Abstract
Background
Infection with any of the 4 related dengue virus serotypes (DENV-1-4) is thought to result in lifelong immunity to homotypic reinfection (ie, reinfection with the same serotype).Methods
Archived serum samples collected as part of an ongoing pediatric dengue cohort study in Nicaragua were tested for DENV by real-time reverse transcription polymerase chain reaction. Samples were collected from 2892 children who presented with an acute febrile illness clinically attributed to a non-DENV cause (hereafter, "C cases"). Test results were added to a database of previously identified symptomatic dengue cases in the cohort to identify repeat infections.Results
Four patients with homotypic DENV reinfections were identified and confirmed among 29 repeat DENV infections (13.8%) with serotype confirmation. Homotypic reinfections with DENV-1, DENV-2, and DENV-3 occurred 325-621 days after the initial infection. Each patient experienced 1 symptomatic dengue case and 1 DENV-positive C case, and 2 patients presented with symptomatic dengue during their second infection. These DENV-positive C cases did not elicit long-lived humoral immune responses, despite viremia levels of up to 6.44 log10 copies per mL of serum.Conclusions
We describe the first set of virologically confirmed homotypic DENV reinfections. Such cases challenge the current understanding of DENV immunity and have important implications for modeling DENV transmission.Free full text

Editor's choice
Homotypic Dengue Virus Reinfections in Nicaraguan Children
Abstract
Background. Infection with any of the 4 related dengue virus serotypes (DENV-1–4) is thought to result in lifelong immunity to homotypic reinfection (ie, reinfection with the same serotype).
Infection with any of the 4 related dengue virus serotypes (DENV-1–4) is thought to result in lifelong immunity to homotypic reinfection (ie, reinfection with the same serotype).
Methods. Archived serum samples collected as part of an ongoing pediatric dengue cohort study in Nicaragua were tested for DENV by real-time reverse transcription polymerase chain reaction. Samples were collected from 2892 children who presented with an acute febrile illness clinically attributed to a non-DENV cause (hereafter, “C cases”). Test results were added to a database of previously identified symptomatic dengue cases in the cohort to identify repeat infections.
Archived serum samples collected as part of an ongoing pediatric dengue cohort study in Nicaragua were tested for DENV by real-time reverse transcription polymerase chain reaction. Samples were collected from 2892 children who presented with an acute febrile illness clinically attributed to a non-DENV cause (hereafter, “C cases”). Test results were added to a database of previously identified symptomatic dengue cases in the cohort to identify repeat infections.
Results. Four patients with homotypic DENV reinfections were identified and confirmed among 29 repeat DENV infections (13.8%) with serotype confirmation. Homotypic reinfections with DENV-1, DENV-2, and DENV-3 occurred 325–621 days after the initial infection. Each patient experienced 1 symptomatic dengue case and 1 DENV-positive C case, and 2 patients presented with symptomatic dengue during their second infection. These DENV-positive C cases did not elicit long-lived humoral immune responses, despite viremia levels of up to 6.44 log10 copies per mL of serum.
Four patients with homotypic DENV reinfections were identified and confirmed among 29 repeat DENV infections (13.8%) with serotype confirmation. Homotypic reinfections with DENV-1, DENV-2, and DENV-3 occurred 325–621 days after the initial infection. Each patient experienced 1 symptomatic dengue case and 1 DENV-positive C case, and 2 patients presented with symptomatic dengue during their second infection. These DENV-positive C cases did not elicit long-lived humoral immune responses, despite viremia levels of up to 6.44 log10 copies per mL of serum.
Conclusions. We describe the first set of virologically confirmed homotypic DENV reinfections. Such cases challenge the current understanding of DENV immunity and have important implications for modeling DENV transmission.
We describe the first set of virologically confirmed homotypic DENV reinfections. Such cases challenge the current understanding of DENV immunity and have important implications for modeling DENV transmission.
(See the editorial commentary by Forshey, Stoddard, and Morrison on pages 979–81.)
Dengue is the most common human arboviral disease worldwide and results from infection with one of the 4 related dengue virus (DENV) serotypes (DENV-1–4) [1]. Following infection with any serotype, patients are thought to develop lifelong immunity to homotypic reinfection (ie, reinfection with the same serotype) but remain at risk for heterotypic infection (ie, infection with another DENV serotype; also known as secondary infection) [1]. The theory of lifelong, serotype-specific protection originated from research performed by Albert Sabin in the 1940s and 1950s. In this work, individuals inoculated with DENV were protected from clinically apparent infections upon challenge with homologous virus, for 18 months [2]. Furthermore, homotypic neutralizing antibodies that developed during other DENV inoculation experiments remained detectable up to 48 years after infection, demonstrating the long-lived nature of immune responses to DENV [3]. Finally, this theory has been supported by epidemiologic studies of sequential outbreaks that occurred during the reemergence of DENV in the Americas [4].
For a number of reasons, it has not been possible to substantiate the dogma of homotypic protective immunity or to discern whether patients are protected from reinfection (ie, whether they experience sterile immunity) or from clinical disease. Animal models do not fully replicate human infection. Nonhuman primates become viremic following DENV inoculation, but they do not develop overt disease [5]. Documenting repeat, serotype-confirmed DENV infections in the same individual can be done in the context of cohort or long-term hospital-based studies [6–9]. This is complicated, however, by the high incidence of asymptomatic or subclinical DENV infections, which represent the majority of all DENV infections [10, 11]. And although some evidence hints at their existence [6, 12–14], homotypic DENV reinfections have never been virologically confirmed.
The Nicaraguan Pediatric Dengue Cohort Study (PDCS) is a community-based prospective cohort study that was established in 2004 to study the natural history of pediatric dengue transmission and viral and host determinants of DENV immunity and pathogenesis [9, 11, 15]. It is now the longest-running continuous cohort in the field of dengue research, with >3500 active participants aged 2–14 years. The majority of symptomatic DENV infections in the PDCS have been caused by DENV-1–3, although the predominant serotype has varied over time. DENV-1 was the predominant serotype in 2004–2005 and 2012–2013. DENV-2 was the predominant serotype from 2005 to 2008, and DENV-3 was dominated from 2009 to 2011 [11, 16]. Since 2007, the PDCS has also included the collection of acute-phase serum samples from febrile patients who received a diagnosis of a nondengue illness. Together, these cases provide a unique sample set in which to identify recurrent DENV infections in patients over time. In the current study, we tested serum from patients with an acute febrile illness and symptoms and/or signs that suggest an alternate diagnosis (hereafter, “C cases”), and by combining these results with all symptomatic dengue cases diagnosed in the PDCS, we confirmed 4 cases of homotypic DENV reinfection.
METHODS
Ethics Statement
The study protocol was reviewed and approved by the institutional review boards of the Nicaraguan Ministry of Health; the University of California (UC), Berkeley; and Stanford University.
Study Design
The design of the PDCS has been described in detail previously [15]. Briefly, children are enrolled at 2 years of age and followed through 14 years of age. Participants receive all primary medical care from study physicians at the Sócrates Flores Vivas Health Center (HCSFV) in District II of Managua, the capital of Nicaragua, and all information is systematically collected onto clinical report forms. Upon presentation to the HCSFV, patient identity is confirmed by fingerprint and a photo identification card, which includes their unique study number. Children who present to the HCSFV with fever are clinically screened for dengue. Acute- and convalescent-phase serum samples are collected from suspected dengue cases (hereafter, “A cases”) and cases of undifferentiated febrile illness (hereafter, “B cases”). Patients with an acute febrile illness and symptoms and/or signs that suggest an alternate diagnosis are called “C cases” and are not routinely tested for DENV. Since 2007, approximately 100 acute-phase samples have been randomly collected from C cases every month and stored at −80°C. Serum samples are labeled with barcoded stickers that identify the participant study number, study year, the case-type (A/B vs C), and the febrile episode. In addition, each year, healthy serum/plasma samples are collected from all study participants.
Clinical Samples
A/B cases were prospectively tested for DENV as part of the PDCS. Archived C-case serum samples were tested retrospectively between 2013 and 2015. C-case samples were selected from each month of 2007 and the months of the peak DENV season in Managua (August–January) during 2008–2012. Up to 75 serum samples per month were randomly selected for testing by real-time reverse-transcription polymerase chain reaction (RT-PCR), for a total of 2892 samples. Real-time RT-PCR results for DENV-positive C cases with quantifiable viremia were added to the database of all A/B dengue cases in the PDCS from 2004–2013. Patients with homotypic DENV re-infections were identified, and clinical information was obtained by chart review.
DENV Testing
For molecular testing of A/B cases, viral RNA was extracted from acute serum using the QIAamp Viral RNA Mini Kit (Qiagen), and samples were tested by a hemi-nested RT-PCR that targets a region of the capsid and premembrane (prM) genes [17]. Quantitation of viremia in A/B cases was performed retrospectively. For this purpose, RNA was reextracted from separate serum aliquots and tested using DENV-specific multiplex real-time RT-PCR analysis, which targets a region of the 5′ untranslated region and capsid gene [18]. Virus isolation was performed for A/B cases at the time of sample acquisition, using C6/36 cells as previously described [19]. Acute- and convalescent-phase serum samples from A/B cases were tested for DENV-specific immunoglobulin M (IgM) by an IgM antibody-capture enzyme-linked immunosorbent assay (MAC-ELISA) and for anti-DENV antibody by an inhibition ELISA [11, 20]. Results of inhibition ELISA were considered positive for acute dengue if seroconversion or a ≥4-fold rise in titer was detected between acute- and convalescent-phase samples. Whether a DENV infection was primary or secondary was determined by the convalescent-phase titer: a titer of <2560 indicated primary infection, and a titer of ≥2560 indicated secondary infection [11].
For C cases, nucleic acids were extracted on an easyMAG instrument, using 140 µL of serum and a 60-µL elution volume. All samples were screened using a multiplex real-time RT-PCR for DENV, Leptospira, and malaria parasites [21]. DENV serotyping and quantitation were performed on positive samples, using a DENV-specific multiplex real-time RT-PCR [18]. C-case samples from individuals with homotypic DENV reinfections were also tested by hemi-nested RT-PCR to confirm DENV RNA detection [22]. Virus isolation, as described above, was attempted from C-case samples over several serial passages after the detection of DENV RNA by real-time RT-PCR (ie, after 2 freeze-thaw cycles).
Annual samples were tested side by side using inhibition ELISA to determine anti-DENV antibody titers [23, 24]. Inhibition ELISA results were considered positive for a DENV infection in the intervening year if a patient had a ≥4-fold rise in titer between preinfection and postinfection annual samples. Additionally, neutralizing antibody titers in annual serum samples were measured in a flow cytometry–based assay, using human Raji-DC-SIGNR cells with reporter virus particles representing the 4 DENV serotypes as previously described [9, 25, 26]. All relevant, available annual serum samples for each case were analyzed side by side. A sigmoidal dose-response curve with a variable slope was generated to determine the 50% neutralization titer (NT50), defined as the serum dilution at which a 50% reduction in infection was observed as compared to the no-antibody (no-serum) control. The assay was performed and interpreted as previously described [9, 25].
DENV testing was performed at the National Virology Laboratory in Managua, the Harris laboratory at UC Berkeley, and the Stanford Clinical Virology Laboratory. All sites maintain 1-way workflow for molecular testing, including the use of separate, dedicated rooms for extraction, master-mix preparation, and nucleic acid amplification. All RT-PCRs were set up with a no-template negative control to monitor for contamination.
Sequencing and Phylogenetic Analysis
The envelope (E) gene was sequenced from C-case serum and A/B-case DENV isolates (patients 1, 3, and 4) or serum (patient 2). Sequencing primers were designed using Primer3 software and an alignment of all whole-genome sequences from Nicaraguan DENV isolates available in GenBank (National Center for Biotechnology Information [NCBI]; 399 sequences). Reverse transcription was performed using SuperScript III Reverse Transcriptase (Life Technologies); long-range PCR was performed using the NEB LongAmp Hot Start Taq 2x Master Mix (New England Biolabs) according to the manufacturer's protocol. Primer sequences and a detailed protocol are available upon request. Sanger sequencing was performed at Elim Biopharmaceuticals (Hayward, California) and the UC Berkeley Sequencing Facility. Phylogenetic trees were generated by ClustalX2 (clustal.org) and MEGA5.2 (megasoftware.net), using neighbor joining analysis and complete E gene sequences. E gene sequences were translated using the ExPASy Translate Tool (available at: http://web.expasy.org/translate/), and protein sequences were aligned using Protein BLAST (NCBI).
RESULTS
Identification of Homotypic DENV Reinfections
Of 2982 C-case samples tested, 130 DENV-positive cases with quantifiable viremia levels were identified (4.5%). Data from these cases were then added to a database of all symptomatic dengue cases identified in the PDCS during 2004–2013 (599 A/B dengue cases). Thirty-two patients with repeat symptomatic DENV infections were identified, including 1 patient with 3 documented infections, and serotype information was available for both infections in 29 infection pairs (87.8%). Among these 29 infection pairs, homotypic DENV reinfections were identified in 4 (13.8%). Clinical histories, examination findings, and clinical diagnoses for each infection from these 4 patients are shown in Table Table1.1. In all cases, patients had a history of fever, although they did not necessarily have documented fever during the visit. Each patient experienced 1 DENV-positive C case and 1 A/B dengue case. In patients 1 and 4, the DENV-positive C case preceded the A/B dengue case. Patient 1 required hospitalization for severe dengue during the second infection (A case); no other patients were hospitalized during these infections.
Table 1.
Clinical Information on 4 Patients Who Developed Homotypic Dengue Virus (DENV) Reinfections
| Patient, Infection Date | Sex | Age at First Infection, y | DENV Serotype | Time Between Infections, d | Category | Day of Fever | Clinical History | Physical Examination Finding(s) | Clinical Diagnosis |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 13 | 1 | 341 | … | … | … | … | … |
| Oct 2011 | … | … | … | … | C | 1 | Sore throat, cough | Temp, 39.2°C | URTI, influenza suspected |
| Sep 2012 | … | … | … | … | A | 2 | Headache, retro-orbital pain, cervical lymphadenopathy, arthralgia, myalgia, rash, dry mouth | Temp, 37.1°C; conjunctival injection; erythematous rash; cold extremities; cyanosis; delayed capillary refill | Severe dengue, hospitalized |
| 2 | F | 4 | 2 | 621 | … | … | … | … | … |
| Sep 2005 | … | … | … | … | A | 1 | Headache, retro-orbital pain, arthralgia | Temp, 38.9°C; HR, 131 beats/min; RR, 35 breaths/min; BP, 90/60 mm Hg | Classic dengue |
| May 2007 | … | … | … | … | C | 1 | Rhinorrhea, cough | Temp, 37.4°C | URTI |
| 3 | F | 4 | 3 | 343 | … | … | … | … | … |
| Oct 2009 | … | … | … | … | A | 2 | Headache, cervical lymphadenopathy, myalgia, loss of appetite, rash | Temp, 39.0°C; conjunctival injection; generalized maculopapular rash; positive tourniquet test | Classic dengue |
| Sep 2010 | … | … | … | … | C | 2 | Headache, cervical lymphadenopathy, cough, nasal congestion, loss of appetite, rhinorrhea, ear ache | Temp, 39.5°C; HR 112 beats/min; RR, 28 breaths/min; pharyngeal erythema | URTI |
| 4 | F | 3 | 3 | 325 | … | … | … | … | … |
| Dec 2009 | … | … | … | … | C | 1 | Diarrhea, sore throat, loss of appetite, hoarseness, cough | Temp, 38.3°C; HR, 98 beats/min; RR, 32 breaths/min; pharyngeal erythema | Diarrhea with dehydration |
| Nov 2010 | … | … | … | … | B | 2 | Intermittent abdominal pain | Temp, 37.1°C | Acute febrile syndrome |
Abbreviations: BP, blood pressure; HR, heart rate; RR, respiratory rate; Temp, temperature; URTI, upper respiratory tract infection.
The results of laboratory testing for DENV are shown in Table Table2.2. Homotypic reinfections involved DENV-1, DENV-2, and DENV-3. The identified serotypes were consistent with the prevalent serotypes in the PDCS during the years in which these infections occurred [11, 16]. All A/B cases were positive by hemi-nested RT-PCR, virus isolation, and serological testing of paired acute-phase (day 1–2) and convalescent-phase (day 15–18) samples. All A/B cases seroconverted anti-DENV IgM, as determined by MAC-ELISA (from negative to positive). Three cases also demonstrated seroconversion of anti-DENV antibody by inhibition ELISA (patients 1, 3, and 4; acute-phase titers, <10; convalescent-phase titers, 26–69), while patient 2 showed a ≥4-fold rise in antibody titer (from 320 to 20480), indicating a secondary DENV infection. DENV-positive C cases were initially identified by a screening real-time RT-PCR [21] and confirmed using the DENV-specific multiplex real-time RT-PCR and hemi-nested RT-PCR [18, 22]. Serotype calls in the DENV-specific multiplex real-time RT-PCR and hemi-nested RT-PCR, which target different regions of the DENV genome, were concordant. To reconfirm DENV detection in C cases, nucleic acids were extracted from a second serum aliquot (made at the time of the C-case presentation, for patients 1, 2, and 4) or reextracted from the only available aliquot (for patient 3) and tested in the DENV-specific multiplex real-time RT-PCR [18]. DENV viremia levels were 2.75–6.44 log10 copies/mL in C cases and 3.70–8.05 log10 copies/mL in A/B cases. DENV-1 was also isolated in culture from the C case of patient 1.
Table 2.
Results of Dengue Virus (DENV) Laboratory Testing
| Patient, Case Category | Hemi-Nested RT-PCR Result | DENV Multiplex RT-PCR Result | Viremia Level, Log10 Copies/mL serum | Virus Isolated | Sequencing Confirmed | IgM Seroconversion | Inhibition ELISA | Neutralizing Antibodies | |
|---|---|---|---|---|---|---|---|---|---|
| Acute- and Convalescent- Phase Seraa | Annual Samplesb | Annual Samplesc | |||||||
| 1 | |||||||||
 C C | DENV-1 | DENV-1 | 2.75 | Yes | Yes | NDd | NDd | Negative | NDe |
 A A | DENV-1 | DENV-1 | 7.59 | Yes | Yes | Yes | Positive | Positive | NDe |
| 2 | |||||||||
 A A | DENV-2 | DENV-2 | 8.00 | Yes | Yes | Yes | Positive | Positive | Positive |
 C C | DENV-2 | DENV-2 | 4.98 | No | Yes | NDd | NDd | Negative | Negativef |
| 3 | |||||||||
 A A | DENV-3 | DENV-3 | 3.70 | Yes | Yes | Yes | Positive | Positive | Positive |
 C C | DENV-3 | DENV-3 | 6.44 | No | Yes | NDd | NDd | NDe | NDe |
| 4 | |||||||||
 C C | DENV-3 | DENV-3 | 3.37 | No | Yes | NDd | NDd | Negative | Negative |
 B B | DENV-3 | DENV-3 | 8.05 | Yes | Yes | Yes | Positive | Positive | Positive |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; IgM, immunoglobulin M; ND, not determined; NT50, 50% neutralization titer; RT-PCR, reverse transcription polymerase chain reaction.
a Positive result defined as a ≥4-fold rise in DENV-specific inhibition ELISA titer in paired acute- and convalescent-phase samples.
b Positive result defined as a ≥4-fold rise in DENV-specific inhibition ELISA titer in paired preinfection and postinfection annual samples [20].
c Positive result defined as a ≥4-fold rise in NT50 in paired preinfection and postinfection annual samples [9].
d Convalescent-phase sample was not collected.
e Sample not available.
f Three-fold rise in NT50 to DENV-2.
Inhibition ELISA of annual samples collected before and after a DENV-positive C case revealed no evidence of recent DENV infection (3 patients had specimens available for analysis), whereas annual samples indicated a DENV infection in the year in which the A/B cases occurred (Table (Table2).2). Three patients developed an A/B dengue case after a DENV-positive C case (secondary infection). In these instances, inhibition ELISA results from acute- and convalescent-phase samples were consistent with primary infection. This analysis includes patient 3, who had 3 separate DENV infections during the study: 2 DENV-3 infections (an A case in 2009 and a C case in 2010), as well as a DENV-1 C case in 2007. Patient 2 was not DENV naive at study entry and experienced a secondary DENV-2 A-case infection in 2005, as well as a DENV-2 C case in 2007.
Annual samples from patients 2–4 were available for neutralizing antibody testing. NT50 and inhibition ELISA results were generally consistent (Table (Table2).2). A/B dengue cases resulted in an appropriate, ≥4-fold rise in NT50 between preinfection and postinfection samples. C-case DENV infections were not captured by neutralization antibody assay for patient 2 (DENV-2 C case in 2007), patient 3 (2007 DENV-1 infection; no sample was available to test for C case in 2010), or patient 4 (DENV-3 C case in 2009). However, patient 2, who experienced a secondary DENV-2 infection in 2005, displayed a 3-fold rise in DENV-2 NT50 following their DENV-2 C case in 2007.
Sequencing and Phylogenetic Analysis
Complete E gene sequences were obtained from both infections for patients 1, 3, and 4 and from the A case for patient 2. The complete E gene sequence could not be obtained from the C case for patient 2; however, the 511–base pair amplicon generated during the first step of the hemi-nested RT-PCR [22], covering portions of the capsid and prM genes, was sequenced for this case. NCBI Nucleotide BLAST results for all sequences matched the serotype detected by RT-PCR analysis.
Further E gene sequence analysis revealed 97% nucleotide identity between paired DENV-1 strains from patient 1 and 99.4% and 99.2% nucleotide identity between paired DENV-3 strains from patients 3 and 4, respectively (Table (Table3).3). Phylogenetic trees generated using these sequences and sequences obtained from contemporaneous Nicaraguan isolates confirmed that the paired strains were nonidentical but closely related (Figure (Figure1).1). In addition, these strains were closely related to the viruses circulating in Nicaragua during the years when the study patients were infected. Translation of the sequences from the strains responsible for homotypic reinfections demonstrated 1–3 amino acid differences in the Envelope protein as compared to the first infecting virus (Table (Table33).
Table 3.
Envelope Nucleotide and Amino Acid Sequence Comparison of C-Case and A/B-Case Dengue Virus Strains
| Patient | Nucleotide Identity, % | Amino Acid Change(s)a |
|---|---|---|
| 1 | 97.0 | I54V, F95L, T181A |
| 3 | 99.4 | A328V, R468K |
| 4 | 99.2 | S184P |
a Amino acids from the C case are displayed first.
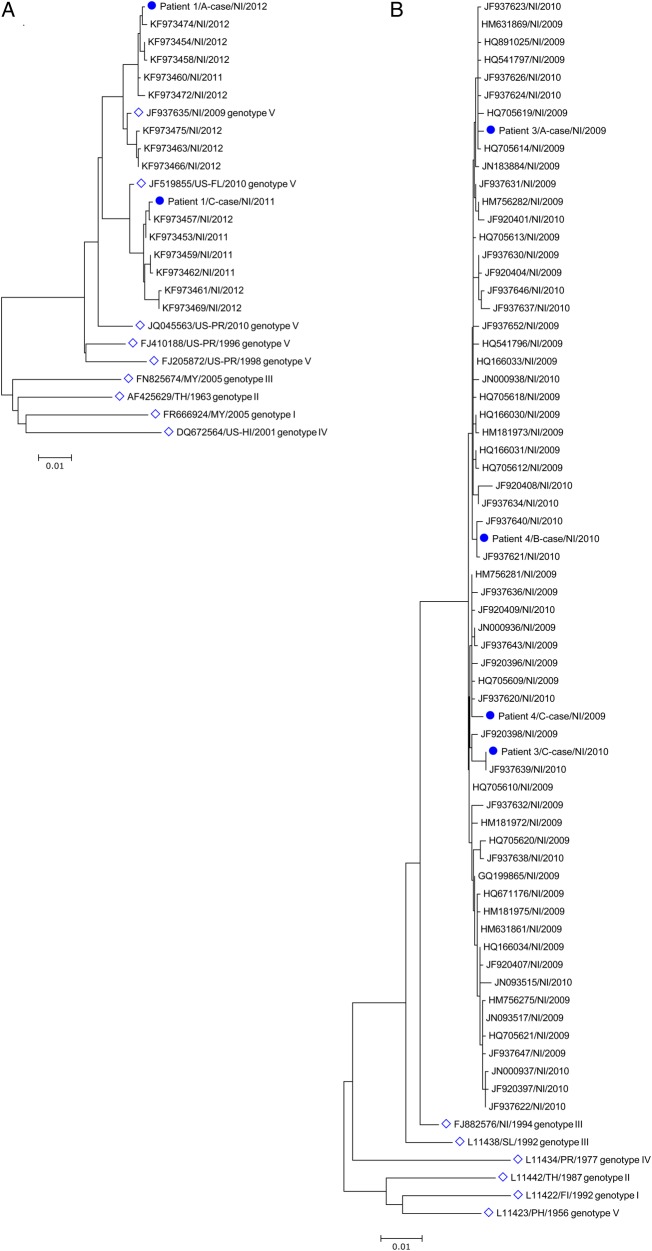
Phylogeny of dengue virus (DENV) strains from patients with homotypic reinfections. A, DENV-1 from patient 1. B, DENV-3 from patients 3 and 4. Phylograms were generated by neighbor joining analysis of all unique envelope (E) gene sequences in the National Center for Biotechnology Information nucleotide database from contemporaneous Nicaraguan isolates of DENV-1 and DENV-3. Patient DENV sequences are labeled with a dot. Reference DENV-1 and DENV-3 E gene sequences representing established genotypes were also included (open diamonds). Sequences are named using the following convention: GenBank accession number/place of origin/year of collection. The scale bars represent the number of nucleotide substitutions per site. Abbreviations: FI, Fiji; MY, Malaysia; NI, Nicaragua; PH, Philippines; SL, Sri Lanka; TH, Thailand; US-FL, Florida; US-HI, Hawaii; US-PR, Puerto Rico.
DISCUSSION
In the current report, we describe the first virologically confirmed cases of homotypic DENV reinfection. Each DENV infection and serotype was detected by ≥2 methods, including viral isolation and distinct RT-PCR assays that target nonoverlapping regions of the DENV genome. Additionally, full-length E gene sequence from both infections for patients 1, 3, and 4 and the A case for patient 2 confirmed the serotypes identified by RT-PCR analysis, and the serotype of the C case for patient 2 was confirmed by sequencing a region of the capsid and prM genes. Phylogenetic analysis of paired E gene sequences further demonstrated that patients were infected with closely related but distinct homotypic DENV strains during their A/B and C infections.
Protective immunity that results from natural DENV infection is presumed to be lifelong and serotype specific, but whether protection results from sterile immunity or the prevention of clinical disease remains unresolved [27]. In the research performed by Albert Sabin, study subjects were protected from clinical disease following rechallenge with DENV-1 at 18 months, but they were not evaluated for viremia [2]. In primate studies, animals typically have not developed viremia upon rechallenge with homologous virus [28–30]. However, much of this research was performed before the routine use of RT-PCR. In a more recent study, Bernardo et al demonstrated low-level viremia only detectable by hemi-nested RT-PCR analysis in 4 of 5 macaques rechallenged with DENV-2 [31]. In the current study, patients developed DENV viremia 1–2 years after a DENV infection of the same serotype. This argues against sterile immunity as the mechanism of protection following natural DENV infection and also demonstrates that, in some cases, serotype-specific immunity to DENV may be short-lived. Furthermore, patients were not protected against clinical disease, as demonstrated by patients 1 and 4. In these patients, an A/B dengue case followed a primary C case of the same serotype, and for patient 1, this resulted in hospitalization with severe dengue.
While findings consistent with homotypic reinfections have been reported in human studies, all lack virologic confirmation and rely primarily on serologic testing [6, 12–14]. Endy et al [12] described one patient with mild dengue fever from DENV-2 who had preexisting monotypic neutralizing antibodies against DENV-2 detected at low levels (50% plaque reduction neutralization titer [PRNT50], 11). Sirivichayakul et al [14] reported 2 cases of patients with monotypic PRNT50 to DENV-2 and -1, respectively, who subsequently developed infections with the same serotype, as determined by mosquito isolation. However, specimens were not available to confirm the first infections in these 3 cases [12, 14]. A possible homotypic DENV-2 reinfection was identified in Puerto Rico. In this case, infections occurred 16 months apart, but the initial infection could not be confirmed, and the patient was removed from further analysis [13]. Finally, Gibbons et al [6] found one possible homotypic DENV-2 reinfection in Thailand, but enzyme immunoassay and RT-PCR analysis yielded discordant serotype results for the first infection [6].
In our patients, homotypic DENV reinfections were identified by testing C cases in the PDCS with real-time RT-PCR assays for DENV that demonstrate improved sensitivity and equivalent specificity as compared to widely-used molecular DENV diagnostic tests [18, 32]. The combination of testing a unique study population with highly sensitive diagnostic tests may have allowed for the detection of homotypic reinfections when they have not been previously identified in large studies of typical, symptomatic dengue cases [6, 9, 10, 13]. When C-case data were added to all symptomatic DENV infections identified in the PDCS, homotypic DENV reinfections accounted for 13.8% of all repeat DENV infections with a confirmed serotype (12.1%, if repeat symptomatic infections without serotype confirmation were included). In the PDCS, symptomatic dengue cases only account for approximately 20% of all DENV infections [11, 33]. Subclinical or asymptomatic DENV infections are identified by inhibition ELISA of annual samples, but the causative serotype in these infections cannot always be accurately determined. Therefore, homotypic DENV reinfections may be more common than indicated by our study. Such repeat infections may have a significant impact on force of infection estimates for DENV, which in turn are used to estimate necessary vaccine coverage and to guide disease control programs [34]. In addition, since the viremia level was detectable and quite high in some of the C cases, these homotypic infections could conceivably contribute to transmission and thus need to be considered in modeling of DENV transmission dynamics.
The viremia level detected at presentation in DENV-positive C cases was as high as 6.44 log10 copies/mL. However, these infections generally did not elicit long-lived humoral immune responses, based on inhibition ELISA of (1) healthy annual samples and (2) samples obtained during A/B dengue cases that followed DENV-positive C cases, as well as analysis of neutralizing antibody titers in healthy annual samples. Inhibition ELISA has demonstrated equivalent sensitivity and specificity to hemagglutination inhibition [23], and testing of annual samples by this method is 80% sensitive for the capture of A/B dengue cases [11]. Taken together, these data are consistent with an altered immune response to C-case DENV infections. Such infections may only elicit a short-lived humoral immune response, or they may be adequately controlled by cellular or innate immune responses without eliciting a detectable humoral response. Interestingly, a 3-fold rise in NT50 was observed following the DENV-2 C case in patient 2, who had experienced a prior secondary A case, indicating that homotypic reinfections may play a role in the maintenance of DENV immunity in endemic regions [25].
Limitations of the current study include the retrospective detection of DENV infections among C cases. Convalescent-phase samples were, therefore, unavailable for serological testing, but annual serum samples were tested for anti-DENV antibodies by inhibition ELISA to evaluate long-lived antibody responses to DENV infection. The dampened immune response to DENV-positive C cases, described earlier, limited our ability to test the possible effect of sequence differences identified in the envelope protein of A/B and C-case DENV strains from patients 1, 3, and 4. Thus, the importance of these amino acid changes in relation to immune escape remains unclear. A final limitation to this study is that DENV was isolated from only 1 C case. However, viral culture is known to be less sensitive than real-time RT-PCR analysis for the detection of DENV. In one prospective method comparison, viral culture was only 27.5% sensitive as compared to RT-PCR analysis [35]. Furthermore, specimens in our study had been stored at −80°C for up to 8 years and had undergone 2 freeze-thaw cycles before attempted viral isolation. These conditions are expected to significantly decrease culture yield, perhaps by as much as 5-fold to >10-fold (E. H., unpublished data) [36, 37].
In conclusion, we present 4 patients with homotypic DENV reinfections, including infections with DENV-1, DENV-2, and DENV-3. To our knowledge, these represent the first virologically confirmed homotypic reinfections in the field of dengue research. Such cases challenge the paradigm of lifelong, serotype-specific DENV immunity following natural infection and have implications for the modeling of DENV transmission.
Notes
Acknowledgments. We thank Yuanyuan Liu, PhD, and Alisha Mohamed-Hadley, PhD, for their contributions in the Pinsky Laboratory during this study; past and present members of the study team based at the Centro de Salud Sócrates Flores Vivas, the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia, and the Sustainable Sciences Institute in Managua, Nicaragua, for their dedication and high-quality work; and the study participants and their families.
We thank Yuanyuan Liu, PhD, and Alisha Mohamed-Hadley, PhD, for their contributions in the Pinsky Laboratory during this study; past and present members of the study team based at the Centro de Salud Sócrates Flores Vivas, the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia, and the Sustainable Sciences Institute in Managua, Nicaragua, for their dedication and high-quality work; and the study participants and their families.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the Thrasher Research Fund (early career award 11979 to J. J. W.); the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants K08AI110528; [to J. J. W.] and R01 AI099631 [to A. B.]); the Pediatric Dengue Vaccine Initiative (grant VE-1 to E. H.); and the Bill and Melinda Gates Foundation and the Instituto Carlos Slim de la Salud (grant to E. H.).
This work was supported by the Thrasher Research Fund (early career award 11979 to J. J. W.); the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants K08AI110528; [to J. J. W.] and R01 AI099631 [to A. B.]); the Pediatric Dengue Vaccine Initiative (grant VE-1 to E. H.); and the Bill and Melinda Gates Foundation and the Instituto Carlos Slim de la Salud (grant to E. H.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
Articles from The Journal of Infectious Diseases are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/infdis/jiw099
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jid/article-pdf/214/7/986/7076857/jiw099.pdf
Citations & impact
Impact metrics
Article citations
Evolving dengue serotype distribution with dominance of dengue virus- 3 in Bangalore: critical insights for vaccine efficacy and implementation.
Lancet Reg Health Southeast Asia, 30:100485, 14 Sep 2024
Cited by: 0 articles | PMID: 39315002 | PMCID: PMC11417586
Dengue Virus Serotype 3 Origins and Genetic Dynamics, Jamaica.
Emerg Infect Dis, 30(10):2149-2154, 27 Aug 2024
Cited by: 0 articles | PMID: 39190550 | PMCID: PMC11431914
Immunologic Crosstalk and Host-Specific Immune Signature Associated with Dengue.
ACS Omega, 9(36):37418-37429, 20 Aug 2024
Cited by: 0 articles | PMID: 39281909 | PMCID: PMC11391553
Review Free full text in Europe PMC
Pooled Cohort Profile: ReCoDID Consortium's Harmonized Acute Febrile Illness Arbovirus Meta-Cohort.
JMIR Public Health Surveill, 10:e54281, 23 Jul 2024
Cited by: 1 article | PMID: 39042429 | PMCID: PMC11288473
Travel surveillance uncovers dengue virus dynamics and introductions in the Caribbean.
Nat Commun, 15(1):3508, 25 Apr 2024
Cited by: 10 articles | PMID: 38664380 | PMCID: PMC11045810
Go to all (74) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of Dengue Virus Infections Among Febrile Children Clinically Diagnosed With a Non-Dengue Illness, Managua, Nicaragua.
J Infect Dis, 215(12):1816-1823, 01 Jun 2017
Cited by: 8 articles | PMID: 28863466 | PMCID: PMC5853235
Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year.
PLoS Negl Trop Dis, 7(8):e2357, 08 Aug 2013
Cited by: 146 articles | PMID: 23951377 | PMCID: PMC3738476
Molecular characterization of dengue viruses circulating during 2009-2012 in Uttar Pradesh, India.
J Med Virol, 87(1):68-75, 02 Jun 2014
Cited by: 27 articles | PMID: 24889214
Immune responses to dengue virus in the skin.
Open Biol, 8(8):180087, 01 Aug 2018
Cited by: 15 articles | PMID: 30135238 | PMCID: PMC6119867
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Bill and Melinda Gates Foundation and the Instituto Carlos Slim de la Salud (grant to E. H.)
NIAID NIH HHS (2)
Grant ID: R01 AI099631
Grant ID: K08 AI110528
National Institute of Allergy and Infectious Diseases
National Institutes of Health (2)
Grant ID: R01 AI099631
Grant ID: K08AI110528





