Abstract
Free full text

Multicenter Evaluation of the Etest Gradient Diffusion Method for Ceftolozane-Tazobactam Susceptibility Testing of Enterobacteriaceae and Pseudomonas aeruginosa
Associated Data
ABSTRACT
Ceftolozane-tazobactam (C/T) is a novel beta-lactam–beta-lactamase inhibitor combination antibiotic approved by the U.S. Food and Drug Administration in 2014 for the treatment of complicated intra-abdominal infections (in combination with metronidazole) and complicated urinary tract infections. In this study, we evaluated the performance of the C/T Etest, a gradient diffusion method. C/T Etest was compared to broth microdilution (BMD) for 51 Enterobacteriaceae challenge isolates and 39 Pseudomonas aeruginosa challenge isolates at three clinical sites. Essential agreement (EA) between the methods ranged from 47 to 49/51 (92.2 to 96.1%) for the Enterobacteriaceae, and categorical agreement (CA) ranged from 49 to 51/51 (96.1 to 100.0%). EA and CA for P. aeruginosa were 100% at all sites. The C/T Etest was also compared to BMD for susceptibility testing on 966 clinical isolates (793 Enterobacteriaceae, including 167 Klebsiella pneumoniae and 159 Escherichia coli isolates, in addition to 173 P. aeruginosa isolates) collected at four clinical sites. EA between Etest and BMD was 96.9% for Enterobacteriaceae isolates and 98.8% for P. aeruginosa isolates. Within the Enterobacteriaceae, isolates from each species examined had >96% CA. For the clinical isolates, no very major errors were identified but two major errors were found (one for K. pneumoniae and one for Providencia rettgeri). By BMD, 47.0% of Enterobacteriaceae and 46.2% of P. aeruginosa challenge strains were nonsusceptible to C/T by CLSI breakpoint criteria; 8.2% of clinical Enterobacteriaceae isolates and 12.1% of clinical P. aeruginosa isolates were nonsusceptible to C/T by CLSI breakpoint criteria. In conclusion, Etest is accurate and reproducible for C/T susceptibility testing of Enterobacteriaceae and P. aeruginosa.
INTRODUCTION
Ceftolozane-tazobactam (C/T) is a new beta-lactam–beta-lactamase inhibitor combination agent that combines a novel oxyimino-cephalosporin (ceftolozane) with an established beta-lactamase inhibitor (tazobactam). C/T was approved by the U.S. Food and Drug Administration in 2014 for the treatment of complicated intra-abdominal infections (cIAI, in combination with metronidazole) and complicated urinary tract infections (cUTI) (1). In clinical trials, C/T demonstrated noninferiority to meropenem when used in combination with metronidazole for the treatment of cIAIs (2, 3). Additionally, C/T was noninferior to levofloxacin for the treatment of cUTIs, including pyelonephritis (4), which was mainly attributable to high rates of levofloxacin-resistant isolates among cUTI-causing bacteria (5, 6). In vitro, C/T inhibits the growth of a high percentage of bacterial isolates from the medically important Enterobacteriaceae family and Pseudomonas aeruginosa, outperforming several current first-line antibiotics for the treatment of multidrug-resistant (MDR) organisms (3, 7,–14). Nevertheless, resistance to C/T has been observed, emphasizing the need for C/T susceptibility testing in clinical microbiology laboratories (15,–17). Here we report a multicenter evaluation of a C/T Etest (bioMérieux, Durham, NC) which was recently cleared by the U.S. FDA for in vitro diagnostic use (18).
MATERIALS AND METHODS
Ethics statement.
Prior to study initiation, each study site acquired approval or waiver from their respective institutional review board.
Quality control.
The following isolates were used for quality control (QC) of the C/T Etest and broth microdilution (BMD) each day of testing: Escherichia coli ATCC 25922, Escherichia coli ATCC 35218, Pseudomonas aeruginosa ATCC 27853, Klebsiella pneumoniae ATCC 700603, and Staphylococcus aureus ATCC 29213 (BMD only), using Clinical and Laboratory Standards Institute (CLSI) QC ranges for these strains. Quality control results were within acceptable limits for 99% of quality control tests performed on each quality control organism, and quality control was verified to be within normal limits on every day that comparative analysis was performed (i.e., challenge and clinical studies). Quality control testing was performed a minimum of 20 times at each site.
Setting.
Testing was performed at the Clinical Microbiology Institute (CMI; Portland, OR), the University of California-Los Angeles (UCLA; Los Angeles, CA) Health System, the University of Indiana (WU; Indianapolis, IN), Washington University in St. Louis School of Medicine (WU; St. Louis, MO), and bioMérieux St. Louis Clinical Affairs (St. Louis, MO).
Clinical isolates and C/T susceptibility testing.
A combination of fresh and previously frozen clinical isolates was evaluated in this study: Enterobacteriaceae (422/793 fresh; 371/793 frozen) and P. aeruginosa (109/173 fresh; 64/173 frozen). Isolates were recovered and identified from patient specimens using standard-of-care culture media and procedures at each study site. For C/T susceptibility testing using the Etest, Mueller-Hinton agar II plates (Remel Inc., San Diego, CA) were inoculated with a sterile cotton swab moistened with a 0.5 McFarland suspension and the Etest strip was placed on the plate with an applicator or forceps. Plates were incubated in ambient air at 35 ± 2°C and read at 16 to 20 h postinoculation. For BMD, 0.01 ml of a 0.5 McFarland solution that had been diluted 1:300 was inoculated into 95 wells of a 96-well plate using a 96-well-plate inoculator. BMD plates containing 0.1 ml of diluted C/T solutions were prepared by bioMérieux according to processes described in CLSI documents M100 and M7 (19, 20) using Becton Dickinson Mueller-Hinton medium and stored at −70°C until use. All BMD plates were read following 16 to 20 h of incubation in ambient air at 35 ± 2°C. Directions for reading the C/T Etest (i.e., the Etest reading guide and the supplemental package insert) were provided to sites. These materials included instruction on interpreting hazes (which were included as growth with the exception of swarming by Proteus species) and colonies within the zone (which were counted as true growth). Magnifying lenses were also supplied to each site, but magnification was not required for reading of Etest results. An example of Etest results for the species used in this study can be found in Fig. 1.
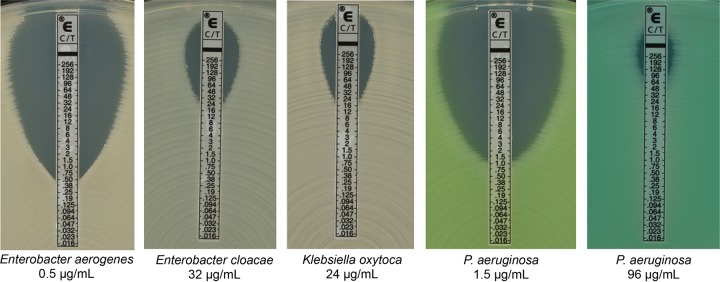
Interpretation of Etest results. C/T Etest strips were placed onto Mueller-Hinton agar plates that had been inoculated using a sterile cotton swab moistened with a 0.5 McFarland suspension. Plates were then read after incubation in ambient air at 35 ± 2°C for 16 to 20 h. The MIC result was read where the zone of inhibition ellipse intersected the Etest strip; if the ellipse intersected between two MIC values, the higher of the two values was reported. Hazy growth and colonies within the ellipse were counted as true growth.
Reproducibility and challenge testing.
Reproducibility testing was performed using a set of 25 on-scale strains (i.e., strains that had expected MICs falling within the measurable range of the Etest device); all strains were tested at three clinical sites using the Etest exclusively. An inoculum density check was performed on all isolates. Challenge testing was performed at three clinical sites using 90 frozen stock organisms provided by bioMérieux. All challenge isolates were tested using Etest and BMD. To derive the MIC for BMD in the challenge study, this trial used a voted reference result as the basis for comparison. The voted reference was derived by comparing the BMD results from all three sites for each isolate. The voted reference result was either a consensus result, the best two of three, or a modal value. In situations where all three reference results were the same, the consensus result was used.
Interpretation of results and data analysis.
Results from all isolates tested for C/T susceptibility using the Etest and BMD methods were included in the analysis. Essential agreement (EA) was defined as agreement between the two methods ± 1 doubling dilution and was calculated by the Biomath team at bioMérieux. Etest results were rounded up to the nearest dilution that was performed for BMD for this calculation. Categorical agreement (CA) between the Etest and BMD, as well as very major, major, and minor error evaluation, was determined using CLSI breakpoint criteria unless otherwise specified.
RESULTS
Reproducibility of C/T Etest.
We used isolates of Enterobacter cloacae (n = 1), Escherichia coli (n = 7), Klebsiella oxytoca (n = 3), Klebsiella pneumoniae (n = 5), and Pseudomonas aeruginosa (n = 9) to evaluate the reproducibility of the Etest at three sites: the Clinical Microbiology Institute (CMI; Portland, OR), the University of California-Los Angeles (UCLA; Los Angeles, CA) Health System, and Washington University in St. Louis School of Medicine (WU). The MICs of C/T required to inhibit growth of each isolate were compared across sites (Table 1), showing that 73/75 (97.3%) isolates tested fell within ±1 doubling dilution of the mode (or the median, if no mode existed).
TABLE 1
Reproducibility of C/T Etest
| Organism | MIC (μg/ml) | DDa from the median/mode | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site A | Site B | Site C | Mode/median | Off-scale | −2 | −1 | 0 | 1 | 2 | Off-scale | |
| E. cloacae | 16 | 16 | 32 | 16 | 2 | 1 | |||||
| E. coli | 1 | 1 | 2 | 1 | 2 | 1 | |||||
| 1 | 2 | 8 | 2b | 1 | 1 | 1 | |||||
| 0.5 | 0.5 | 1 | 0.5 | 2 | 1 | ||||||
| 0.5 | 0.5 | 1 | 0.5 | 2 | 1 | ||||||
| 0.125 | 0.125 | 0.125 | 0.125 | 3 | |||||||
| 2 | 2 | 2 | 2 | 3 | |||||||
| 0.125 | 0.125 | 0.125 | 0.125 | 3 | |||||||
| K. oxytoca | 4 | 2 | 4 | 4 | 1 | 2 | |||||
| 0.5 | 0.5 | 0.5 | 0.5 | 3 | |||||||
| 1 | 0.5 | 2 | 1b | 1 | 1 | 1 | |||||
| K. pneumoniae | 64 | 256 | 128 | 128b | 1 | 1 | 1 | ||||
| 1 | 2 | 1 | 1 | 2 | 1 | ||||||
| 32 | 256 | 64 | 64b | 1 | 1 | 1 | |||||
| 2 | 1 | 1 | 1 | 2 | 1 | ||||||
| 0.5 | 0.25 | 0.25 | 0.25 | 2 | 1 | ||||||
| P. aeruginosa | 128 | 128 | 256 | 128 | 2 | 1 | |||||
| 64 | 64 | 128 | 64 | 2 | 1 | ||||||
| 1 | 1 | 2 | 1 | 2 | 1 | ||||||
| 0.5 | 0.5 | 1 | 0.5 | 2 | 1 | ||||||
| 2 | 2 | 4 | 2 | 2 | 1 | ||||||
| 1 | 1 | 1 | 1 | 3 | |||||||
| 1 | 1 | 1 | 1 | 3 | |||||||
| 0.5 | 0.5 | 0.5 | 0.5 | 3 | |||||||
| 0.5 | 0.5 | 0.5 | 0.5 | 3 | |||||||
| Total | 0 | 0 | 5 | 54 | 14 | 2 | 0 | ||||
C/T Etest challenge study.
To examine the accuracy of the Etest, we compared MICs derived by C/T Etest to MICs derived by BMD for 51 Enterobacteriaceae and 39 P. aeruginosa strains at CMI, UCLA, and WU (see Table S1 in the supplemental material for details on each isolate). Among the Enterobacteriaceae isolates evaluated, 23/51 (45%) were resistant to C/T by BMD using the CLSI breakpoints (susceptible [S], ≤2/4 μg/ml; intermediate [I], 4/4 μg/ml; resistant [R], ≥8/4 μg/ml), while 27/51 (53%) were susceptible and 1/51 was intermediate (Table 2). Similarly, 18/39 (46%) P. aeruginosa strains evaluated were resistant and 21/39 (54%) were susceptible by BMD using CLSI breakpoint criteria (S, ≤4/4 μg/ml; I, 8/4 μg/ml; R, ≥16/4 μg/ml). Essential agreement (EA) between the C/T Etest and BMD methods ranged from 47/51 (92.2%) to 49/51 (96.1%) for the Enterobacteriaceae species; categorical agreement (CA) ranged from 49/51 (96.1) to 51/51 (100%), with 1 very major error, 1 major error, and 2 minor errors identified. EA and CA for P. aeruginosa were 100% at all sites.
TABLE 2
Challenge study of C/T Etesta
| Organism(s) | Site | No. of isolates | No. with indicated result by BMD | Performance, no. (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | EA | CA | VMEs | MEs | mEs | |||
| Enterobacteriaceae | A | 51 | 27 | 1 | 23 | 47 (92.2) | 51 (100) | 0 (0) | 0 (0) | 0 (0) |
| B | 51 | 27 | 1 | 23 | 49 (96.1) | 49 (96.1) | 0 (0) | 1 (3.7) | 1 (2.0) | |
| C | 51 | 27 | 1 | 23 | 48 (94.1) | 49 (96.1) | 1 (4.3) | 0 (0) | 1 (2.0) | |
| P. aeruginosa | A | 39 | 21 | 0 | 18 | 39 (100) | 39 (100) | 0 (0) | 0 (0) | 0 (0) |
| B | 39 | 21 | 0 | 18 | 39 (100) | 39 (100) | 0 (0) | 0 (0) | 0 (0) | |
| C | 39 | 21 | 0 | 18 | 39 (100) | 39 (100) | 0 (0) | 0 (0) | 0 (0) | |
Clinical performance of C/T Etest.
We examined the performance of the C/T Etest at five sites: CMI, UCLA, WU, Indiana University School of Medicine (Indianapolis, IN), and bioMérieux St. Louis Clinical Affairs (St. Louis, MO). A combined total of 966 clinical isolates were evaluated across the five study sites, including 793 strains of Enterobacteriaceae and 173 isolates of P. aeruginosa. The majority of Enterobacteriaceae (428/793 [54.0%]) were isolated from urine, while respiratory specimens were the most common source of P. aeruginosa isolates (52/173 [30.1%]) (see Table S2 for the specimen type from which each isolate was recovered and the corresponding MIC of each isolate). Per CLSI breakpoints for the Enterobacteriaceae, 56/793 (7.1%) of patient isolates were resistant to C/T by BMD, 728/793 (91.8%) were susceptible, and 9/793 (1.1%) had MICs that fell within the intermediate category (Table 3). The species with the highest percentage of resistant isolates were E. cloacae (8/53 [15.1%]) and K. oxytoca (7/58 [12.1%]). For P. aeruginosa, 17/173 (9.8%) of patient isolates were resistant by BMD using CLSI breakpoints, while 152/173 (87.9%) of isolates were susceptible and 4/173 (2.3%) were of intermediate susceptibility. EA between BMD and Etest was 768/793 (98.5%) for Enterobacteriaceae collectively (Fig. 2A), with the EA for each species within the Enterobacteriaceae ranging from 93.4% (K. pneumoniae) to 100%. CA for Enterobacteriaceae isolates collectively was 781/793 (98.5%), with the CA for each species within the Enterobacteriaceae ranging from 96.2% (E. cloacae) to 100%. For P. aeruginosa, EA and CA of the C/T Etest versus BMD were 171/173 (98.8%) and 172/173 (99.4%), respectively (Table 3; Fig. 2B). No very major errors were identified in our clinical study. However, two major errors were found: one K. pneumoniae isolate with an Etest MIC of 64 μg/ml and a BMD MIC of 0.5 μg/ml and one P. rettgeri isolate with an Etest MIC of 8 μg/ml and a BMD MIC of 2 μg/ml. Eleven minor errors were also identified: 10 from Enterobacteriaceae isolates and 1 from a P. aeruginosa isolate, with no obvious bias toward one pattern of minor error.
TABLE 3
Clinical performance of C/T Etest
| Organism(s) | No. of isolates | No. (%) matching CLSI breakpoint criteria | No. (%) matching EUCAST breakpoint criteria | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Result by BMD | Performance | Result by BMD | Performance | ||||||||||||
| S | I | R | EA | CA | VMEs | MEs | mEs | S | R | EA | CA | VMEs | MEs | ||
| Enterobacteriaceae (total) | 793 | 728 (91.8) | 9 (1.1) | 56 (7.1) | 768 (96.8) | 781 (98.5) | 0 (0) | 2 (0.3) | 10 (1.3) | 718 (90.5) | 75 (9.5) | 768 (96.8) | 785 (99.0) | 3 (4.0) | 5 (0.7) |
| Citrobacter koseri | 48 | 48 (100) | 0 (0) | 0 (0) | 48 (100) | 48 (100) | 0 (NDa) | 0 (0) | 0 (0) | 47 (97.9) | 1 (2.1) | 48 (100) | 48 (100) | 0 (0) | 0 (0) |
| Enterobacter cloacae | 53 | 44 (83.0) | 1 (1.9) | 8 (15.1) | 51 (96.2) | 51 (96.2) | 0 (0) | 0 (0) | 2 (3.8) | 44 (83.0) | 9 (17.0) | 51 (96.2) | 52 (98.1) | 0 (0) | 1 (2.3) |
| Escherichia coli | 159 | 146 (91.8) | 1 (0.6) | 12 (7.5) | 153 (96.2) | 159 (100) | 0 (0) | 0 (0) | 2 (1.3) | 146 (91.8) | 14 (8.8) | 153 (96.2) | 158 (99.4) | 0 (0) | 1 (0.7) |
| Klebsiella oxytoca | 58 | 52 (89.7) | 1 (1.7) | 7 (12.1) | 56 (96.6) | 58 (100) | 0 (0) | 0 (0) | 0 (0) | 50 (86.2) | 9 (15.5) | 56 (96.6) | 57 (98.3) | 0 (0) | 1 (2.0) |
| Klebsiella pneumoniae | 167 | 153 (91.6) | 1 (0.6) | 13 (7.8) | 156 (93.4) | 164 (98.2) | 0 (0) | 1 (0.7) | 2 (1.2) | 151 (90.4) | 16 (9.6) | 156 (93.4) | 165 (98.8) | 1 (6.3) | 1 (0.7) |
| Morganella morganii | 50 | 45 (90.0) | 0 (0) | 5 (10.0) | 50 (100) | 50 (100) | 0 (0) | 0 (0) | 0 (0) | 45 (90.0) | 5 (10.0) | 50 (100) | 49 (98.0) | 0 (0) | 1 (2.2) |
| Proteus mirabilis | 65 | 62 (95.4) | 1 (1.5) | 2 (3.1) | 64 (98.5) | 64 (98.5) | 0 (0) | 0 (0) | 1 (1.5) | 61 (93.8) | 4 (6.2) | 64 (98.5) | 64 (98.5) | 0 (0) | 0 (0) |
| Proteus vulgaris | 49 | 48 (98.0) | 1 (2.0) | 0 (0) | 48 (98.0) | 48 (98.0) | 0 (ND) | 0 (0) | 1 (2.0) | 3 (6.1) | 46 (93.9) | 48 (98.0) | 47 (95.9) | 2 (4.3) | 0 (0) |
| Providencia rettgeri | 40 | 39 (97.5) | 0 (0) | 1 (2.5) | 39 (97.5) | 39 (97.5) | 0 (0) | 1 (2.6) | 0 (0) | 38 (95.0) | 2 (5.0) | 39 (97.5) | 40 (100) | 0 (0) | 0 (0) |
| Providencia stuartii | 42 | 34 (81.0) | 3 (7.1) | 5 (11.9) | 40 (95.2) | 41 (97.6) | 0 (0) | 0 (0) | 1 (2.4) | 34 (81.0) | 8 (19.0) | 40 (95.2) | 42 (100) | 0 (0) | 0 (0) |
| Serratia liquefaciens | 10 | 10 (100) | 0 (0) | 0 (0) | 10 (100) | 10 (100) | 0 (ND) | 0 (0) | 0 (0) | 10 (100) | 0 (0) | 10 (100) | 10 (100) | 0 (ND) | 0 (0) |
| Serratia marcescens | 52 | 47 (90.4) | 0 (0) | 5 (9.6) | 52 (100) | 51 (98.1) | 0 (0) | 0 (0) | 1 (1.9) | 46 (88.5) | 6 (11.5) | 52 (100) | 52 (100) | 0 (0) | 0 (0) |
| Pseudomonas aeruginosa | 173 | 152 (87.9) | 4 (2.3) | 17 (9.8) | 171 (98.8) | 172 (99.4) | 0 (0) | 0 (0) | 1 (0.6) | 152 (87.9) | 21 (12.1) | 171 (98.8) | 173 (100) | 0 (0) | 0 (0) |
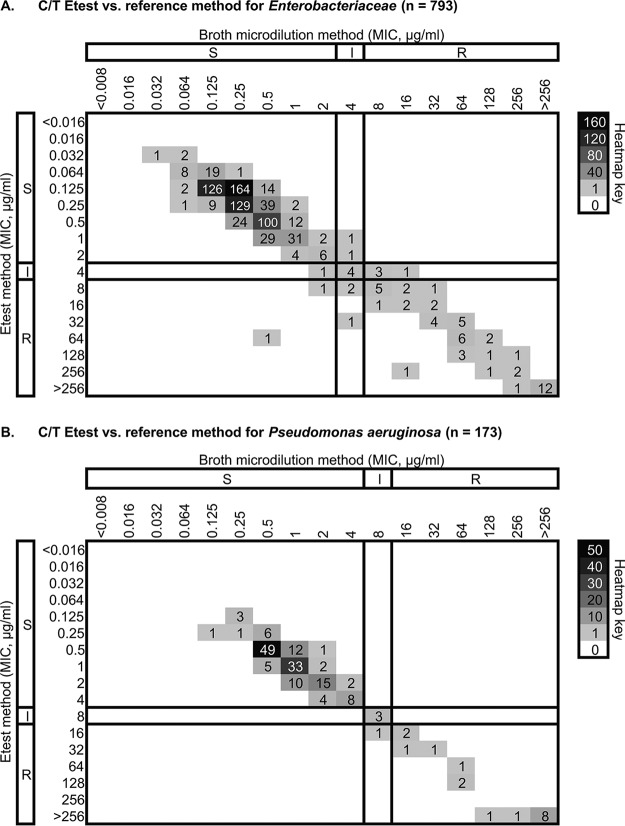
Correlation of C/T MICs from Etest and BMD. Clinical isolates identified as Enterobacteriaceae (n = 793) (A) or P. aeruginosa (B) (n = 173) were subjected to C/T susceptibility testing by Etest and BMD. The numbers of isolates with MICs corresponding to each test method are shown, with darker squares corresponding to MIC values displayed by a larger number of isolates. CLSI breakpoints are shown as dark solid lines, with their respective interpretation for each method shown to the left and top of the plot.
We also analyzed the categorical interpretation of our clinical data using European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints, which are more conservative than CLSI breakpoints for Enterobacteriaceae and do not include an intermediate category (for Enterobacteriaceae, S, ≤1/4 μg/ml, and R, >1/4 μg/ml; for P. aeruginosa, S, ≤4/4 μg/ml, and R, >4/4 μg/ml). Using EUCAST breakpoints, CA was higher for both Enterobacteriaceae (785/793 [99.0%]) and P. aeruginosa (173/173 [100%]) than CLSI breakpoints. However, more very major (n = 3) and major errors (n = 5) were found when using EUCAST criteria (Table 3).
DISCUSSION
C/T is a novel beta-lactam–beta-lactamase inhibitor combination that was developed for the treatment of infections with multidrug-resistant Gram-negative bacteria. In this study, C/T susceptibility testing was performed on a large number of Gram-negative bacterial isolates at multiple study sites across a large geographic distribution in the continental United States. We found that the MIC values derived by the Etest for C/T correlated well with MIC values obtained by BMD for both Enterobacteriaceae and P. aeruginosa, exceeding the >90% threshold of EA required by the FDA. Additionally, we identified very few categorical and very major errors using CLSI breakpoint criteria for Enterobacteriaceae and P. aeruginosa.
Despite growing interest in the use of C/T by physicians (21), only a limited number of studies have evaluated the performance of alternatives to BMD for C/T susceptibility testing. Among these studies, a considerable range of performance has been observed for a variety of C/T susceptibility testing devices. For example, a research-use-only (RUO) version of the Etest performed poorly compared to BMD for susceptibility testing of a small number of meropenem-nonsusceptible P. aeruginosa isolates in Michigan (22), but these data were later questioned when a follow-up to this study suggested favorable performance characteristics for the C/T Etest (<96% EA and CA) (23). Although our study did not produce any very major errors in the clinical isolate portion of the study, one very major error was identified in the challenge set study. Interestingly, the isolate for which this error occurred had Etest and BMD results that were internally consistent at each site; however, this isolate had MICs in the susceptible range at one site but MICs that were resistant at the two other sites. Therefore, when the voted BMD reference value was generated, this created a very major error for the one site at which the isolate tested as susceptible. Given the consistency of test results for this isolate within each site, it appears that this error was not due to a failure of the test device but rather an artifact of the data analysis in combination with the possibility of the loss of a resistance plasmid.
Other devices that use gradient diffusion (e.g., Liofilchem MIC test strip) for C/T testing have been shown to have unacceptably high error rates (23). These discrepancies could reflect the fact that interpretation of MIC results from gradient diffusion devices can be more subjective than BMD results. In addition, the population distribution of many Gram-negative species falls near the breakpoint for this antimicrobial agent, which can be challenging for accurate categorical interpretation. However, various degrees of performance have also been reported for disk diffusion assays for C/T disks (23,–25). Despite these limitations, in most clinical laboratory settings, C/T susceptibility testing options like gradient diffusion that are more accessible than BMD are critically needed because approximately 10% of infections for which C/T therapy is indicated are caused by a C/T-resistant organism and therefore susceptibility testing is needed to optimize therapy and antimicrobial stewardship (6, 7, 9,–17, 24, 26, 27). Indeed, C/T resistance and treatment failures have already been described and appear to be acquired/mediated via a variety of mechanisms, emphasizing the need for widely accessible phenotypic C/T susceptibility testing (8, 11, 24, 28,–30).
This study has limitations, including the small number of isolates for some of the specific species evaluated and the lack of resistant isolates for some species tested. The strengths of this study include reference BMD as a reference method, standardization of preparation of the BMD panels, the large number of isolates evaluated, and the multicenter nature of the study.
In conclusion, testing a large number of clinical strains from different geographic regions within the United States, we found that the bioMérieux C/T Etest is a reproducible and accurate method for C/T susceptibility testing of Enterobacteriaceae and P. aeruginosa.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by bioMérieux. We thank Meghan Wallace, Sheri Garret, and Paul Magnano for excellent technical assistance with this study.
Funding Statement
This study was funded by bioMerieux.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00717-18.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.00717-18
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/56/9/e00717-18.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jcm.00717-18
Article citations
Approaches to Testing Novel β-Lactam and β-Lactam Combination Agents in the Clinical Laboratory.
Antibiotics (Basel), 12(12):1700, 05 Dec 2023
Cited by: 1 article | PMID: 38136734 | PMCID: PMC10740869
Review Free full text in Europe PMC
High risk of bloodstream infection of carbapenem-resistant enterobacteriaceae carriers in neutropenic children with hematological diseases.
Antimicrob Resist Infect Control, 12(1):66, 08 Jul 2023
Cited by: 1 article | PMID: 37422680 | PMCID: PMC10329308
Antimicrobial Activity of Ceftolozane-Tazobactam, Ceftazidime-Avibactam, and Cefiderocol against Multidrug-Resistant Pseudomonas aeruginosa Recovered at a German University Hospital.
Microbiol Spectr, 10(5):e0169722, 03 Oct 2022
Cited by: 9 articles | PMID: 36190424 | PMCID: PMC9603231
Susceptibility of Meropenem-Resistant and/or Carbapenemase-Producing Clinical Isolates of Enterobacterales (Enterobacteriaceae) and Pseudomonas aeruginosa to Ceftazidime-Avibactam and Ceftolozane-Tazobactam as Assessed by In Vitro Testing Methods.
Antibiotics (Basel), 11(8):1023, 29 Jul 2022
Cited by: 2 articles | PMID: 36009892 | PMCID: PMC9405240
Comparative Evaluation of Vitek 2 and Etest versus Broth Microdilution for Ceftazidime/Avibactam and Ceftolozane/Tazobactam Susceptibility Testing of Enterobacterales and Pseudomonas aeruginosa.
Antibiotics (Basel), 11(7):865, 27 Jun 2022
Cited by: 4 articles | PMID: 35884118 | PMCID: PMC9312067
Go to all (10) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Multicenter Evaluation of the New Etest Gradient Diffusion Method for Piperacillin-Tazobactam Susceptibility Testing of Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii Complex.
J Clin Microbiol, 58(2):e01042-19, 28 Jan 2020
Cited by: 9 articles | PMID: 31597745 | PMCID: PMC6989064
Verification of Ceftazidime-Avibactam and Ceftolozane-Tazobactam Susceptibility Testing Methods against Carbapenem-Resistant Enterobacteriaceae and Pseudomonas aeruginosa.
J Clin Microbiol, 56(2):e01093-17, 24 Jan 2018
Cited by: 27 articles | PMID: 29167294 | PMCID: PMC5786715
Ceftolozane-tazobactam activity against drug-resistant Enterobacteriaceae and Pseudomonas aeruginosa causing healthcare-associated infections in Australia and New Zealand: Report from an Antimicrobial Surveillance Program (2013-2015).
J Glob Antimicrob Resist, 10:186-194, 19 Jul 2017
Cited by: 12 articles | PMID: 28735046
Ceftolozane/tazobactam: place in therapy.
Expert Rev Anti Infect Ther, 16(4):307-320, 09 Mar 2018
Cited by: 64 articles | PMID: 29493397
Review
 a
a




