confirmed by primary transcript in situ hybridization analysis (Wijgerde et al. 1995; data not shown). In contrast, the ε- and γ-globin gene expression levels were the same as in the wild-type locus at the embryonic stage in primitive cells. At this stage, the β-globin gene is not expressed and hence the competitive effect is absent.
Interestingly, the expression of the γ-globin genes in the double mutant Δ3Δβp is completely absent in early fetal liver (Fig. 4B, confirmed by in situ primary transcript analysis, not shown). However, they are expressed at the embryonic stage (Fig. 4C), although the level has been decreased by ~40%. An even bigger decrease is observed for ε-globin gene expression. This result shows that the locus can be expressed in primitive, but not definitive, cells and suggests that HS3 interacts with sequences other than the basic promoter of the β-globin gene, for example, the more upstream part of the promoter, the enhancer just downstream of the gene, or sequences much farther away from the β-globin gene (for review, see Stamatoyannopoulos and Grosveld 2001). The result also suggests that the Δ3Δβp double mutation may completely destabilize the normal chromatin structure of the locus in definitive cells. We therefore carried out DNaseI sensitivity assays of the double- and single-mutant transgenic lines.
DNaseI hypersensitive site analysis
Globin gene transcription is normally correlated with the presence of DNaseI HSs at the promoter of the expressed genes and the LCR (Tuan et al. 1985; Grosveld et al. 1987). In order to determine the effect of the targeted deletions in the β-globin promoter alone and in combination with HS2 or HS3, we analyzed DNaseI hypersensitivity of the γ-globin promoter and of the individual HSs of the LCR in isolated nuclei from wild-type, Δβp, Δ2Δβp, Δ3, and Δ3Δβp E12.5 fetal liver cells, using DNaseI PCR (Fig. 5A,B; McMorrow et al. 2000) and Southern blots (Fig. 5C). The results show that the γ-globin gene remains DNaseI sensitive in all of the lines with the exception of the Δ3Δβp line. In this double mutant, sensitivity has become the same as observed in the control murine Zfp37 gene. The latter has a low DNaseI sensitivity (i.e., it is not insensitive, McMorrow et al. 2000) in the early fetal liver (Fig. 5A). The hypersensitivity of all of the other HSs has been maintained in all of the lines with the exception of the Δ3Δβp line (Fig. 5B–D). Surprisingly, in this line all hypersensitivity to DNaseI digestion has been lost. We conclude that this is due to the double mutation and not just the absence of HS3, because in the Δ3 single mutation line, HS5, HS4, HS2, and HS1 are still present (Fig. 5B [green line], D). The loss of all hypersensitivity suggests that the chromatin structure of the Δ3Δβp double mutant locus has been severely affected and that the entire ACH structure might have been lost in this double mutant.
Histone acetylation and 3C analysis of the mutant β-globin loci
In order to link our DNaseI hypersensitivity mapping data to the chromatin acetylation status, we performed chromatin immunoprecipitation (ChIP) assays in formaldehyde-fixed chromatin, isolated from E12.5 nuclei from the same transgenic lines analyzed for DNaseI hypersensitivity. The assays were performed using antibodies specific for acetylated histones H3 (Ac-H3) and H4 (Ac-H4) and quantitative PCR at different sites throughout the locus (Fig. 6A). As would be expected from the DNaseI sensitivity analysis, the histone acetylation status follows the gene expression and DNaseI sensitivity patterns. There was a significant enrichment of the acetylated histones H3 (Fig. 6) and H4 (data not shown) in all of the expressing lines throughout the locus in erythroid cells, but not in the brain. In contrast, the enrichment of acetylated histones was much less in the Δ3Δβp locus, indeed suggesting that the chromatin structure has been severely affected.
We therefore carried out a 3C analysis of the double mutant Δ3Δβp locus versus the wild type, the single mutant Δ3, and Δβp loci. Figure 7A shows one of the triplicate PCR amplifications of the ligation after cross-linking of the locus in E12.5 fetal liver cells, using the XPB gene as the reference and an EcoRI-restriction-digested β-globin PAC as the PCR control (Palstra et al. 2003). Figure 7B summarizes the quantitation data of all of the 3C experiments. Two points stand out. First, when the β-globin gene is used as the fixed fragment and measured against the other sites forming the ACH, the interaction of the gene with the fragment containing the most active sites of the LCR (HS2 and HS3) in the single mutant Δ3 is decreased to a similar level as that observed for the Δβp mice (as discussed earlier). Its interaction with the other sites in the ACH has remained the same (after normalization; see the legend for Fig. 7). This suggests that HS3 significantly contributes to the stability of the ACH. Second and importantly, all interactions, including the HS5 interactions, are lost in the double mutant Δ3Δβp line, correlating loss of ACH formation with the lack of γ- and δ-globin gene expression in definitive cells, the decrease in histone acetylation, and the loss of HS5. The same analysis on embryonic blood of the double mutant shows that the ACH can be formed at the stage when the ε- and γ-genes are expressed (Fig. 8), which suggests that there is no intrinsic defect preventing ACH formation.
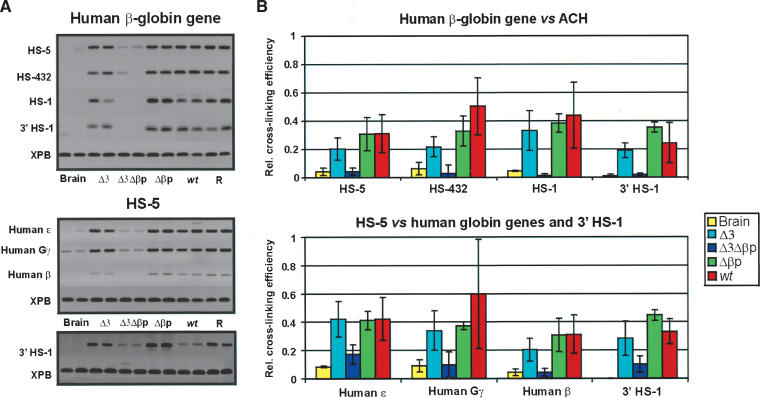
Spatial organization of the mutant transgenic globin gene loci in definitive fetal erythroid cells. (A) Agarose gels, showing the PCR products as a result of 3C ligations representing the various interactions between the β-globin gene and the HS5, relative to the HSs of the ACH and the globin genes, respectively. (R) Random control (Palstra et al. 2003). (B) Calculation of the relative cross-linking efficiency of the fragment containing the human β-globin gene relative to the HSs forming the ACH and of an HS5 fragment, relative to the globin genes and 3′ HS1 (based on at least three experiments in duplicate per primer set). Relative cross-linking efficiencies were normalized for the number of positive embryos analyzed, that is, the number of human transgenic loci relative to the mouse control locus. Note that the contacts between HS5 and 3′HS1 are maintained in the Δ3Δβp transgenic line, albeit weaker when compared with the remaining transgenic lines, indicative of the CH complex formation in erythroid cells.
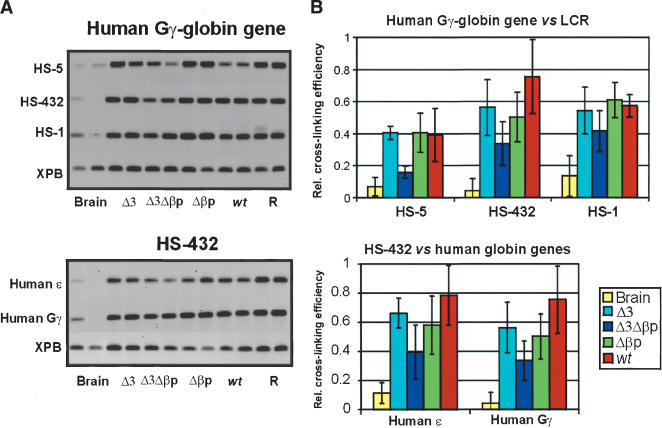
Spatial organization of the mutant transgenic globin gene loci in primitive E10.5 erythroid cells. (A) Agarose gels, showing the PCR products as a result of 3C ligations representing the various interactions between the Gγ-globin gene (upper panel) or the HS432 fragment (lower panel), relative to the HSs of the LCR and the globin genes. (R) Random control (Palstra et al. 2003). (B) Calculation of the relative cross-linking efficiency shown in A.
Discussion
We have analyzed the role of the most active sites of the LCR, HS2 and HS3, and the β-globin promoter on the expression, the chromatin structure, and the 3-D configuration of the human β-globin locus, using a 185-kb human β-globin PAC inserted in transgenic mice. This locus contains the LCR, all of the genes, and the 3′HS1 and forms, just like the endogenous mouse locus, an ACH with active transcription (Fig. 1; Palstra et al. 2003). The results are the first functional validation of the role of the LCR and the genes in the formation/maintenance of the ACH.
We propose at this stage of the 3C work to use two terms for the complex of interactions we have analyzed previously and in this paper. The complex formed by hypersensitive sites, such as that seen by Palstra et al. (2003) in precursor cells, we propose to name a chromatin hub (CH, rather than pre-ACH), whereas the complex involving hypersensitive sites and active genes (i.e., the flanking sites, the LCR, and the genes) is named the active chromatin hub (ACH) as originally proposed by Tolhuis et al. (2002).
Deletion of the β-globin gene promoter
A human β-globin promoter deletion was generated in the context of the full LCR to explore the interactions between the LCR and the β-globin gene and whether or not the interaction of the gene with the ACH is the result of transcription. We have recently shown for the mouse locus that committed precursor cells, which do not yet transcribe the β-globin genes, already show an interaction between the most 5′ part of the LCR and the HSs flanking the locus (Tolhuis et al. 2002; Palstra et al. 2003). Interestingly, all of these elements contain CTCF-binding sites (Farrell et al. 2002; Bulger et al. 2003). When the cells differentiate, the full LCR and the expressed genes participate in the interaction, forming the ACH and the active transcriptosome. However, those results do not show whether the interaction of the globin genes with the ACH is required for transcription or whether the association with the ACH is simply the result of transcription.
The deletion of the β-globin promoter does not lead to any appreciable change in the expression pattern of the ε- and γ-globin genes in primitive cells when the β-globin gene is normally not transcribed. However, in definitive cells, when the β-globin gene is normally transcribed, it results in an increase of transcription of the γ- and δ-globin genes, in agreement with the earlier results, showing that the genes are transcribed competitively with a single gene expressed at any particular moment (Wijgerde et al. 1995 and references therein), and the total output of the locus remains the same when another β-globin gene is added (Dillon et al. 1997; Wai et al. 2003). Such a mechanism suggests that the ACH would only be able to bind a single gene for transcription. Deletion of the β-globin promoter leads to a reduction in the frequency of interaction of the β-globin gene with the LCR in the ACH (~30%), but importantly, a substantial interaction remains. Unfortunately, our experiments do not show to what extent the remaining interaction is due to the interactions of the δ- and γ-genes or whether the remaining interaction of the β-globin gene still competes with those of the γ- or δ-globin genes with the ACH. In any case, the results of the deletion of the β-globin promoter strongly suggest that other (nonpromoter) interactions take place between the ACH and the β-globin gene (see following). In addition, the results suggest that the interactions with the ACH or the stability of the ACH is not dependent on transcription of the β-globin gene per se and that the association of the β-globin gene with the ACH is also not the result of its transcription. Instead, we suggest that this association may be a requisite for transcription and could provide an explanation for proposals like the LCR being involved in loading the polymerase on the promoter (Johnson et al. 2001).
Deletion of the β-globin promoter combined with LCR HS2 and HS3 deletion
The β-globin promoter deletion was subsequently combined with the deletion of either HS2 or HS3 in cis, because previous data have implicated these HSs as functionally the most important in the LCR; that is, HS2 has the greatest enhancing activity and HS3 the most efficient chromatin opening activity (Ellis et al. 1996; Milot et al. 1996).
There was no significant difference in γ-globin gene expression between the wild-type and the Δβp and Δ2Δβp transgenic lines during primitive erythropoiesis. In contrast, in the absence of HS3, that is, in the Δ3 and Δ3Δβp lines, γ-globin expression is reduced, when compared with the wild-type PAC transgenic line. This decrease in γ- (and more severe decrease in ε-) globin gene expression in primitive cells of the Δ3Δβp lines is very similar to what is seen here and published previously for deletion of HS3 only (Milot et al. 1996; Navas et al. 1998). The fact that there is no difference between the Δ3 and Δ3Δβp lines confirms that HS3 is important in the primitive expression pattern.
The situation is very different in definitive erythroid cells. The absence of the β-globin promoter in conjunction with the HS2 deletion leads to very similar levels of expression of the γ- and δ-globin genes when compared with the β-globin promoter deletion alone. This suggests that HS2 normally interacts primarily with the β-globin globin gene promoter. If HS2 were to primarily interact with sequences other than the β-globin promoter (e.g., the remainder of the gene or its flanking sequences or the γ- or δ-globin gene promoter or yet other sequences), these “other” interactions would be lost with the HS2 deletion and would probably result in a decrease of transcription of the γ- and δ-globin genes. Although it has been suggested that HS2 and the β-globin promoter interact on the basis of the RNA TRAP analysis (Carter et al. 2002) or the 3C analysis (Palstra et al. 2003), these techniques currently do not have sufficient resolution to distinguish the different parts of the LCR and/or the globin genes to actually confirm an HS2/β-globin promoter interaction.
In contrast, the absence of the β-globin promoter in conjunction with the absence of HS3 has a dramatic effect on the expression of the γ- and δ-globin genes in definitive cells. The transcripts are no longer detectable and all DNaseI sensitivity is lost. This suggests that HS3 normally interacts with sequences other than the β-globin promoter and that this interaction is important for transcription of the globin genes and maintenance of the ACH. For the same resolution reasons as described for HS2, it is presently not clear what these sequences would be, although in the case of the β-globin gene, its 3′ enhancer would be a candidate for such an interaction (Liu et al. 1997).
The 3-D structure of the locus
Three aspects of the double deletion Δ3Δβp mutant are most interesting, namely, the observation that the ACH is destabilized, all DNaseI hypersensitivity (including HS5) is lost, and histone acetylation throughout the locus is severely affected. We have previously shown that the region comparable to HS5 in the murine locus (the 5′ side of the LCR) already interacts with the hypersensitive sites flanking the region, 3′ HS1 and the HS-62.5/HS-60 in murine erythroid precursors before the globin genes are transcribed (Palstra et al. 2003). We do not know why the hypersensitivity at HS5, HS2, and HS1 is lost and whether this loss is the result of ACH destabilization. A number of labs have previously shown that HS1–4 are involved in the efficiency of β-globin gene expression, and the rather weak hypersensitive site HS5 is not required for the expression of the human β-globin locus in mice and that its only role would be a border function in embryonic, but not fetal/adult red cells (Wai et al. 2003 and references therein). However, it is present in the globin CH at all stages analyzed to date. In addition, it is also known that the upstream HS-111 (the equivalent of mouse HS-62.5/HS-60, Bulger et al. 2003; also not present in all of the lines reported here) and downstream 3′ HS1 are not required for a high level of expression of the β-globin locus when introduced in transgenic mice (Strouboulis et al. 1992) and that such lines express at the same level as lines that contain the 5′ and 3′ flanking sites (Bungert et al. 1995; Peterson et al. 1996; Chang et al. 1998; Calzolari et al. 1999). At first sight, these data would suggest that the HSs involved in the CH complex formation are not very important, but why then would there be a CH? We have suggested a general “organizational” role for these flanking elements (Palstra et al. 2003). They are all binding sites for CTCF (Farrell et al. 2002; Bulger et al. 2003), of which there are many throughout the genome. Erythroid-specific factors such as EKLF and chromatin remodeling factors (Armstrong et al. 1998) may make these binding sites accessible to CTCF or similar factors (they are erythroid-specific hypersensitive sites). Alternatively, CTCF may bind first. CTCF and associated proteins would then act as a self-organizer to provide a CH complex to enhance further looping to form the complete erythroid-specific ACH (Fig. 1B) through the action of additional factors present on the LCR and genes. We postulate that the exact position of the flanking HS sites relative to the LCR and genes is not very important (they are not required for normal expression) and that CTCF (or similar protein) sites scattered throughout the genome could take over the function. Thus, in transgenic mouse experiments in the absence of the flanking HSs (e.g., the HS-111 and 3′ HS1; Fig. 1A; as in Strouboulis et al. 1992), other CTCF sites at the site of integration of the transgene would take over the initial CH organization step. When multiple interactions of the ACH and the gene are disrupted, the ACH becomes destabilized, and hence the high concentration of histone modification enzymes (postulated in de Laat and Grosveld 2003) would diminish, resulting in a loss of acetylation, as observed in the Δ3Δβp mice.
In conclusion, we postulate that the active organization of the locus would use similar principles (but on a larger scale) as the folding of an enzyme to create an active site or “pocket”. Initial folding of the locus would be nonspecific in terms of exact position or site but would provide the template to allow the proper and precise folding of the LCR (which would be like creating an active site of an enzyme) and allow interactions with the genes (which in the analogy is the substrate). In other words, the function of the CH complex lies in the fact that it would cause a spatial restriction by forming a loop containing the genes and LCR and hence stimulate ACH formation. When different sets of genes are present in a locus, they may use the same or different flanking sites to initiate the “folding” process, followed by the interaction of specific sequences (like the globin LCR HS1–4) to create a compartment, the ACH, to transcribe the different genes in different tissues (de Laat and Grosveld 2003).
Materials and methods
Generation of human β-globin locus deletion constructs
A PAC clone containing the entire human β-globin locus was modified by homologous recombination in E. coli DH10B cells, as described previously (Imam et al. 2000).
For the generation of the β-globin promoter deletion (Δβp), a 483-bp fragment was PCR amplified using Vent DNA polymerase (NEBiolabs) and cloned into pBluescript (Stratagene). Subsequently, a β-globin promoter deletion was generated, spanning from position -139 to +49 relative to the Cap site of the gene, using StyI/NcoI restriction endonucleases, respectively. DNA sequencing was performed to confirm the deletion breakpoint and to exclude the possibility of sequence alterations introduced by Vent polymerase during the PCR step. A HincII/SmaI fragment was then subcloned into the NaeI site of pDF-25 recombination vector and used for the subsequent targeting procedure.
The generation of the HS2 (Δ2) and HS3 deletion (Δ3) has been described elsewhere (Milot et al. 1996). A 1.2-kb and a 1.5-kb HindIII fragment containing the HS2 and HS3 deletions, respectively, have been subcloned into the HindIII site of the pDF-25 recombination vector for the targeting procedure into a β-globin gene promoter deletion-containing PAC (Δβp).
Following modifications, the resulting PAC clones were mapped extensively to ensure that only the desired modification was present and no other alteration had occurred during the targeting procedure.
Transgenic mice
Purified 185-kb NotI fragments were microinjected into the pronuclei of FVB fertilized mouse oocytes and transferred into the oviducts of pseudopregnant F1 (CABxC57B1) female mice. Transgenic founders were identified by Southern blotting of DNA isolated from tail biopsies and probed with cosmids (cos) LCRε and cosγγδβ, containing the 5′ and 3′ parts of the human globin locus, respectively (Strouboulis et al. 1992). In addition, PCR reactions were performed to verify the deletion junction points. Those founders that contained all of the characteristic EcoRI fragments for the entire locus were processed for breeding and further analysis. Copy numbers were determined by quantitation of Southern blots via PhosphorImager analysis. Internal probes were also used to verify the structure of the transgenes.
RNA isolation and S1 nuclease protection assay
RNA was isolated from frozen transgenic E10.5 embryonic yolk sacs, E12.5 fetal livers, and adult blood and subjected to S1 nuclease protection assays as previously described (Wijgerde et al. 1996). S1 probes were end-labeled with T4 polynucleotide kinase and equimolar amounts were used per reaction. Signal quantitation was performed using a PhosphorImager (Molecular Dynamics).
DNA and primary transcript fluorescence in situ hybridization (FISH)
Peripheral blood cells were cultivated for 24 h in RPMI 1640 medium (GIBCO-BRL) and chromosome preparations were made according to standard procedures. DNA FISH was carried out as described in Mulder et al. (1995), using the human LCR probe labeled with digoxigenin and immunohistochemically detected with fluorescein. DNA was counterstained with DAPI.
Primary transcript in situ hybridization to detect transcription of the human and mouse globin genes in the yolk sac and fetal liver cells was performed as described previously (Wijgerde et al. 1995). Quantitation of transcriptional signals was done by counting at least 300 cells from each slide with a fluorescence microscope.
Isolation of nuclei and DNaseI hypersensitive site analysis
Nuclei suspension from six transgenic E12.5 fetal livers for each line was performed by 20 strokes of a Dounce pestle (type B) and 100-μL aliquots were digested with increasing amounts of DNaseI at 37°C for 3 min. Reactions were stopped and treated with proteinase K and extracted with phenol/chloroform. After ethanol precipitation, DNA was resuspended in water in a concentration of 10 ng/μL.
PCR: Radioactive PCR was performed to determine DNaseI sensitivity of human γ- and β-globin gene promoter and the individual LCR HSs, using 10 ng of template DNA as previously described (McMorrow et al. 2000). Primer sets were as follows: human γ-globin gene promoter (forward, 5′-GCCTTGACCAATAGCCTTGACA-3′; reverse, 5′-CTGGACTAGGAGCTTATTGATAAC-3′; fragment size, 171 bp), HS5 (forward, 5′-CTGACCTATATCTGGCAGGACTC-3′; reverse, 5′-GGAAACTGGAGAAACTGGGA-3′; fragment size, 205 bp), HS3 (forward, 5′-ACTTTGCGAGCTGGTGTGC-3′; reverse, 5′-ATCTCTCCTGGCTCCTGGGA-3′; fragment size, 180 bp), HS2 (forward, 5′-CCCTCTATCCCTTCCAGCATC-3′; reverse, 5′-TTGAGCCAGAAGGTTTGCTTA-3′; fragment size, 200 bp), HS1 (forward, 5′-TTCTGTGCTTTCCTGGCTG-3′; reverse, 5′-ATGACGGGTTGATGGGTG-3′; fragment size, 210 bp). Primer sets and product sizes for HS4, human β-globin promoter, mouse βmaj, and Zfp37 and PCR conditions have been published previously (McMorrow et al. 2000). Each reaction has been done in triplicate and band intensity has been quantitated using Phosphor-Imager analysis and normalized against either mouse Zfp37 or βmaj signal.
Southern blots: DNaseI-treated DNA was digested with HindIII or PstI, run on 0.7% agarose gels, blotted, and hybridized with PCR-generated probes from the ends of the restriction fragments corresponding to the different hypersensitive sites.
ChIP
Five transgenic E12.5 fetal livers and brains for each line were incubated in DMEM, containing 1% formaldehyde and 10% fetal calf serum at 37°C for 10 min. Following washing with PBS, the cells were pelleted and resuspended in 200 μL SDS lysis buffer (Upstate Biotechnology) for 10 min on ice. Cell lysate was then sonicated to reduce DNA length to between 200 bp and 1 kb and debris was removed by centrifugation. ChIP was performed using the acetyl-histone H3 immunoprecipitation assay kit (Upstate Biotechnology) and acetyl-histone H4 immunoprecipitation assay kit (Upstate Biotechnology), according to the manufacturer's instructions. Enrichment for specific immunoprecipitated sequences, including HS5, ε-, γ-, Ψβ-, and δ-globin genes promoters, were determined by radioactive PCR, using primer sets and conditions as described (McMorrow et al. 2000 and this study). Input and unbound DNA samples were included as negative controls. Each reaction has been done in triplicate and band intensity has been quantitated using PhosphorImager analysis and normalized against mouse Zfp37 signal.
Chromosome conformation capture (3C) analysis
Isolation and formaldehyde fixation of embryonic blood (from E10.5 embryos) and fetal liver cells (from E12.5 embryos), EcoRI restriction enzyme digestion of cross-linked DNA in the nucleus, intramolecular ligation, reversal of cross-links, PCR analysis of ligation products, and calculation of relative cross-linking frequencies were carried out, as previously described (Tolhuis et al. 2002; Palstra et al. 2003). In order to correct for differences in quality and quantity of template, we normalized the ligation frequencies between globin site pairs to those detected between two restriction fragments (with the sites analyzed 8.3 kb apart) in the XPB gene locus (encoding for a subunit of the basal transcription factor TFIIH), assuming that expression levels and spatial conformation of this gene were similar in all analyzed tissues. Also, to compare signal intensities obtained with different primer sets in a quantitative manner, we included a control template containing all possible ligation products in equimolar amounts to correct for the PCR amplification efficiency of each primer set. For this purpose, we used a 185-kb PAC clone, spanning the complete transgenic locus and a 60–70-kb PAC containing the control mouse XPB locus (PAC Clone #443-C18, MRC gene service).
Acknowledgments
We are indebted to Patrick Molenbeek, Vincent Maes, and Agnes Boer for animal care; to John Kong-a-San for microinjection; to Nynke Gillemans, Bas Tolhuis, Miyata Masato, and Petros Papadopoulos for technical assistance; and to Drs. and Sjaak Philipsen for critically reading of this manuscript. This work was supported by a Marie Curie long-term fellowship (IHPMFCT199900175) to G.P.P. and by the European Commission and the Nederlandse Organisatie voor Weteuschappelijh Orderzoek (The Netherlands) grants to F.G.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Notes
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.289704.
References
- Armstrong J.A., Bieker, J.J., and Emerson, B.M. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95: 93-104. [Abstract] [Google Scholar]
- Bulger M., Schubeler, D., Bender, M.A., Hamilton, J., Farrell, C.M., Hardison, R.C., and Groudine, M. 2003. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 23: 5234-5244. [Europe PMC free article] [Abstract] [Google Scholar]
- Bungert J., Dave, U., Lim, K.C., Lieuw, K.H., Shavit, J.A., Liu, Q., and Engel, J.D. 1995. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes & Dev. 9: 3083-3096. [Abstract] [Google Scholar]
- Calzolari R., McMorrow, T., Yannoutsos, N., Langeveld, A., Grosveld, F. 1999. Deletion of a region that is a candidate for the difference between the deletion forms of hereditary persistence of fetal hemoglobin and δβ-thalassemia affects β- but not γ-globin gene expression. EMBO J. 18: 949-958. [Europe PMC free article] [Abstract] [Google Scholar]
- Carter D., Chakalova, L., Osborne, C.S., Dai, Y.F., and Fraser, P. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32: 623-626. [Abstract] [Google Scholar]
- Chang J., Lu, R., Lin, C., Xu, S.M., Kan, Y.W., Porcu, S., Carlson, E., Kitamura, M., Yang, S., Flebbe-Rehwaldt, L., et al. 1998. Transgenic knockout mice exclusively expressing human hemoglobin S after transfer of a 240-kb β-globin yeast artificial chromosome: A mouse model of sickle cell anemia. Proc. Natl. Acad. Sci. 95: 14886-14890. [Europe PMC free article] [Abstract] [Google Scholar]
- de Laat W. and Grosveld, F. 2003. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 11: 447-459. [Abstract] [Google Scholar]
- Dekker J., Rippe, K., Dekker, M., and Kleckner, N. 2002. Capturing chromosome conformation. Science 295: 1306-1311. [Abstract] [Google Scholar]
- Dillon N., Trimborn, T., Strouboulis, J., Fraser, P., and Grosveld, F. 1997. The effect of distance on long-range chromatin interactions. Mol. Cell 1: 131-139. [Abstract] [Google Scholar]
- Ellis J., Talbot, D., Dillon, N., and Grosveld, F. 1993. Synthetic human β-globin 5′HS2 constructs function as locus control regions only in multicopy transgene concatamers. EMBO J. 12: 127-134. [Europe PMC free article] [Abstract] [Google Scholar]
- Ellis J., Tan-Un, K.C., Harper, A., Michalovich, D., Yannoutsos, N., Philipsen, S., and Grosveld, F. 1996. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 15: 562-568. [Europe PMC free article] [Abstract] [Google Scholar]
- Farrell C.M., West, A.G., and Felsenfeld, G. 2002. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol. 22: 3820-3831. [Europe PMC free article] [Abstract] [Google Scholar]
- Fiering S., Epner, E., Robinson, K., Zhuang, Y., Telling, A., Hu, M., Martin, D.I., Enver, T., Ley, T.J., and Groudine, M. 1995. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-globin locus. Genes & Dev. 9: 2203-2213. [Abstract] [Google Scholar]
- Forrester W.C., Epner, E., Driscoll, M.C., Enver, T., Brice, M., Papayannopoulou, T., and Groudine, M. 1990. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes & Dev. 4: 1637-1649. [Abstract] [Google Scholar]
- Fraser P., Pruzina, S., Antoniou, M., and Grosveld, F. 1993. Each hypersensitive site of the human β-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes & Dev. 7: 106-113. [Abstract] [Google Scholar]
- Grosveld F., van Assendelft, G.B., Greaves, D.R., and Kollias, G. 1987. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51: 975-985. [Abstract] [Google Scholar]
- Hanscombe O., Whyatt, D., Fraser, P., Yannoutsos, N., Greaves, D., Dillon, N., and Grosveld, F. 1991. Importance of globin gene order for correct developmental expression. Genes & Dev. 5: 1387-1394. [Abstract] [Google Scholar]
- Hardison R.C., Chui, D.H., Giardine, B., Riemer, C., Patrinos, G.P., Anagnou, N., Miller, W., and Wajcman, H. 2002. Hb-Var: A relational database of human hemoglobin variants and thalassemia mutations at the globin gene server. Hum. Mutat. 19: 225-233. [Abstract] [Google Scholar]
- Hug B.A., Wesselschmidt, R.L., Fiering, S., Bender, M.A., Epner, E., Groudine, M., and Ley, T.J. 1996. Analysis of mice containing a targeted deletion of β-globin locus control region 5′ hypersensitive site 3. Mol. Cell. Biol. 16: 2906-2912. [Europe PMC free article] [Abstract] [Google Scholar]
- Imam A.M., Patrinos, G.P., de Krom, M., Bottardi, S., Janssens, R.J., Katsantoni, E., Wai, A.W., Sherratt, D.J., and Grosveld, F.G. 2000. Modification of human β-globin locus PAC clones by homologous recombination in Escherichia coli. Nucleic Acids Res. 28: E65. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson K.D., Christensen, H.M., Zhao, B., and Bresnick, E.H. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8: 465-471. [Abstract] [Google Scholar]
- Kioussis D., Vanin, E., deLange, T., Flavell, R.A., and Grosveld, F.G. 1983. β-Globin gene inactivation by DNA translocation in gamma β-thalassaemia. Nature 306: 662-666. [Abstract] [Google Scholar]
- Liu Q., Bungert, J., and Engel, J.D. 1997. Mutation of geneproximal regulatory elements disrupts human ε-, γ-, and β-globin expression in yeast artificial chromosome transgenic mice. Proc. Natl. Acad. Sci. 94: 169-174. [Europe PMC free article] [Abstract] [Google Scholar]
- McMorrow T., van den Wijngaard, A., Wollenschlaeger, A., van de Corput, M., Monkhorst, K., Trimborn, T., Fraser, P., van Lohuizen, M., Jenuwein, T., Djabali, M., et al. 2000. Activation of the β globin locus by transcription factors and chromatin modifiers. EMBO J. 19: 4986-4996. [Europe PMC free article] [Abstract] [Google Scholar]
- Milot E., Strouboulis, J., Trimborn, T., Wijgerde, M., de Boer, E., Langeveld, A., Tan-Un, K., Vergeer, W., Yannoutsos, N., Grosveld, F., et al. 1996. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87: 105-114. [Abstract] [Google Scholar]
- Mulder M.P., Wilke, M., Langeveld, A., Wilming, L.G., Hagemeijer, A., van Drunen, E., Zwarthoff, E.C., Riegman, P.H., Deelen, W.H., van den Ouweland, A.M., et al. 1995. Positional mapping of loci in the DiGeorge critical region at chromosome 22q11 using a new marker (D22S183). Hum. Genet. 96: 133-141. [Abstract] [Google Scholar]
- Navas P.A., Peterson, K.R., Li, Q., Skarpidi, E., Rohde, A., Shaw, S.E., Clegg, C.H., Asano, H., and Stamatoyannopoulos, G. 1998. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol. Cell. Biol. 18: 4188-4196. [Europe PMC free article] [Abstract] [Google Scholar]
- Palstra R.J., Tolhuis, B., Splinter, E., Nijmeijer, R., Grosveld, F., and de Laat, W. 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35: 190-194. [Abstract] [Google Scholar]
- Peterson K.R., Clegg, C.H., Navas, P.A., Norton, E.J., Kimbrough, T.G., and Stamatoyannopoulos, G. 1996. Effect of deletion of 5′HS3 or 5′HS2 of the human β-globin locus control region on the developmental regulation of globin gene expression in β-globin locus yeast artificial chromosome transgenic mice. Proc. Natl. Acad. Sci. 93: 6605-6609. [Europe PMC free article] [Abstract] [Google Scholar]
- Philipsen S., Talbot, D., Fraser, P., and Grosveld, F. 1990. The β-globin dominant control region: Hypersensitive site 2. EMBO J. 9: 2159-2167. [Europe PMC free article] [Abstract] [Google Scholar]
- Splinter E., Grosveld, F., and de Laat, W., 2004. 3C technology: Analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 375: 493-507. [Abstract] [Google Scholar]
- Stamatoyannopoulos G. and Grosveld, F. 2001. Hemoglobin switching. In The molecular basis of blood diseases, 3rd ed. (eds. G. Stamatoyannopoulos et al.), pp. 135-182. WB Saunders, Philadelphia.
- Strouboulis J., Dillon, N., and Grosveld, F. 1992. Developmental regulation of a complete 70-kb human β-globin locus in transgenic mice. Genes & Dev. 6: 1857-1864. [Abstract] [Google Scholar]
- Talbot D., Philipsen, S., Fraser, P., and Grosveld, F. 1990. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 9: 2169-2177. [Europe PMC free article] [Abstract] [Google Scholar]
- Tolhuis B., Palstra, R.J., Splinter, E., Grosveld, F., and de Laat, W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10: 1453-1465. [Abstract] [Google Scholar]
- Tuan D., Solomon, W., Li, Q., and London, I.M. 1985. The `β-like-globin' gene domain in human erythroid cells. Proc. Natl. Acad. Sci. 82: 6384-6388. [Europe PMC free article] [Abstract] [Google Scholar]
- Urnov F.D. and Wolffe, A.P. 2001. Chromatin remodeling and transcriptional activation: The cast (in order of appearance). Oncogene 20: 2991-3006. [Abstract] [Google Scholar]
- Wai A.W.K., Gillemans, N., Raguz-Bolognesi, S., Pruzina, S., Zafarana, G., Meijer, D., Philipsen, S., and Grosveld, F. 2003. HS5 of the human β-globin locus control region: A developmental stage-specific border in erythroid cells. EMBO J. 22: 4489-4500. [Europe PMC free article] [Abstract] [Google Scholar]
- Wijgerde M., Grosveld, F., and Fraser, P. 1995. Transcription complex stability and chromatin dynamics in vivo. Nature 377: 209-213. [Abstract] [Google Scholar]
- Wijgerde M., Gribnau, J., Trimborn, T., Nuez, B., Philipsen, S., Grosveld, F., and Fraser, P. 1996. The role of EKLF in human β-globin gene competition. Genes & Dev. 10: 2894-2902. [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.289704
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/18/12/1495.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Enhancer Function in the 3D Genome.
Genes (Basel), 14(6):1277, 16 Jun 2023
Cited by: 3 articles | PMID: 37372457 | PMCID: PMC10298269
Review Free full text in Europe PMC
Forced enhancer-promoter rewiring to alter gene expression in animal models.
Mol Ther Nucleic Acids, 31:452-465, 31 Jan 2023
Cited by: 1 article | PMID: 36852088 | PMCID: PMC9958407
Chromatin Hubs: A biological and computational outlook.
Comput Struct Biotechnol J, 20:3796-3813, 05 Jul 2022
Cited by: 4 articles | PMID: 35891791 | PMCID: PMC9304431
Review Free full text in Europe PMC
Deficiencies in the DNA Binding Protein ARID3a Alter Chromatin Structures Important for Early Human Erythropoiesis.
Immunohorizons, 5(10):802-817, 18 Oct 2021
Cited by: 0 articles | PMID: 34663594 | PMCID: PMC8900713
Transcriptional enhancers and their communication with gene promoters.
Cell Mol Life Sci, 78(19-20):6453-6485, 19 Aug 2021
Cited by: 20 articles | PMID: 34414474 | PMCID: PMC8558291
Review Free full text in Europe PMC
Go to all (115) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Hypersensitive site 2 specifies a unique function within the human beta-globin locus control region to stimulate globin gene transcription.
Mol Cell Biol, 19(4):3062-3072, 01 Apr 1999
Cited by: 60 articles | PMID: 10082573 | PMCID: PMC84100
Cooperativeness of the higher chromatin structure of the beta-globin locus revealed by the deletion mutations of DNase I hypersensitive site 3 of the LCR.
J Mol Biol, 365(1):31-37, 03 Oct 2006
Cited by: 17 articles | PMID: 17056066 | PMCID: PMC2826273
Effects of human locus control region elements HS2 and HS3 on human beta-globin gene expression in transgenic mouse.
Blood Cells Mol Dis, 31(3):360-369, 01 Nov 2003
Cited by: 9 articles | PMID: 14636653
Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights.
Gene, 205(1-2):73-94, 01 Dec 1997
Cited by: 147 articles | PMID: 9461381
Review





