Abstract
Free full text

Inactivation of Bap1 Cooperates with Losses of Nf2 and Cdkn2a to Drive the Development of Pleural Malignant Mesothelioma in Conditional Mouse Models
Associated Data
Abstract
Pleural malignant mesothelioma (MM) is a therapy-resistant cancer affecting the serosal lining of the thoracic cavity. Mutations/deletions of BAP1, CDKN2A, and NF2 are the most frequent genetic lesions in human MM. We introduced various combinations of these deletions in the pleura of conditional knockout (CKO) mice, focusing on the contribution of Bap1 loss. While homozygous CKO of Bap1, Cdkn2a, or Nf2 alone gave rise to few or no MMs, inactivation of Bap1 cooperated with loss of either Nf2 or Cdkn2a to drive development of MM in ~20% of double-CKO mice, and a high incidence (22/26, 85%) of MMs was observed in Bap1;Nf2;Cdkn2a (triple)-CKO mice. MM onset was rapid in triple-CKO mice, with a median survival of only 12 weeks, and MMs from these mice were consistently high-grade and invasive. Adenoviral-Cre treatment of normal mesothelial cells from Bap1;Nf2;Cdkn2a CKO mice, but not from mice with knockout of one or any two of these genes, resulted in robust spheroid formation in vitro, suggesting that mesothelial cells from Bap1;Nf2;Cdkn2a mice have stem cell-like potential. RNA-seq analysis of MMs from triple-CKO mice revealed enrichment of genes transcriptionally regulated by the polycomb repressive complex 2 (PRC2) and others previously implicated in known Bap1-related cellular processes. These data demonstrate that somatic inactivation of Bap1, Nf2, and Cdkn2a results in rapid, aggressive MMs, and that deletion of Bap1 contributes to tumor development, in part, by loss of PRC2-mediated gene repression of tumorigenic target genes and by acquisition of stem-cell potential, suggesting a potential avenue for therapeutic intervention.
Introduction
Pleural malignant mesothelioma (MM) is an aggressive, treatment-resistant cancer, which is causally related to asbestos exposure. Early cytogenetic and deletion mapping studies had uncovered several prominent sites of chromosomal loss in human pleural MM, including 3p21, 9p21 and 22q12, and these recurrent abnormalities often occurred in combination in a given tumor, suggesting a multi-step pathogenetic process (1). Alterations in the target tumor suppressor genes at two of these sites, i.e., CDKN2A at 9p21 and NF2 at 22q12, were first discovered more than two decades ago (2–6), whereas the identity of the crucial 3p21 gene in MM remained a mystery until 2011, when BAP1 was identified as the critical MM tumor suppressor gene at this location (7,8). In a recent comprehensive study as part of The Cancer Genome Atlas, somatic genetic alterations of BAP1, NF2 and CDKN2A were observed in combination in 25/74 (34%) pleural MMs (9).
The alterations of CDKN2A involve heterozygous or homozygous deletions of all or part of the locus (2). Abnormalities of NF2 in MM frequently are biallelic, consisting of both an inactivating mutation and loss of the second allele (10). Biallelic losses of CDKN2A have been documented in ~40% of primary MMs (7) and up to 90% of MM cell lines (2), whereas inactivating NF2 mutations have been reported in 20-55% of cases (5–7,10). Using Sanger sequencing, somatic inactivating mutations of BAP1 were initially reported in 20-25% of sporadic MMs (7,8), but more recent studies with other sequencing platforms such as targeted next generation sequencing and multiplex ligation-dependent probe amplification have uncovered BAP1 alterations in ~60% of cases, with a preponderance of exonic deletions (11).
The CDKN2A locus encodes the tumor suppressors p16INK4A and p14ARF, which regulate the Rb and p53 cell cycle pathways, respectively. In MM, the deleted region of CDKN2A typically includes exon 2 (2), which encodes portions of p16INK and p14ARF, and is thus predicted to affect both Rb and p53 pathways. Re-expression of p16INK4A in MM cells results in cell cycle arrest and tumor suppression/regression (12), while re-expression of p14ARF in MM cells induces G1 arrest/apoptosis (13). Underscoring the relevance of Cdkn2a to MM pathogenesis, heterozygous Cdkn2a knockout mice treated with asbestos develop MM at a significantly accelerated rate compared to asbestos-treated wild-type (WT) littermates (14). Additionally, mice deficient for both p16Ink4a and p19Arf exhibit enhanced asbestos-induced MM formation relative to mice deficient for either 16Ink4a or 19Arf alone (14).
Loss of the NF2 product, Merlin, in MM leads to cell cycle progression by upregulation of cyclin D1 both transcriptionally (15) and post-transcriptionally via activation of mTORC1 (16). NF2 also inhibits Pak and FAK signaling, which play key roles in cell migration and spreading, respectively, and inactivation of NF2 in MM cells promotes invasiveness and spreading (17,18). Nf2+/− mice treated with asbestos develop MM at an accelerated rate compared to asbestos-treated WT mice, suggesting that merlin inactivation contributes significantly to MM development (4,19,20). Moreover, mice with heterozygous losses of both Nf2 and Cdkn2a show further acceleration of asbestos-induced MM, with resulting tumors exhibiting increased cancer stem cells and enhanced tumor spreading capability compared to that observed in mice with losses of only one or the other of these genes (21). Similarly, conditional knockout (CKO) mice harboring homozygous deletions of both Nf2 and p16Ink4a/19Arf in the mesothelial lining of the thoracic cavity developed a high incidence of pleural MM that showed increased pleural invasion compared to Nf2;Tp53 CKO mice (20).
BRCA1-associated protein-1 (BAP1) was discovered as an ubiquitin hydrolase that associates with the RING finger domain of BRCA1 and enhances BRCA1-mediated inhibition of breast cancer cell growth (22). BAP1 interacts with ASXL family members to form the polycomb group (PcG) repressive deubiquitinase (PR-DUB) complex involved in stem cell pluripotency and other developmental processes (23). BAP1 also interacts with and deubiquitinates the transcriptional regulator host cell factor 1 (HCF1) (24). Importantly, BAP1 has been shown to form complexes with numerous transcription factors and cofactors, including transcription factor YY1 (25). The BAP1 ubiquitin carboxyl hydrolase activates transcription in an enzyme-dependent manner and regulates the expression of a variety of genes involved in numerous processes, including cell proliferation, DNA damage and inflammatory responses, metabolism, and various mechanisms of cell death (24,26–29). In animal models, asbestos exposure induces a significant increase in the incidence of MM in heterozygous Bap1-mutant mice as compared to asbestos-exposed WT littermates (30,31).
MM patients with germline BAP1 mutations have an improved long-term survival compared to MM patients without such heritable variants (32,33). However, it remains unclear whether somatic BAP1 mutations/deletions are associated with a poor prognosis in sporadic MM, as is the case in uveal melanoma and renal cell carcinoma (34,35). While most human MMs exhibit somatic alterations of BAP1, NF2 and/or CDKN2A, currently it is not known if inactivation of BAP1 cooperates with loss of NF2 and/or CDKN2A to drive a more aggressive MM phenotype. Here we address these questions experimentally using CKO models.
Materials and Methods
Mouse strains and genotyping
All mouse studies were performed in accordance with a protocol approved by the Fox Chase Cancer Center (FCCC) IACUC. Nf2f/f;Cdknaf/f mice in FVB/N genetic background (20), a gift of Anton Berns, were maintained in our laboratory in a mixed FVB/N × 129/Sv background. The LoxP sites in the Cdkn2a locus of these mice permit excision of exon 2, which results in inactivation of both p16Ink4a and p19Arf.
Bap1f/f mice in FVB/N background were developed in our laboratory using zinc finger nuclease (ZFN) technology, with the assistance of the FCCC Transgenic Mouse Facility. Custom ZFNs targeting Bap1 were designed and validated in mammalian cells by Sigma-Aldrich (St. Louis, MO). A pair of ZFNs was identified that binds to and cuts within a site in intron 5 of Bap1 with high efficiency and specificity. We then designed a donor DNA construct containing LoxP sites in introns 6 and 7 (Figure 1A), such that upon adenovirus-mediated expression of Cre recombinase there is deletion of exon 7 of Bap1. The ZFN mRNAs and donor DNA were injected into the pronucleus of one-cell embryos of FVB/N mice, and embryos were then transferred into pseudo-pregnant females. DNA from pups was analyzed for correct targeting by PCR amplification of the gene and sequencing. Three founder mice with LoxP sites integrated in the Bap1 locus were identified, one of which was used for experiments reported here. Representative examples of genotyping of DNA from Bap1 CKO mice are shown in Figure 1B.
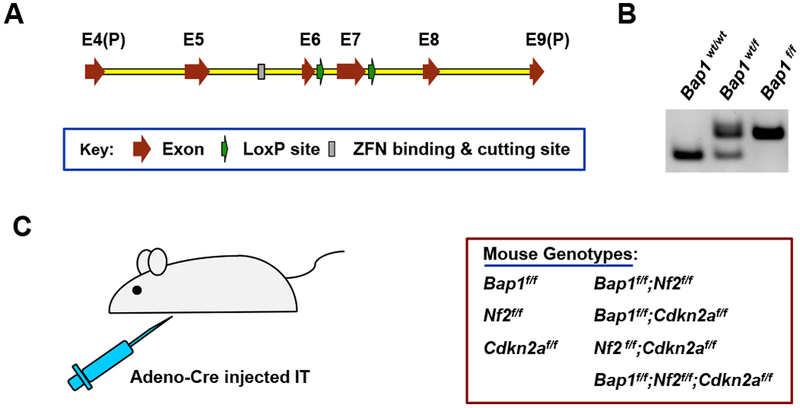
Generation of Bap1 conditional knockout (CKO) mouse with zinc finger nuclease (ZFN) technology, and experimental schema used for crosses with other CKO mice. A, Schematic drawing of Bap1 LoxP donor DNA sequence. Donor DNA contains region spanning part of exon 4, E4(P), to part of exon 9, E9(P). LoxP sites were inserted into introns 6 and 7. A pair of ZFNs was identified that binds to and cuts within a site in intron 5 of Bap1 with high efficiency and specificity. A donor DNA construct was then designed that contained two LoxP sites, one in intron 6 and one in intron 7, as shown, such that expression of Adeno-Cre in the mouse mesothelial cell lining the pleural cavity excises exon 7 of Bap1. B, Genotyping of DNA from wild type (wt) mouse as well as heterozygous (wt/f) and homozygous (f/f) Bap1 CKO mice. C, Strategy used to homozygously excise Bap1, Nf2, and/or Cdkn2a via intrathoracic (IT) injection of Adeno-Cre virus in mice with the various genotypes shown.
In addition to Bap1f/f, Nf2f/f, and Cdkn2af/f mice, these CKO animals were crossed to generate cohorts with the following genotypes: Bap1f/f;Nf2f/f, Bap1f/f;Cdkn2af/f, Nf2f/f;Cdkn2af/f and Bap1f/f;Nf2f/f;Cdkn2af/f. The following primers and annealing temperatures were used for genotyping: Bap1f/f 1063: 5’-CCC TGA GAC CCA GAA AAT CA-3’, Tm = 54.3°C; Bap1f/f 1228: 5’-GGG AGG CTC TTT GAA TTG GA-3’, Tm = 54.9°C; Nf2f/f 1048: 5’-CTT CCC AGA CAA GCA GGG TTC-3’, Tm = 58.0°C; Nf2f/f 1049: 5’-GAA GGC AGC TTC TTC CTT AAG TC-3’, Tm = 53.2°C; Cdkn2af/f 1025: 5’-GCA GTG TTG CAG TTT GAA CCC-3’, Tm = 57.4°C; and Cdkn2af/f 1026: 5’-TGT GGC AAC TGA TTC AGT TGG-3’, Tm = 55.8°C.
Adeno-Cre injections
Ad5CMVCre (Adeno-Cre) virus was obtained from the Viral Vector Core of the University of Iowa. Mice 8-12-weeks of age were injected intrathoracically (20), specifically intrapleurally, with Adeno-Cre virus (50 μl of 3-6 × 1010 PFU), with approximately equal numbers of mice of each gender in each cohort. Mice were monitored daily and were euthanized upon visible signs of distress, including extreme fatigue, labored breathing, or when mice exhibited a 10% change in body weight. Tissues of all organs of the pleural and peritoneal cavities were collected from sacrificed mice, and tumor specimens were subjected to histopathological assessment. Portions of tumors were also saved in both O.C.T. Compound and RNAlater Solution (Thermo Fisher, Waltham, MA) and immediately frozen at −80°C.
Tumor histopathology, immunohistochemistry and RT-PCR analysis
The histopathological procedures used are essentially the same as previously described (23). In summary, after paraffin embedding, sectioning and deparaffinization, hematoxylin and eosin (H&E)-stained sections were used for histopathologic evaluation, and unstained sections were used for immunohistochemistry (IHC). For IHC, sections were subjected to heat-induced epitope retrieval using citrate buffer (pH 6.0) for 20 min.
MMs were diagnosed by two independent pathologists (A.J.K.-S. and K.Q.C.). To confirm the diagnosis of MM, IHC was performed for various MM markers, including mesothelin, detected with D-16 antibody (Santa Cruz Biotechnology, Dallas, TX), and cytokeratin 8, detected with TROMA-1 antibody (DSHB, University of Iowa, Iowa City, IA). In addition, reverse transcription-PCR (RT-PCR) analysis was also performed to analyze MM markers, including Wt1, Msln (mesothelin), and Krt18/19 (cytokeratin 18/19). RT-PCR analysis for Msln used primers 5′-ATCAAGACATTCCTGGGTGGG-3′ and 5′-CGGTTAAAGCTGGGAGCAGAG-3′. Primers for other MM markers have been previously described (4). To assess cell proliferation, IHC staining was performed with antibodies for Ki-67 (Dako/Agilent, Santa Clara, CA).
Immunoblot analysis and antibodies
Immunoblots were prepared with 30-50 μg of protein lysate per sample, as previously described (21). Anti-Bap1 antibody (A302-243, 1:2000) was purchased from Bethyl Laboratories (Montgomery, TX). Anti-Bap1 (C-4, sc-28383, 1:2000), anti-p19Arf (5-C3-1, sc-32748 1:1000), anti-Gapdh (6C5, sc-32233, 1:50,000), and anti-β-actin (C4, sc-47778, 1:50,000) were from Santa Cruz Biotechnology; anti-Nf2 (D1D8, #6995S, 1:1000), anti-total-Akt (#9272S, 1:1000), and anti-P-Akt, Ser473 (D9E XP, #4060, 1:1000) were from Cell Signaling Technology (Danvers, MA); and anti-p16 (ab189034 1:1000) was from Abcam (Cambridge, MA). Immunoblots were imaged using Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Ontario Canada; P90720 Cat. No. WBKLS0500).
Spheroid growth assay
Primary normal mesothelial cells were isolated from individual CKO mice, 10-12 weeks of age, with each of the following genotypes: Bap1f/f, Nf2f/f, Cdkn2af/f, Bap1f/f;Nf2f/f, Bap1f/f;Cdkn2af/f, Nf2f/f;Cdkn2af/f, and Bap1f/f;Nf2f/f;Cdkn2af/f. The mesothelial cells were isolated by sacrificing animals and adding trypsin (0.25% Trypsin-EDTA, Gibco, 25200-056) to the peritoneal cavity, according to the method of Bot et al. (36). Normal mesothelial cells at passage 2 were then seeded in 6-well plates (Thermo Fisher BioLite 6-well Multidish, Cat. #130184) and treated with 4×1010 PFU/ml of either control Ad5CMVempty (Adeno-CMV) or Adeno-Cre virus for 1 h. Cells were then washed in PBS, cultured in normal mesothelial media (34) for 72 h, and trypsinized. Next, 5,000 mesothelial cells were seeded in each well of a 6-well non-adherent plate (Corning, Costar) in serum-free DMEM/F12 media supplemented with B27, EGF (10 ng/mL), basic FGF (10 ng/mL) and penicillin/streptomycin, as described (21). The experiment was performed in triplicate for each cell genotype. Spheroids were photographed using light microscopy after 9 days of culture.
RNA-seq analysis
Tumor RNA was isolated in Trizol and purified with RNeasy columns (Qiagen, Germantown, MD). RNA-seq was performed with a HiSeq Sequencer and the following reagents: TruSeq Stranded mRNA Library, HiSeq Rapid SR Cluster, and HiSeq Rapid SBS v2 kits (Illumina, San Diego, CA). Stranded mRNA-seq libraries were prepared according to Illumina’s product guide. First-strand cDNA was synthesized using SuperScript II reverse transcriptase (Thermo Fisher) and random primers at 42°C for 15 min, followed by second strand synthesis at 16°C for 1 h. Adapters with Illumina P5/P7 sequences and indices were ligated to cDNA fragments, and libraries were pooled and loaded onto the sequencer. Fastq files were aligned to the mm10 mouse genome using TopHat2. Raw sequence counts for each gene were produced with HTseq (https://htseq.readthedocs.io) and differentially expressed genes were identified by DESeq2 (37). RNA-seq heatmaps were constructed from log2(x+1)-transformed counts/million, standardized across rows, using the “heatmap.2” function from the R gplots library. Raw sequencing data have been deposited in the GEO repository (accession number GSE131942).
Gene set enrichment analysis
For functional enrichment analysis, genes identified as differentially expressed by DESeq2 with nominal p-value < 0.5 were ranked by fold-change and mapped to human genes using R biomaRt (38). These genes were then analyzed using the GSEAPreranked method of Gene Set Enrichment Analysis (GSEA) (39) (with “classic” enrichment statistic) applied to the curated (c2) and Gene Ontology gene sets from the Molecular Signature Database (MSigDB). Confirmation of differentially expressed genes was performed by quantitative RT-PCR analysis; Taqman (Thermo Fisher) assay IDs used for 13 genes validated by RT-PCR analysis are shown in Supplemental Table 1.
Results
Temporal intrathoracic (IT) inactivation of Bap1 together with Nf2 and/or Cdkn2a results in MM and other tumors
Injection of Adeno-Cre virus into the pleural space of Rosa26 LacZ reporter (R26R) mice has been previously reported to result in efficient β-galactosidase expression in the mesothelial cell lining of the chest cavity (20). Locotemporal expression of Cre recombinase by IT injection of Adeno-Cre was used to induce mesothelial cell-specific loss of Bap1, Nf2, and/or Cdkn2a in homozygous single-gene CKO mice and in homozygous compound CKO mice. The injection scheme used for the CKO mice is depicted in Figure 1C. Tumors that arose in most mouse cohorts were mainly MMs, although other neoplasms were also observed (Table 1).
Table 1.
Summary of tumors observed in homozygous conditional knockout (CKO) mice arising after intrathoracic (IT) injection of Adeno-Cre.
| Genotypea | Bap1f/f | Bap1f/f; Nf2f/f | Bap1f/f; Cdkn2af/f | Nf2f/f; Cdkn2af/f | Bap1f/f;Nf2f/f; Cdkn2af/f |
|---|---|---|---|---|---|
| No. mice injected | 32 | 42 | 27 | 24 | 26 |
| MMb | 1 (3.1%) | 7c (16.6%) | 6 (22.2%) | 15d (62.5%) | 22 (84.6%) |
| Epithelioid | 0 | 0 | 0 | 0 | 0 |
| Mixed (Biphasic) | 0 | 6 | 1 | 4 | 8 |
| Sarcomatoid | 1 | 1 | 5 | 11 | 14 |
| Lymphoma | 1 | 0 | 0 | 0 | 0 |
| Adenocarcinoma | 2 | 0 | 2 | 0 | 0 |
| HCC / ICC | 0 | 28c,e | 0 | 2d | 0 |
| Hemangiocarcinoma | 0 | 0 | 2 | 0 | 0 |
| Other causes of deathf | 29 | 10 | 17 | 9 | 4 |
In the cohorts of mice with homozygous knockout of a single gene only, few MMs were seen. Among 32 Bap1f/f CKO mice injected IT with Adeno-Cre, a slow growing, well differentiated MM was identified in a single animal 45 weeks after injection. Interestingly, this MM acquired loss of Nf2 and p16Ink4a expression during tumor formation (Supplemental Figure 1A). As expected, this tumor showed Bap1 loss-related deficient deubiquitination activity; and consistent with the acquired loss of Nf2/Merlin, downstream Yap was not phosphorylated (Supplemental Fig. 1B). PCR analysis confirmed that losses of Nf2 and Cdkn2a are acquired (Supplemental Figure 1C). MMs were observed in 2 of 15 Nf2f/f mice and 0 of 12 Cdkn2af/f mice injected with Adeno-Cre, results comparable to those reported in an earlier study (20).
Among the IT-injected mice with homozygous inactivation of two genes, MMs were observed in 7 of 42 (16.6%) Bap1;Nf2 CKO mice, 6 of 27 (22.2%) Bap1;Cdkn2a CKO mice, and 15 of 24 (62.5%) Nf2;Cdkn2a CKO mice. The highest incidence of MM (22 of 26, 84.6%) was observed in Bap1;Nf2;Cdkn2a triple-CKO mice. A summary of MM incidence among Bap1 CKO mice and mice with the various compound CKO genotypes is shown in Figure 2A.
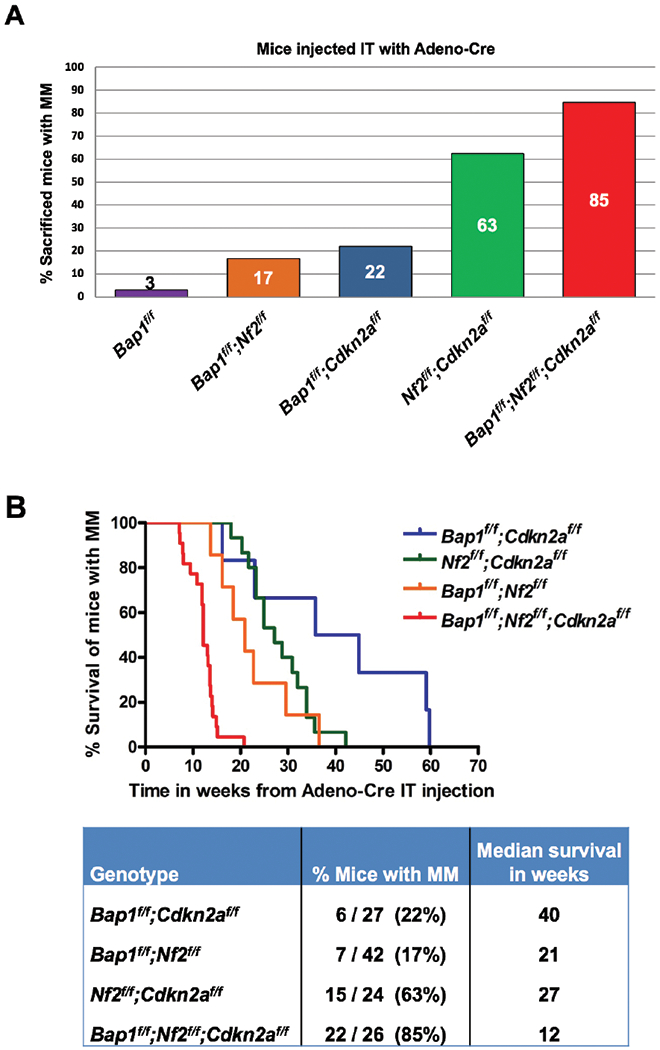
Incidence and latency of pleural malignant mesothelioma (MM) in mouse cohorts with different genotypes following intrathoracic (IT) injections with Adeno-Cre virus. A, Incidence of MM in the various CKO mouse groups. B, Tumor latency curves for MMs arising after Adeno-Cre injections in mice with CKO of different combinations of Bap1, Nf2 and/or Cdkn2a. Kaplan-Meier curves depict survival of mice succumbing to MM arising after IT injections with Adeno-Cre. Note rapid development of MM in triple-CKO mice, with the median survival being only 12 weeks, which was significantly different (p<0.0001) from that of any of the other compound CKO mice. The survival differences between the other three double knockout (DKO) mouse cohorts were not statistically significant (Supplemental Table 2).
With regard to histological subtype, notably no epithelioid MMs were observed with any of the mouse genotypes. Sarcomatoid MMs predominated in most cohorts, with the exception being Bap1;Nf2 CKO mice, in which 6 of 7 MMs showed mixed (biphasic) histology.
MM latency varied between 21 and 40 weeks among the different compound double-CKO mice. A markedly shorter MM latency, 12 weeks, was observed in Bap1;Nf2;Cdkn2a triple-CKO mice, a highly significant difference (log rank test, p<0.0001) from the 27 weeks observed for Nf2;Cdkn2a CKO mice or for any of the other compound double-CKO (DKO) mice. The MM survival differences between the three cohorts with different DKO combinations were not statistically significant (Supplemental Table 2). Kaplan-Meier survival curves of IT-injected mice that developed MM are shown in Figure 2B. Additionally, MMs from Bap1;Nf2;Cdkn2a CKO mice were consistently high-grade, very invasive, and highly proliferative tumors (Figure 3A,,B),B), whereas MM morphology was generally more variable and less hypercellular with the other mouse genotypes. Immunoblotting confirmed the absence of expression of proteins encoded by conditionally knocked out genes Bap1, Nf2 and Cdkn2a (p16Ink4a and p19Arf) in 12 of 12 MMs tested, and interestingly there was also acquired loss of expression of Bap1 in 4 of 6 MMs from Nf2;Cdkn2a CKO mice (Figure 4). We also assessed the status of Akt in these tumors, because Akt activation occurs frequently in human and murine MMs (4). Akt activation was seen in all MMs tested, as indicated by expression of phosphorylated (active) Akt (Figure 4).
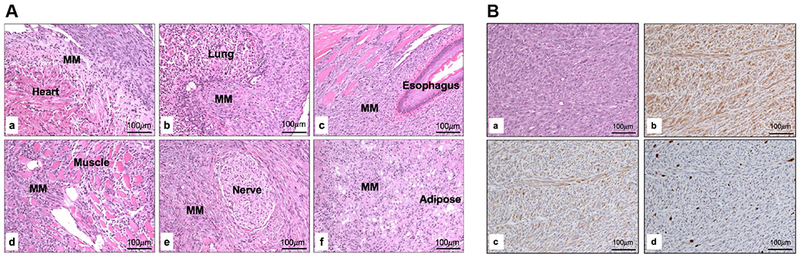
Histopathological assessment of representative high-grade MMs from Bap1;Nf2;Cdkn2a CKO mice. A, Histopathology of highly invasive, disseminated MM that arose in a Bap1;Nf2;Cdkn2a CKO mouse following IT injection of Adeno-Cre. Note MM invasion affecting multiple organs including the heart (a), lung (b), esophagus (c), skeletal muscle (d), nerve (e), and mediastinal adipose tissue (f). B, Serial sections of a MM from a Bap1;Nf2;Cdkn2a CKO mouse showing H&E staining (a), and IHC for mesothelin (b), cytokeratin 8 marker TROMA-1 (c), and nuclear proliferation marker Ki-67 (d).
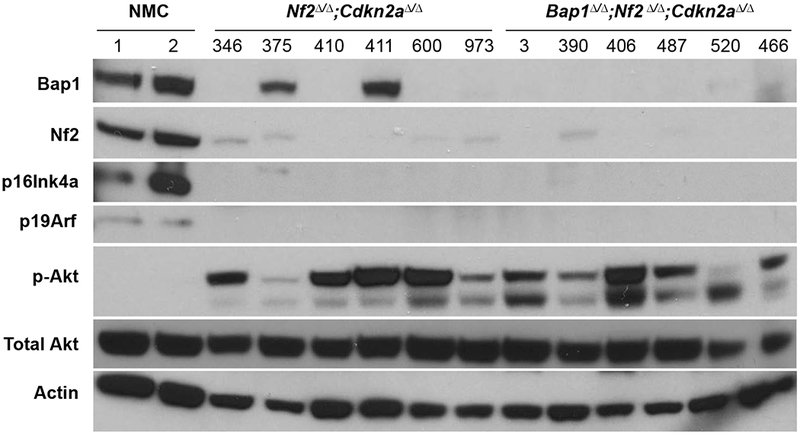
Western blot analysis of normal mesothelial cells (NMC) and MMs from Nf2;Cdkn2a double-knockout (DKO) and Bap1;Nf2;Cdkn2a triple-knockout (TKO) mice. Immunoblots confirm loss of expression of proteins encoded by conditionally knocked out genes Bap1, Nf2 and Cdkn2a (p16Ink4a and p19Arf) in all tumors. Acquired loss of expression of Bap1 was also observed in 4 of 6 MMs from Nf2;Cdkn2a DKO mice. Interestingly, this acquired loss of Bap1 expression appears to have occurred at the post-translational level, as these tumors showed no Bap1 mutations, deletions, or loss of mRNA expression. Phosphorylated Akt, indicative of Akt activation, is seen in all MMs shown.
Besides MM, various other tumor types were also seen in mice injected IT with Adeno-Cre (Table 1). Among mice with homozygous inactivation of Bap1 alone in the thoracic cavity, 2 lung adenocarcinomas and 1 lymphoma were seen. The remaining mice died of other age-related diseases, including bladder obstructions or lymphoproliferative diseases. Among the compound CKO mice, most notable was the cohort with inactivation of both Bap1 and Nf2, which showed a high incidence (28 of 42, 66.7%) of hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) (Supplemental Figure 2A and 2B, respectively), with these tumors generally arising later than MMs in this cohort (median 35 weeks vs. 21 weeks, respectively). Altogether, 28 Bap1;Nf2 CKO mice developed hepatic cancers, including 7 with HCC alone, 4 with ICC alone, 14 with both HCC and ICC, 1 with HCC and MM, and 2 with HCC, ICC, and MM.
Inactivation of three MM driver tumor suppressor genes is sufficient to induce a stem cell-like phenotype in mesothelial cells
To assess the cooperativity of Bap1, Nf2, and Cdkn2a losses in vitro, we next used a spheroid growth assay. Primary normal mesothelial cells were isolated from Bap1f/f, Nf2f/f, Cdkn2af/f, Bap1f/f;Nf2f/f, Bap1f/f;Cdkn2af/f, Nf2f/f;Cdkn2af/f, and Bap1f/f;Nf2f/f;Cdkn2af/f mice. The mesothelial cells were then exposed to either control Adeno-CMV or Adeno-Cre virus for 1 h, seeded in non-adherent plates in serum-free stem cell medium (21), and photographed after 9 days. There was little or no evidence of spheroid formation in cultured WT mesothelial cells or mesothelial cells with Adeno-Cre-induced homozygous inactivation of one or various combinations of two tumor suppressor genes (representative examples shown in Figure 5). In contrast, Adeno-Cre-treated mesothelial cells from Bap1f/f;Nf2f/f;Cdkn2af/f mice, i.e., with inactivation of all three MM driver genes, showed multiple large spheroids, indicative of stem cell features (Figure 5).
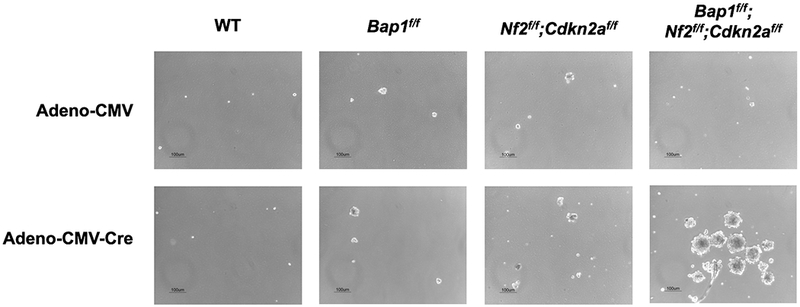
Ad5CMVCre virus-exposed normal mesothelial cells (NMC) from Bap1f/f;Nf2f/f;Cdkn2af/f mice harbor more and much larger stem cell-like spheroids than do those from wild-type (WT), Bap1f/f, and Nf2f/f;Cdkn2af/f mice. Equal numbers of NMC from mice with the various cited genotypes were seeded on non-adherent 6-well plates in stem cell media, and spheroids were photographed after 9 days.
RNA-seq analysis of MMs from triple-CKO mice reveals enrichment of genes transcriptionally regulated by the polycomb repressive complex 2 (PRC2)
Given the striking shift in MM latency between Bap1;Nf2;Cdkn2a CKO mice and Nf2;Cdkn2a CKO mice, as well as the consistently more aggressive nature of MMs from triple-CKO (TKO) mice, we performed RNA-seq analysis on MMs from these two cohorts to shed light on the contribution of Bap1 inactivation in this experimental setting. This analysis was carried out on all 6 MMs from Bap1;Nf2;Cdkn2a CKO mice shown in Figure 4 versus 2 MMs from Nf2;Cdkn2a CKO mice in which Bap1 expression was retained (tumors 375 and 411 in Figure 4). Among 1,425 genes identified as differentially expressed with fold-change of at least 2 and false discovery rate (FDR) <1%, significantly fewer genes (628, 44%) were upregulated than downregulated (p-value = 8.4 × 10−6, exact binomial test) in MMs from Bap1-deficient TKO mice versus Bap1-expressing MMs from DKO mice. Functional enrichment analysis with GSEA identified several highly significant (family-wise error rate < 0.0005) curated gene sets. Prominent among the positively enriched gene sets were those related to writers of histone H3K27Me3, including PRC2 and SUZ12 (Figure 6A), as well as genes participating in cell proliferation and transcription regulation involving ubiquitin signaling mediated by the Bap1-HCF1-YY1 complex, EED-related stem cell pluripotency, cytokine signaling, and cell adhesion; 100 of the top pathways differentially affected in MMs from TKO versus DKO mice are presented in Supplemental Table 3. Considering that Bap1 plays an important role in chromatin remodeling and transcriptional regulation (23–25), and the fact that many of the top differentially expressed genes were significantly enriched for PRC2 target genes (40), we decided to focus further on PRC2 targets here. A heatmap of genes differentially expressed in MMs from triple-CKO mice versus MMs from Nf2;Cdkn2a double-CKO mice is shown in Figure 6B. Thirteen cancer-related genes were validated by RT-PCR analysis, 6 of which (e.g., Ptchd1 and Diras1) are depicted in Figure 6C and Supplemental Figure 3. Mmp9 protein was also shown to be overexpressed and activated in MMs from TKO mice versus those from DKO mice (Figure 6D).
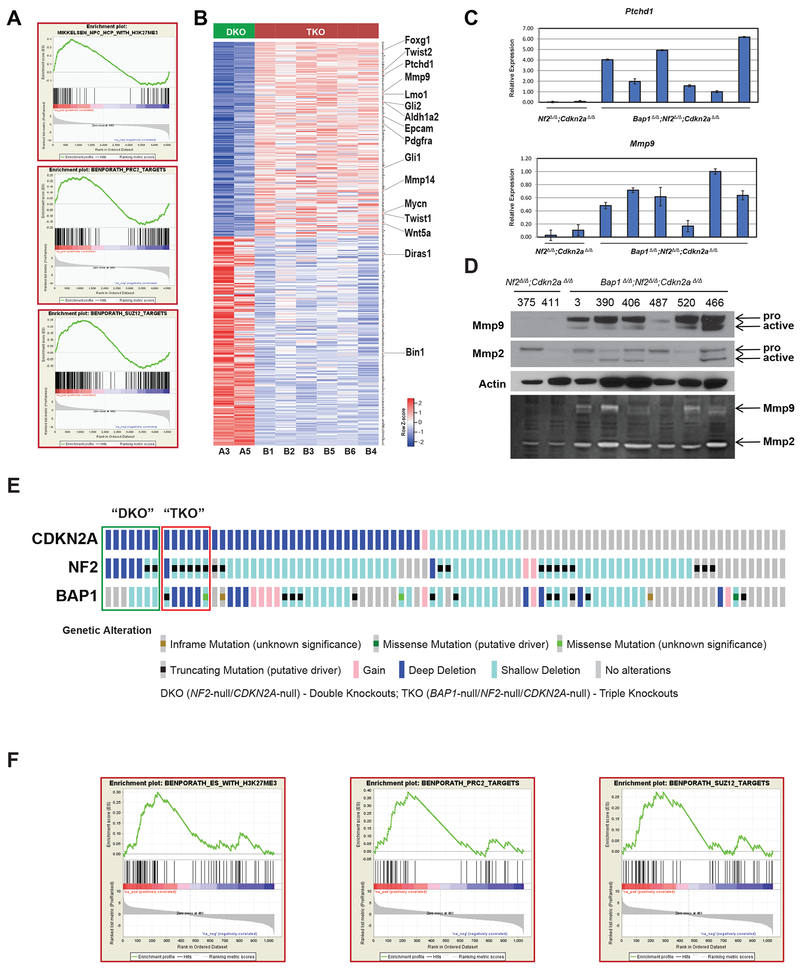
Heatmap and GSEA of same MMs (B1-B6) from Bap1;Nf2;Cdkn2a triple-knockout (TKO) mice shown in Figure 4 versus two MMs, A3 and A5 (375 and 411, respectively), from Nf2;Cdkn2a double knockout (DKO) mice retaining expression of Bap1. A, Gene enrichment plots of several gene sets differentially expressed in MMs from TKO versus DKO mice. B, Top 400 PRC2 target genes differentially expressed with at least 2-fold change and FDR of <1%. Genes are ranked by significance, with most significantly upregulated PRC2 target at top and most significantly downregulated at bottom. A number of the indicated upregulated genes have been implicated in oncogenesis, cancer cell stemness, pro-invasion/metastasis, adhesion, and cytokine and/or hedgehog signaling, and two downregulated genes (Bin1, Diras1) encode tumor suppressors. C, RT-PCR analysis was performed on 13 of the 16 indicated genes (2 shown here), validating the RNA-seq data in each instance. D, Immunoblot demonstrating overexpression of Mmp9 and Mmp2 in MMs from TKO mice (upper panel). Upper bands indicate respective pro-Mmp9 and pro-Mmp2; lower bands represent cleaved, active forms. Lysates were run on Novex 10% Zymogram gel and stained using colloidal blue staining kit to visualize Mmp2/9 activity (lower panel). E, Assessment of BAP1, CDKN2A and NF2 in 87 human MMs from TCGA. Seven MMs (green open rectangle, designated DKO here) were null for both CDKN2A and NF2, and 6 (red open rectangle, designated TKO) were null for BAP1, CDKN2A and NF2. F, Gene enrichment plots of several gene sets differentially expressed in TKO versus DKO MMs.
Discussion
In this investigation, our aim was to induce mesothelial cell-specific homozygous deletions of three tumor suppressor genes (Bap1, Nf2, and Cdkn2a) that have been implicated as major drivers of human MM pathogenesis. While the majority of malignant tumors observed were MMs, additional tumor types also resulted, possibly due to infection of other intrathoracic tissues or by passage of virus via blood vessels to the liver. Notably, HCCs and ICCs were very common later in life in Bap1;Nf2 CKO mice but were observed in only two CKO mice with any of the other genotypes. Notably, liver-specific deletion of Nf2 in the developing or adult mouse has been reported to promote a marked, progressive expansion of progenitor cells throughout the liver, with all surviving mice eventually developing both HCC and ICC (41). Interestingly, HCC and ICC were not seen in our Bap1;Nf2;Cdkn2a CKO cohort, perhaps because these mice succumbed to MM before hepatic tumors could be detected.
Overall, the MMs observed in our GEM cohorts generally showed morphological features, immunohistochemical staining, and invasiveness similar to that of human sarcomatoid MMs, with the exception being those observed in Bap1;Nf2 CKO mice, in which 6 of 7 MMs showed mixed (biphasic) histology that also was similar histologically to their human biphasic counterparts.
The Kaplan-Meier curves (Figure 2B) indicate that homozygous inactivation of different combinations of Bap1, Nf2, and Cdkn2a affect MM latency differently, with losses of all three genes profoundly accelerating tumor development. Specifically, Bap1;Nf2;Cdkn2a CKO mice bearing MM showed much shorter tumor latency than mice in any compound double-CKO cohort. Moreover, MMs from triple-CKO mice consistently appeared to be high grade, very invasive, hypercellular tumors whereas MMs from mice with other genotypes were more variable phenotypically. All MMs observed in our study were sarcomatoid or biphasic, similar to what was reported by Jongsma et al. for mice with homozygously floxed conditional compound alleles that were injected IT with Adeno-Cre (20). For example, in their Nf2;Cdkn2a CKO mice, 31 MMs were sarcomatoid, 13 had mixed histology, and only 1 was epithelioid (20). These investigators had speculated that the predominant epithelioid histology seen in human MMs, as opposed to mice, may be the result of species-specific differences or route of induction, including the long latency and exposure to asbestos in the human disease counterpart (20). It is also likely that the particular mouse strain can influence the phenotype observed. For example, in our previous asbestos carcinogenesis work, Nf2+/− mice in a 129Sv/Jae genetic background injected intraperitoneally (IP) with asbestos developed roughly equivalent percentages of epithelioid, mixed, and sarcomatoid MMs (4), whereas asbestos-injected Nf2+/− mice in a FVB/N background mostly developed sarcomatoid MMs (21).
Heterozygous knockout (+/−) and heterozygous mutant (+/mut) Bap1 mice injected IP with asbestos exhibit a highly significant increase in the incidence of MMs as compared to asbestos-injected WT littermates, providing in vivo evidence that Bap1 inactivation plays an important role in MM tumorigenesis (30,31). Similarly, heterozygous Nf2 mice injected with asbestos develop MM with a markedly shorter latency than asbestos-treated WT littermates (4,19). Heterozygous Cdkn2a knockout mice treated with asbestos also develop MM at an accelerated rate compared to asbestos-treated WT mice (14). Furthermore, asbestos-exposed compound Nf2+/−;Cdkn2a+/− mice show further acceleration of asbestos-induced MM and increased presence of cancer stem cells (21). These carcinogenesis studies, in combination with the gene knockout model studies reported here strongly support a working hypothesis that Bap1, Nf2, and Cdkn2a are key drivers of MM pathogenesis and that the collective losses of these three tumor suppressors is sufficient to drive rapidly disseminated, lethal disease, similar to the poor clinical outcome of human pleural MM.
In one study, CKO mice in which only one of these tumor suppressor genes was homozygously deleted developed MM at a low rate, with spontaneous MM-like tumors being identified in 5 of 30 Nf2f/f mice and 0/17 Cdkn2af/f mice injected IT with Adeno-Cre (20), supporting the notion that MM development requires the combined involvement of multiple tumor suppressor gene alterations (42). Similarly, in our Bap1 CKO cohort, MM was seen in only 1 of 32 mice. The fact that MM arises spontaneously in a considerable number of CKO mice made deficient for both alleles of two or more tumor suppressor genes, provides compelling support for the role of genetics in MM causation and the need for accumulated genetic and epigenetic alterations for tumor formation (1). As an example, even in the one Bap1 CKO mouse that did develop MM, it is intriguing that there was acquired loss of Nf2 and p16Ink4a expression during tumorigenesis.
The in vivo findings presented here with CKO mice closely mimic those found in the human disease counterpart, in which losses of BAP1, CDKN2A, and NF2 are frequently seen in various combinations (7,43). Their frequent occurrence in MM and their individual roles both in various cancer predisposition syndromes and in tumor progression strongly suggest that they are key drivers in MM pathogenesis. Our initial work on BAP1 provided the first demonstration that genetics influences the risk of MM, a cancer linked to mineral fiber carcinogenesis (8). However, there is evidence that germline mutations of CDKN2A and NF2 may also modulate susceptibility to mineral fiber carcinogens. For example, Betti et al. recently described a family in which both MM and cutaneous melanoma cases were found to have a deleterious germline missense mutation in the CDKN2A gene, and the family member with MM was determined to be of the low exposure category (44). Additionally, germline mutations in NF2 have been reported in patients who developed MM. In one case, an individual with neurofibromatosis, type II (NF2) developed peritoneal MM at the relatively young age of 40 (45). Comparative genomic hybridization analysis and IHC staining of the MM tissue revealed loss of the NF2 gene and protein product, respectively. A second NF2 patient developed a pleural MM at age 75 (46). Both patients were reported to have occupational asbestos exposure.
Our observation that CKO mice with losses of all three tumor suppressor genes had much higher MM penetrance and markedly shorter survival than mice with loss of one or two of these genes fits with the model put forward by Vogelstein and Kinzler (47). They proposed that in solid tumors of adults, alterations in a minimum of three mutated driver genes are needed for a cell to evolve into an invasive (malignant) tumor. In our CKO models, loss of one or two tumor suppressors was not consistently sufficient to result in a high incidence of MMs. The exception was the Nf2;Cdkn2a CKO model, but even in these mice tumor latency was much longer than in the triple-CKO mouse model. One possibility is that in Nf2;Cdkn2a CKO mice, significantly more time is needed in order to accumulate additional genetic and epigenetic alterations sufficient to result in an invasive cancer. In fact, 4 of 6 MMs tested from Nf2;Cdkn2a CKO mice acquired loss of Bap1 expression (Figure 4), suggesting that Bap1 loss may be favored in the progression of these tumors. In human MM, tumors are characterized by the presence of multiple somatic genetic and cell signaling alterations. The fact that large spheroids can be reproducibly formed in vitro in normal mesothelial cells following Adeno-Cre-induced excision of Bap1, Cdkn2a and Nf2, but not by the excision of only one or two of these genes, also provides experimental support for cooperativity between the inactivation of these three tumor suppressor genes in MM tumorigenesis.
Our RNA-seq analysis revealed that MMs from Bap1;Nf2;Cdkn2a CKO mice exhibit positively enriched gene sets consistent with cellular processes previously implicated in Bap1 function, such as genes mediated by the Bap1-HCF1-YY1 complex, which play a role in cellular proliferation and transcription regulation involving ubiquitin signaling (25). In addition, in the experimental context used here, we have uncovered a connection between Bap1 and PRC2 that has been less well established than other previously reported Bap1-related cellular activities.
Interestingly, a considerable number of upregulated genes in MMs from triple-CKO mice have been implicated as oncogenes or pro-invasion/metastasis genes, whereas several tumor suppressor genes were downregulated. Lafave et al. demonstrated that BAP1 inactivation leads to EZH2-dependent transformation (48). Although their report did not include RNA-seq analysis on human MM tumors for comparison with our murine data, they demonstrated that BAP1-deficient MM cells are sensitive to EZH2 pharmacologic inhibition, and our findings add support to their conclusion that such an approach may have clinical efficacy. It is noteworthy that PRC2 subunits such as Ezh2 have been shown to have context-dependent oncogenic or tumor suppressive roles in different cancers (49).
To investigate if our CKO mouse model findings are recapitulated in human MMs with biallelic inactivation of BAP1, CDKN2A and NF2 versus MMs with biallelic inactivation of CDKN2A and NF2 only, we analyzed data from The Cancer Genome Atlas (TCGA) (Supplemental Materials and Methods; Figure 6E, ,F).F). GSEA analysis of the TCGA data revealed several positively enriched gene sets associated with SUZ12, PRC2, as well as EZH2 mediated tri-methylation of H3K27 in human MMs with biallelic inactivation of all three tumor suppressor genes (Figure 6F; Supplemental Table 4), similar to the situation in MMs from our TKO mice.
Mechanistically, our findings suggest that losses of Bap1, Nf2 and Cdkn2a cooperate by disrupting multiple cellular pathways that lead to the transformation of normal mesothelial cells to MM cells. Thus, Cdkn2a loss would result in insensitivity to antigrowth signals (via p16Ink4a loss) and evasion of apoptosis (p19Arf loss), whereas Nf2 loss would be expected to result in evasion of apoptosis and tissue invasion/metastasis (Hippo, Pak) and TAZ-related MM cell transformation and transcriptional induction of distinct pro-oncogenic genes, including cytokines (50). Based on the upregulated genes identified in MMs from TKO mice, other possible effects such as Lmo1-induced ECM and integrin-related self-sufficiency in growth signals, Aldh1a2- and Gli1-related maintenance of cancer stem cells, and Mmp9-induced invasiveness (Figure 6D) may be mechanistically connected with Bap1 inactivation and will be explored in future studies.
Collectively, Bap1f/f;Nf2f/f;Cdkn2af/f mice injected IT with Adeno-Cre appear to be useful for modeling an aggressive form of MM. These MMs develop at a high incidence after a short latency period. Moreover, our finding that loss of Bap1 contributes to MM pathogenesis, in part, via loss of PRC2-mediated gene repression of certain oncogenic target genes, suggests a potential avenue for therapeutic intervention that may be rapidly tested in this CKO model.
Supplementary Material
1
2
3
4
5
6
7
8
Acknowledgments
Grant Support
This work was supported by NCI grants CA175691 (to J.R. Testa and F.J. Rauscher III) and CA06927 (to FCCC) and an appropriation from the Commonwealth of Pennsylvania to FCCC. Other support was provided by the Local #14 Mesothelioma Fund of the International Association of Heat and Frost Insulators and Allied Workers. We thank Emmanuelle Nicolas for RT-PCR analyses, Eric Ross for statistical assistance, Alfonso Bellacosa for critical reading of the manuscript, and Raza Zaidi, Xavier Graña, Siddharth Balachandran, and Richard Pomerantz for scientific advice and technical suggestions. The following FCCC core services assisted this project: Laboratory Animal, Transgenic Mouse, Genomics, Cell Culture, DNA Sequencing, Histopathology, and Biostatistics and Bioinformatics Facilities.
Footnotes
Conflict of Interest Statement
JRT and MC have a pending patent application on BAP1 mutation testing. JRT has provided legal consultation regarding genetic aspects of mesothelioma. The remaining authors have no potential conflicts of interest with regard to the publication of this work.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-18-4093
Read article for free, from open access legal sources, via Unpaywall:
https://cancerres.aacrjournals.org/content/canres/79/16/4113.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1158/0008-5472.can-18-4093
Article citations
Contemporary management of mesothelioma.
Breathe (Sheff), 20(2):230175, 01 Jun 2024
Cited by: 1 article | PMID: 39015660 | PMCID: PMC11250169
Review Free full text in Europe PMC
Pharmacological inhibition of CDK4/6 impairs diffuse pleural mesothelioma 3D spheroid growth and reduces viability of cisplatin-resistant cells.
Front Oncol, 14:1418951, 01 Jul 2024
Cited by: 0 articles | PMID: 39011477 | PMCID: PMC11246887
NF2: An underestimated player in cancer metabolic reprogramming and tumor immunity.
NPJ Precis Oncol, 8(1):133, 15 Jun 2024
Cited by: 0 articles | PMID: 38879686 | PMCID: PMC11180135
Review Free full text in Europe PMC
Characterization of KLHL14 anti-oncogenic action in malignant mesothelioma.
Heliyon, 10(6):e27731, 09 Mar 2024
Cited by: 2 articles | PMID: 38509883 | PMCID: PMC10950656
Comprehensive bioinformatics analysis of the role of VWF in the tumor microenvironment of malignant mesothelioma.
Medicine (Baltimore), 102(41):e35579, 01 Oct 2023
Cited by: 0 articles | PMID: 37832118 | PMCID: PMC10578691
Go to all (30) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE131942
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Fluorescence in situ hybridization (FISH) provides estimates of minute and interstitial BAP1, CDKN2A, and NF2 gene deletions in peritoneal mesothelioma.
Mod Pathol, 33(2):217-227, 30 Sep 2019
Cited by: 10 articles | PMID: 31570769
The prognostic significance of BAP1, NF2, and CDKN2A in malignant peritoneal mesothelioma.
Mod Pathol, 29(1):14-24, 23 Oct 2015
Cited by: 62 articles | PMID: 26493618
Usefulness of NF2 hemizygous loss detected by fluorescence in situ hybridization in diagnosing pleural mesothelioma in tissue and cytology material: A multi-institutional study.
Lung Cancer, 175:27-35, 19 Nov 2022
Cited by: 2 articles | PMID: 36442384
The significance of BAP1 and MTAP/CDKN2A expression in well-differentiated papillary mesothelial tumour: a series of 21 cases and a review of the literature.
Pathology, 56(5):662-670, 09 May 2024
Cited by: 0 articles | PMID: 38789301
Review
Funding
Funders who supported this work.
NCI (1)
Grant ID: CA175691
NCI NIH HHS (4)
Grant ID: P30 CA006927
Grant ID: R01 CA148805
Grant ID: R01 CA175691
Grant ID: P30 CA010815





