Abstract
Purpose
The epithelial-to-mesenchymal transition (EMT) is a cell development-regulated process in which noncoding RNAs act as crucial modulators. Recent studies have implied that EMT may contribute to resistance to epidermal growth factor receptor (EGFR)-directed therapy. The aims of this study were to determine the potential role of microRNAs (miRNA) in controlling EMT and the role of EMT in inducing the sensitivity of human bladder cancer cells to the inhibitory effects of the anti-EGFR therapy.Experimental design
miRNA array screening and real-time reverse transcription-PCR were used to identify and validate the differential expression of miRNAs involved in EMT in nine bladder cancer cell lines. A list of potential miR-200 direct targets was identified through the TargetScan database. The precursor of miR-200b and miR-200c was expressed in UMUC3 and T24 cells using a retrovirus or a lentivirus construct, respectively. Protein expression and signaling pathway modulation, as well as intracellular distribution of EGFR and ERRFI-1, were validated through Western blot analysis and confocal microscopy, whereas ERRFI-1 direct target of miR-200 members was validated by using the wild-type and mutant 3'-untranslated region/ERRFI-1/luciferse reporters.Results
We identified a tight association between the expression of miRNAs of the miR-200 family, epithelial phenotype, and sensitivity to EGFR inhibitors-induced growth inhibition in bladder carcinoma cell lines. Stable expression of miR-200 in mesenchymal UMUC3 cells increased E-cadherin levels, decreased expression of ZEB1, ZEB2, ERRFI-1, and cell migration, and increased sensitivity to EGFR-blocking agents. The changes in EGFR sensitivity by silencing or forced expression of ERRFI-1 or by miR-200 expression have also been validated in additional cell lines, UMUC5 and T24. Finally, luciferase assays using 3'-untranslated region/ERRFI-1/luciferase and miR-200 cotransfections showed that the direct down-regulation of ERRFI-1 was miR-200-dependent because mutations in the two putative miR-200-binding sites have rescued the inhibitory effect.Conclusions
Members of the miR-200 family appear to control the EMT process and sensitivity to EGFR therapy in bladder cancer cells and the expression of miR-200 is sufficient to restore EGFR dependency at least in some of the mesenchymal bladder cancer cells. The targets of miR-200 include ERRFI-1, which is a novel regulator of EGFR-independent growth.Free full text

miR-200 Expression Regulates Epithelial to Mesenchymal Transition in Bladder Cancer Cells and Reverses Resistance to EGFR Therapy
Abstract
Purpose
The epithelial to mesenchyme transition (EMT) is a cell development-regulated process in which noncoding RNAs act as crucial modulators. Recent studies have implied that EMT may contribute to resistance to epidermal growth factor receptor (EGFR)-directed therapy. The aims of this study were to determine the potential role of microRNAs (miRNAs) in controlling EMT and the role of EMT in inducing the sensitivity of human bladder cancer cells to the inhibitory effects of the anti-EGFR therapy.
Experimental Design
miRNA array screening and real-time reverse transcription-polymerase chain reaction were used to identify and validate the differential expression of miRNAs involved in EMT in 9 bladder cancer cell lines. A list of potential miR-200 direct targets was identified through the TargetScan database. The precursor of miR-200b and c were expressed in UMUC3 and T24 cells using a retrovirus or a lentivirus construct, respectively. Protein expression and signaling pathway modulation as well as intracellular distribution of EGFR and ERRFI-1 were validated through western blot analysis and confocal microscopy, whereas ERRFI-1 direct target of miR-200 members was validated by using the wild-type and mutanty 3′UTR/ERRFI-1/Luciferse reporters.
Results
We identified a tight association between the expression of miRNAs of the miR-200 family, epithelial phenotype, and sensitivity to EGFR inhibitors-induced growth inhibition in bladder carcinoma cell lines. Stable expression of miR-200 in mesenchymal UMUC3 cells increased E-cadherin levels, decreased expression of ZEB-1, ZEB-2, ERRFI-1, and cell migration, and increased sensitivity to EGFR blocking agents. The changes in EGFR sensitivity by silencing or forced expression of ERRFI-1 or by miR-200 expression have also been validated in additional cell lines, UMUC5 and T24. Finally, luciferase assays using 3′UTR/ERRFI-1/Luc and miR-200 co-transfections demonstrated that the direct down-regulation of ERRFI-1 was miR-200-dependent since mutations in the two putative miR-200-binding sites have rescued the inhibitory effect.
Conclusions
Members of the miR-200 family appear to control the EMT process and sensitivity to EGFR therapy, in bladder cancer cells and that expression of miR-200 is sufficient to restore EGFR dependency, at least in some of the mesenchymal bladder cancer cells. The targets of miR-200 include ERRFI-1, which is a novel regulator of EGFR-independent growth.
Introduction
Bladder cancer is a common malignancy characterized by frequent recurrences and a poor clinical outcome when tumors progress to invasive disease (1). A multi-step accumulation of genetic and epigenetic factors result in uncontrolled cellular proliferation, invasion, increased cell survival and metastatic spread. Recent advances in the use of high-throughput methods of molecular analysis have made it possible to identify genetic profiles characteristic of distinct tumor types and to identify targets and pathways that may underlie a particular clinical behavior; however, systemic chemotherapy remains palliative and only modestly effective (2).
Targeted therapies, including inhibitors of receptor tyrosine kinases, appear to be most effective within the context of documented patient selection and using algorithms that take into account target expression and validation of molecular pathways. Growth factor receptors, such as the epidermal growth factor receptor (EGFR), fibroblast growth factor receptor (FGFR) or platelet-derived growth factor receptor (PDGFR) are differentially expressed by bladder cancer cell lines or tumors, making inhibitors to these molecules attractive therapeutic tools (3-5). Debate is ongoing as to whether EGFR expression or mutation in the tyrosine kinase domain correlates well with response to EGFR blockade by tyrosine kinase inhibitors (TKIs) and to survival. In previous studies, we have demonstrated that mutations in the EGFR protein or gene amplifications are not as frequent in bladder cancer as in other cancers, for example lung cancer (6, 7), suggesting that other predictors remain to be identified. Very recently, several groups reported that sensitivity to EGFR-directed therapy is associated with expression of E-cadherin and other properties of a typical “epithelial” tumor phenotype (8-13).
A phenomenon known as epithelial to mesenchyme transition (EMT) has been implicated as having a role in tumor invasion/migration and metastasis (14). Loss of E-cadherin expression is a hallmark of the EMT process, which is probably required for enhanced tumor-cell motility. EMT is essential during mesoderm development or neural plate formation in vertebrate and it is thought to be important for wound healing and tumor metastasis. During EMT, downregulation of E-cadherin allows epithelial cells to undergo changes in cell morphology and motility so that they adopt mesenchymal characteristics. Expression of E-cadherin is controlled by several transcriptional repressors, including Twist, Snail1, Snail2/Slug, E47, ZEB1/TCF8, and ZEB2/SIP1, which bind to E-boxes in the E-cadherin promoter (15).
Recent studies have demonstrated that ZEB1 and ZEB2 are direct targets of miRNAs from the miR-200 family in human breast cancer cells (MDA-MB231), canine kidney cells (MDCK), murine models and a NCI-60 panel of cell lines (16-19). miRNAs are short noncoding RNAs that regulate protein production by pairing with appropriate complementary stretches in mRNAs (20,21). Hourteau et al (16), Gregory et al (17), and Park et al (19) all identified a new role for miR-200c, miR-141, miR-200b, and miR-205, all members of the miR-200 family of miRNAs, in EMT through the direct regulation of ZEB1 and ZEB2. Korpal et al (18) also showed that ectopic expression of these miRNAs leads to re-expression of E-cadherin and epithelial phenotypes in NMuMG murine mammary model that undergone EMT upon TGF-β stimulation (18) and mesenchymal MDAMB231 cells (16). Hundreds of miRNAs are encoded in the human genome, with an estimated 30% of mRNAs possessing conserved miRNA binding sites, suggesting that miRNA-based regulation is an integral component of the global gene expression program (22). The presence of diverse and often strong phenotypes, such as EMT or diseases associated with mutations or altered expression of miRNAs, suggests their importance in cell development processes and in the biology of cancers (23, 24).
The overall goal of this study was to investigate the mechanisms underlying the EMT process and EMT-induced resistance to EGFR-directed therapy in human bladder cancer cells. Our results revealed a close, direct association between expression of miRNAs of the miR-200 family, E-cadherin expression, and sensitivity to the EGFR antagonists. We also demonstrated that ZEB1 and ZEB2 are involved in the EMT process and identified a new target (ERRFI-1/Mig6) that also plays an important role in mediating EGFR-inhibitor resistance. These results should help us to prospectively identify bladder tumors that will be most susceptible to EGFR-directed therapy.
Materials and Methods
Cell lines and culture conditions
The UMUC series of urothelial carcinomas and 253J BV cells were isolated and fingerprinted and genotyped by the specimen core at the M. D. Anderson Genitourinary Specialized Program of Research Excellence in Bladder Cancer. KU7 cells were supplied by W. Benedict at M. D. Anderson. The cell lines were maintained at 37ْC in modified Eagle's minimum essential medium (MEM) supplemented with 10% fetal bovine serum, vitamins, sodium pyruvate, L-glutamine, penicillin, streptomycin, and nonessential amino acids.
Reagents and chemicals
ImClone Systems, Inc. (New York, NY) generously provided cetuximab (C225), a selective antihuman EGFR antagonist monoclonal antibody in a stock concentration of 2 mg/mL. Iressa (Astra-Zeneca) and Erlotinib/Tarceva (Genentech), which are specific EGFR kinase inhibitors have been generously provided by the manufacturer or purchased from the UT MD Anderson Pharmacy. The following antibodies were purchased from the manufacturer's list and were used for western blot analysis: EGFR (Lab Vision Corp., Fremont, CA); E-cadherin and P-cadherin (BD Transduction Laboratories, Lexington, KY); actin, and vinculin (Sigma-Aldrich Corporation, St. Louis, MO); ErbB receptor inhibitor-1 (ERRFI-1) and pEGFR (Tyr1068; Cell Signaling Technology, Danvers, MA). Pre-miRNAs and antagomirs were purchased from Ambion, Inc. (Austin, TX). The TaqMan gene expression assays, miRNA expression assays, and ERRFI-1. The short hairpin RNA (shRNA) against ERRFI-1 and non-targeting controls, as well as miR-200b RFP lentiviral system were all obtained from Open Biosystems-Thermo Scientific.
Cell proliferation assay
Cells (5 × 103) were plated in 96-well plates for 24 hours, serum-starved for 24 hours in 2% fetal calf serum supplemented with MEM, and then treated with or without EGFR inhibitors at increasing concentrations in an EGF-nonstimulated environment for 30 hours. We measured the effect of cetuximab (C225), Erlotinib (Tarceva) and Iressa on DNA synthesis by pulse-labeling cells with [3H]thymidine (MP Biomedicals, Santa Ana, CA) for 2 hours followed by lysis in 100 μL of 0.1 mol/L KOH. Cells were harvested onto fiberglass filters and incorporated tritium was quantified by β-counter.
Wound-healing assay
UMUC3 cells (4 × 106) were plated in 6-well plates for 24 to 48 hours (until they reached confluence). A diametric scratch was performed using a pipette tip followed by 2 culture media changes. Cells were photographed in several pre-marked spots as time zero. Multiple photographs were then taken at 24 and 48 hours in the same spots for comparison.
Plasmid construction and luciferase reporter assays
3′UTR reporter plasmids for miR-200c were constructed via insertion of miR-200c seed sequence into the XbaI restriction site 3′ to luciferase gene in the pGL3-control plasmid (Promega Corp., Madison, WI). An ERRFI-1 3′ UTR segment of 648bp was amplified by PCR from human cDNA and inserted into the pMIR-REPOR (Ambion) with MluI and SpeI sites. For point mutations we used the QuikChange® II Site-Directed Mutagenesis Kit (Qiagen) following manufactory's instruction. The primers for point mutations (U:C) are Mutant 1-F: 5′-ccttgtgttgctggttcctattcagtacctcctggggattgttt-3′; Mutant 1-R: 5′-aaacaatccccaggaggtactgaataggaaccagcaacacaagg-3′; Mutant 2-F: 5′-cactgatttctgcattatgtgtacagtaccggacaaaggattttattcattttgtt-3′; Mutant 2-R: 5′-aacaaaatgaataaaatcctttgtccggtactgtacacataatgcagaaatcagtg-3′. The sequences of the recombinant plasmids were confirmed by DNA sequencing.
Reporter vector transfection was performed using Lipofectamine-2000 reagent (Invitrogen) as described previously (25). miRNA transfections were performed using 20 nmol/L Lipofectamine-2000. Plasmid transfections were carried out similarly but with 50nM/L of reporter plasmid in 24-well plates plus 0.02 μg cytomegalovirus-renilla.
Real-time reverse transcription-polymerase chain reaction (RT-PCR) to quantify mature, miRNAs
Total RNA was extracted using a Mirvana extraction reverse transcription kit (Applied Biosystems) and 10 ng total RNA along with miR-specific primer miRNA were used for expression analysis. cDNA was synthesized using Taqman miRNA specific kit (Ambion-Applied Biosystems).
Immunoblots
Cells were treated with cetuximab (C225) for 3 hours, harvested at approximately 75% to 80%, and lysed. Protein concentration was assayed using the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA). To prepare cell extracts, cells were washed 3 times with phosphate-buffered saline, and then lysed with RIPA buffer [50 mM Tris–HCl, pH 7.5; 150 mM NaCl; 0.5% NP-40; 0.1% sodium dodecyl sulfate; 0.1% sodium deoxycholate; protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN); and 1 mM sodium ortho-vanadate] for 20 minutes on ice. To measure the activity (phosphorylation) of EGFR, we used an anti-Tyr-1068--specific antibody (Cell Signaling Technololgy) raised against the kinase, which measures the level of autophosphorylated EGFR. Western blot analyses were performed as previously described (25). Antibodies anti-phosphorylated p42/44 MAPKinase and pp42/44 were purchased from Cell Signaling Technology. All chemicals were purchased from Sigma Immunochemicals.
Indirect immunofluorescence staining
Cellular localization of EGFR and ERRFI1 was determined using indirect immunofluorescence as previously described (24). Briefly, cells grown on glass coverslips were fixed (without permeabilization) in 3.7% paraformaldehyde at room temperature for 10 minutes and then extracted with ice-cold acetone. Cells were treated with or without anti-EGFR (LabVision, Inc.) and ERRFI-1 antibodies (Cell Signaling Technology) and then treated with Alexa-488--labeled goat anti-rabbit and Alexa-546--labeled goat anti-mouse secondary antibodies (Molecular Probes, Inc., Eugene, OR). Confocal analysis was carried out using a Zeiss laser-scanning confocal microscope and established methods involving processing of the same section for each detector (2 excitations corresponding to 546 and 488). Co-localization of the 2 proteins (EGFR and ERRFI-1) was indicated by the presence of yellow color as a result of overlapping red and green pixels.
RNA isolation, microarray platform, and statistics
All transcriptome data were generated from duplicates of the cell lines. Cells were plated and total RNA was isolated independently using Trizol reagent (Roche), followed by clean-up with RNeasy® Mini kit (Qiagen, Valencia, CA). RNA was used for the synthesis of biotin-labeled cRNA, which was prepared using the Illumina RNA amplification kit (Ambion, Inc.). Briefly, 500 ng of total RNA from each specimen was converted to cDNA then to cRNA by in vitro transcription. After purification, 1.5 μg of cRNA was fragmented and hybridized to Illumina human-6 v 2 (Illumina, Inc., San Diego, CA) chip. After being washed, the slides were scanned with Bead Station 500X (Illumina, Inc.), and the signal intensities were quantified with Bead Studio (Illumina, Inc.). Quantile normalization was used to normalize the data.
Statistics
Each experiment was performed at least twice and at least one duplicate. MiRNA data normalization was performed by VSN. All calculations including statistical analysis were performed by one-way or multi-way ANOVA. miRNA target prediction and associated mRNA pathway analysis were performed using Ingenuity Pathway Analysis and TargetScan (www.targetscan.org).
Results
miRNAs of the miR-200 family are expressed in bladder cancer cell lines that display an “epithelial” phenotype
To identify miRNAs that might be important for regulating E-cadherin expression, we compared global miRNA expression patterns in 2 bladder cell lines: an EGFR-sensitive, E-cadherin-positive cell line (UMUC5) and an EGFR-resistant, E-cadherin negative, and vimentin-positive cell line (KU7). We used miRNA array analysis that contained a dataset composed of all known miRNAs biologically demonstrated or predicted through bioinformatics (Assuragen, MiRInform, Dec 2007). Triplicate samples of untreated and cetuximab (C225)-treated UMUC5 and KU7 cells were analyzed using this platform. A diagram demonstrating the total number or miRNA species expressed by the 2 cells without treatment is shown in Fig. 1A (left panel). All miRNAs were then ranked according to the differences in expression between the untreated epithelial and mesenchymal cells with a cut-off of a two-fold difference between the 3 replicate samples of the cells (Fig. 1A, right panel, red dots). We then performed a cluster analysis in which we grouped the known (validated) and the predicted miRNAs by their level of differential expression. One example of such a cluster is shown in Fig. 1B. Six miRNAs were selectively expressed at very high levels in the epithelial UMUC5 cells compared to mesenchymal KU7 cells. These miRNAs were miR-200a/141, miR-200b/c/429, and miR-205, all of which are part of the miR-200 family.

A) Volcano plots showing all miRNAs detected in the 2 cell lines (left panel) or only the miRNA with significantly different expression (right panel, red dots) as opposed to non significant levels of differential exression (right panel, blue dots). Bayesian log odds of differential expression are plotted against log2 (expression in UMUC5 divided by expression in KU7 cells). B) Heatmap of differentially expressed miRNA clustered by cell type (left, low-expressed miRNAs in KU7 cells; right, high expression of the same miRNAs in UMUC5 cells). C-G) Quantification of miRNAs in a panel of 9 bladder cancer cell lines as measured by TaqMan real-time RT-PCR. Data are means of triplicate RT-PCR assays.
We then evaluated the expression of the miR-200 family of miRNAs in a panel of 9 bladder cancer cells that display varying sensitivities to EGFR inhibitor-induced growth arrest. The panel contained 4 epithelial cell lines (UMUC6, UMUC9, UMUC16, UMUC5 and UMUC6), 5 mesenchymal cell lines (UMUC2, T24, UMUC3, UMUC13, and KU7), and 1 cell line that contained markers of both phenotypes (253J B-V). Importantly, the latter cell line was derived from 253J cells through 5 cycles of in vivo selection as previously described (26) with the goal of producing a more invasive phenotype. The resultant 253J B-V cells were highly invasive in vitro and in vivo and some of the epithelial characteristics of the mother cell were lost (12). We observed increased expression of miR-200b/c, miR-205, miR-429, and miR-141 in all “epithelial” cells (E-cadherin positive), including the UMUC5 cells, whereas expression levels were very low in the “mesenchymal” cells, including KU7 cells (Fig. 1C-G). The 253J BV cells, which express low levels of E-cadherin and some vimentin (12) and are considered to be part of the EGFR-sensitive group, displayed co-expression of 3 of the miR-200 family members (Fig. 1D-G), suggesting that miR-200 family members are important regulators of EMT in bladder cancer cells. miR-141 was co-expressed with miR-205 in the UMUC5, UMUC6, UMUC9, and UMUC16 cells; however, the epithelial UMUC9 cell line lacked miR-205, whereas 253J BV cells lacked miR-141 (Fig, 1 E and G). A comparison of miRNA expression patterns in UMUC5 and KU7 cells incubated with or without the EGFR antagonist C225 revealed that miR-200c was the only miRNA species induced substantially in the EGFR-sensitive UMUC5 cells, whereas no miRNA species, known or predicted was modulated substantially by treatment with cetuximab (C225) in the EGFR-resistant KU7 cells (data not shown). These observations, corroborated by our personal observation that C225 treatment induced upregulation of E-cadherin in UMUC5 cells, indicated that miR-200 is the best candidate to use in a study of the relationship between the EMT and EGFR signaling.
Because miR-200c, miR-200b, and miR-429 target the same sequences in their target mRNAs and because miR-200c seemed to be associated with the epithelial phenotype and with responsiveness to the EGFR inhibition by C225, we next decided to use a luciferase-based reporter system to measure the functionality of miR-200 family in our panel of “epithelial” bladder cancer cell lines. These constructs had the “seed” sequence of miR-200c inserted downstream of the luciferase gene of the pGL3 vector. As an example, the UMUC5 epithelial cell line showed low baseline luciferase activity as opposed to the mesenchymal cell lines in which the baseline luciferase activity was very high although further suppressed by the transient expression of a miR-200c precursor (Fig. 2, A), and not by the unrelated miR-26b, demonstrating that miR-200b/c/429 expressed by E-cadherin-positive cells are also functional. Further real-time RT-PCR analysis in the same panel of bladder cancer cell lines revealed that the expression of miR-200 family members was inversely correlated with the expression of ZEB1 and ZEB2 (Fig. 2, B and C, respectively). The expression of E-cadherin suppressors ZEB1 and ZEB2 was inversely correlated with the E-cadherin expression (Fig. 2D), suggesting that the epithelial phenotype of bladder cancer cells may also be regulated by miR-200 family members as it has been recently described in breast cancer cells (16), NCI-60 cell lines (19), and colon cancer cells (17) or in amurine model of EMT (18). We looked for other classical cadherins that are involved in homotypic interactions and found that P-cadherin was also associated with the expression of miRNAs of the miR-200 family (Fig. 2, E). However, P-cadherin did not correlate with EGFR sensitivity because it was not expressed by 253J-BV (Fig. 2, D compared to E).
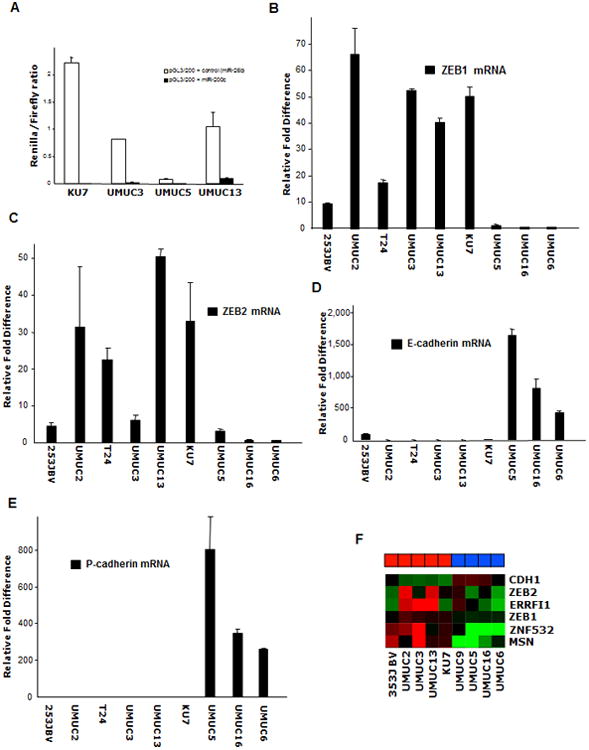
A) The Renilla reporter plasmid harboring a miR-200c site with miR-200c precursor or the unrelated miR-26b (control) precursor were transiently co-transfected into KU7, UMUC3, UMUC5, and UMUC13 cells along with a firefly luciferase reporter (pGL3 control) for normalization. The data are mean ± standard error of mean of separate transfections (n=6), and are shown as the ratio of Renilla activity to firefly. B-E) Measurement by real-time RT-PCR of epithelial and mesenchymal markers: ZEB1, ZEB2, E-cadherin, and P-cadherin. The data are means of measurements from triplicate experiments. F) Heat-map of 6 differentially expressed putative targets of miR-200c in a panlel of 9 bladder cancer cell lines using the Illumina gene grofilling array platform.
ZEB1, ZEB2, and ERRFI-1 are inversely associated with miR-200c expression
We inquired the TargetScan and PictarVert databases for potential direct targets of miR-200b/c/429 that would associate to an EMT phenotype and targets involved in the EGFR pathway. Several targets were identified according to their ranking in terms of content of conserved miR-200 sites and their expression pattern in our panel of bladder cancer cell lines as a result of gene expression profiling of the 9 cell lines. This list included ZEB1, ZEB2, ERRFI-1, MSN (a modulator of the cytoskeleton), and an unknown Zn finger transcription factor (ZNF532). A heat map showing the relative differences between the cell lines with respect to these potential miR-200 direct targets is shown in Fig. 2F. Further scrutiny for known direct modulators of EGFR-related signaling eliminated all of the targets except ERRFI-1. Real-time RT-PCR analysis revealed that ERRFI-1 mRNA levels were very low in 3 of 5 epithelial cell lines and high in all 3 resistant cell lines (Fig. 3A), suggesting that it is a good candidate target of miR-200 and that its higher baseline expression may be a component of EGFR-resistance.

A) Measurement by real-time westernblot of ERRFI-1 and E-cadherin. B) Measurement by real-time RT-PCR of mRNA relative levels of TGF-α. The data are means of measurements from triplicate experiments. C) Cell proliferation measurement (DNA index) of 9 bladder cancer cell lines in the absence or presence of different cetuximab (C225) concentrations. Data are means of at least 3 triplicate experiments.
Analysis of the 3′UTR region of ERRFI-1 and ZEB1/ZEB2 revealed that although ZEB1 and ZEB2 displayed at least 5 potential binding sites for the miR-200 family members (miR200a/b/c/429/141 and 205), ERRFI-1 displayed 2 such conserved sites (http:TargetScan.org), with 1 additional miRNA conserved site for miR-148/152, which was not found in either ZEB1 or ZEB2 3′UTRs. We analyzed the expression pattern of the miR-148 in all 9 cell lines and found no correlation with an epithelial or a mesenchymal phenotype, which underscored its role as a primary ERRFI-1 regulator (data not shown).
Because an intriguing property of ERRFI-1 is its ability to bind more tightly to activated EGFR than to the unligated receptor, we next determined which EGFR ligands are expressed by our cell lines. Gene expression array analysis revealed that three of the E-cadherin-positive cell lines, 253JBV, UMUC16, and UMUC9 also expressed TGF-α; whereas none of the mesenchymal cell lines expressed this gene; (Fig. 3B). In addition, these “autocrine-positive” and E-cadherin positive cells displayed slightly higher levels of the ERRFI-1 protein (Fig. 3A), suggesting that the “feed-back” mechanism of ligand-induced ERRFI-1 up-regulation is present in these cells, as previously described (27). Importantly, UMUC5, UMUC5 which are E-cadherin positives but do not express the TGF-α have almost undetectable levels of ERRFI-1 protein (Fig. 3A and B). An evaluation of cell growth inhibition by the cetuximab (C225) treatment and expression of ERRFI-1 and TGF-α revealed that KU7, UMUC2, UMUC13, and UMUC3 (high expressers of ERRFI-1) and E-cadherin-negative, were resistant for up to 1 μM cetuximab (C225) concentrations; whereas UMUC9, 253J BV, and UMUC16 (moderate ERRFI-1 expressers and high TGF-α expressers) or UMUC5 and UMUC6 (no detectable expression of ERRFI-1 protein or TFG-α mRNA) cell proliferation was inhibited up to 40% by 1 μM cetuximab (C225) (Fig. 3C). Importantly, these assays were done in EGF-unstimulated or “baseline” growth conditions, in the presence of Dulbecco's MEM supplemented with 2% fetal calf serum and reflect the “lower inhibitory efficiencies” for the ligand antagonist cetuximab (C225).
miR-200c expression induced a mesenchymal to epithelial transition (MET) phenotype
To more directly define the role of miR-200c in maintaining the epithelial phenotype and EGFR dependency in bladder cancer cells, we stably transduced the mesenchymal UMUC3 cells with a retroviral vector containing the miR-200c precursor as shown previously (16). Stable expression of miR-200c in fibroblast-like mesenchymal UMUC3 cells resulted in morphological changes suggestive of epithelial differentiation in that the cells grew in a more compact fashion and displayed cobblestone-like cell morphologies (Fig. 4A). These changes were associated with a reduced invasive phenotype as measured by in vitro wound-healing assays, where the miR-200c--expressing clones showed a significantly reduced wound-closing efficiency at 48 hours (Fig. 4, B). The high miR-200c levels expressed by the UMUC3 clones (Fig. 4C) were associated with the appearance of E-cadherin mRNA transcripts and reduced mRNA transcripts for ZEB1 and ZEB2 (Fig. 4D). These differences were also true at the protein levels, as shown for ZEB1 and ZEB2 using immunofluorescence microscopy and confocal analysis (Fig. 4E) and for E-cadherin by western blot analysis (Fig. 4F). These changes were also associated with a reduced ERRFI-1 protein expression (Fig. 4F). Based on morphological criteria as well as biochemical and biological behavior, we concluded that the UMUC3/200c clones demonstrated a MET phenotype and reduced ERRFI-1 expression.

A) Phase contrast microscopy of UMUC3 cells transduced with a miR-200c containing retrovirus (UMUC3/200c) or with an empty, control, retroviral construct (UMUC3-E). Scale bars represent 100 μm. B) Measurement of in vitro cell migration by “wound healing” assay. Representative pictures for same single spot are shown. The experiment was performed twice in triplicate experiments. Scale bars represent 400 μm. C) Quantification of miR-200c in empty virus transduced (UMUC3-E) and miR-200c-transduced (UMUC3/200c) cells. The levels of miR-200c in UMUC5 cells are plotted for comparison. D) Measurement by real-time RT-PCR of the epithelial and mesenchymal markers E-cadherin, ZEB1 and ZEB2 in empty virus-transduced cells and miR-200c-transduced UMUC3 cells. E) Confocal microscopy analysis of the UMUC3 series co-stained for ZEB1 or ZEB2 (red pixels) and nuclear DNA (green pixels). Note downregulation of nuclear ZEB1 and ZEB2 in UMUC3 clones expressing the miR-200c. F) Measurement of ERRFI-1 and E-cadherin protein levels by western blot. Actin reprobing served as internal (loading) control. Lower panel, OD relative values expressed as ratios between actin (internal control) and ERRFI-1. Note up-regulation of E-cadherin and down-regulation of ERRFI-1 protein levels upon miR-200c expression.
miR-200c expression reversed the EGFR resistant phenotype
Expression of miR-200c in UMUC3 cells constantly but modestly reduced the mRNA transcripts of ERRFI-1 (data not shown), whereas the ERRFI-1 protein levels were significantly reduced, suggesting an additional post-translational mechanism involved in ERRFI-1 downregulation in MET clones (Fig. 4F). It is well documented that the EGFR is subject to ligand-induced downregulation mediated by receptor internalization and subsequent degradation by lysosome and the ubiquitin/proteasome system (27-29). Thus, it has recently been suggested that ERRFI-1 is able to bind EGFR molecules, including the trafficking EGFR molecules and that ERRFI-1 becomes expressed at a time in which cells contain a sizable population of activated EGFRs that are being routed through the endosomal compartment. The blockade of kinase activity imposed by ERRFI-1 to trafficking EGFR molecules is expected to shift the kinase/phosphatase equilibrium in favor of phosphatases, therefore leading to a decline of phosphorylated EGFR levels and concomitant loss of signaling competence (28).
To interrogate the location of these EGFR/ERRFI-1 complexes we performed immunofluorescence staining and confocal microscopy studies of UMUC3 cells co-stained with antibodies specific for EGFR (Fig. 5A, green pixels) and ERRFI-1 (Fig. 5A, red pixels). The results revealed significant co-localized signals (yellow pixels) inside the cytoplasm, corresponding to high levels of EGFR trafficking, as well as on the cell membrane edges, such as ruffles. To our surprise, the distribution of EGFR molecules in the UMUC3/200c clones was very different, with most of the EGFR localized on the cell membrane, suggesting a higher rate of the surface-recycled EGFR. ERRFI-1 expression (red pixels) was also lower in UMUC3/200c cells compared to UMUC3 controls (Fig. 5 A, middle and right panels, compared to left panel) confirming our immunoblot analysis.

A) Confocal microscopy analysis of the UMUC3 series costained for ERRFI-1 (red pixels) and EGFR (green pixels) and a DNA dye, showing the nucleus (blue pixels). Note the yellow pixels (left panel) as a result of red and green pixels co-localization. B) Immunoblot of autophosphorylated EGFR, total EGFR, and total ERRFI-1 in cells transduced with miR-200c from the experiment above. In this experiment, actin served as the internal control. C) Upper panels, Immunoblot of phosphorylated MAPKinase, total MAPKinase and total EGFR of the UMUC3 series. Cells grown in 2% serum-supplemented MEM were left untreated or were treated with increased concentrations of cetuximab (C225) for 3 hours. Lower panel, OD relative values expressed as ratios between EGFR (internal control) and pMAPKinase. D) Cell proliferation measurement of the UMUC3 series using radioactive thymidine incorporation. Each experiment was done in at least two different triplicates.
EGFR downstream mitogenic signaling is affected by miR-200c expression
Feedback inhibition is an important mechanism of signaling pathway regulation and is crucial in modulating the intensity and duration of signals generated by receptors. Thus, sustained activation of the signal-regulated kinase group of MAPKinases is necessary for agonist-induced cell proliferation (30). EGF-stimulated ERK activation is often relatively transient, and in fibroblasts, EGF is a less effective mitogen than PDGF. A whole genome expression analysis on two of the UMUC3/200c clones revealed that they express moderate levels of mRNA/TGF-α, which may be responsible for autocrine loop–driven baseline EGFR activation, although the overall levels of ERRFI-1 prtotein are lower than the empty-vector-transfected UMUC3 cells, as previously described. Given that TGF-α uncouples internalized EGFR molecules from degradation by driving their rapid recycling to the cell surface (31, 32), ERRFI-1 may represent the major, if not the only, means of restraining the recursive positive feedback loop generated by autocrine stimulation of the EGFR via TGF-α. Thus, a reduction of the ERRFI-1 baseline levels in the stable UMUC3/200c clones would result in higher baseline EGFR activation. Western blot analysis demonstrated that UMUC3/200c clones have indeed higher baseline EGFR autophosphorylation associated with reduced levels of ERRFI-1 as compared to UMUC3 controls (Fig. 5B). Furthermore, concentrations of cetuximab (C225) as low as 10 nM were sufficient to block the baseline MAPKinase activity in UMUC3/200c clones, whereas 1 μM of cetuximab (C225) had no effect on MAPKinase activity in UMUC3/E (Fig. 5C). To determine whether morphological and biochemical changes were relevant for the mitogenic response to anti-EGFR therapy, we treated UMUC3 controls and miR-200c clones with increased concentrations of C225 for 36 hours and measured their proliferation index (Fig. 5D). We found that expression of miR-200c in the resistant UMUC3 cells resulted in a significantly higher cell growth inhibition to anti-EGFR therapy using cetuximab (C225) (Fig. 5D) as well as other anti-EGFR agents, such as erlotinib/Tarceva (data not shown). Thus, cetuximab (C225) concentrations of up to 1 μM-produced cell-cycle inhibition ranging from 40% to 60% translated into similar or even higher efficiency in cell growth inhibition than the rest of epithelial bladder cancer cells tested in baseline conditions. The importance of ERRFI-1 in modulating the cell-cycle response to EGFR-targeted therapy was further demonstrated by specific ERRFI-1 silencing in UMUC3 and in T24 cells, another E-cadherin-negative urothelial cell (Fig. 6A) where a significant reduction of ERRFI-1 levels was associated with a 30% cell-cycle inhibition of cells treated with 10 nM cetuximab (Fig. 6B). In addition, transfection of the full-length myc-tagged ERRFI-1 into sensitive, E-cadherin-positive UMUC5 cells resulted in the loss of EGFR sensitivity to relevant EGFR inhibitor concentrations (Fig. 6C). As we have previously mentioned, and according to the computer-based prediction programs, the 3′UTR of the ERRFI-1 gene contains two putative miR-200 binding sites (Fig. 6D, red rectangles). To demonstrate that ERRFI-1 is indeed a direct target of miR-200 family members, we constructed four constructs, encompassing the 650bp region of the 3′UTR shown in Fig. 6D. We also modified the two binding sites one by one as single mutants or both sites as double mutants. We engineered a two-base-pair site-directed mutagenesis which allowed us to identify binding differences of miR-200b/c/429 translated to modulation of the luciferase reporter's activity (Fig. 6E). Because cotransfection with either miR-200c (Fig. 6E), miR-200b or miR-429 (data not shown) and wild-type 3′UTR/ERRFI-1 significantly reduced the luciferase activity in UMUC3 and T24 and because the double mutant constructs were able to rescue this activity, we concluded that ERRFI-1 is a direct target of the miR-200 in these cells (Fig. 6E). Because we used M1 and M2 point mutations and not deletions of the miRNA binding sites and because the sequence complementarities between the miR-200 and the M2 site is expanding beyond the “seed” region we cannot conclude that M2 site is less important than the M1 site, as it may also be the result of a residual binding despite the point mutations that we introduced at this site. To finally demonstrate that expression of miR-200 into T24 results in modulation of cell proliferation response using EGFR inhibitors, we transfected a lentiviral construct with or without the miR-200b sequence into T24, an EGFR-resistant cells (Fig. 6F). Treatment of these cells with various concentrations of the EGFR inhibitor Iressa resulted in an increase in cell's response to this inhibitor as measured by DNA replication (Fig. 6G). We have included in this experiment the highly-sensitive UMUC9 cells for comparison (Fig. 6G). In addition, miR-200b expression resulted into expression of E-cadherin, a 18 fold reduction of ERRFI-1/mRNA (data not shown), suggesting a similar phenotype to the UMUC3/200c clones above.

ERRFI-1 is a direct target of miR-200 and implicated in response to EGFR inhibitors. A). Western blot measurement of ERRFI-1 protein in UMUC3 and T24 cells after transfection with a non-targeting sh construct, a GAPDH or a ERRFI-1sh construct. Vinculin served as internal control. B) Cell proliferation measurement of the UMUC3 and T24 series using radioactive thymidine incorporation after stransfections described in the previous panel. Each experiment was done in at least two triplicates. C) Cell proliferation measurement of the UMUC5 series using radioactive thymidine incorporation after ERRFI-1 transfection. The level of ERRFI-1 protein is shown in the right panel. Vinculin served as internal control. D) Schematic representation of the 3′UTR region of ERRFI-1 displaying the miRNA potential binding sites as predicted by TargetScan. Red boxes represent miR-200b/c/429 potential binding sites. E) measurement of miR-200 repressive activity on wild-type and mutant 3′UTR/ERRFFI-1 reporters. F) Real-time RT-PCR of T24 series after miR-200b transduction using a lentiviral system. PEV, empty vector-transduced cells served as negative controls. G) Cell proliferation measurement of the T24 series using radioactive thymidine incorporation after miR-200b lentiviral transduction. Each experiment was done in at least two different triplicates. UMUC9 served as positive controls being highly-sensitive responders to EGFR blockers, in this case, Iressa.
Discussion
We found that expression of miRNAs members of the 200-mRNA family, in particular miR-200c and b, are sufficient to reverse the biology of a mesenchymal cell line, not only by its morphological appearance but also by its EGFR signaling. Patterns of miRNA expression are thought to coordinately regulate a specific biological function, not just a gene involved in a specific pathway, so we examined whether the miRNA expression patterns found in bladder cancer cell lines might reflect potential markers of an epithelial phenotype in tight connection with EGFR signaling, which might actually represent a signature of an EGFR-sensitive phenotype in bladder cancer. ZEB1, ZEB2 and ERRFI-1 are three examples of miR-200 direct targets, their downregulation by miR-200 being associated with an EGFR sensitive phenotype. What would be the ERRFI-1 specific role to this change? We have recently reported that an “uninterrupted flow” from an active EGFR and its downstream PI-3-Kinase and MAPKinase signaling pathways towards GSK-3β inactivation seem to play central roles in defining the EGFR sensitivity or the EGFR-dependency in bladder cancer cells (25). Thus, we conclude that a higher baseline expression of ERRFI-1 (as found in many mesenchymal cell lines) that is not the resultant of an autocrine loop, might impair the EGFR-MAPKinase signaling flow and switch the dependence for cell proliferation from EGFR-driven to another signaling or growth factor receptor signaling pathway.
ERRFI-1, also known as RALT, Gene 33, or Mig6, is a widely expressed adaptor protein that can be transcriptionally induced by a variety of external stimuli, including various growth factors, cytokines, and stress factors (33-36). Because several studies have reported that loss of ERRFI-1 signaling mediated by RNAi enhances the mitogenic potency of suboptimal doses of EGF (37, 38) with paralleled ERK/MAPKinase and PI3-Kinase/AKT activation, accumulation of cyclins, crucial in the G1→S transition, we reasoned that low baseline expression of ERRFI-1 might define a cell line's “addiction” for ligand/EGFR signaling loop, promoting cell proliferation and finally defining an “EGFR-dependent” phenotype. An intriguing property of ERRFI-1 is its ability to bind more tightly to activated EGFR than to the unliganded receptor (38, 39) by using a double-headed mechanism for inhibiting EGFR to the cyclin-like face of the kinase domain of EGFR, a region that is conserved between all four EGFR family members (40). Based on these observations, we reasoned that the higher the ratio between TGF-α expression and ERRFI-1 expression in a cell line, the higher would be the chances that that cell line would be dependent on EGFR-driven mitogenic signals and would be mostly sensitive to anti-EGFR therapy in vitro. Two of the cell lines expressing miR-200 displayed very low levels of ERRFI-1 protein whereas other three lines expressing miR-200 and TGF-α displayed slightly higher levels of ERRFI-1 protein, although all five were defined as EGFR sensitive. These cell lines were indeed some of the most sensitive to EGFR inhibitors, including cetuximab (C225) treatment (12). Of particular interest are 2 recent studies that demonstrate that secretion of EGFR agonists facilitate disease control in patients with metastatic colorectal cancer treated with cetuximab (C225) as well as in experimental metastatic lung model in mice (41, 42). Based on these reports and the present work, we conclude that high expression of miR-200 would facilitate the best EGFR functionality, hence most efficiently targeted by anti-EGFR therapy and that additional TGF-α expression through autocrine or paracrine secretion would probably serve to fine tune the EGFR dependency in vivo.
Particularly intriguing was our previous finding that bladder cancer cells displaying an EMT are also less responsive to anti-EGFR targeted therapy (12), suggesting that alterations in the EGFR signaling are associated with such a phenotype. We reasoned that the association between E-cadherin expression and high anti-EGFR sensitivity could be the result of 2 situations: (1) EMT-regulating miRNAs might co-regulate additional targets involved in EGFR signaling or (2) there are additional miRNAs that primarily target components of the EGFR pathway and showed similar expression patterns to those that regulate an EMT phenotype. In that study, we demonstrated the importance of E-cadherin expression in an epithelial background where the silencing of E-cadherin substantially modulated EGFR response to therapy in 253J BV cell lines from a sensitive to a resistant phenotype (12). In the present study, we were able to completely reverse the EGFR responsiveness to therapy using a mesenchymal background (e.g., UMUC3 and T24) by the introduction of miR-200 and modulation of its direct or indirect target genes, including E-cadherin and ERRFI-1. The 2 studies support each other by placing emphasis on the importance of an epithelial phenotype and its relation to EGFR responsiveness. Although the exact mechanistic explanation of how E-cadherin expression affects EGFR responsiveness in an epithelial background is still hypothetical at this point, based on previous reports and our findings, we can assume a scenario in which EGFR spatial distribution may play a role in the EGFR responsiveness. Thus, in an epithelial background with low expression of ERRFI-1, EGFR is rapidly recycled to the cell membrane and E-cadherin may serve as a scaffold for EGFR, which would result in increased stability and exposure on the cell surface for optimal accessibility of EGFR-targeted therapy. This was also illustrated by findings in our current study that in MET UMUC3/200 clones in which E-cadherin expression was induced by miR-200c expression and ERRFI-1 was down regulated, the EGFR was mostly exposed on the cell membrane and the responsiveness to its inhibitors was optimal. In this case, either down regulation of ERRFI-1 or E-cadherin might affect the EGFR spatial distribution that would impact its signaling. It would be important to know whether E-cadherin is required for EGFR to be accessible by ERRFI-1 or whether E-cadherin downregulation triggers feed-forward upregulation of ERRFI-1 (43). In contrast, in a mesenchymal background, there is higher baseline expression of ERRFI-1, which binds EGFR to its kinase domain, maintaining the EGFR/agonist loop in a rather inhibited state. Again, this finding was supported by our confocal microscopy analysis, which showed that the cell-surface pool and most of the internalized, trafficking EGFR were bound to ERRFI-1, and by western blot, which showed that baseline EGFR autophosphorylation was lower in the mesenchymal background as compared to the epithelial background of UMUC3 cells and the expression of miR-200c facilitates the EGFR downstream signaling pathways towards MAPKinase modulation. Importantly, the “switch” in EGFR responsiveness was demonstrated for two different mesenchymal cell lines and using two members of the miR-200 family.
Conclusions
Recent therapeutic advances have identify targeted therapy, such as growth factor inhibition, as being more effective in the context of a documented patient selection and using algorithms that take into account targeted expression and molecular pathways validation. Growth factor receptors, such EGFR, fibroblast growth factor receptor, and PDGFR, are differentially expressed by bladder cancer cell lines or tumors, making the use of inhibitors to these molecules an attractive therapeutic strategy. By exploring the mechanistic relationship between miRNAs-driven gene re-programming and the implications to the therapeutic response, our studies open a new avenue to the understanding the mechanism of acquired resistance to the EGFR-targeted therapy in the context of bladder cancer ontogeny and unravel additional markers to evaluate for a future informed therapeutic decision.
Acknowledgments
We thank GJ Brock for his generosity in providing the retrovirus constructs for miR-200c and J. Kuriyan for providing the full-length myc-tagged ERRFI-1 construct. This work was supported by the NCI-Bladder SPORE and the MD Anderson CORE grant to C.D and by the Alliance for Nanohealth seed grants to L.A. Dr. Calin is supported in part, as a Fellow of The University of Texas M. D. Anderson Research Trust, and University of Texas System Regents Research Scholar and as a Ladjevardian Regents Research Scholar Fund.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/1078-0432.ccr-08-2245
Read article for free, from open access legal sources, via Unpaywall:
https://clincancerres.aacrjournals.org/content/clincanres/15/16/5060.full.pdf
Free to read at clincancerres.aacrjournals.org
http://clincancerres.aacrjournals.org/cgi/content/abstract/15/16/5060
Free after 12 months at clincancerres.aacrjournals.org
http://clincancerres.aacrjournals.org/cgi/reprint/15/16/5060.pdf
Free after 12 months at clincancerres.aacrjournals.org
http://clincancerres.aacrjournals.org/cgi/content/full/15/16/5060
Citations & impact
Impact metrics
Article citations
A bibliometric analysis of bladder cancer and microRNA research: Trends and advances from 2008 to 2022.
Medicine (Baltimore), 103(43):e40289, 01 Oct 2024
Cited by: 0 articles | PMID: 39470484 | PMCID: PMC11521070
miR-200c targeting GLI3 inhibits cell proliferation and promotes apoptosis in non-small cell lung cancer cells.
Medicine (Baltimore), 103(38):e39658, 01 Sep 2024
Cited by: 0 articles | PMID: 39312343 | PMCID: PMC11419521
Urinary miRNAs: Technical Updates.
Microrna, 13(2):110-123, 01 Jan 2024
Cited by: 0 articles | PMID: 38778602
Review
A New Approach to Melanoma Treatment: microRNAs.
Curr Top Med Chem, 24(16):1362-1376, 01 Jan 2024
Cited by: 0 articles | PMID: 38676490
Review
Investigating genomic, proteomic, and post-transcriptional regulation profiles in colorectal cancer: a comparative study between primary tumors and associated metastases.
Cancer Cell Int, 23(1):192, 05 Sep 2023
Cited by: 0 articles | PMID: 37670299 | PMCID: PMC10478430
Go to all (299) article citations
Other citations
Wikipedia
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
miR-200c inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3.
J Transl Med, 12:305, 04 Nov 2014
Cited by: 77 articles | PMID: 25367080 | PMCID: PMC4226852
MiR-200c-3p suppression is associated with development of acquired resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors in EGFR mutant non-small cell lung cancer via a mediating epithelial-to-mesenchymal transition (EMT) process.
Cancer Biomark, 28(3):351-363, 01 Jan 2020
Cited by: 19 articles | PMID: 32417760
The TGFβ-miR-499a-SHKBP1 pathway induces resistance to EGFR inhibitors in osteosarcoma cancer stem cell-like cells.
J Exp Clin Cancer Res, 38(1):226, 28 May 2019
Cited by: 28 articles | PMID: 31138318 | PMCID: PMC6540516
Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer.
Cancer Metastasis Rev, 28(3-4):335-344, 01 Dec 2009
Cited by: 243 articles | PMID: 20012924 | PMCID: PMC5915353
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P30 CA006973
Grant ID: P30 CA016672






