Abstract
Free full text

Dynamics of the actin cytoskeleton mediates receptor cross talk: An emerging concept in tuning receptor signaling
Abstract
Recent evidence implicates the actin cytoskeleton in the control of receptor signaling. This may be of particular importance in the context of immune receptors, such as the B cell receptor, where dysregulated signaling can result in autoimmunity and malignancy. Here, we discuss the role of the actin cytoskeleton in controlling receptor compartmentalization, dynamics, and clustering as a means to regulate receptor signaling through controlling the interactions with protein partners. We propose that the actin cytoskeleton is a point of integration for receptor cross talk through modulation of protein dynamics and clustering. We discuss the implication of this cross talk via the cytoskeleton for both ligand-induced and low-level constitutive (tonic) signaling necessary for immune cell survival.
Introduction
Adaptive immunity is a highly differentiated biological system composed of B and T lymphocytes that play a critical role in the fight against infections and malignancies. Appropriate function of the immune system critically depends on the fine-tuned control of signals from cell-surface receptors and their activation threshold governing downstream cellular events such as cell differentiation. The signaling threshold is vital for ensuring efficient cell activation, but only when required, such as upon pathogen invasion. Diminished lymphocyte activation leads to a variety of immune deficiencies, whereas a lowered activation threshold can lead to autoimmune disorders and cancerogenesis, emphasizing the importance of refined mechanisms ensuring correct threshold for cell activation (Tsubata, 1999; Hermiston et al., 2003).
Emerging evidence implicates the actin cytoskeleton as a key regulator of cell signaling. The cellular cytoskeleton is a continuously remodeled dynamic network that provides force and support to cellular structures and also serves as tracks for vesicle and organelle movement. The actin cortex underlies the plasma membrane, where it has been traditionally thought to mainly act as support to the membrane. However, the cortical actin cytoskeleton is in intimate interaction with both lipid and protein components of the plasma membrane. Indeed, cortical actin participates in various cellular events occurring proximal to the membrane, such as endocytosis, focal adhesions, and regulation of membrane protein diffusion and organization (Le Clainche and Carlier, 2008; Mooren et al., 2012; Trimble and Grinstein, 2015). The multifunctional and dynamic quality of actin impart it with characteristics that make it a potent mechanism for controlling and/or tuning receptor signaling; this may be particularly important for receptors that need to be tightly regulated, such as immune receptors. Indeed, mutations in genes that encode actin regulatory proteins are associated with human immunodeficiencies, such as Wiskott-Aldrich syndrome, underscoring the importance of the actin cytoskeleton in immune cell activation (Derry et al., 1994; Machesky and Insall, 1998).
In this review, we concentrate on recent studies examining the role of the actin cytoskeleton in controlling receptor triggering and cell signaling. We focus on the role of the cytoskeleton in regulating receptor compartmentalization, dynamics, and clustering. These parameters play a key role in defining the function of a protein, because they may facilitate or inhibit interactions with protein partners and thus regulate the cellular outcome of receptor engagement. We propose that actin serves as a critical point of integration of receptor signaling such that changes in the cytoskeleton induced by one signal can readily influence the function of other receptors. We frame this discussion largely in terms of the B cell receptor (BCR), for which recent studies have highlighted a striking role for actin in constraining BCR signaling. We discuss the implication of receptor cross talk with the cytoskeleton not only in the context of the low-level constitutive (tonic) signal necessary for B cell survival, but also as a mechanism to influence the threshold of activation. We posit that modulation of the actin cytoskeleton is a general mechanism for integrating cross talk between different signaling pathways. Understanding the mechanisms how the cytoskeleton can control protein–protein interactions to consequently regulate cell signaling may have broad implications for understanding human health and disease.
Actin cortex and other submembranous cytoskeletal structures
As this review is focused on the role of the actin cytoskeleton in controlling receptor signaling, it is prudent to briefly introduce some of the key structures of the cytoskeleton and the mechanisms that regulate these structures, although this is by no means meant to be a comprehensive review of the cytoskeleton; for this, we refer the reader to several excellent reviews (Bezanilla et al., 2015; Carlier et al., 2015; Case and Waterman, 2015; De La Cruz and Gardel, 2015). What we aim to highlight, and hope the reader will consider throughout this review, is that different actin-based structures may differentially modulate the steady-state organization and dynamics of membrane proteins, and although few of the studies discussed herein specifically define the actin features in relation to their data, we believe that this in an important consideration that may have significant implications for both the interpretation of data and the functional implications of the findings.
The actin cytoskeleton broadly refers to actin-based cytoskeletal structures, encompassing a variety of functionally distinct structures of different actin organization and regulated by different actin regulatory proteins (Fig. 1). The actin cortex is composed of a dense mesh-like array of F-actin estimated to be between 50 nm and 2 µm in thickness tangent to the plasma membrane and anchored to the cell membrane through interaction with both membrane proteins and lipids (Bray and White, 1988; Charras et al., 2006). This structure provides the core “skeleton” of the cell, functioning to define cell shape and provide resistance to mechanical stress. Lamellipodia refers to the sheet-like protrusive structure composed of branched F-actin and primarily involved in cell locomotion (Small et al., 2002). The finger-like protrusions known as filopodia consist of long, straight, bundled F-actin and are thought to function as exploratory fingers for the cell (Mattila and Lappalainen, 2008). To fulfill these various functions in a multitude of cellular events, both the dynamics and structural properties of the actin cytoskeleton must be tightly regulated. This is achieved by a plethora of actin-binding proteins that orchestrate different aspects of actin dynamics in a highly spatiotemporally controlled manner. Many actin-binding proteins are recruited to and activated, or inactivated, at the cell membrane, regulating the formation of these submembranous cytoskeletal structures (Saarikangas et al., 2010). Each of these structures involves a distinct balance of actin polymerization, depolymerization, and filament capping and bundling and share several factors conveying these properties (Fig. 1). All actin-based structures require actin nucleation and polymerization that converts monomeric actin into F-actin. Two main actin nucleators are the Arp2/3 complex, which forms branched actin networks, and the formin family proteins that induce formation of long and straight filaments. The sheet-like lamellipodia is considered chiefly nucleated by the Arp2/3 complex (Welch et al., 1997), whereas the activity of formins is involved in inducing linear F-actin in filopodia (Peng et al., 2003). Although all actin-based structures in cells require a set of actin-regulatory factors, the balance of the activity of these regulatory factors varies in different actin-based structures. Actin depolymerization by the cofilin family of proteins, for example, is important in lamellipodia, where filaments undergo highly dynamic treadmilling (Bugyi and Carlier, 2010), whereas cross-linking of filaments by actin-bundling proteins, such as fascin, is critical to provide the required stability to filopodia (Kureishy et al., 2002).
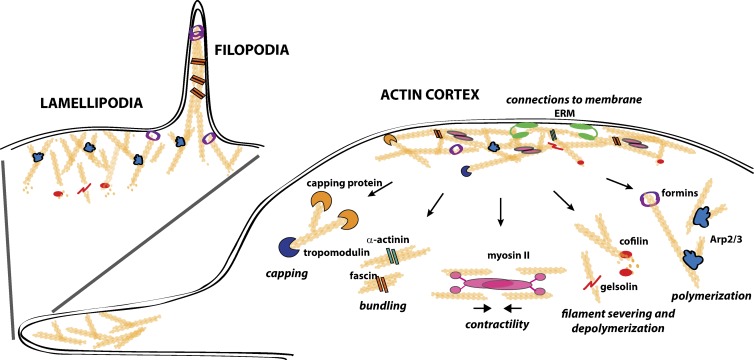
General principles of the regulation of submembranous actin structures by various actin-binding proteins. Carefully regulated balance of filament polymerization, depolymerization, bundling, and capping together with contractility and regulated connections to the plasma membrane lead to different properties of actin-based structures, such as actin cortex, lamellipodia, and filopodia. Classical examples of actin-binding proteins that convey these activities are illustrated. The actin cortex is a relatively stable mesh-like network of interconnected and contractile filaments that are physically linked with the membrane and aligned under it in a juxtaposed manner. At the leading edge of the cell, lamellipodia are highly dynamic structures where branched filaments are aligned perpendicular to the membrane and push it forward. Filopodia, also found in the leading edge, are finger-like structures, where long and straight filaments are bundled together to protrude the membrane outward.
The actin cortex is the least studied of these submembranous actin structures, although it is present in virtually every cell. De novo formation of the F-actin network under the membrane of expanded blebs has been used as a model system for actin cortex generation (Charras et al., 2006). The cortex seems to be generated by cooperation of two actin nucleator machineries: Arp2/3 complex and the formin mDia1. The balance of these two nucleators, together with filament cross-linkers and the molecular motor myosin II, which provides contractility, contributes to the relatively stable but still rapidly adjustable cortical meshwork of F-actin (Fritzsche et al., 2013; Bovellan et al., 2014). The flexibility of mature cortex is illustrated by the existence of both short and long filaments with differential dynamic behavior, as demonstrated by single-particle tracking (Gowrishankar et al., 2012) and photobleaching studies (Fritzsche et al., 2013).
The F-actin network of the cortex is involved in various cellular events at the plasma membrane. Cortex remodeling is required upon endo- and exocytosis, for instance, and it serves as the origin or building material for other actin structures like lamellipodia and filopodia. Located right beneath the plasma membrane, the actin cortex also physically connects to it by various membrane–cytoskeleton linker proteins (Fig. 2). It is hardly surprising that such a dense filamentous mesh close to and intimately connected with the plasma membrane can affect the behavior and function of membrane proteins. Interestingly, already the ancient bacterial actin homolog MreB is able to organize bacterial cell membrane and membrane protein diffusion (Strahl et al., 2014), underlining the significance of the cortical cytoskeleton in the regulation of membrane protein function. It should also be kept in mind that the actin cytoskeleton further works together with the microtubule cytoskeleton as well as various intermediate filaments. Whereas microtubules provide polarity and intracellular organelle organization, for instance, the cytoplasmic intermediate filaments are considered critical for cell resistance against mechanical stress (Huber et al., 2015).

Examples of proteins connecting the plasma membrane to the actin cytoskeleton. (top left) ERM proteins (Neisch and Fehon, 2011). (bottom left) Protein 4.1-ankyrin-spectrin network (Baines, 2010; Baines et al., 2014). (top right) Septins (Gilden et al., 2012; Bridges and Gladfelter, 2015). (middle right) Filamins (Lin et al., 2001; Stossel et al., 2001; Beekman et al., 2008; Zhou et al., 2010). (bottom right) Myosin 1 (McConnell and Tyska, 2010).
In the immune system, the critical role of the actin cytoskeleton is demonstrated by various human pathologies and mouse models. For example, the causal gene product for Wiskott-Aldrich syndrome, an X-linked recessive disease characterized by immune dysfunction and recurrent infections as well as predisposition to develop lymphomas and leukemias, is Wiskott-Aldrich syndrome protein, WASP, an immune cell–specific activator of the Arp2/3 complex (Derry et al., 1994). The same syndrome is also caused by mutations in WASP-interacting protein, WIP (Lanzi et al., 2012), which forms a complex with and stabilizes WASP (de la Fuente et al., 2007). A recent study in T cells demonstrated that WASP-dependent Arp2/3 activation is required for actin polymerization foci localized at sites of T cell receptor (TCR) signaling and promotion of PLCγ activation and subsequent calcium signaling. However, T cell immunological synapse is able to form upon inhibition of Arp2/3 activity by the inhibitor CK666, suggesting that formin-based actin polymerization plays the major structural role (Kumari et al., 2015). Moreover, Rho family GTPases including Cdc42 and Rac, which are considered master regulators of the actin cytoskeleton capable of activating multiple cytoskeletal regulators (Heasman and Ridley, 2008), play an important role in B cell activation. B cell–specific deletion of Cdc42 was recently shown to lead to a dramatic impairment in the generation of antibody producing plasma cells (Burbage et al., 2015). In addition, inactivation of Rac1 and Rac2 in a mouse model leads to a complete lack of B cells, possibly because of deficient BCR signaling during development, as has been shown in mature B cells (Walmsley et al., 2003; Brezski and Monroe, 2007; Arana et al., 2008). Notably, impaired activity of G-nucleotide exchange factors such as Vav and DOCK8, which regulate the activity of RhoGTPases, has profound effects on B cell activity and the humoral immune response, through defective formation of the immunological synapse (Doody et al., 2001; Weber et al., 2008; Randall et al., 2009). Although it is currently not well understood how much of these disease phenotypes are caused by abnormalities in the actin cortex or membrane protein functions, a recent study has shown that in B cells, absence of the actin regulatory protein, WIP, affects signaling capabilities, likely because of altered organization and dynamics of BCR and the coreceptor CD19 (Keppler et al., 2015), providing strong genetic evidence that the actin cytoskeleton plays a key role in receptor signaling. Here, we discuss means by which the actin cytoskeleton can regulate membrane receptor signaling thresholds and integration of different signals through controlling molecular organization and dynamics of cell surface proteins.
Constitutive dynamic molecular assemblies in the plasma membrane
The plasma membrane is densely packed with each of the thousands of surface proteins represented by up to hundreds of thousands of molecules. For example, mature B cells express two isoforms of the BCR, with between 20,000 and 150,000 molecules of IgM and 250,000 and 300,000 molecules of IgD (Mattila et al., 2013). How these thousands of molecules are organized within the membrane is an important question for understanding the mechanism of receptor triggering and the initiation of signaling. Although plasma membrane proteins were once considered to be largely randomly distributed on the surface of cells (Singer and Nicolson, 1972), research over the last 40 years has consistently demonstrated that the plasma membrane is compartmentalized. Indeed, using a variety of experimental techniques to probe the topography of membrane proteins, research has uncovered a smorgasbord of protein organizations: some proteins appear to be monomers; some are dimers, trimers, or higher-order oligomers; and some are found as discrete clusters of tens of molecules of different molecular species, which have been variably called “nanoclusters” or “protein islands.” Such varied organization has been noted not only for different proteins but also for proteins of the same molecular type, including the BCR, and this can be rather confusing for the uninitiated. Further complicating the matter, many cell surface proteins undergo clustering upon activation, and these structures also need to be named; in the case of immune cells, the term “microclustering” has become common nomenclature. What is emerging is a dynamic view of the plasma membrane, where cell surface proteins are likely found on a continuum from single proteins to clusters of a few to tens of molecules. In the following paragraphs, we discuss some of the work that has led to this modified view of the plasma membrane, potential mechanisms that regulate these dynamic assemblies, and importantly, the functional significance for intercellular communication.
Researchers at the interface between immunology and biophysics made many of these important observations of the nonrandom distribution of cell surface proteins in lymphocytes. Early studies used flow cytometry–based Förster resonance energy transfer (FRET) experiments, which are sensitive to intermolecular distances less than 10 nm, the reasoning being that if plasma membrane proteins are homogeneously distributed, then the likelihood of two proteins being within such close proximity would be highly unlikely and FRET would not occur. Instead, FRET experiments revealed homo- and heteroassociations of numerous membrane proteins including major histocompatability complex (MHC) class I and II (Chakrabarti et al., 1992; Matko et al., 1994; Szöllósi et al., 1996), epidermal growth factor receptor (Gadella and Jovin, 1995), interleukin-2 receptor α-subunit and ICAM-1 (Szöllösi et al., 1987; Burton et al., 1990), and MHC class I and II, CD20, and tetraspanins (Szöllósi et al., 1996). Although these studies established that, in contrast to the fluid mosaic model, many plasma membrane proteins were nonrandomly distributed, they could not provide spatial information about protein distribution or the number of molecules that are closely associated. To address this, researchers turned to transmission EM (TEM), which revealed nanometer-scale “islands” of proteins including MHC class I (Damjanovich et al., 1995), MHC class II (Jenei et al., 1997), and the interleukin-2 receptor α-subunit and transferrin receptor (Vereb et al., 2000). Interestingly, these studies also revealed higher hierarchical level receptor clustering in which these protein “islands” organized into “island groups” with mean sizes between 400 and 800 nm, raising the question of whether there was functional significance to the codistribution of these proteins. Wilson et al. also used TEM to visualize the distribution of several cell surface proteins in B cells, mast cells, and fibroblasts using a technique to generate plasma membrane sheets, which exposes the inner surface of the plasma membrane for immunostaining. By counting gold particles they found that approximately one-third of particles marking FcεR1 are distributed as singlets and the rest are found in clusters of two or three particles and occasionally up to nine particles (Wilson et al., 2000). In contrast to the observation of the codistribution of other proteins mentioned previously (Damjanovich et al., 1995; Jenei et al., 1997; Vereb et al., 2000), Wilson et al. (2001) found that linker for activation of T cells (LAT), found primarily in clusters of <20 molecules, was segregated from FcεR1. Using this technique of plasma membrane sheets, but instead labeling membrane proteins via biotinylation of sulfhydryl or carboxyl groups, Lillemeier et al. provided evidence that the nonrandom distribution of cell surface proteins is not specific to only some classes of proteins but rather all membrane-associated proteins are clustered in cholesterol-rich islands in a sea of protein-free and cholesterol-low membrane (Lillemeier et al., 2006). Although these EM studies further support the model of the compartmentalization of membrane proteins, they may be underestimating the density of these domains because of extensive sample processing and potential loss of antibody epitopes, illustrated by often very sparse immunogold decoration of the sample.
Because these constitutive assemblies are smaller than the diffraction-limited resolution of confocal and epifluorescence microscopy, defined as half the wavelength of light (λ/2) or ~250 nm, it has been difficult to resolve these structures using light microscopy, which would allow for less sample processing than TEM studies. Recently, super-resolution imaging techniques including near-field scanning optical microscopy (Betzig et al., 1986), stochastic optical reconstruction microscopy (STORM; Rust et al., 2006), photo-activated localization microscopy (PALM; Betzig et al., 2006), and BLINK microscopy (Steinhauer et al., 2008) have been applied to the study of receptor organization on the nanoscale. For example, MHC class I, visualized by near-field scanning optical microscopy, was found in islets with radii of ~70–600 nm and contained on the order of 25–125 molecules each (Hwang et al., 1998). Nanoclusters of the lectin DC-SIGN were ~80 nm in diameter and contained a mean of 12 molecules but in some cases upward of 65 molecules (Itano et al., 2012). Using STORM, we found that both IgM and IgD BCR are found in clusters with a mean radius of ~60–80 nm and estimated to contain between 20 and 120 molecules per cluster (Mattila et al., 2013). IgD was found more often in nanoclusters and these clusters had significantly higher mean density of molecules (Fig. 3). This is quite consistent with a recent two-color STORM study of IgM and IgD, which also suggested that these proteins are not found coclustered but instead are localized on separate nanoclusters (Maity et al., 2015). It is perhaps also not surprising that previous FRET studies found IgM to be largely monomeric on the surface of resting B cells (Tolar et al., 2005), given the much lower proportion of IgM in clusters compared with IgD (38% compared with 70%, respectively; Mattila et al., 2013) Moreover, based on the mean radius of a nanocluster seen in our STORM data, one can appreciate that if a cluster has a low density of receptors, then it is likely that they will be >10 nm apart (the maximum intermolecular distance for FRET measurements) while clearly still having a nonrandom or compartmentalized distribution (Fig. 3). We also found that a key regulator of BCR signaling, the positive coreceptor CD19, also exists in nanoclusters, and the proportion of molecules in clusters and density of clusters was within the range of IgM and IgD (Mattila et al., 2013). It is important to note that it is not only transmembrane proteins that display a nonrandom distribution; proteins bound to the outer or inner leaflet of the plasma membrane exhibit constitutive clustering on the nanoscale. For example, the Ras family of small G proteins, which are bound to the inner leaflet of the plasma membrane, are found in nanoclusters containing approximately seven proteins and radii of roughly 9 nm, notably smaller than the nanoclusters discussed above. Approximately 30% of K-ras is organized in clusters, and the remaining 70% is found as monomers (Plowman et al., 2005). Interestingly, these authors report that this ratio of monomer to cluster is independent of expression level, suggesting a mechanism that actively regulates the extent of clustering. More recently, Gaus and colleagues report that the nature of the membrane anchor impacts protein clustering, such that proteins anchored to the inner leaflet of the plasma membrane exhibit significantly different clustering parameters than proteins anchored to the outer leaflet (Magenau et al., 2015).
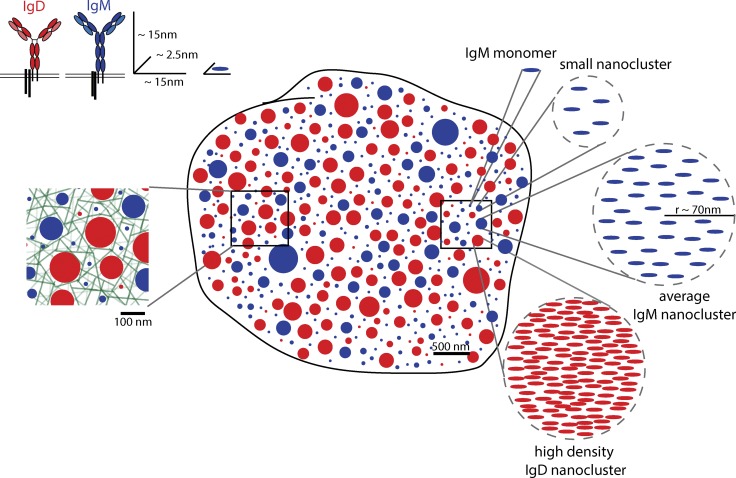
Model of the nanoscale organization of IgM and IgD at the B cell surface. Schematic diagram illustrating IgM (blue) and IgD (red) nanoclusters at the surface of B cells based on super-resolution STORM analysis reported in Mattila et al. (2013). A heterogeneity of clusters is present from monomers to small clusters of varying size and number of molecules. Dashed circles on the right show zoomed-in view of nanoclusters in box outlined. Based on a mean cluster size with radius of ~70 nm and considering the surface area of the cell and the number of BCR at the cell surface, we calculated a theoretical estimate of receptor density within nanoclusters and depict an average IgM cluster and a high-density IgD cluster, as IgD was found to be significantly more densely packed than IgM nanoclusters. Inset box on left depicts a model of the actin cytoskeleton (green) in relation to nanoclusters based on electron tomography data from Morone et al. (2006). Diagram is to scale.
Collectively, these data firmly establish the nonrandom distribution of cell surface proteins as a general phenomenon. Of note, proteins are found as monomers to clusters ranging from a few molecules to tens of molecules, which are typically called either protein islands or nanoclusters. However, caution should be taken in assigning absolutes to the number and proportion of molecules within these constitutive assemblies as calculations based on super-resolution localizations and protein density may be subject to inaccuracies. For example, a recent two-color PALM study suggests that LAT is predominantly found in very small nanoclusters (two to three molecules) and that previous studies noting larger LAT clusters (Lillemeier et al., 2010) may have overestimated LAT cluster sizes because of differences in data analysis and statistical techniques (Sherman et al., 2011). It is, however, also likely that the range of nanocluster size and density reflects the dynamic nature of these assemblies, instead of a view where these entities are static. The biophysical mechanisms that regulate the size and temporal lifetimes of these assemblies largely have yet to be elucidated. Gheber and Edidin (1999) proposed that these dynamic assemblies are dependent on diffusional barriers and the trafficking of proteins to the cell surface in vesicles. Thus, the population of clusters might be heterogeneous in size and concentration at any given time because clusters are born at different moments of exocytic vesicle fusion, after which their size increases, but concentration decreases over time. Using both mathematical modeling and experimental testing, they provide evidence that clusters are born by the delivery of vesicles to the plasma membrane and that the size and lifetime of these assemblies are dependent on diffusional barriers mediated by the actin cytoskeleton. Stabilizing the actin cytoskeleton increases the lifetime of clusters and conversely depolymerizing the actin network increases the size of clusters as molecules diffuse away, ultimately decreasing the lifetime of clusters (Lavi et al., 2007, 2012).
The formation of nanoclusters via vesicle delivery to the plasma membrane may not be the only mechanism of formation, as mathematical modeling supported by experimental data suggests that even transient protein–actin interactions coupled with actin dynamics/treadmilling and myosin contractility can lead to protein nanoclustering, even for glycophosphatidylinositol (GPI)-anchored proteins, which are themselves not directly linked to actin (Goswami et al., 2008; Gowrishankar et al., 2012). These nanoclusters of GPI-anchored proteins should not be mistaken for ‘raft domains’ described by Suzuki et al., in which various GPI-anchored proteins are largely mobile and form transient homodimers based on ectodomain protein–protein interactions (Suzuki et al., 2012). However, the assembly of greater raft domains, or “nanoclusters” of rafts, likely involves actin filaments (Goswami et al., 2008; Gowrishankar et al., 2012; Suzuki et al., 2012). A role for the actin cytoskeleton in the formation and/or maintenance of nanoclusters is consistent with data showing actin staining associated with protein islands in T cells visualized by TEM and a dramatic reduction in the density of islands upon actin disruption (Lillemeier et al., 2006). In contrast, our data suggest that the organization of both the BCR and CD19 is not altered upon disruption of the actin cytoskeleton (Mattila et al., 2013), although it should be noted that the length of drug treatment in these two studies differed dramatically; Lillemeier and colleagues treated cells with actin inhibitors for 150 min, whereas we used an acute treatment (5 min), and thus it could be that the remaining cortical actin network is sufficient for maintenance of BCR and CD19 nanoclusters. Interestingly, the size of many receptor nanoclusters is consistent with actin-defined compartments (~50–200 nm) revealed by electron tomography (Morone et al., 2006) and super-resolution imaging showing the mean “hole” in the actin mesh is <100 nm (Brown et al., 2012), suggesting that the actin meshwork may indeed limit the size of these clusters.
Another mechanism for nanoclusters of plasma membrane proteins are protein–protein and protein–lipid interactions. This has been shown for the adaptor protein LAT, which exhibits constitutive nanoscale clustering that is dependent on two juxtamembrane cysteine residues as well as four distal tyrosines, indicative of a role for palmitoylation and protein–lipid interactions, and protein–protein interactions, respectively (Sherman et al., 2011). In the case of BCR, Reth and colleagues have shown that the size of IgD complexes isolated biochemically was dependent on class-specific amino acids in the transmembrane region (Schamel and Reth, 2000), suggesting that protein–protein interactions might be particularly important for IgD–BCR nanoclusters. We found that CD19 nanoclusters were dependent on the tetraspanin CD81 (Mattila et al., 2013). Tetraspanins are a large family of small transmembrane proteins that form homo- and multimers that further interact in cis with a multitude of membrane proteins. So-called tetraspanin-enriched microdomains within the plasma membrane laterally organize the membrane and strongly influence organization of their interaction partners, including immune receptors and integrins (Charrin et al., 2014). Interestingly, the role of CD81 in CD19 nanoclustering does not appear to be holding the clusters together, as deficiency of CD81 did not lead to loss of nanoclusters, but rather in organizing CD19 within the clusters, as deficiency of CD81 led to an 80% increase in the density of CD19 within nanoclusters (Mattila et al., 2013). A recent study using dual-color stimulated emission depletion microscopy to gain super-resolution imaging of various tetraspanin-enriched microdomain structures in membrane sheets of B cells and dendritic cells found that tetraspanins CD37, CD53, CD81, and CD82 formed individual nanoclusters (<120 nm) that showed little overlap with each other (Zuidscherwoude et al., 2015). This is surprising considering earlier studies of tetraspanin–tetraspanin heterophilic interactions (Yáñez-Mó et al., 2009); however, specific cell types and tetraspanins might differ in this respect.
Tetraspanin-enriched microdomains are one example of membrane-organizing microdomains, but lipids themselves are also capable of provoking membrane order. Lipid raft membrane domains were first characterized by Simons and Ikonen (1997) and suggested to promote compartmentalization of membrane proteins for membrane trafficking or serve as signaling platforms. Initial studies on these sphingolipid and cholesterol-rich assemblies relied on biochemical methods using detergent extraction that led to notable controversy in the field. However, technological advances have now provided ample evidence of the existence of separated lipid domains that function in the organization of membrane proteins (Lingwood and Simons, 2010). Recent work combining super-resolution imaging and fluorescence lifetime measurements of membrane dyes that report lipid packing indeed implicates lipid microdomains in the nonrandom distribution of proteins (Owen et al., 2012).
An often overlooked mechanism for membrane protein organization may be the interaction of these glycosylated cell surface proteins with members of the secreted family of lectins, called galectins, which can bind and cross-link cell surface proteins, creating glycan-based domains (Brewer et al., 2002). Indeed, the galectin lattice has been implicated in the compartmentalization of cell surface proteins (Pace et al., 1999; Chen et al., 2007) and the regulation of epidermal growth factor receptor diffusion (Lajoie et al., 2009), providing compelling evidence for a possible role for these proteins in the nonrandom organization of cell surface proteins. Interestingly, galectins may also influence the nanoscale clustering of proteins through domains other than the carbohydrate recognition domain, as has been suggested for H-ras and K-ras (Belanis et al., 2008; Shalom-Feuerstein et al., 2008). It may also be that interactions with the extracellular matrix influence domain formation. In support of this, truncation of the carbohydrate recognition domain of the transmembrane lectin DC-SIGN altered the formation and stability of constitutive DC-SIGN microdomains (Liu et al., 2012). Taking these observations together, it is likely that a single mechanism does not control the nanoscale organization of proteins, but rather a combination of these mechanisms is at work either in isolation or in combination, depending on the specific protein, resulting in the variety of protein islands of different size and density observed at the surface of cells.
A compelling question is whether there is functional significance for the constitutive nanoscale clustering of cell surface proteins. One of the earliest proposals for the function of constitutive clustering was that the clustering of MHC class I and class II, which present pathogen-derived peptides to T cells, would facilitate antigen presentation. To test this, MHC class I cluster size was altered and the functional effect of this alteration examined. By incubating cells with exogenous β-2 microglobulin, a constituent of MHC class I, the small clusters of MHC class I could be dispersed and a corresponding reduction in T cell responses was observed (Bodnár et al., 2003). In contrast, stabilizing the actin-based membrane skeleton resulted in larger than normal clusters and enhanced T cell responses (Kwik et al., 2003; Fooksman et al., 2006). Thus, the size and stability of constitutive nanoclustering may modulate intercellular communication. In the case of MHC, this clustering may serve as a mechanism to enhance TCR recognition of rare peptide–MHC complexes. Conversely, the constitutive nanoscale clustering of K-Ras and H-Ras proteins is proposed to act as a functional platform for signaling as abrogation of nanoclustering inhibits signal transduction (Tian et al., 2007). Another possibility is that the nanoscale compartmentalization of the plasma membrane serves to segregate functional units of signaling complexes. This notion is consistent with TEM data showing TCR and FcεR1 islands are segregated from the adaptor protein LAT (Wilson et al., 2001; Lillemeier et al., 2010) in the steady state, although recent two-color PALM data of TCR and LAT challenge the view that these molecular species are present in separate nanodomains (Sherman et al., 2011). Conversely, it has been suggested that the coclustering of IL2R and ICAM-1 may help to promote the directed delivery of cytokines to target cells (Burton et al., 1990).
In the case of the BCR, an interesting question is why the proportion and density of IgM in constitutive clusters is less than IgD. Do differences in the clustering state of the receptor regulate its properties, such as mechanism of triggering or requirements for ligand valency? Indeed, a mutant of IgD unable to induce BiFC and thus presumably monomeric was found to be more active (Yang and Reth, 2010). Moreover, monovalent engagement readily activated IgG1-BCRs expressed on memory B cells (Avalos et al., 2014), and this isotype has an enhanced capacity to oligomerize/cluster upon ligand engagement (Liu et al., 2010). Although the enhanced response of memory B cells is no doubt linked to increased affinity of these BCRs, it may also be that the nanoscale organization (Avalos et al., 2014) of this isotype alters triggering properties leading to enhanced sensitivity. Interestingly, a recent study suggests that memory T cells have increased antigen sensitivity through increased number and size of TCR oligomers at the cell surface (Kumar et al., 2011). Because TCRs do not undergo somatic hypermutation leading to increased affinity, as BCRs do, the increased prevalence of oligomeric TCRs may be reflective of a process of avidity maturation, as suggested by the authors. Intriguingly, altered nanoscale organization of the adhesion molecule LFA-1 and the lectin DC-SIGN has been noted in the differentiation of monocytes to dendritic cells, and these changes in receptor organization have important functional consequences for ligand binding (Cambi et al., 2004, 2006). It will be interesting to determine the mechanisms for these differentiation-specific organizations and whether these alterations are mediated by changes in the actin cytoskeleton.
The actin cytoskeleton regulates receptor mobility
Extensive research in membrane protein biology has demonstrated that many cell surface proteins exhibit restricted diffusion on both the micrometer scale over milliseconds as well as nanometer and microsecond scales (Kusumi et al., 2012). Consequently, it was proposed in the membrane-skeleton fence model that transmembrane proteins protrude into the cytoplasm and collide with the underlying cortical actin cytoskeleton (membrane skeleton) resulting in temporary confinement of membrane proteins (Kusumi et al., 2005). Confined diffusion is observed not only for transmembrane proteins that protrude into the cytoplasm but also for lipids and GPI-anchored proteins, which are only linked to the outer layer of the plasma membrane, and thus, the anchored transmembrane protein picket model was proposed; in this model, transmembrane proteins are anchored to and aligned along the membrane skeleton, effectively acting as rows of pickets to the free diffusion of GPI-anchored proteins and lipids (Kusumi et al., 2005, 2012). Molecules within these membrane compartments delineated by fences and pickets undergo short-term confined diffusion and long-term “hop” diffusion between compartments. (Fujiwara et al., 2002; Suzuki et al., 2005). Many studies have provided evidence that support the model that the actin cytoskeleton defines diffusion barriers for membrane proteins and lipids by disrupting the actin cytoskeleton either genetically or pharmacologically (Sheetz et al., 1980; Haggie et al., 2004; Charrier et al., 2006; Lenne et al., 2006; Wheeler et al., 2007). The first study to directly visualize the actin cytoskeleton and an immune receptor simultaneously, in this case FcεR, demonstrated that actin filaments defined micron-sized domains that confine receptors (Andrews et al., 2008). We took a similar approach to examine the role of the actin cytoskeleton on BCR mobility. Using dual-view total internal reflection fluorescence microscopy and single-particle tracking, we provided evidence that the actin cytoskeleton defines diffusion barriers for the BCR (Treanor et al., 2010). Interestingly, the diffusion of the two BCR isotypes expressed by mature naive B cells, IgM and IgD, showed a high degree of restricted diffusion with ~50% of IgM and almost 90% of IgD nearly immobile. This observation is in striking contrast to the nearly entirely mobile state reported for FcεR (Andrews et al., 2008). It should be noted that because of the low labeling density required for single-particle tracking experiments, it is not possible to verify if the tracked molecules were BCR monomers or organized in nanoclusters on the cell surface. It is tempting to speculate that perhaps protein dynamics reflect the state of clustering to some extent (at least on this macroscopic timescale). Indeed, the proportion of IgM–BCR versus IgD–BCR found in clusters (Mattila et al., 2013) is similar to the proportion of very slow diffusing BCRs for these two isotypes (Treanor et al., 2010). Consistent with this notion, GPI-AP nanoclusters are less mobile than GPI-AP monomers (Goswami et al., 2008). Interestingly, the diffusion of both IgM and IgD increased upon treatment with actin depolymerizing agents, although not to the same extent, suggesting additional mechanisms may regulate BCR diffusion, particularly in the case of IgD.
An intriguing question is whether membrane protein diffusion dynamics are spatially modulated within the cell depending on the local actin architecture. For example, membrane protein diffusion may be different in the relatively stable cross-linked and membrane-linked network of actin filaments within the cortex, compared with the highly dynamic organization of actin within lamellipodia, which is undergoing continuous remodeling. In our study, we primarily focused on BCR movement within the cortex and only rather crudely defined actin-rich versus actin-poor regions within this area. We did, however, observe linear BCR diffusion at the cell periphery within actin-rich filopodia (Treanor et al., 2010), suggesting that indeed, membrane protein diffusion may be regulated by different F-actin–based structures. The mechanism for this linear diffusion of BCR within filopodia has yet to be investigated, as well as how this might modulate BCR triggering within these exploratory fingers. Linear diffusion of a population of CD36 has also been reported, although in this case it was within “troughs” radiating from the perinuclear region and aligned along microtubules where actin was locally displaced from the membrane (Jaqaman et al., 2011). Indeed, the actin cytoskeleton does not work in isolation from other cytoskeletal networks, and thus, the proximity and connections between the actin network and the plasma membrane are critical with regard to the effect on membrane proteins. In mammalian cells, various linkers modulate the connection between the plasma membrane and the cytoskeleton. These linkers include ezrin-radixin-moesin (ERM) proteins, 4.1 protein, ankyrin, spectrin, septin, filamin, and myosin 1, which we have highlighted in Fig. 2. In B cells, we found that not only actin, but also the ERM family protein ezrin defined barriers to BCR diffusion. The binding of ERM proteins to both membranous targets and actin is regulated by phosphorylation (Bretscher et al., 2002), thus providing a mechanism to modify membrane protein diffusion in highly dynamic manner. Indeed, in B cells, ezrin underwent dynamic reorganization, which appeared to allow BCR to transition from immobile to mobile states, acting as a kind of “gate” to plasma membrane compartments (Treanor et al., 2010). Interestingly, in these studies we noted that the ERM network is much more dynamic than actin reorganization in the cortex, suggesting that indeed, the ERM proteins may be a key mechanism to rapidly regulate membrane protein diffusion. Because both TCR and BCR stimulation induce a transient dephosphorylation of ERM proteins (Delon et al., 2001; Faure et al., 2004; Gupta et al., 2006; Ilani et al., 2007), these proteins may provide a more general mechanism to tune receptor diffusion in response to ligand binding.
In contrast to BCR, the mobility of the key coreceptor CD19 was less affected by disruption of the actin cytoskeleton in naive unstimulated B cells, despite showing highly limited mobility (Mattila et al., 2013). These observations were supported by simultaneous visualization of the actin cytoskeleton and single-particle tracking experiments, which showed that CD19 diffusion was independent of the density of actin. Instead, immobilization of CD19 was largely mediated by the tetraspanin CD81; CD19 diffusion was markedly increased in CD81-deficient B cells. Interestingly, deficiency of CD81 rendered CD19 mobility more sensitive to disruption of the actin cytoskeleton, suggesting some degree of cross talk between protein–protein mechanisms controlling receptor movement and the underlying actin cytoskeleton. Similar to CD81, other tetraspanins, like CD151 and CD82 for instance, have been found to regulate the diffusion of their interacting proteins, such as α6 integrins and EGF receptor, respectively (Danglot et al., 2010; Yang et al., 2012). Thus, tetraspanin microdomains within the plasma membrane may play an important role in regulating diffusion of several membrane proteins. Only a few studies report on the dynamics of tetraspanins themselves. A relatively slow half-life of 10–15 s was demonstrated for the bulk of CD63 and CD82 in photobleaching studies (Hoorn et al., 2012). A more detailed study of CD9 tetraspanin by single particle tracking revealed high mobility for the majority of CD9 molecules, whereas a minority appeared confined in CD9 tetraspanin microdomains. Interestingly CD9 tetraspanin microdomains were very stable in both position and shape over an imaging period of 10 min and resistant to latrunculin B treatment (Espenel et al., 2008), in line with our data on tetraspanin-dependent but largely actin-independent mobility of CD19 (Mattila et al., 2013).
Actin controls signaling
The BCR presents an interesting model for the role of actin in regulating signaling, with differential requirements for actin depolymerization and polymerization spatially and temporally modulated. Given the potential role for actin in organizing receptor nanoclusters and membrane protein diffusion, and the importance of these parameters in signaling, it is likely that the findings on the role of actin in regulating BCR signaling may be broadly relevant to many cell surface receptors. We found that the cytoskeleton plays a critical role in controlling BCR signaling and that treatment with agents disrupting actin organization was sufficient to induce BCR signaling in the absence of any ligand (Treanor et al., 2010; Mattila et al., 2013). We hypothesized that by restricting the diffusion of BCR, the cytoskeleton regulates collaboration with the activatory coreceptor CD19; disruption of the diffusion barrier defined by the actin cytoskeleton increases BCR (BCR nanocluster) diffusion and thus increases the likelihood that the BCR will encounter the coreceptor CD19 (Treanor and Batista, 2010; Treanor, 2012). This idea is consistent with the “collision coupling” or “mobile receptor” hypothesis (Jans, 1992; Jans and Pavo, 1995), which postulated that lateral diffusion of membrane proteins was required to facilitate protein-protein interactions necessary for signal transduction. In support of this hypothesis, we found that the spontaneous BCR signal triggered by disruption of the cytoskeleton is dependent on CD19 (Mattila et al., 2013). These data suggest that release of BCR nanoclusters allows for either BCR collisions that activate CD19 or direct BCR access to CD19 that then facilitates signaling through the BCR (Fig. 4). The coming together of nanoclusters of BCR and CD19 is reminiscent of the concatenation of protein islands of TCR and the coreceptor LAT (Lillemeier et al., 2010; Sherman et al., 2011). Thus, it seems likely that by restricting the diffusion of the BCR, the cytoskeleton is capable of regulating the interactions between the BCR and CD19. Although our data suggest an “opening up” of the actin cytoskeleton is necessary to facilitate protein–protein collisions, actin-void structures such as troughs aligned along microtubules can create channels for the linear diffusion of CD36 that promotes receptor interaction and clustering (Jaqaman et al., 2011).

BCR nanoclusters are brought together to form signaling BCR microclusters. (A) En-face view of the cell–cell contact showing nanoclusters of BCR and CD19 are brought together to form signaling microclusters. Some microclusters contain only IgM, some contain only IgD, and some contain both IgM and IgD as shown in Depoil et al. (2008). Note that not all BCRs form microclusters; monomers and nanoclusters of BCR not in microclusters are shown as semitransparent. (B) Side view of cell–cell contact showing actin and ERM proteins are reorganized to form a fence around BCR microclusters as shown in Treanor et al. (2011). These signaling microclusters lead to localized reorganization of actin that will release unengaged monomers/nanoclusters of BCR from the actin-defined diffusion barrier providing a positive feedback loop to amplify BCR signaling.
If the actin cytoskeleton restricts the interaction between BCR and CD19 by limiting BCR mobility, then alteration of the actin cytoskeleton must be a critical step in the initiation of BCR signaling in response to ligand. Indeed, several lines of evidence support an opening up of the actin network upon BCR stimulation. Hao and colleagues have shown that BCR stimulation is accompanied by rapid depolymerization of the actin cytoskeleton (Hao and August, 2005). Moreover, ERM proteins are rapidly dephosphorylated upon BCR stimulation, which would lead to a conformational folding of the protein and release of the membrane–actin connection, allowing for increased mobility of BCR (Gupta et al., 2006; Treanor et al., 2011). We also found that BCR stimulation was accompanied by a transient increase in the overall mobility of the BCR (Treanor et al., 2011), supporting the idea that signaling induced alteration in actin modifies BCR diffusion dynamics, thus creating a positive feedback loop whereby the increased BCR mobility increases the likelihood of interaction between BCR and CD19 (Mattila et al., 2013), further amplifying BCR signaling. In line with this, B cells deficient in key cytoskeleton regulators such as Cdc42 and WIP result in altered cytoskeleton and defective BCR signaling. Alteration of the actin cytoskeleton and release from diffusional barriers may not only be important for BCR signaling; indeed, the first study to demonstrate a functional outcome from release of a receptor from actin-defined confinement showed that alterations in the actin cytoskeleton induced by phorbol ester increases the mobility of LFA-1 and consequently LFA-1 inside-out signaling (Kucik et al., 1996). Moreover, destabilization of actin was reported to increase the diffusion of serotonin1A receptor, and this increased diffusion was strongly correlated with the efficiency of ligand-mediated signaling (Ganguly et al., 2008). Collectively, these data indicate that depolymerization of the actin cytoskeleton and release of receptors from diffusional barriers is an important step in the initiation of signaling. Interestingly, a recent study has demonstrated that actin-defined confinement regions sequester the transmembrane chemokine CX3CL1 from the membrane metalloprotease ADAM10 and disruption of the cytoskeleton increases association and consequently cleavage and release of soluble chemokine (Wong et al., 2014). Thus, actin-defined membrane compartmentalization may be a general mechanism for regulating a variety of cellular processes.
Although alteration of actin and release of receptor diffusion boundaries is important for BCR signaling, it has also been shown that the actin cytoskeleton plays an important role in morphological changes that accompany B cell activation (Kuokkanen et al., 2015). Upon encountering ligand, the B cell rapidly spreads out over the ligand-containing membrane and then slowly contracts back (Fleire et al., 2006). Although this response clearly requires actin polymerization, it is not necessarily in contradiction to the depolymerization of actin discussed above. In fact, it is likely that the redistribution and polymerization of actin at the periphery of the cell during cell spreading further facilitates the breakdown of the cortical actin meshwork, which was immobilizing BCR nanoclusters. The polymerization of actin occurs not only at the lamellipodia but also at BCR microclusters (Treanor et al., 2011), the micron-scale signaling active clusters that, according to our STORM data, are multiple nanoclusters of BCR and CD19 brought together (Fig. 4; Mattila et al., 2013). The formation and integrity of BCR and TCR microclusters is dependent on the actin cytoskeleton as pretreatment of cells with actin depolymerizing agents abrogates microcluster formation (Campi et al., 2005; Fleire et al., 2006; Varma et al., 2006) and treatment after microcluster formation results in the breakup of microclusters and loss of both ligand and signaling proximal kinase from within clusters (Treanor et al., 2011). The ERM-mediated linkage of the actin-cytoskeleton was also important for maintaining BCR microcluster integrity and downstream signaling (Treanor et al., 2011). In contrast, the large central accumulation of TCRs known as the central supramolecular activation cluster is not affected by disruption of F-actin, likely because these TCRs are on the surface of extracellular microvesicles that bud from the center of the immunological synapse (Choudhuri et al., 2014).
Clearly, the actin cytoskeleton has multifaceted roles in regulating BCR signaling; these roles are spatially and temporally modulated to both permit interaction of receptor nanoclusters as well as support the interaction and thus sustained signaling. This is not to imply that actin is the only player; no doubt additional mechanisms are important for the formation and maintenance of signaling BCR microclusters. It has been suggested that ligand engagement by the BCR induces a conformational change in the Fc portion to reveal an interface in the Cμ4 domain that promotes BCR clustering (Tolar et al., 2009). In addition, we recently found that the tetraspanin molecule CD81, which governs the mobility and the molecular organization of CD19 within the membrane as discussed above, also functions as an amplifier of BCR signaling by facilitating the collaboration of CD19 with the BCR (Mattila et al., 2013). In B cells deficient in CD81, B cell spreading and the number of microclusters are reduced. The molecular mechanism by which CD81 facilitates BCR activation is not clear, but it might involve supporting the optimal conformation or membrane localization of CD19.
Actin dynamics as a mechanism of integrating receptor cross talk
Clearly, the actin cytoskeleton plays a critical role in controlling and regulating receptor signaling. This raises the question of whether alterations in the actin cytoskeleton induced through one receptor signaling pathway alter diffusion dynamics of another receptor, thus facilitating the interaction between functional protein partners. Does the actin cytoskeleton provide a mechanism for mediating cross talk between cell surface receptors and thus the integration of various environmental stimuli? In support of this idea, a recent study has shown that in Toll-like receptor (TLR)-activated B cells, cofilin-mediated increase in actin turnover leads to faster BCR diffusion as well as stronger cell activation by suboptimal BCR stimulation (Freeman et al., 2015). In the in vivo context, infection detected via innate immune receptors would modulate actin regulation of BCR-CD19 interactions resulting in “priming” of BCR and increased BCR sensitivity for ligand (Fig. 5). Moreover, in the in vivo context, cells are not static but rather constantly migrating within lymphoid tissue in response to chemokine signaling, and thus, the actin cytoskeleton is undergoing constant dynamic remodeling. How this effects receptor diffusion within the various F-actin–based structures is a key question. For example, extension of lamellipodia in a migrating cell could either lead to release of receptors from constrains formed by F-actin or induce rapid linear motion with rearward actin flow for receptors that remain bound to actin. How these altered diffusion dynamics impact on receptor signaling is an important area to investigate.

Actin dynamics as a mechanism of integrating receptor cross talk. We propose that the actin cytoskeleton provides a mechanism to integrate environmental stimuli through modification of receptor organization and dynamics and, consequently, protein–protein interactions regulating receptor signaling. In the context of B cells, alterations in the actin cytoskeleton induced through non–BCR-signaling pathways are able to tune BCR activation through increased BCR diffusion dynamics and thus the probability that the BCR will encounter CD19. Changes in actin dynamics could be induced, for example, through innate immune receptors such as TLRs, as recently shown by Freeman et al. (2015), or through other environmental stimuli such as cytokines and chemokines. A recent study showed that BAFFR signaling leads to phosphorylation of Igα/β subunits and subsequent activation of the kinase Syk (Schweighoffer et al., 2013); perhaps this is caused by BAFFR-induced changes in the cytoskeleton and increased interaction between BCR and CD19. This integration of environmental stimuli via actin dynamics could result in priming of BCR and increased BCR sensitivity for ligand. It could also offer perspective on tonic BCR signaling as periodic spatiotemporal changes in the cytoskeleton could modulate BCR–CD19 interactions and initiate localized ligand-independent signaling.
Release of receptors from cytoskeletal constraints caused by altered actin dynamics or organization could offer a novel perspective on BCR tonic signaling necessary for B cell survival (Lam et al., 1997). The mechanism for this low-level ligand-independent constitutive signal, which critically depends on cell surface expression of BCR, is not well understood. We speculate that periodic spatiotemporal changes in the cytoskeleton, caused for example by cell migration, interactions with other cells, or integration of other environmental stimuli, could modulate BCR-CD19 collisions and initiation of localized “tonic” BCR signaling. Indeed, in the study by Freeman et al. (2015), activation of B cells with TLR ligands and the resultant alteration in actin dynamics led to increased BCR diffusion and increased basal levels of phosphorylated ERK and Akt, likely corresponding to tonic signaling. In addition to BCR, tonic signaling also depends on B cell activating factor (BAFF), a cytokine of the TNF family, which binds to BAFF receptor (BAFFR) on B cells. A recent study showed that BAFFR signaling leads to phosphorylation of BCR Igα/β subunits and subsequent activation of the kinase Syk (Schweighoffer et al., 2013). Interestingly, a recent study has shown that Syk is important for actin cytoskeleton reorganization and simply inhibiting this kinase is sufficient to alter actin organization and mobility of FcγR in macrophages (Jaumouillé et al., 2014). A role for Syk in actin dynamics is supported by our observations that Syk-deficient B cells have altered F-actin organization (Treanor et al., 2010). Moreover, B cells deficient in other signaling molecules known to be involved in actin organization, including PLCγ2 and Vav, show decreased BCR diffusion (Treanor et al., 2010) consistent with the idea that altered actin dynamics modulates receptor mobility. Thus, the important collaboration between BCR and BAFFR in supporting B cell survival may be caused by BAFFR-induced changes in the cytoskeleton leading to the activation of the BCR-CD19 arm of signaling. In the in vivo context, it is unlikely that receptors work in isolation, but rather collaborate to integrate the multitude of environmental signals. Interestingly, in B cells, CD19 appears to act as a hub for PI3K signaling and consequently, defects in CD19 signaling in WIP-deficient B cells as a consequence of altered CD19 organization and dynamics results in PI3K activation defects by BCR, CD40, BAFFR, and CXCR5 (Keppler et al., 2015). The actin cytoskeleton could provide a mechanism to integrate these signals through modification of receptor organization and dynamics and, consequently, protein–protein interactions regulating receptor signaling. In further support of this idea, a recent study suggested that remodeling of the cytoskeleton initiated by costimulatory signals can regulate TCR signaling (Tan et al., 2014). Furthermore, an overactive phenotype in dendritic cells with disrupted integrin–actin linkage was recently demonstrated (Morrison et al., 2014). Whether these phenotypes correlate with increased receptor diffusion and altered intermolecular interactions remains to be shown.
Conclusion and perspectives
A new view of the plasma membrane is emerging, one in which membrane proteins are organized in constitutive dynamic molecular assemblies, regulated by a combination of protein–protein and protein–lipid interactions and interactions with the underlying actin cytoskeleton. The mobility of many of these membrane constituents is restricted by the actin cytoskeleton, and this may play a key role in controlling receptor signaling through limiting the interactions with functional protein partners. This role of the cytoskeleton of regulating protein–protein interactions and thus signaling might also provide an explanation for ligand-independent receptor signaling, such as tonic BCR signaling, which is absolutely required for B cell survival but mechanistically enigmatic. However, many questions remain with respect to how this critical cytoskeletal regulation of receptors is achieved and how cells maintain optimal signal thresholds for various continually changing conditions. Environmental stimuli in the in vivo context may modulate the actin cytoskeleton and thus tune the threshold of receptor activation, either by increasing actin dynamics and reorganization or by stabilizing the cortical actin cytoskeleton. Future research will need to identify how the milieu of environmental stimuli influences the cytoskeleton and consequently BCR triggering. Such knowledge may help in our understanding of autoimmune disease and B cell malignancies as well as inform effective vaccination strategies. Furthermore, this information may also prove vital for understanding cytoskeletal regulation of signaling and cross talk of various signal pathways in several other cell types.
Acknowledgments
We apologize to the many scientists whose work was not cited owing to space limitations.
P.K. Mattila is supported by the Academy of Finland (grant 257000), Turku Collegium for Science and Medicine, and the Sigrid Juselius and Jane and Aatos Erkko foundations. F.D. Batista is supported by Cancer Research UK and the Francis Crick Institute. B. Treanor is supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN418756) and the Canadian Institutes of Health Research (FRN136808).
The authors declare no competing financial interests.
References
- Andrews N.L., Lidke K.A., Pfeiffer J.R., Burns A.R., Wilson B.S., Oliver J.M., and Lidke D.S.. 2008. Actin restricts FcepsilonRI diffusion and facilitates antigen-induced receptor immobilization. Nat. Cell Biol. 10:955–963. 10.1038/ncb1755 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Arana E., Vehlow A., Harwood N.E., Vigorito E., Henderson R., Turner M., Tybulewicz V.L., and Batista F.D.. 2008. Activation of the small GTPase Rac2 via the B cell receptor regulates B cell adhesion and immunological-synapse formation. Immunity. 28:88–99. 10.1016/j.immuni.2007.12.003 [Abstract] [CrossRef] [Google Scholar]
- Avalos A.M., Bilate A.M., Witte M.D., Tai A.K., He J., Frushicheva M.P., Thill P.D., Meyer-Wentrup F., Theile C.S., Chakraborty A.K., et al. . 2014. Monovalent engagement of the BCR activates ovalbumin-specific transnuclear B cells. J. Exp. Med. 211:365–379. 10.1084/jem.20131603 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Baines A.J. 2010. The spectrin-ankyrin-4.1-adducin membrane skeleton: adapting eukaryotic cells to the demands of animal life. Protoplasma. 244:99–131. 10.1007/s00709-010-0181-1 [Abstract] [CrossRef] [Google Scholar]
- Baines A.J., Lu H.C., and Bennett P.M.. 2014. The Protein 4.1 family: hub proteins in animals for organizing membrane proteins. Biochim. Biophys. Acta. 1838:605–619. 10.1016/j.bbamem.2013.05.030 [Abstract] [CrossRef] [Google Scholar]
- Beekman J.M., van der Poel C.E., van der Linden J.A., van den Berg D.L., van den Berghe P.V., van de Winkel J.G., and Leusen J.H.. 2008. Filamin A stabilizes Fc gamma RI surface expression and prevents its lysosomal routing. J. Immunol. 180:3938–3945. 10.4049/jimmunol.180.6.3938 [Abstract] [CrossRef] [Google Scholar]
- Belanis L., Plowman S.J., Rotblat B., Hancock J.F., and Kloog Y.. 2008. Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Mol. Biol. Cell. 19:1404–1414. 10.1091/mbc.E07-10-1053 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Betzig E., Lewis A., Harootunian A., Isaacson M., and Kratschmer E.. 1986. Near Field Scanning Optical Microscopy (NSOM): Development and Biophysical Applications. Biophys. J. 49:269–279. 10.1016/S0006-3495(86)83640-2 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., and Hess H.F.. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313:1642–1645. 10.1126/science.1127344 [Abstract] [CrossRef] [Google Scholar]
- Bezanilla M., Gladfelter A.S., Kovar D.R., and Lee W.L.. 2015. Cytoskeletal dynamics: a view from the membrane. J. Cell Biol. 209:329–337. 10.1083/jcb.201502062 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bodnár A., Bacsó Z., Jenei A., Jovin T.M., Edidin M., Damjanovich S., and Matkó J.. 2003. Class I HLA oligomerization at the surface of B cells is controlled by exogenous beta(2)-microglobulin: implications in activation of cytotoxic T lymphocytes. Int. Immunol. 15:331–339. 10.1093/intimm/dxg042 [Abstract] [CrossRef] [Google Scholar]
- Bovellan M., Romeo Y., Biro M., Boden A., Chugh P., Yonis A., Vaghela M., Fritzsche M., Moulding D., Thorogate R., et al. . 2014. Cellular control of cortical actin nucleation. Curr. Biol. 24:1628–1635. 10.1016/j.cub.2014.05.069 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bray D., and White J.G.. 1988. Cortical flow in animal cells. Science. 239:883–888. 10.1126/science.3277283 [Abstract] [CrossRef] [Google Scholar]
- Bretscher A., Edwards K., and Fehon R.G.. 2002. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3:586–599. 10.1038/nrm882 [Abstract] [CrossRef] [Google Scholar]
- Brewer C.F., Miceli M.C., and Baum L.G.. 2002. Clusters, bundles, arrays and lattices: novel mechanisms for lectin-saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol. 12:616–623. 10.1016/S0959-440X(02)00364-0 [Abstract] [CrossRef] [Google Scholar]
- Brezski R.J., and Monroe J.G.. 2007. B cell antigen receptor-induced Rac1 activation and Rac1-dependent spreading are impaired in transitional immature B cells due to levels of membrane cholesterol. J. Immunol. 179:4464–4472. 10.4049/jimmunol.179.7.4464 [Abstract] [CrossRef] [Google Scholar]
- Bridges A.A., and Gladfelter A.S.. 2015. Septin form and function at the cell cortex. J. Biol. Chem. 290:17173–17180. 10.1074/jbc.R114.634444 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Brown A.C., Dobbie I.M., Alakoskela J.M., Davis I., and Davis D.M.. 2012. Super-resolution imaging of remodeled synaptic actin reveals different synergies between NK cell receptors and integrins. Blood. 120:3729–3740. 10.1182/blood-2012-05-429977 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bugyi B., and Carlier M.F.. 2010. Control of actin filament treadmilling in cell motility. Annu. Rev. Biophys. 39:449–470. 10.1146/annurev-biophys-051309-103849 [Abstract] [CrossRef] [Google Scholar]
- Burbage M., Keppler S.J., Gasparrini F., Martínez-Martín N., Gaya M., Feest C., Domart M.C., Brakebusch C., Collinson L., Bruckbauer A., and Batista F.D.. 2015. Cdc42 is a key regulator of B cell differentiation and is required for antiviral humoral immunity. J. Exp. Med. 212:53–72. 10.1084/jem.20141143 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Burton J., Goldman C.K., Rao P., Moos M., and Waldmann T.A.. 1990. Association of intercellular adhesion molecule 1 with the multichain high-affinity interleukin 2 receptor. Proc. Natl. Acad. Sci. USA. 87:7329–7333. 10.1073/pnas.87.18.7329 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cambi A., de Lange F., van Maarseveen N.M., Nijhuis M., Joosten B., van Dijk E.M., de Bakker B.I., Fransen J.A., Bovee-Geurts P.H., van Leeuwen F.N., et al. . 2004. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J. Cell Biol. 164:145–155. 10.1083/jcb.200306112 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cambi A., Joosten B., Koopman M., de Lange F., Beeren I., Torensma R., Fransen J.A., Garcia-Parajó M., van Leeuwen F.N., and Figdor C.G.. 2006. Organization of the integrin LFA-1 in nanoclusters regulates its activity. Mol. Biol. Cell. 17:4270–4281. 10.1091/mbc.E05-12-1098 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Campi G., Varma R., and Dustin M.L.. 2005. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202:1031–1036. 10.1084/jem.20051182 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Carlier M.F., Pernier J., Montaville P., Shekhar S., and Kühn S.. Cytoskeleton Dynamics and Motility group . 2015. Control of polarized assembly of actin filaments in cell motility. Cell. Mol. Life Sci. 72:3051–3067. 10.1007/s00018-015-1914-2 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Case L.B., and Waterman C.M.. 2015. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17:955–963. [Europe PMC free article] [Abstract] [Google Scholar]
- Chakrabarti A., Matko J., Rahman N.A., Barisas B.G., and Edidin M.. 1992. Self-association of class I major histocompatibility complex molecules in liposome and cell surface membranes. Biochemistry. 31:7182–7189. 10.1021/bi00146a022 [Abstract] [CrossRef] [Google Scholar]
- Charras G.T., Hu C.K., Coughlin M., and Mitchison T.J.. 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175:477–490. 10.1083/jcb.200602085 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Charrier C., Ehrensperger M.V., Dahan M., Lévi S., and Triller A.. 2006. Cytoskeleton regulation of glycine receptor number at synapses and diffusion in the plasma membrane. J. Neurosci. 26:8502–8511. 10.1523/JNEUROSCI.1758-06.2006 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Charrin S., Jouannet S., Boucheix C., and Rubinstein E.. 2014. Tetraspanins at a glance. J. Cell Sci. 127:3641–3648. 10.1242/jcs.154906 [Abstract] [CrossRef] [Google Scholar]
- Chen I.J., Chen H.L., and Demetriou M.. 2007. Lateral compartmentalization of T cell receptor versus CD45 by galectin-N-glycan binding and microfilaments coordinate basal and activation signaling. J. Biol. Chem. 282:35361–35372. 10.1074/jbc.M706923200 [Abstract] [CrossRef] [Google Scholar]
- Choudhuri K., Llodrá J., Roth E.W., Tsai J., Gordo S., Wucherpfennig K.W., Kam L.C., Stokes D.L., and Dustin M.L.. 2014. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 507:118–123. 10.1038/nature12951 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Damjanovich S., Vereb G., Schaper A., Jenei A., Matkó J., Starink J.P., Fox G.Q., Arndt-Jovin D.J., and Jovin T.M.. 1995. Structural hierarchy in the clustering of HLA class I molecules in the plasma membrane of human lymphoblastoid cells. Proc. Natl. Acad. Sci. USA. 92:1122–1126. 10.1073/pnas.92.4.1122 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Danglot L., Chaineau M., Dahan M., Gendron M.C., Boggetto N., Perez F., and Galli T.. 2010. Role of TI-VAMP and CD82 in EGFR cell-surface dynamics and signaling. J. Cell Sci. 123:723–735. 10.1242/jcs.062497 [Abstract] [CrossRef] [Google Scholar]
- De La Cruz E.M., and Gardel M.L.. 2015. Actin mechanics and fragmentation. J. Biol. Chem. 290:17137–17144. 10.1074/jbc.R115.636472 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- de la Fuente M.A., Sasahara Y., Calamito M., Antón I.M., Elkhal A., Gallego M.D., Suresh K., Siminovitch K., Ochs H.D., Anderson K.C., et al. . 2007. WIP is a chaperone for Wiskott-Aldrich syndrome protein (WASP). Proc. Natl. Acad. Sci. USA. 104:926–931. 10.1073/pnas.0610275104 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Delon J., Kaibuchi K., and Germain R.N.. 2001. Exclusion of CD43 from the immunological synapse is mediated by phosphorylation-regulated relocation of the cytoskeletal adaptor moesin. Immunity. 15:691–701. 10.1016/S1074-7613(01)00231-X [Abstract] [CrossRef] [Google Scholar]
- Depoil D., Fleire S., Treanor B.L., Weber M., Harwood N.E., Marchbank K.L., Tybulewicz V.L., and Batista F.D.. 2008. CD19 is essential for B cell activation by promoting B cell receptor-antigen microcluster formation in response to membrane-bound ligand. Nat. Immunol. 9:63–72. 10.1038/ni1547 [Abstract] [CrossRef] [Google Scholar]
- Derry J.M., Ochs H.D., and Francke U.. 1994. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 79:922. [Abstract] [Google Scholar]
- Doody G.M., Bell S.E., Vigorito E., Clayton E., McAdam S., Tooze R., Fernandez C., Lee I.J., and Turner M.. 2001. Signal transduction through Vav-2 participates in humoral immune responses and B cell maturation. Nat. Immunol. 2:542–547. 10.1038/88748 [Abstract] [CrossRef] [Google Scholar]
- Espenel C., Margeat E., Dosset P., Arduise C., Le Grimellec C., Royer C.A., Boucheix C., Rubinstein E., and Milhiet P.E.. 2008. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J. Cell Biol. 182:765–776. 10.1083/jcb.200803010 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Faure S., Salazar-Fontana L.I., Semichon M., Tybulewicz V.L., Bismuth G., Trautmann A., Germain R.N., and Delon J.. 2004. ERM proteins regulate cytoskeleton relaxation promoting T cell-APC conjugation. Nat. Immunol. 5:272–279. 10.1038/ni1039 [Abstract] [CrossRef] [Google Scholar]
- Fleire S.J., Goldman J.P., Carrasco Y.R., Weber M., Bray D., and Batista F.D.. 2006. B cell ligand discrimination through a spreading and contraction response. Science. 312:738–741. 10.1126/science.1123940 [Abstract] [CrossRef] [Google Scholar]
- Fooksman D.R., Grönvall G.K., Tang Q., and Edidin M.. 2006. Clustering class I MHC modulates sensitivity of T cell recognition. J. Immunol. 176:6673–6680. 10.4049/jimmunol.176.11.6673 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Freeman S.A., Jaumouillé V., Choi K., Hsu B.E., Wong H.S., Abraham L., Graves M.L., Coombs D., Roskelley C.D., Das R., et al. . 2015. Toll-like receptor ligands sensitize B-cell receptor signalling by reducing actin-dependent spatial confinement of the receptor. Nat. Commun. 6:6168 10.1038/ncomms7168 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fritzsche M., Lewalle A., Duke T., Kruse K., and Charras G.. 2013. Analysis of turnover dynamics of the submembranous actin cortex. Mol. Biol. Cell. 24:757–767. 10.1091/mbc.E12-06-0485 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., and Kusumi A.. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157:1071–1081. 10.1083/jcb.200202050 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gadella T.W. Jr., and Jovin T.M.. 1995. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J. Cell Biol. 129:1543–1558. 10.1083/jcb.129.6.1543 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ganguly S., Pucadyil T.J., and Chattopadhyay A.. 2008. Actin cytoskeleton-dependent dynamics of the human serotonin1A receptor correlates with receptor signaling. Biophys. J. 95:451–463. 10.1529/biophysj.107.125732 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gheber L.A., and Edidin M.. 1999. A model for membrane patchiness: lateral diffusion in the presence of barriers and vesicle traffic. Biophys. J. 77:3163–3175. 10.1016/S0006-3495(99)77147-X [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gilden J.K., Peck S., Chen Y.C., and Krummel M.F.. 2012. The septin cytoskeleton facilitates membrane retraction during motility and blebbing. J. Cell Biol. 196:103–114. 10.1083/jcb.201105127 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., and Mayor S.. 2008. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 135:1085–1097. 10.1016/j.cell.2008.11.032 [Abstract] [CrossRef] [Google Scholar]
- Gowrishankar K., Ghosh S., Saha S., C R., Mayor S., and Rao M.. 2012. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 149:1353–1367. 10.1016/j.cell.2012.05.008 [Abstract] [CrossRef] [Google Scholar]
- Gupta N., Wollscheid B., Watts J.D., Scheer B., Aebersold R., and DeFranco A.L.. 2006. Quantitative proteomic analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid raft dynamics. Nat. Immunol. 7:625–633. 10.1038/ni1337 [Abstract] [CrossRef] [Google Scholar]
- Haggie P.M., Stanton B.A., and Verkman A.S.. 2004. Increased diffusional mobility of CFTR at the plasma membrane after deletion of its C-terminal PDZ binding motif. J. Biol. Chem. 279:5494–5500. 10.1074/jbc.M312445200 [Abstract] [CrossRef] [Google Scholar]
- Hao S., and August A.. 2005. Actin depolymerization transduces the strength of B-cell receptor stimulation. Mol. Biol. Cell. 16:2275–2284. 10.1091/mbc.E04-10-0881 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Heasman S.J., and Ridley A.J.. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701. 10.1038/nrm2476 [Abstract] [CrossRef] [Google Scholar]
- Hermiston M.L., Xu Z., and Weiss A.. 2003. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 21:107–137. 10.1146/annurev.immunol.21.120601.140946 [Abstract] [CrossRef] [Google Scholar]
- Hoorn T., Paul P., Janssen L., Janssen H., and Neefjes J.. 2012. Dynamics within tetraspanin pairs affect MHC class II expression. J. Cell Sci. 125:328–339. 10.1242/jcs.088047 [Abstract] [CrossRef] [Google Scholar]
- Huber F., Boire A., López M.P., and Koenderink G.H.. 2015. Cytoskeletal crosstalk: when three different personalities team up. Curr. Opin. Cell Biol. 32:39–47. 10.1016/j.ceb.2014.10.005 [Abstract] [CrossRef] [Google Scholar]
- Hwang J., Gheber L.A., Margolis L., and Edidin M.. 1998. Domains in cell plasma membranes investigated by near-field scanning optical microscopy. Biophys. J. 74:2184–2190. 10.1016/S0006-3495(98)77927-5 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ilani T., Khanna C., Zhou M., Veenstra T.D., and Bretscher A.. 2007. Immune synapse formation requires ZAP-70 recruitment by ezrin and CD43 removal by moesin. J. Cell Biol. 179:733–746. 10.1083/jcb.200707199 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Itano M.S., Steinhauer C., Schmied J.J., Forthmann C., Liu P., Neumann A.K., Thompson N.L., Tinnefeld P., and Jacobson K.. 2012. Super-resolution imaging of C-type lectin and influenza hemagglutinin nanodomains on plasma membranes using blink microscopy. Biophys. J. 102:1534–1542. 10.1016/j.bpj.2012.02.022 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jans D.A. 1992. The mobile receptor hypothesis revisited: a mechanistic role for hormone receptor lateral mobility in signal transduction. Biochim. Biophys. Acta. 1113:271–276. 10.1016/0304-4157(92)90001-Q [Abstract] [CrossRef] [Google Scholar]
- Jans D.A., and Pavo I.. 1995. A mechanistic role for polypeptide hormone receptor lateral mobility in signal transduction. Amino Acids. 9:93–109. [Abstract] [Google Scholar]
- Jaqaman K., Kuwata H., Touret N., Collins R., Trimble W.S., Danuser G., and Grinstein S.. 2011. Cytoskeletal control of CD36 diffusion promotes its receptor and signaling function. Cell. 146:593–606. 10.1016/j.cell.2011.06.049 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jaumouillé V., Farkash Y., Jaqaman K., Das R., Lowell C.A., and Grinstein S.. 2014. Actin cytoskeleton reorganization by Syk regulates Fcγ receptor responsiveness by increasing its lateral mobility and clustering. Dev. Cell. 29:534–546. 10.1016/j.devcel.2014.04.031 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jenei A., Varga S., Bene L., Mátyus L., Bodnár A., Bacsó Z., Pieri C., Gáspár R. Jr., Farkas T., and Damjanovich S.. 1997. HLA class I and II antigens are partially co-clustered in the plasma membrane of human lymphoblastoid cells. Proc. Natl. Acad. Sci. USA. 94:7269–7274. 10.1073/pnas.94.14.7269 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Keppler S.J., Gasparrini F., Burbage M., Aggarwal S., Frederico B., Geha R.S., Way M., Bruckbauer A., and Batista F.D.. 2015. Wiskott-Aldrich Syndrome Interacting Protein Deficiency Uncovers the Role of the Co-receptor CD19 as a Generic Hub for PI3 Kinase Signaling in B Cells. Immunity. 43:660–673. 10.1016/j.immuni.2015.09.004 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kucik D.F., Dustin M.L., Miller J.M., and Brown E.J.. 1996. Adhesion-activating phorbol ester increases the mobility of leukocyte integrin LFA-1 in cultured lymphocytes. J. Clin. Invest. 97:2139–2144. 10.1172/JCI118651 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kumar R., Ferez M., Swamy M., Arechaga I., Rejas M.T., Valpuesta J.M., Schamel W.W., Alarcon B., and van Santen H.M.. 2011. Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity. 35:375–387. 10.1016/j.immuni.2011.08.010 [Abstract] [CrossRef] [Google Scholar]
- Kumari S., Depoil D., Martinelli R., Judokusumo E., Carmona G., Gertler F.B., Kam L.C., Carman C.V., Burkhardt J.K., Irvine D.J., and Dustin M.L.. 2015. Actin foci facilitate activation of the phospholipase C-γ in primary T lymphocytes via the WASP pathway. eLife. 4:4. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuokkanen E., Šuštar V., and Mattila P.K.. 2015. Molecular control of B cell activation and immunological synapse formation. Traffic. 16:311–326. 10.1111/tra.12257 [Abstract] [CrossRef] [Google Scholar]
- Kureishy N., Sapountzi V., Prag S., Anilkumar N., and Adams J.C.. 2002. Fascins, and their roles in cell structure and function. BioEssays. 24:350–361. 10.1002/bies.10070 [Abstract] [CrossRef] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R.S., Kondo J., and Fujiwara T.. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. 10.1146/annurev.biophys.34.040204.144637 [Abstract] [CrossRef] [Google Scholar]
- Kusumi A., Fujiwara T.K., Chadda R., Xie M., Tsunoyama T.A., Kalay Z., Kasai R.S., and Suzuki K.G.. 2012. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 28:215–250. 10.1146/annurev-cellbio-100809-151736 [Abstract] [CrossRef] [Google Scholar]
- Kwik J., Boyle S., Fooksman D., Margolis L., Sheetz M.P., and Edidin M.. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 100:13964–13969. 10.1073/pnas.2336102100 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lajoie P., Goetz J.G., Dennis J.W., and Nabi I.R.. 2009. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J. Cell Biol. 185:381–385. 10.1083/jcb.200811059 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lam K.P., Kühn R., and Rajewsky K.. 1997. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 90:1073–1083. 10.1016/S0092-8674(00)80373-6 [Abstract] [CrossRef] [Google Scholar]
- Lanzi G., Moratto D., Vairo D., Masneri S., Delmonte O., Paganini T., Parolini S., Tabellini G., Mazza C., Savoldi G., et al. . 2012. A novel primary human immunodeficiency due to deficiency in the WASP-interacting protein WIP. J. Exp. Med. 209:29–34. 10.1084/jem.20110896 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lavi Y., Edidin M.A., and Gheber L.A.. 2007. Dynamic patches of membrane proteins. Biophys. J. 93:L35–L37. 10.1529/biophysj.107.111567 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lavi Y., Gov N., Edidin M., and Gheber L.A.. 2012. Lifetime of major histocompatibility complex class-I membrane clusters is controlled by the actin cytoskeleton. Biophys. J. 102:1543–1550. 10.1016/j.bpj.2012.01.042 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Le Clainche C., and Carlier M.F.. 2008. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 88:489–513. 10.1152/physrev.00021.2007 [Abstract] [CrossRef] [Google Scholar]
- Lenne P.F., Wawrezinieck L., Conchonaud F., Wurtz O., Boned A., Guo X.J., Rigneault H., He H.T., and Marguet D.. 2006. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 25:3245–3256. 10.1038/sj.emboj.7601214 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lillemeier B.F., Pfeiffer J.R., Surviladze Z., Wilson B.S., and Davis M.M.. 2006. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. USA. 103:18992–18997. 10.1073/pnas.0609009103 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lillemeier B.F., Mörtelmaier M.A., Forstner M.B., Huppa J.B., Groves J.T., and Davis M.M.. 2010. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11:90–96. 10.1038/ni.1832 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lin R., Karpa K., Kabbani N., Goldman-Rakic P., and Levenson R.. 2001. Dopamine D2 and D3 receptors are linked to the actin cytoskeleton via interaction with filamin A. Proc. Natl. Acad. Sci. USA. 98:5258–5263. 10.1073/pnas.011538198 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lingwood D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327:46–50. 10.1126/science.1174621 [Abstract] [CrossRef] [Google Scholar]
- Liu P., Wang X., Itano M.S., Neumann A.K., Jacobson K., and Thompson N.L.. 2012. The formation and stability of DC-SIGN microdomains require its extracellular moiety. Traffic. 13:715–726. 10.1111/j.1600-0854.2012.01337.x [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Liu W., Meckel T., Tolar P., Sohn H.W., and Pierce S.K.. 2010. Intrinsic properties of immunoglobulin IgG1 isotype-switched B cell receptors promote microclustering and the initiation of signaling. Immunity. 32:778–789. 10.1016/j.immuni.2010.06.006 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Machesky L.M., and Insall R.H.. 1998. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8:1347–1356. 10.1016/S0960-9822(98)00015-3 [Abstract] [CrossRef] [Google Scholar]
- Magenau A., Owen D.M., Yamamoto Y., Tran J., Kwiatek J.M., Parton R.G., and Gaus K.. 2015. Discreet and distinct clustering of five model membrane proteins revealed by single molecule localization microscopy. Mol. Membr. Biol. 32:11–18. [Abstract] [Google Scholar]
- Maity P.C., Blount A., Jumaa H., Ronneberger O., Lillemeier B.F., and Reth M.. 2015. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci. Signal. 8:ra93 10.1126/scisignal.2005887 [Abstract] [CrossRef] [Google Scholar]
- Matko J., Bushkin Y., Wei T., and Edidin M.. 1994. Clustering of class I HLA molecules on the surfaces of activated and transformed human cells. J. Immunol. 152:3353–3360. [Abstract] [Google Scholar]
- Mattila P.K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454. 10.1038/nrm2406 [Abstract] [CrossRef] [Google Scholar]
- Mattila P.K., Feest C., Depoil D., Treanor B., Montaner B., Otipoby K.L., Carter R., Justement L.B., Bruckbauer A., and Batista F.D.. 2013. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 38:461–474. 10.1016/j.immuni.2012.11.019 [Abstract] [CrossRef] [Google Scholar]
- McConnell R.E., and Tyska M.J.. 2010. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 20:418–426. 10.1016/j.tcb.2010.04.004 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mooren O.L., Galletta B.J., and Cooper J.A.. 2012. Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 81:661–686. 10.1146/annurev-biochem-060910-094416 [Abstract] [CrossRef] [Google Scholar]
- Morone N., Fujiwara T., Murase K., Kasai R.S., Ike H., Yuasa S., Usukura J., and Kusumi A.. 2006. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J. Cell Biol. 174:851–862. 10.1083/jcb.200606007 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Morrison V.L., James M.J., Grzes K., Cook P., Glass D.G., Savinko T., Lek H.S., Gawden-Bone C., Watts C., Millington O.R., et al. . 2014. Loss of beta2-integrin-mediated cytoskeletal linkage reprogrammes dendritic cells to a mature migratory phenotype. Nat. Commun. 5:5359 10.1038/ncomms6359 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Neisch A.L., and Fehon R.G.. 2011. Ezrin, Radixin and Moesin: key regulators of membrane-cortex interactions and signaling. Curr. Opin. Cell Biol. 23:377–382. 10.1016/j.ceb.2011.04.011 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Owen D.M., Williamson D.J., Magenau A., and Gaus K.. 2012. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat. Commun. 3:1256 10.1038/ncomms2273 [Abstract] [CrossRef] [Google Scholar]
- Pace K.E., Lee C., Stewart P.L., and Baum L.G.. 1999. Restricted receptor segregation into membrane microdomains occurs on human T cells during apoptosis induced by galectin-1. J. Immunol. 163:3801–3811. [Abstract] [Google Scholar]
- Peng J., Wallar B.J., Flanders A., Swiatek P.J., and Alberts A.S.. 2003. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13:534–545. 10.1016/S0960-9822(03)00170-2 [Abstract] [CrossRef] [Google Scholar]
- Plowman S.J., Muncke C., Parton R.G., and Hancock J.F.. 2005. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 102:15500–15505. 10.1073/pnas.0504114102 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Randall K.L., Lambe T., Johnson A.L., Treanor B., Kucharska E., Domaschenz H., Whittle B., Tze L.E., Enders A., Crockford T.L., et al. . 2009. Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat. Immunol. 10:1283–1291. 10.1038/ni.1820 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rust M.J., Bates M., and Zhuang X.. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–795. 10.1038/nmeth929 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Saarikangas J., Zhao H., and Lappalainen P.. 2010. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90:259–289. 10.1152/physrev.00036.2009 [Abstract] [CrossRef] [Google Scholar]
- Schamel W.W., and Reth M.. 2000. Monomeric and oligomeric complexes of the B cell antigen receptor. Immunity. 13:5–14. 10.1016/S1074-7613(00)00003-0 [Abstract] [CrossRef] [Google Scholar]
- Schweighoffer E., Vanes L., Nys J., Cantrell D., McCleary S., Smithers N., and Tybulewicz V.L.. 2013. The BAFF receptor transduces survival signals by co-opting the B cell receptor signaling pathway. Immunity. 38:475–488. 10.1016/j.immuni.2012.11.015 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Shalom-Feuerstein R., Plowman S.J., Rotblat B., Ariotti N., Tian T., Hancock J.F., and Kloog Y.. 2008. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 68:6608–6616. 10.1158/0008-5472.CAN-08-1117 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sheetz M.P., Schindler M., and Koppel D.E.. 1980. Lateral mobility of integral membrane proteins is increased in spherocytic erythrocytes. Nature. 285:510–511. 10.1038/285510a0 [Abstract] [CrossRef] [Google Scholar]
- Sherman E., Barr V., Manley S., Patterson G., Balagopalan L., Akpan I., Regan C.K., Merrill R.K., Sommers C.L., Lippincott-Schwartz J., and Samelson L.E.. 2011. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 35:705–720. 10.1016/j.immuni.2011.10.004 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Simons K., and Ikonen E.. 1997. Functional rafts in cell membranes. Nature. 387:569–572. 10.1038/42408 [Abstract] [CrossRef] [Google Scholar]
- Singer S.J., and Nicolson G.L.. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175:720–731. 10.1126/science.175.4023.720 [Abstract] [CrossRef] [Google Scholar]
- Small J.V., Stradal T., Vignal E., and Rottner K.. 2002. The lamellipodium: where motility begins. Trends Cell Biol. 12:112–120. 10.1016/S0962-8924(01)02237-1 [Abstract] [CrossRef] [Google Scholar]
- Steinhauer C., Forthmann C., Vogelsang J., and Tinnefeld P.. 2008. Superresolution microscopy on the basis of engineered dark states. J. Am. Chem. Soc. 130:16840–16841. 10.1021/ja806590m [Abstract] [CrossRef] [Google Scholar]
- Stossel T.P., Condeelis J., Cooley L., Hartwig J.H., Noegel A., Schleicher M., and Shapiro S.S.. 2001. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2:138–145. 10.1038/35052082 [Abstract] [CrossRef] [Google Scholar]
- Strahl H., Bürmann F., and Hamoen L.W.. 2014. The actin homologue MreB organizes the bacterial cell membrane. Nat. Commun. 5:3442 10.1038/ncomms4442 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Suzuki K., Ritchie K., Kajikawa E., Fujiwara T., and Kusumi A.. 2005. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88:3659–3680. 10.1529/biophysj.104.048538 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Suzuki K.G., Kasai R.S., Hirosawa K.M., Nemoto Y.L., Ishibashi M., Miwa Y., Fujiwara T.K., and Kusumi A.. 2012. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat. Chem. Biol. 8:774–783. 10.1038/nchembio.1028 [Abstract] [CrossRef] [Google Scholar]
- Szöllósi J., Horejsí V., Bene L., Angelisová P., and Damjanovich S.. 1996. Supramolecular complexes of MHC class I, MHC class II, CD20, and tetraspan molecules (CD53, CD81, and CD82) at the surface of a B cell line JY. J. Immunol. 157:2939–2946. [Abstract] [Google Scholar]
- Szöllösi J., Damjanovich S., Goldman C.K., Fulwyler M.J., Aszalos A.A., Goldstein G., Rao P., Talle M.A., and Waldmann T.A.. 1987. Flow cytometric resonance energy transfer measurements support the association of a 95-kDa peptide termed T27 with the 55-kDa Tac peptide. Proc. Natl. Acad. Sci. USA. 84:7246–7250. 10.1073/pnas.84.20.7246 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tan Y.X., Manz B.N., Freedman T.S., Zhang C., Shokat K.M., and Weiss A.. 2014. Inhibition of the kinase Csk in thymocytes reveals a requirement for actin remodeling in the initiation of full TCR signaling. Nat. Immunol. 15:186–194. 10.1038/ni.2772 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tian T., Harding A., Inder K., Plowman S., Parton R.G., and Hancock J.F.. 2007. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol. 9:905–914. 10.1038/ncb1615 [Abstract] [CrossRef] [Google Scholar]
- Tolar P., Sohn H.W., and Pierce S.K.. 2005. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat. Immunol. 6:1168–1176. 10.1038/ni1262 [Abstract] [CrossRef] [Google Scholar]
- Tolar P., Hanna J., Krueger P.D., and Pierce S.K.. 2009. The constant region of the membrane immunoglobulin mediates B cell-receptor clustering and signaling in response to membrane antigens. Immunity. 30:44–55. 10.1016/j.immuni.2008.11.007 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Treanor B. 2012. B-cell receptor: from resting state to activate. Immunology. 136:21–27. 10.1111/j.1365-2567.2012.03564.x [Abstract] [CrossRef] [Google Scholar]
- Treanor B., and Batista F.D.. 2010. Organisation and dynamics of antigen receptors: implications for lymphocyte signalling. Curr. Opin. Immunol. 22:299–307. 10.1016/j.coi.2010.03.009 [Abstract] [CrossRef] [Google Scholar]
- Treanor B., Depoil D., Gonzalez-Granja A., Barral P., Weber M., Dushek O., Bruckbauer A., and Batista F.D.. 2010. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity. 32:187–199. 10.1016/j.immuni.2009.12.005 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Treanor B., Depoil D., Bruckbauer A., and Batista F.D.. 2011. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. J. Exp. Med. 208:1055–1068. 10.1084/jem.20101125 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Trimble W.S., and Grinstein S.. 2015. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol. 208:259–271. 10.1083/jcb.201410071 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tsubata T. 1999. Co-receptors on B lymphocytes. Curr. Opin. Immunol. 11:249–255. 10.1016/S0952-7915(99)80041-7 [Abstract] [CrossRef] [Google Scholar]
- Varma R., Campi G., Yokosuka T., Saito T., and Dustin M.L.. 2006. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 25:117–127. 10.1016/j.immuni.2006.04.010 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vereb G., Matkó J., Vámosi G., Ibrahim S.M., Magyar E., Varga S., Szöllosi J., Jenei A., Gáspár R. Jr., Waldmann T.A., and Damjanovich S.. 2000. Cholesterol-dependent clustering of IL-2Ralpha and its colocalization with HLA and CD48 on T lymphoma cells suggest their functional association with lipid rafts. Proc. Natl. Acad. Sci. USA. 97:6013–6018. 10.1073/pnas.97.11.6013 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Walmsley M.J., Ooi S.K., Reynolds L.F., Smith S.H., Ruf S., Mathiot A., Vanes L., Williams D.A., Cancro M.P., and Tybulewicz V.L.. 2003. Critical roles for Rac1 and Rac2 GTPases in B cell development and signaling. Science. 302:459–462. 10.1126/science.1089709 [Abstract] [CrossRef] [Google Scholar]
- Weber M., Treanor B., Depoil D., Shinohara H., Harwood N.E., Hikida M., Kurosaki T., and Batista F.D.. 2008. Phospholipase C-gamma2 and Vav cooperate within signaling microclusters to propagate B cell spreading in response to membrane-bound antigen. J. Exp. Med. 205:853–868. 10.1084/jem.20072619 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Welch M.D., DePace A.H., Verma S., Iwamatsu A., and Mitchison T.J.. 1997. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 138:375–384. 10.1083/jcb.138.2.375 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wheeler D., Sneddon W.B., Wang B., Friedman P.A., and Romero G.. 2007. NHERF-1 and the cytoskeleton regulate the traffic and membrane dynamics of G protein-coupled receptors. J. Biol. Chem. 282:25076–25087. 10.1074/jbc.M701544200 [Abstract] [CrossRef] [Google Scholar]
- Wilson B.S., Pfeiffer J.R., and Oliver J.M.. 2000. Observing FcepsilonRI signaling from the inside of the mast cell membrane. J. Cell Biol. 149:1131–1142. 10.1083/jcb.149.5.1131 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wilson B.S., Pfeiffer J.R., Surviladze Z., Gaudet E.A., and Oliver J.M.. 2001. High resolution mapping of mast cell membranes reveals primary and secondary domains of Fc(epsilon)RI and LAT. J. Cell Biol. 154:645–658. 10.1083/jcb.200104049 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wong H.S., Jaumouillé V., Heit B., Doodnauth S.A., Patel S., Huang Y.W., Grinstein S., and Robinson L.A.. 2014. Cytoskeletal confinement of CX3CL1 limits its susceptibility to proteolytic cleavage by ADAM10. Mol. Biol. Cell. 25:3884–3899. 10.1091/mbc.E13-11-0633 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., and Sánchez-Madrid F.. 2009. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19:434–446. 10.1016/j.tcb.2009.06.004 [Abstract] [CrossRef] [Google Scholar]
- Yang J., and Reth M.. 2010. Oligomeric organization of the B-cell antigen receptor on resting cells. Nature. 467:465–469. 10.1038/nature09357 [Abstract] [CrossRef] [Google Scholar]
- Yang X.H., Mirchev R., Deng X., Yacono P., Yang H.L., Golan D.E., and Hemler M.E.. 2012. CD151 restricts the α6 integrin diffusion mode. J. Cell Sci. 125:1478–1487. 10.1242/jcs.093963 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zhou A.X., Hartwig J.H., and Akyürek L.M.. 2010. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 20:113–123. 10.1016/j.tcb.2009.12.001 [Abstract] [CrossRef] [Google Scholar]
- Zuidscherwoude M., Göttfert F., Dunlock V.M., Figdor C.G., van den Bogaart G., and van Spriel A.B.. 2015. The tetraspanin web revisited by super-resolution microscopy. Sci. Rep. 5:12201 10.1038/srep12201 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.201504137
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jcb/article-pdf/212/3/267/1370756/jcb_201504137.pdf
Citations & impact
Impact metrics
Article citations
Aspartyl proteases target host actin nucleator complex protein to limit epithelial innate immunity.
EMBO Rep, 25(11):4846-4875, 30 Sep 2024
Cited by: 0 articles | PMID: 39349750 | PMCID: PMC11549443
DDR1 Drives Malignant Progression of Gastric Cancer by Suppressing HIF-1α Ubiquitination and Degradation.
Adv Sci (Weinh), 11(35):e2308395, 18 Jul 2024
Cited by: 1 article | PMID: 39024501 | PMCID: PMC11425230
The method for assessing the specificity of developing CAR therapies.
Biophys Rep (N Y), 4(3):100172, 16 Jul 2024
Cited by: 0 articles | PMID: 39025235 | PMCID: PMC11344002
Hem1 inborn errors of immunity: waving goodbye to coordinated immunity in mice and humans.
Front Immunol, 15:1402139, 04 Jul 2024
Cited by: 0 articles | PMID: 39026677 | PMCID: PMC11254771
Review Free full text in Europe PMC
Control of germinal center B cell survival and IgE production by an adaptor molecule containing PH and SH2 domains, Aps/Sh2b2.
Sci Rep, 14(1):17767, 01 Aug 2024
Cited by: 0 articles | PMID: 39090233 | PMCID: PMC11294469
Go to all (75) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
GPCRs and actin-cytoskeleton dynamics.
Methods Cell Biol, 132:165-188, 10 Feb 2016
Cited by: 17 articles | PMID: 26928544
Phagocytosis: receptors, signal integration, and the cytoskeleton.
Immunol Rev, 262(1):193-215, 01 Nov 2014
Cited by: 270 articles | PMID: 25319336
Review
B cell receptor-induced Ca2+ mobilization mediates F-actin rearrangements and is indispensable for adhesion and spreading of B lymphocytes.
J Leukoc Biol, 93(4):537-547, 29 Jan 2013
Cited by: 21 articles | PMID: 23362305
Signaling to Rho GTPases.
Exp Cell Res, 253(1):166-179, 01 Nov 1999
Cited by: 220 articles | PMID: 10579921
Review
Funding
Funders who supported this work.
Academy of Finland (1)
Grant ID: 257000
Canadian Institutes of Health Research (1)
Grant ID: FRN136808
Cancer Research UK
Natural Sciences and Engineering Research Council of Canada (1)
Grant ID: RGPIN418756

 3
3



