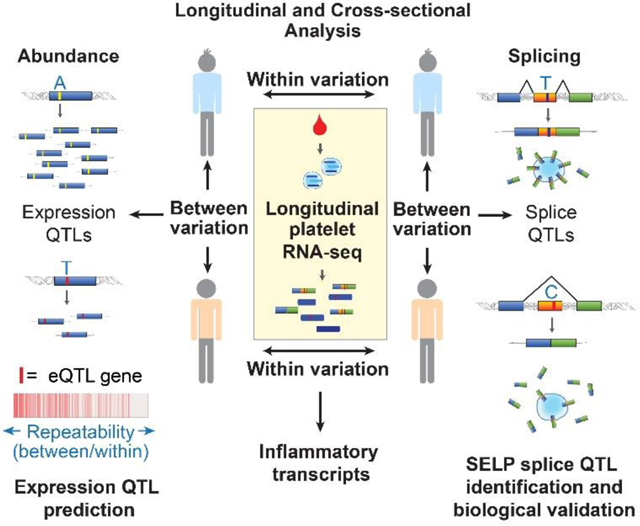
Abstract
Rationale
Longitudinal studies are required to distinguish within versus between-individual variation and repeatability of gene expression. They are uniquely positioned to decipher genetic signal from environmental noise, with potential application to gene variant and expression studies. However, longitudinal analyses of gene expression in healthy individuals-especially with regards to alternative splicing-are lacking for most primary cell types, including platelets.Objective
To assess repeatability of gene expression and splicing in platelets and use repeatability to identify novel platelet expression quantitative trait loci (QTLs) and splice QTLs.Methods and results
We sequenced the transcriptome of platelets isolated repeatedly up to 4 years from healthy individuals. We examined within and between individual variation and repeatability of platelet RNA expression and exon skipping, a readily measured alternative splicing event. We find that platelet gene expression is generally stable between and within-individuals over time-with the exception of a subset of genes enriched for the inflammation gene ontology. We show an enrichment among repeatable genes for associations with heritable traits, including known and novel platelet expression QTLs. Several exon skipping events were also highly repeatable, suggesting heritable patterns of splicing in platelets. One of the most repeatable was exon 14 skipping of SELP. Accordingly, we identify rs6128 as a platelet splice QTL and define an rs6128-dependent association between SELP exon 14 skipping and race. In vitro experiments demonstrate that this single nucleotide variant directly affects exon 14 skipping and changes the ratio of transmembrane versus soluble P-selectin protein production.Conclusions
We conclude that the platelet transcriptome is generally stable over 4 years. We demonstrate the use of repeatability of gene expression and splicing to identify novel platelet expression QTLs and splice QTLs. rs6128 is a platelet splice QTL that alters SELP exon 14 skipping and soluble versus transmembrane P-selectin protein production.Free full text

Longitudinal RNA-seq Analysis of the Repeatability of Gene Expression and Splicing in Human Platelets Identifies A Platelet SELP Splice QTL
Abstract
Rationale:
Longitudinal studies are required to distinguish within versus between-individual variation, and repeatability of gene expression. They are uniquely positioned to decipher genetic signal from environmental noise, with potential application to gene variant and expression studies. However, longitudinal analyses of gene expression in healthy individuals—especially with regards to alternative splicing—are lacking for most primary cell types, including platelets.
Objective:
To assess repeatability of gene expression and splicing in platelets and use repeatability to identify novel platelet eQTLs and sQTLs.
Methods and Results:
We sequenced the transcriptome of platelets isolated repeatedly up to 4 years from healthy individuals. We examined within and between-individual variation and repeatability of platelet RNA-expression and exon skipping, a readily measured alternative splicing event. We find that platelet gene expression is generally stable between and within individuals over time—with the exception of a subset of genes enriched for the inflammation gene ontology. We show an enrichment among repeatable genes for associations with heritable traits, including known and novel platelet eQTLs. Several exon skipping events were also highly repeatable, suggesting heritable patterns of splicing in platelets. One of the most repeatable was exon 14 skipping of SELP. Accordingly, we identify rs6128 as a platelet sQTL and define an rs6128-dependent association between SELP exon 14 skipping and race. In vitro experiments demonstrate that this single nucleotide variant directly affects exon 14 skipping and changes the ratio of transmembrane versus soluble P-selectin protein production.
Conclusions:
We conclude that the platelet transcriptome is generally stable over 4-years. We demonstrate the use of repeatability of gene expression and splicing to identify novel platelet eQTLs and sQTLs. rs6128 is a platelet sQTL that alters SELP exon 14 skipping and soluble versus transmembrane P-selectin protein production.
INTRODUCTION
Platelets are abundant, accessible blood cells that are increasingly used in gene expression studies. Like nucleated cells, platelets possess a diverse portfolio of RNAs, including coding mRNAs, small non-coding RNAs, lncRNAs, and others1–3. Yet their anucleate nature offers unique advantages over nucleated cells for studying gene expression. For one, ex vivo handling (cell isolation method, processing time, buffers, etc.) of nucleated cells can immediately affect the expression of thousands of transcripts4–6. On the other hand, platelets are transcriptionally unaffected by isolation7,8, allowing capture of the native in vivo gene expression signature. These attractive features render platelets an excellent choice for RNA diagnostics and gene expression studies.
Platelets have been used in GWAS, gene-phenotype9,10, diagnostic11, and differential expression studies with an emphasis on RNA abundance to elucidate mediators of platelet reactivity in health and disease12. Genetic modifiers of RNA abundance in platelets, called expression quantitative trait loci (eQTL), have also been described13,14. eQTLs are DNA sequence variants associated with gene expression that affect nearby (cis-) or remote (trans-) genes in a cell type specific manner. eQTLs are particularly important in genetic studies because they provide an intermediate and mechanistic link between a phenotype and gene association.
Beyond RNA abundance, it is now known that platelets and megakaryocytes harbor alternative structural features of RNA, including alternative start and stop sites, and alternative splicing12, that diversify the transcriptome and proteome15,16 and alter cellular function. In platelets, activation induces RNA splicing, and thereby modulates functional protein expression15,17. It is probable that genetic variants called splice QTLs (sQTLs) also influence basal and activation dependent RNA splicing levels in platelets. However, sQTLs for platelets have not yet been described.
Other major knowledge gaps exist regarding RNA abundance and structure in platelets. Nearly all platelet studies have been cross-sectional, examining gene expression at a single time point. Yet, gene expression can vary both between-individuals and within-individuals over time. Hormonal changes, circadian rhythm, inflammation, diet, and aging are examples of environmental cues that might alter gene expression within healthy individuals18–20. Such normal changes in gene expression can mask the ability to detect signal in differential gene expression, diagnostic, and genetic studies, and confound their analysis. Thus, understanding within individual versus between individual variation in gene expression is important for the design and interpretation of gene expression studies, and can be used to prioritize candidates in genetic studies.
With regards to genetic studies, several reports have suggested using repeatability to identify eQTL genes21–23. In vivo repeatability can only be calculated from multiple samplings from the same individual, and refers to the proportion of variation attributed to between-individual versus within-individual variation24. In the straightforward view, repeatability sets an upper-bound to broad sense heritability25: if between individual differences are not repeatable because of low between, and/or high within-individual variation, there will be insufficient power to detect heritable, genetic signal. For this reason, it has been recommended to measure the repeatability of a trait before performing GWAS21. Carlborg et. al.22 found that censoring mouse eQTL data on repeatability was an effective method for prioritizing transcripts with a high a priori likelihood of successful eQTL identification. Hoffman et. al.23 also demonstrated the potential use of within-individual technical variation to narrow candidates and facilitate eQTL prediction, although this study employed only single time point replicates. Together, these studies imply that longitudinal analysis of gene expression may facilitate prospective eQTL (and sQTL) gene discovery. Surprisingly, longitudinal analyses of gene expression are scarce in primary cells from healthy individuals, and absent for platelets. To our knowledge, the repeatability of alternative splicing over time has not been established for any primary cell type.
In this study, we use longitudinal RNA-seq analysis to examine within (intra-) and between (inter-) individual variability of the human platelet transcriptome. We examine repeatability of gene abundance and exon skipping, a readily measured alternative splicing event. We retrospectively demonstrate the use of repeatability to decipher heritable signal from environmental noise, and identify eQTL genes. We prospectively use repeatability to prioritize eQTL and sQTL gene candidates for novel platelet eQTL and sQTL discovery.
METHODS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Human subjects and Platelet isolation.
Subjects were independently recruited from the University of Utah and Duke University (Table 1).
Table 1.
Characteristics of cohorts 1 and 2 and timeline of platelet collection.
| Cohort 1 (=31) | Cohort 2 (n=7) | |
|---|---|---|
| Center | Duke University | University of Utah |
| Age (yrs) | 42 ± 11 | 47 ± 9.1 |
| Male Gender, n (%) | 10 (32%) | 5 (71%) |
| Race/Ethnicity, n (%) | ||
 Black or African American Black or African American | 13 (42%) | 0% |
 Caucasian Caucasian | 15 (48%) | 7 (100%) |
 Unknown or Not Reported Unknown or Not Reported | 3 (10%) | 0% |
| Platelet RNA-sequencing | ||
  | ||
All subjects provided informed consent and this study was IRB approved by each institution. Subjects were healthy and without active medical conditions. No subjects had undergone surgery for the past 4 months. Any illness required resolution of symptoms for at least 7 days prior to sampling. Cohort 2 subjects were prospectively recruited. Cohort 1 subjects were part of a clinical study where they were previously exposed to aspirin, however no subjects were on aspirin for at least 4 weeks prior to each blood sampling. Blood was drawn by venipuncture into citrate tubes (cohort 1) or acid-citrate-dextrose (cohort 2) and sample processing initiated within 30 minutes of phlebotomy. Platelets were isolated via magnetic leukocyte depletion using CD45 microbeads (Miltenyi) as we have previously described1,2,26.
RNA isolation and sequencing.
RNA was isolated using phenol-chloroform extraction (cohort 1) as previously described1 or DirectZol kit (cohort 2, Zymogen). More details on RNA isolation are found in the Online Supplemental Methods. Sequencing libraries for cohorts 1 and 2 were barcoded and prepared using kits: KAPA Stranded mRNA-Seq Kit (Roche #KK8421) and TruSeq unstranded v2 with poly(A) selection (Illumina #RS-122) respectively. Libraries were sequenced 50 cycles, single end, on Illumina HiSeq 4000 (cohort 1) or Illumina HiSeq 2000 (cohort 2) to a depth of ~20–40 million mapped reads per sample. Fastq files have been deposited in the NIH Sequence Read Archive PRJNA531691.
RNA-seq analysis.
For analysis of expression variation, reads were aligned to GRCh38/hg38 using Novoalign (Novocraft) as we have previously described1. Reads were assigned to flattened Ensembl gene annotations using the USeq analysis package27. Read counts were normalized separately for each cohort using the DESeq2 analysis package28. Non-coding RNAs were selected according to Ensembl transcript biotype. Heatmaps, clustering (complete linkage), density plots, boxplots, and scatterplots, were generated in R29. Read distribution plots were generated using integrative genomics viewer (IGV)30. Gene ontology enrichment was analyzed using DAVID31.
Analysis of individual transcript variation.
RNA-seq data has a strong mean-variance relationship. The DESeq2 regularized log transformation (RLD)28 was applied to counts which preferentially shrinks the overall variance among low abundant transcripts (while retaining outliers), thus allowing a more straightforward comparison of transcript variation across all expression levels. Lowest abundant transcripts were also arbitrarily removed. Within-individual variation was calculated as the standard deviation of all samples from the same individual. Total variation was calculated as the standard deviation of samples across all individuals in each cohort, thus within-individual variation is a sub-component of total individual variation. Sources of variance were quantified with a linear mixed model using the R package variancePartition23. Repeatability was calculated with the formula σb2/( σw2 + σb2) in the R package ‘heritability’32, including correction for sex in repeatability calculations for eQTL enrichment analysis and gene prioritization for eQTL and sQTL discovery.
Exon skipping analysis.
Exon skipping events were identified from triads of Exon/Exon and Exon/Intron junctions with > 5 reads per junction and the calculated percent spliced in (PSI; see Figure 5D) > .05 and < .95 in > 30% of samples. Junctions were excluded where the flanking Exon/Intron junction pairs varied by more than 10 fold in > 70% of samples. By this strategy we identified 245 exons (from 194 different transcripts) that were skipped in a significant fraction of the transcripts in some but not all samples.
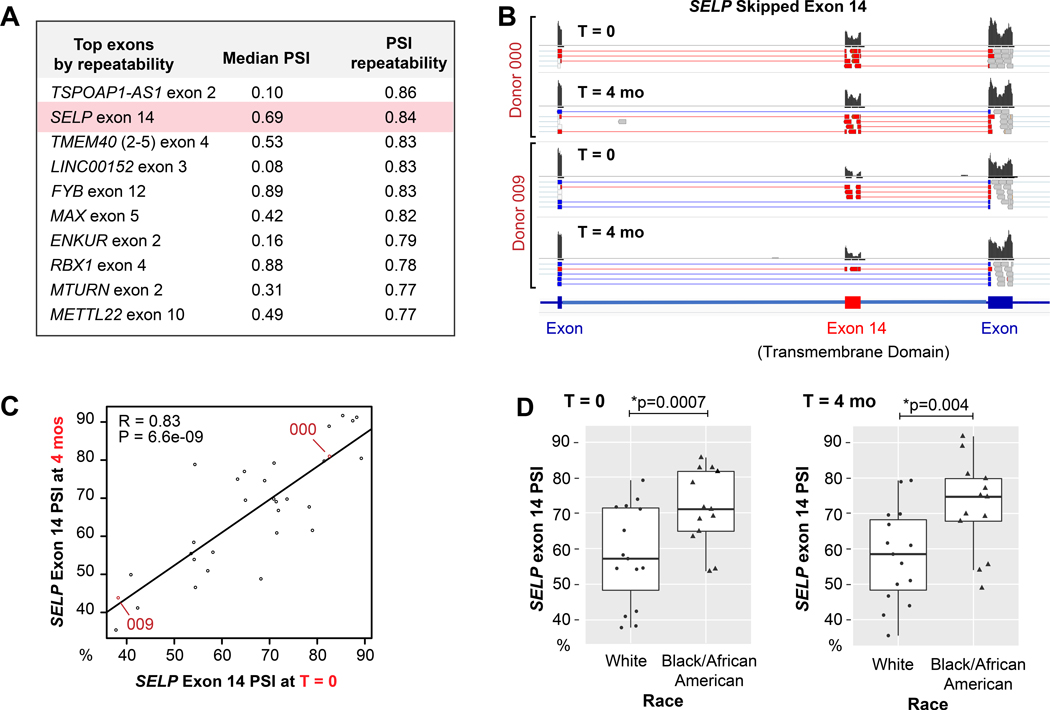
A: Table of the most repeatable exon skipping events in platelets. B: Representative IGV plots of sequencing reads from two different individuals at time 0 and 4 months, showing the differential distribution of reads between individuals that align to or skip exon 14 of SELP. The histograms indicate the cumulative abundance of reads that aligned to each exon. A subset of individual reads is shown below each histogram that indicate split splice junction reads by thin lines (absent in read) that connect to thick lines (mapped portion of read). Red and blue reads are splice junction reads that align to or skip exon 14 respectively. C: Correlation plot of SELP exon 14 PSI. Each point represents the PSI for an individual donor at time 0 (x-axis) and 4 months (y-axis). Donors represented in the IGV plots in B are labeled in red text. D) Boxplot of SELP exon 14 mean PSI according to race at T=0 and T=4 months.
SELP mini-gene.
The complete open reading frame for SELP transcript ENST00000263686.10 with a c-terminal DYK tag, and introns 13 and 14 that flank exon 14, were cloned into PCDNA-CMV and pCDH-MSCV-GFP vectors. A single nucleotide change C->T was made at rs6128. 293T cells (HEK 293T/17, ATCC CRL-11268) were maintained according to ATCC recommendations and used between passages 10–20. 293T cells were transfected with lipofectamine 2000. 24 hours after transfection, SELP RNA splicing was analyzed by PCR at cycles 25 and 30 using primers flanking exon 14 (5’-gtcaactaccgtgccaacct; 5’-taaggactcgggtcaaatgc). For flow cytometry experiments, cells were either co-transfected with a GFP plasmid (PCDNA-CMV) or GFP was contained within the same backbone as SELP (PCDH-MSCV). Surface expression of P-selectin was assessed by staining with Psel.KO2.3 APC antibody (ThermoFisher #17–0626-82) and analyzed by flow cytometry on a CytoFLEX analyzer (Beckman Coulter). MFI of P-selectin was normalized to GFP expression as assessed on live/transfected cells gated according to forward/side scatter (live) and FL-1 (GFP+) intensity. Soluble P-selectin was measured using a Quantikine ELISA kit (R&D Systems #DPSE00). For western blots, cells were lysed with RIPA, lysates denatured and reduced, proteins separated using an 10% SDS-PAGE, transferred onto a PVDF membrane, and blotted for tagged P-selectin with anti-DYK antibody (Cell Signaling Tech. #2368S), followed by anti-rabbit HRP secondary antibody (Rockland #18–8816-33) and chemiluminescent detection (ThermoFisher #34580).
Novel platelet eQTL and sQTL analysis.
RNA-seq fastq files for 234 previously published samples (Best et. al.8,33, Netherlands cohort; hereafter referred to as NL cohort) were retrieved from NCBI short read archive PRJNA35358833. These, and fastq files for cohort 1, were aligned with STAR34 to human reference genome (build HG38) in a splice-aware manner, and variants were called and filtered using the workflow built from the Genome Analysis Toolkit (GATK)35 best practices for variant calling on RNA-seq. Additional details on variant calls, filters used, and a note on the caveats and limitations of RNA-seq based variant analysis are included in the Online Supplemental Methods. Variants tested for eQTLs were limited to within 2 kb of all genes not identified as eQTLs by the previously published PRAX113 dataset, and with repeatability > 0.9 (238 genes) in the cohort 1 dataset. For comparison, the equivalent number of genes with lowest repeatability were also included. Combined filtering resulted in 641 variants across 181 genes that were tested for gene abundance–variant association. Gene-variant association was tested in the R package SNPassoc36 using an additive model of variant-allele dosage (0,1,2), while controlling for the covariates sex, age, and population structure37,38 (see Online Figure X and Online Supplemental Methods for details). Benjamini and Hochberg FDR correction for multiple testing (641 gene-variant tests) is reported. However, a conservative significance threshold of p < 1e-6 was used to filter novel eQTLs as if genome wide analysis had been performed13. Allelic imbalance of each significant eQTL was evaluated with the Wilcoxon rank sum test on the proportion of reference variant reads to total reads in each heterozygote individual.
A generalized linear model was used for logistic regression analysis in R to test the association of SELP exon 14 splicing and self-reported race or rs6128 variant-allele dosage. For this, SELP exon 14 inclusion counts versus total inclusion + exclusion counts were used as the binomial response variable. Where specified, models controlled for potential covariates including race or population structure, rs6128 genotype, sex, and age.
Statistics.
Significance for multimodality was calculated according to Hartigan’s dip test statistic. Wilcoxon test with adjustment for multiple comparisons was used to test for similarity in distributions of between and within sample correlations. Kolmogorov–Smirnov test was used to test for enrichment in the rank of genes associated with genetic/heritable traits. Gene Set Enrichment Analysis (GSEA)39 pre-ranked was used to evaluate enrichment among genes tested for the presence of significant eQTLs as identified in the PRAX1 cohort. For this, only the subset of genes tested by PRAX1 were included. The association between eQTL presence and different repeatability thresholds was estimated with odds ratio (odds at each threshold compared to no threshold) and significance evaluated with Chi-square test of independence. Two sided T-tests and correlation tests for significance (alpha = 0.05) were performed in R29 using functions cor.test and t.test, which sets a lower p value limit of 2.2e-16.
RESULTS
To assess within and between individual variation of the platelet transcriptome, we performed RNA-seq analysis of leuko-depleted platelets isolated longitudinally from two independent cohorts of healthy individuals. For cohort 1, we analyzed platelets from 31 individuals at an initial visit (T0) and 4 months later. For cohort 2 we analyzed platelets from 7 individuals at T0 and then longitudinally over 4 years. The characteristics of the two cohorts are detailed in Table 1.
Platelets contain a stable within-individual gene expression signature.
Unsupervised clustering analysis of all pair-wise distances within and between individual transcriptomes in cohort 1 indicated a robust within-individual (self) RNA expression signature (Figure 1A), with most self-pairs clustering as nearest neighbor pairs. As depicted in Figures 1B--C,C, the mean within-individual correlation of platelet transcriptomes isolated 4 months apart was very high (r mean±sd=0.987±0.012). Between-individual correlations of the global platelet transcriptome were also high (0.947±0.024), but significantly lower (p < 2.2e-16) than within-individual correlations. Grouping samples by race, age, and sex only partly corrected the difference (Figure 1C). Within individual clustering of non-coding RNAs was also robust (Online Figure I A), with a mean within-individual correlation of 0.984±0.013. The mean between individual correlation for non-protein coding transcripts (0.905±0.031, Online Figure I B-C) was significantly weaker compared to protein coding transcripts (p < 2.2e-16), which is consistent with previous cross-sectional studies40. Raw and normalized counts for each transcript in cohort 1 are found in Online Datasets I and II.
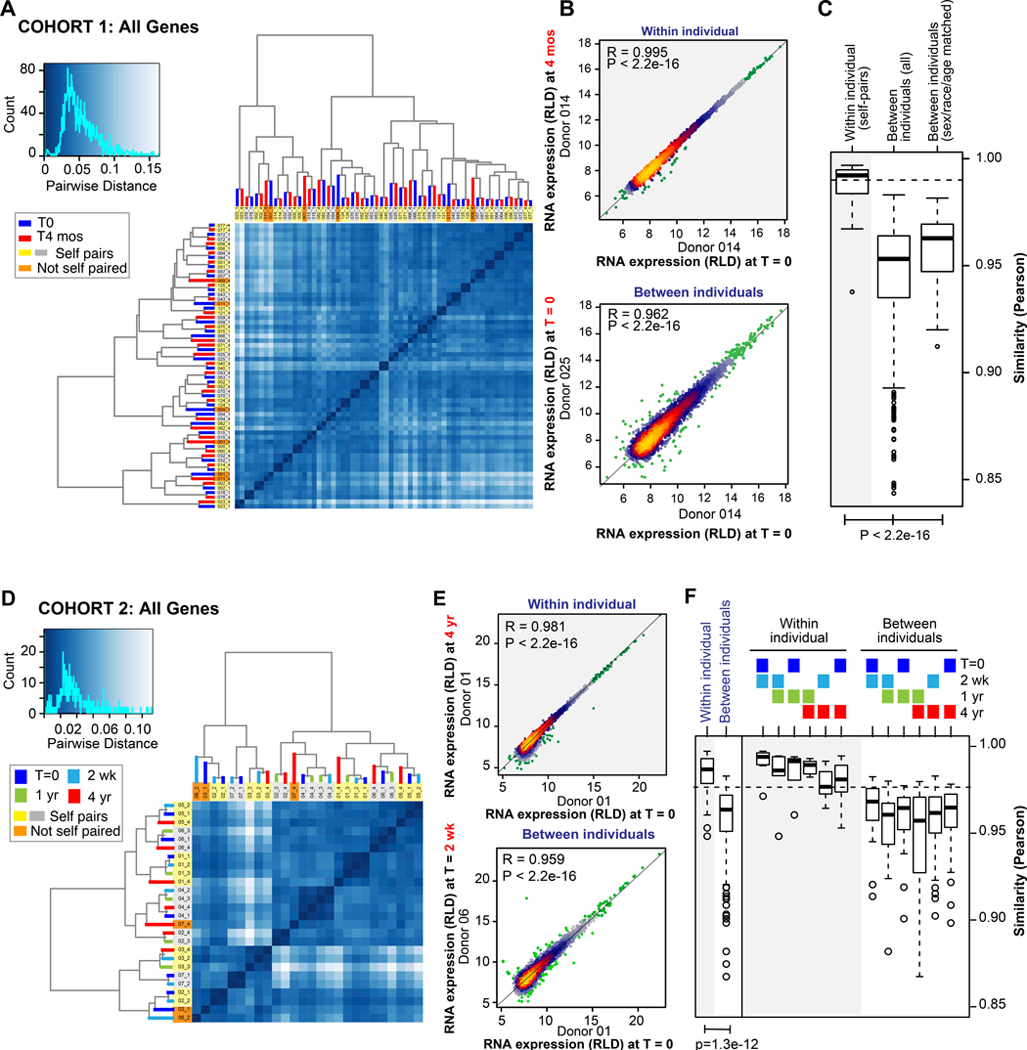
A and D: Unsupervised clustering and heatmaps of total RNA expression in platelets from all samples in A) cohort 1 and D) cohort 2. The histograms to the left of each heatmap show the distribution of distances between all pairs of samples, and the darkness of blue indicates the degree of similarity between pairs of samples. Samples that cluster as neighbors in the heatmap dendrograms reflect transcriptomes with the highest similarity. Nearest neighbor self-pairs are highlighted in yellow and gray, whereas nearest neighbor non-self pairs are highlighted in orange. B and E: Example individual correlation plots of all transcripts in B) cohort 1 or E) cohort 2. Each data point represents the regularized, log-transformed expression level (RLD) of a single transcript from the specified donor at time 0 (x axis) versus 0, 2 wk, 4 months, or 4 years (y axis) within the same individual (top panels) or a different individual (bottom panels). Points are heat-colored according to density. P values are from Pearson correlation. C and F: Boxplots summarizing the RNA expression Pearson correlation between all within versus between-individual pairs at C) time 0 and 4 months or F) in aggregate at all time points (left) or at the individually specified time points (right). With regards to specified time points in F, note that the average within-individual correlation did not significantly decrease as samples taken farther apart were compared. For example, there was not a significant difference when comparing the average within-individual correlation of T0 versus 2 weeks with the average within-individual correlation of T0 versus 4 years. Boxplots for cohort 1 (C) are shown before and after adjusting for age, sex, and race, whereas they are not adjusted for cohort 2 (F), because of the smaller sample size. P values are from Wilcox test, adjusted.
Unsupervised clustering of total RNA transcriptomes in cohort 2 resulted in robust self-clustering and suggested minimal transcriptional drift over 4 years (Figure 1D). Within-individual correlations for samples isolated 4 years apart remained comparable to within-individual correlations for samples isolated only 2 weeks apart, and significantly higher than between-individual correlations regardless of time-point (Figure 1E--F).F). As shown in Online Figure I D, the within-self non-coding RNA signature was also robust, and uniquely identified individuals at all time points over the 4 years without exception—a reflection of the significantly higher within-self correlations in non-coding RNA expression compared to those between-individuals (Online Figure I E-F). Raw and normalized counts for each transcript in cohort 2 are found in Online Datasets III and IV.
Within and between individual transcript variation in platelets is reproducible across cohorts.
Together, the data in Figure 1 indicate similar patterns of gene expression variation between cohorts 1 and 2: the average within-individual correlations were similar (0.983±0.13 vs 0.987±0.12) for each cohort, as were the average between-individual correlations (0.958±0.021 vs 0.947±0.024). We further defined the specific transcripts in each cohort that displayed the least and most overall (total) variation compared to within individual variation (see Online Datasets V-VI). As depicted in Figure 2A and Online Figure II A-B, high within individual variation was limited to a small number of moderately expressed transcripts that were consistently variable in both cohorts 1 and 2. Transcripts that varied the most within individuals in both cohorts were enriched for those within the inflammatory and defense response gene ontologies (GO; Figure 2B)18,41. As shown in Figure 2C and Online Figure II C-D, transcripts with high total variation spanned a broader range of expression levels, yet the extent of variation was still consistent between cohorts 1 and 2. The transcripts with the highest total variation predominantly overlapped those that varied the most within-individuals (Online Figure II E-F) leading to an enrichment in the inflammatory and defense response GO (Figure 2D). In addition to inflammatory transcripts, we noted several genes with high total variation that were previously associated with sex, race, or platelet eQTLs10,13,42. However, unlike the inflammatory transcripts, most of these did not demonstrate high within individual variation (see bottom right quadrants of Online Figure II E-F).

The mean within and total individual variation (standard deviation, SD) was calculated from the regularized log transformed expression (RLD) for each transcript. A and C: the A) within or C) total individual variation of each transcript in cohort 1 (x-axis) plotted against the respective variation of each transcript in cohort 2 (y-axis). The horizontal and vertical lines at 0.5 marks an arbitrary threshold of variation used for the Venn diagrams in B and D. B and D: Venn diagrams of the overlap in the transcripts with highest B) within and D) total variation. Listed below each Venn are the significantly enriched GO terms for the transcripts overlapping both cohorts. FDR = Benjamini False Discovery Rate calculated by David65.
Variance partition analysis23 was used to further partition and quantify for each gene the amount of variation attributable to sex, race, and other covariates, and to decouple between from within and total variation. As shown in Online Figure III, for more than half of all genes, between individual variation was responsible for the majority (>50%) of gene expression variation, followed by residual within individual variation. On the other hand, sex and race affected only a small number of genes. Other known covariates including age and sample processing contributed to only a minor portion of variation for each gene.
Repeatability defines heritable platelet gene expression and predicts eQTL genes.
As discussed in the introduction, we hypothesized that repeatability, which captures the within and between individual variation in a single index (see methods), could be used to prioritize genes for eQTL analysis. Therefore, we calculated repeatability for each gene (Online Datasets V-VI), and retrospectively tested whether repeatability is associated with genetic and heritable regulation of gene expression in platelets. To do this we utilized the published PRAX113 dataset as an independent (no overlap with the current cohorts) and cross-platform (microarray) validation dataset. PRAX1 previously associated platelet transcripts with sex and race, and identified 612 platelet eQTL genes. We used cohort 1 for the analysis because it was larger in size than cohort 2 and better matched the diversity and demographics of PRAX1. We ranked all genes in the PRAX1 microarray according to the repeatability calculated by RNA-seq in cohort 1. Ranking by repeatability resulted in a significant enrichment for genes associated with sex, race, or eQTLs (p < 2e-16), that was significantly greater than ranking genes by abundance, within-individual, or total variation (p < 2e-6; also see Online Figure IV). As shown in Figure 3A, 100% of the top 15 repeatability ranked genes are significantly associated in expression with sex, race, or a cis-eQTL. As an example, MFN2 is an established platelet eQTL gene13 that was among the most repeatable genes (repeatability = 0.98). This is because MFN2 expression varies according to eQTL genotype13 by more than 8 fold between individuals, while remaining relatively constant within individuals over time (Figure 3B).
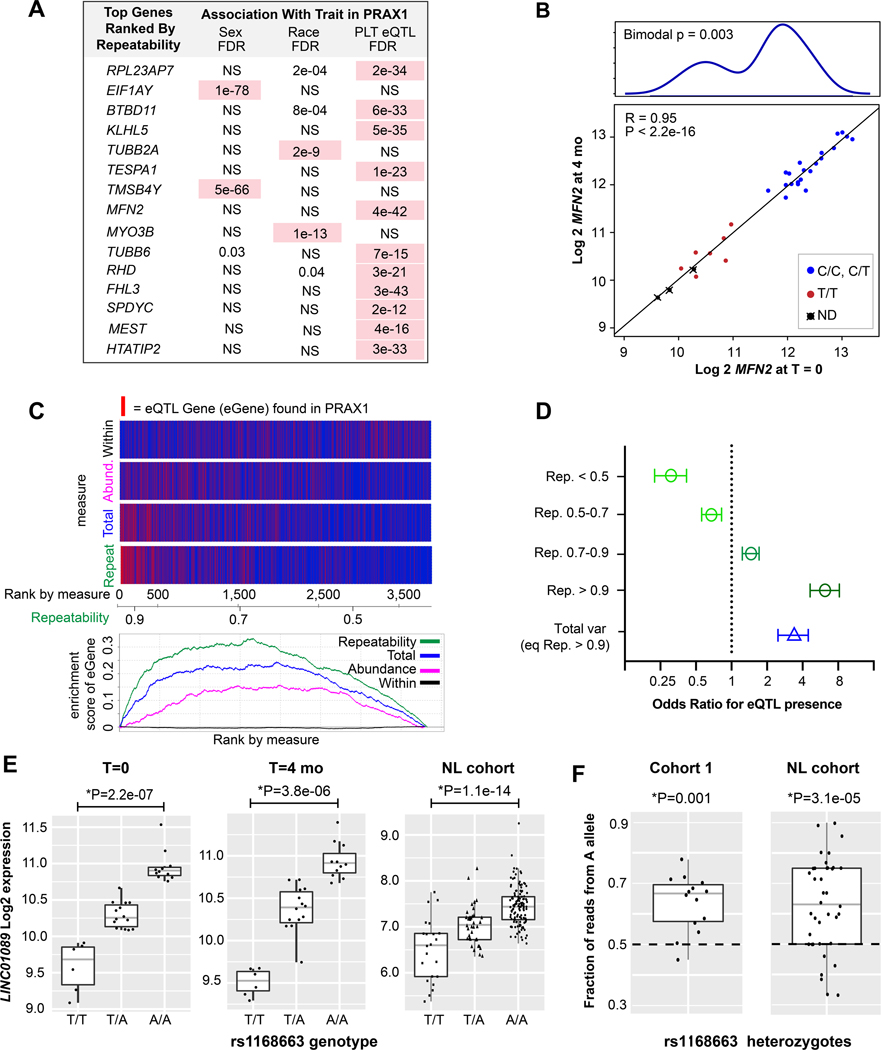
A) Table of transcripts with the highest repeatability in cohort 1 RNA-seq data, and their reported association with race, sex, or eQTLs in PRAX113,42 microarray data. Associations with FDR < 1e-4 are highlighted in pink. NS = not significant. B) Correlation plot of RNA expression (log normalized) of MFN2 at time 0 (x-axis) and 4 months (y-axis). Points are colored according to rs1474868 genotype (ND = not determined). Above is a density histogram showing a bimodal distribution according to genotype. Bimodal P value from Hartigan’s diptest for multi-modality. C) Top: enrichment plots for the presence of eQTLs ranked according to different measures: within variation, mean expression abundance, total variation, or repeatability. The axis below the plot indicates the gene rank according to each measure, and indicates the value of the repeatability measure (the values of the other measures are not noted on the axis). Genes with a known eQTL are in red, those without are in blue. Thus, genes with the highest repeatability are nearly 100% eQTL genes, whereas those with the lowest repeatability are nearly 0%. Bottom: plot of cumulative enrichment scores for each metric. D) Odds ratios for the likelihood of identifying an eQTL for genes at the indicated repeatability thresholds compared to the same number of genes ranked by total variation. E) Boxplots of LINC01089 expression according to rs1168863 genotype in cohort 1 at time 0 and 4 months and in the NL cohort33. *P values adjusted for age, sex, and cohort 1) race or cohort 2) population structure (inferred genetic ancestry37,38). F) Boxplots demonstrating allelic imbalance of rs1168863 within heterozygotes in cohort 1 and the NL cohort. The proportion of RNA-seq reads with A nucleotide versus T nucleotide was calculated and plotted for each heterozygote individual.
When assessing specifically the enrichment for eQTL genes, GSEA indicated a significant enrichment (p = 0) of known eQTLs as repeatability increased, with those with repeatability >0.68 (leading edge) accounting for the enrichment (Figure 3C). Significant enrichment was also observed when ranking by mean expression abundance or total variation, although the enrichment score for these measures was lower than for repeatability. According to binned analysis of odds ratios, the odds of identifying an eQTL for a gene with a repeatability <0.5 is 3.2 fold lower (p = 5e-15) than testing a gene at random (Figure 3D). The odds of identifying an eQTL for a gene with a repeatability > 0.9 is 6.2 fold higher (p = 6e-45) than random and 1.8 fold higher (p=0.006, adjusted) than ranking according to total variation. Furthermore, for known platelet cis-eQTLs, there was a significant correlation between the eQTL FDR and the repeatability of its associated transcript (Online Figure V). Thus, repeatability indicates an enrichment for and strength of cis-eQTL signal in platelets and may be a useful filtering and prioritization strategy to identify genes with an eQTL signal.
Microarrays differ from RNA-seq in accuracy, sensitivity, and comprehensiveness, and some eQTL genes identifiable by RNA-seq may have been missed by PRAX1. Therefore, we used RNA-seq data to re-interrogate 238 genes with high repeatability (>0.9), yet with no cis-eQTLs previously found. We tested these for cis-eQTLs using a publicly available dataset13,42 collected in the Netherlands, of platelet RNA-seq from 234 healthy individuals (NL cohort). Genetic variants were called from RNA-seq reads across each gene, and tested for association with RNA-seq abundance. Despite the known limitations of RNA-seq in calling genetic variants (e.g. most variants are located in promoters and introns), 11 new probable platelet eQTL genes (Online Dataset VII) were identified. In contrast, when analyzing the same number of genes with the lowest repeatability, we did not identify any additional eQTLs—a difference which was statistically significant (11/238 vs 0/238, p=0.0009, Fisher Exact). Allele Specific Expression (ASE) analysis of allelic imbalance, which measures the ratio of read counts coming from each allele within heterozygotes, confirmed a significant and directionally consistent within-sample eQTL effect on allelic imbalance for 8 of the 11 genes (Online Dataset VII). An example of one of the novel platelet eQTL genes is long non-coding RNA LINC01089—one of the most repeatable (0.95) and abundant (top 10% by RNA-seq) transcripts in platelets. As shown in Figure 3E, there is a strong additive allele dosage effect of rs1168663 on LINC01089 expression among cohort 1 individuals at both time points, and among individuals in the NL cohort. As shown in Figure 3F, LINC01089 expression demonstrates significant allelic imbalance in cohort 1 and in the NL cohort. Together these data define several novel platelet cis-eQTLs, demonstrating the utility of repeatability from longitudinal expression data to predict heritable gene expression variation and prioritize targets for prospective identification of cis-eQTL genes.
Alternative exon skipping in platelets is maintained within-individuals over time.
To assess within-versus between-individual stability of alternative splicing, we focused on an alternative splicing event that is readily measured in RNA-seq data: exon skipping. We stringently identified exon skipping events in cohort 1 (see methods), and calculated Percent of exon Spliced In (PSI; Figure 4A) for each (Online Datasets VIII-XI). As shown in Figure 4B, there was a broad range of exon skipping levels among the different exon skipping events that was mostly consistent between individuals and within-individuals over time. Unsupervised clustering analysis using PSI resulted in a preference for within-individual clustering compared to between-individual clustering (Online figure VI), although this was not as robust as clustering based on expression. Nonetheless, the within-individual correlation of PSI was significantly higher than between-individual correlation, independent of age, race, or sex (Figure 4B--C),C), suggesting a heritable component of exon skipping levels in platelets.
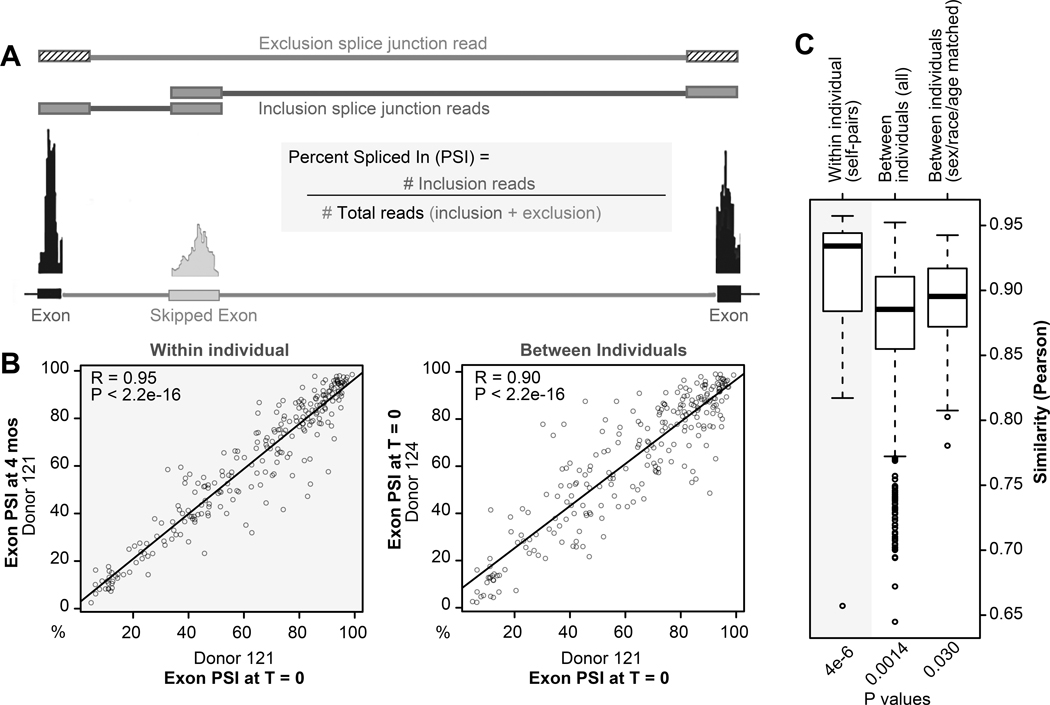
A) Schematic of how exon skipping events are defined. Percent exon Spliced In (PSI) is calculated using splice junction reads and is the ratio of exon inclusion junction reads over total junction reads. B) Correlation plots the PSI for all exon skipping events within (left panel) and between (right panel) individuals. Each point represents a single exon skipping event from the specified donor at time 0 (x axis) versus 0 or 4 months (y axis). C) Boxplots summarizing Pearson correlations of within versus between-individual pairs when analyzing PSI of all exon skipping events at time 0 and 4 months, and after adjusting for age, sex, and race. *Wilcox test, adjusted.
Identification of a platelet splice QTL associated with race that affects exon 14 skipping in SELP.
Unlike eQTLs, platelet sQTLs have not previously been identified. We therefore used repeatability to prioritize exon skipping events, with the goal of identifying novel, robust, and physiologically relevant platelet cis-sQTLs. To this end, we assessed the within/between individual variation of PSI for each exon skipping event, and ranked each by repeatability (Online Dataset VIII for full table). As shown in Figure 5A, Exon 14 of SELP, which codes the leukocyte adhesion and platelet activation marker P-selectin, ranked second among repeatable exon skipping events. The difference in exon 14 exon skipping between donors, but stability within individuals is illustrated by the alignment plots in Figure 5B and the correlation plot in Figure 5C.
Exon 14 skipping predicts an in-frame deletion of the transmembrane domain of P-selectin. An exon 14 deficient isoform of P-selectin was previously detected in endothelial cells and platelets43–45 and at significant levels in the human circulation44–46. PCR (Online Figure VII), cloning, and Sanger sequencing (data not shown) verified that the RNA isoform predicted by RNA-seq in our own cohorts matches the previously described soluble protein isoform in plasma. Together, this implicates exon 14 skipping as a heritable source of variability in P-selectin protein cell surface and soluble plasma levels between individuals.
Previous studies have associated soluble P-selectin in the plasma with a variety of clinical and genetic factors including race and single nucleotide polymorphisms (SNPs)47–50. As shown in Figure 5D, we observed a significant increase of SELP exon 14 inclusion among blacks/African Americans compared to whites at both time points. A search for the most likely responsible genetic variants identified a SNP, rs6128, within exon 14 of SELP with a homozygous MAF (T/T) that is much higher among Africans (0.29) compared to Europeans (0.04)51. Intriguingly, rs6128 has been associated with plasma P-selectin52 levels and diabetic retinopathy, especially among African Americans50. However, the direct relationship of rs6128 to soluble P-selectin is unclear since the C to T transition does not change the protein sequence or modify canonical splice sites. Bioinformatic analysis53 of the sequence surrounding rs6128 predicted a net loss of an exonic splicing silencer (ESS) motif and a net gain of 2 exonic splicing enhancer motifs (ESE) (Figure 6A). To determine whether rs6128 is associated with SELP exon 14 splicing in platelets, we inferred rs6128 genotypes from RNA-seq reads in cohort 1 and the NL cohort33 (variant details are in Online Dataset XII). As shown in Figure 6B--C,C, the rs6128 SNP is significantly associated in both cohorts with SELP exon 14 skipping in platelets. The association of rs6128 with SELP exon 14 skipping was independent of age, sex, race, or population structure.
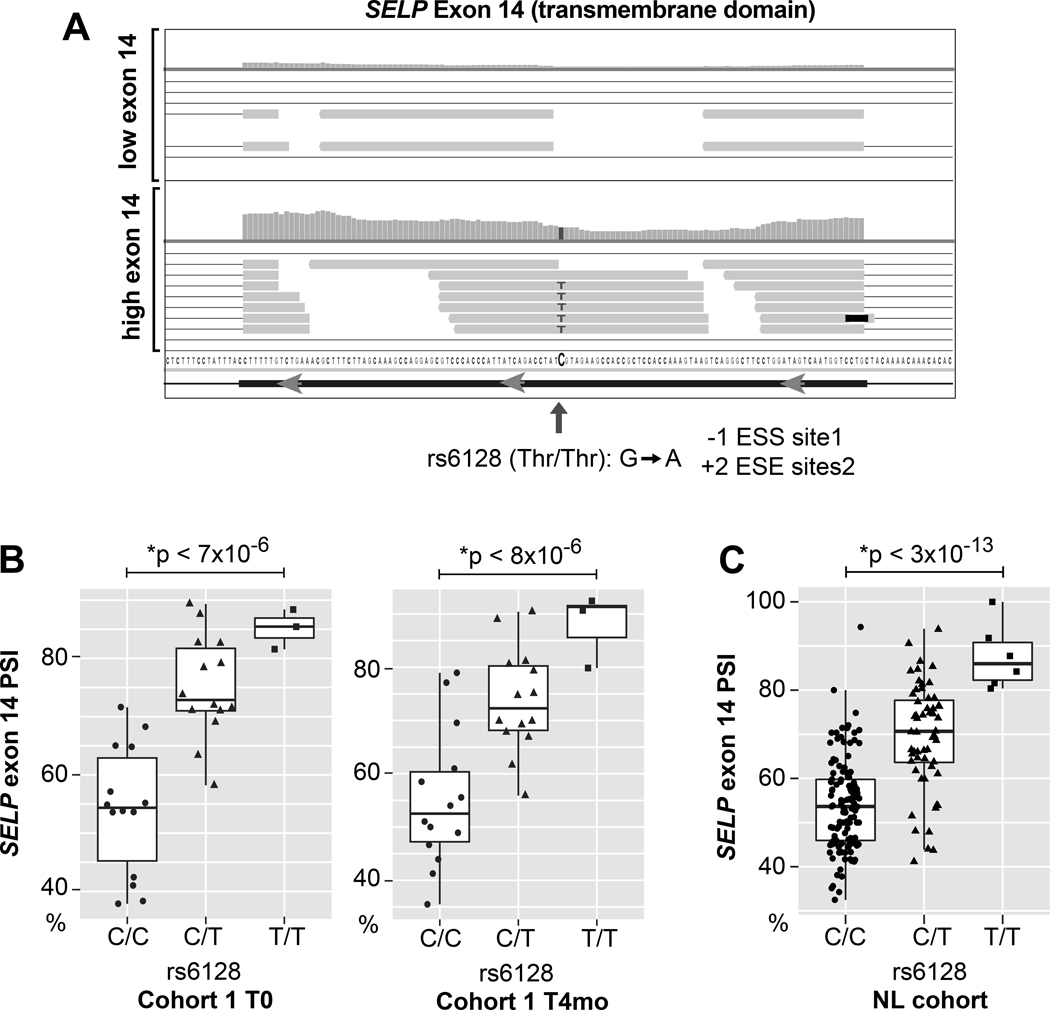
A) Close up IGV plot showing read distribution across SELP exon 14 for Top) an individual with rs6128 A/A and relatively high levels of exon skipping reads or Bottom) an individual with rs6128 (T/T) variant. The C->T change does not change amino acid sequence, but alters exonic splicing silencer and enhancer sites as predicted by Ex-Skip53. B) Boxplot of SELP exon 14 mean PSI according to rs6128 genotype inferred from RNA-seq in cohort 1 at time 0 and 4 months. C) Boxplot of SELP exon 14 mean PSI according to rs6128 genotype inferred from RNA-seq data published in the NL cohort. *P values adjusted for age, sex, and B) race or C) population structure (inferred genetic ancestry37,38).
We tested whether the difference in rs6128 MAF between Africans and Europeans might account for the association of exon 14 skipping with race indicated in Figure 5D. Consistent with this, there was no difference in exon 14 skipping levels between blacks/African Americans and whites after correcting for rs6128 (p = .4 and .3 for T=0 and T=4 months respectively).
Given the importance of P-selectin in disease, we extended the SELP exon 14 splicing analysis to additional diseases also available through Best et. al.8,33. As shown in Online Figure VIII, none of the diseases examined (non-small cell lung cancer, multiple sclerosis, or pulmonary hypertension) was significantly associated with SELP exon 14 splicing, or the effect of rs6128 on splicing. This indicates that the levels of exon 14 skipping are very stable within the individual, even in the context of environmental stressors which have been shown to trigger changes in platelet transcript abundance11,33.
Rs6128 directly affects SELP exon 14 skipping and the proportion of soluble to surface P-selectin in vitro.
Non-causal markers are commonly falsely identified in genetic association studies because of linkage with other unobserved variables54. To specifically test a causative effect of rs6128 on SELP exon 14 splicing, we generated mini-gene constructs of the SELP ORF with rs6128 C/C or T/T, and included introns flanking exon 14 (Figure 7A). Since it is known that promoter differences can influence splicing55, two different promoters (CMV or MSCV) were tested for each minigene construct. Constructs were expressed in HEK 293 cells, which lack endogenous P-selectin. Following transfection, RT-PCR analysis confirmed that the single nucleotide change from C/C to T/T resulted in a significant shift in the ratio of SELP RNA isoforms (Figure 7B), in the direction consistent with RNA-seq results. This occurred for both promoters, but with a more pronounced shift observed for the CMV promoter. Western blot analysis indicated that the T/T variant resulted in a shift toward exon 14 inclusion in P-selectin protein (Online Figure IX). As shown in Figure 7C, and consistent with inclusion of the transmembrane domain, the single nucleotide change from C/C to T/T significantly increased (2 fold) the amount of surface P-selectin on HEK 293 cells. In contrast, the T/T variant significantly decreased the amount of soluble P-selectin in the supernatant as measured by ELISA (Figure 7D). Together this data demonstrates a causal relationship between rs6128 genotype, the amount of exon 14 inclusion in SELP RNA, and the proportion of soluble to surface P-selectin expression.

A) Schematic of mini-gene constructs of SELP that include the ORF of SELP, and the introns flanking exon 14. The C/C and T/T constructs vary by a single nucleotide at rs6128. Constructs were cloned into vectors with 2 different promoters (CMV or MSCV). After transfection into HEK 293 cells, the introns are spliced out and exon 14 is variably spliced out (skipped). The extent of exon 14 skipping is measured by PCR via exon 14 flanking primers that generate two PCR products of different sizes. B) RT-PCR analysis of SELP exon 14 skipping following transfection of HEK 293 cells with rs6128 C/C or T/T vectors. Shown is a representative result from 5 independent experiments. Below are bar graphs and standard error summary of PSI calculated according to densitometry analysis of the exon 14 inclusion band (upper band) divided by the sum of the upper and lower bands (total).*paired t-test, n=5 independent experiments. C) Flow cytometry analysis of P-selectin surface expression following transfection of HEK 293 cells with rs6128 C/C or T/T vectors. Top is a representative histogram overlay of P-selectin surface expression 24 hours after transfection with CMV promoter empty vector, rs6128 C/C, or T/T. Below are bar graph and standard error summaries of the fold change (normalized to transfection) of surface P-selectin MFI following transfection. *paired T test, n=5–6 pairs per group. D) ELISA analysis of soluble P-selectin in supernatants of HEK 293 cells following transfection with rs6128 C/C or T/T vectors. *paired T test, n=12–14 pairs per group.
DISCUSSION
Analysis of within-individual versus between individual variation in two independent cohorts indicated that the human platelet transcriptome is highly stable in healthy individuals for up to 4 years. There are very few longitudinal studies available on primary nucleated cells for comparison. One study with similar design by Radich56 et. al. observed a 30% within-individual misclassification rate for leukocyte transcriptomes even after selecting for a gene signature that maximized variation between individuals. Although differences exist between this published study and our current work, we identified that platelets had a lower within-individual misclassification rate (10–12%) without signature selection. We speculate that their anucleate nature and 7–10 day lifespan moderate in vitro and in vivo RNA changes in platelets, promoting a stable and defined in vivo healthy gene expression signature.
Platelet gene expression profiling via RNA-sequencing is emerging as a relevant tool for platelet function studies, for defining the consequences and causes of disease, and for disease diagnostics8–10,12,14,33,57. However, almost all published studies to date, have relied on single-time point comparisons of the platelet transcriptome between a disease cohort and healthy subjects. As healthy subjects are often used as the “baseline” or “control” condition in these studies, understanding whether the platelet transcriptome is durable – or not – in health is critical to understanding the robustness of these comparisons. Our finding that the platelet transcriptome is generally stable over 4 years in healthy individuals lends validity to these comparisons. In depth analysis of the individual transcripts that do vary within and between individuals also suggest some limitations and caveats.
Although most transcripts were stable, a few transcripts varied substantially within individuals. Most within-variable transcripts were related to inflammation. These may inform studies evaluating the effects of inflammation on platelet gene expression, and may be of relevance to clinical findings that inflammatory stress is associated with platelet counts and function58. The range of variation for inflammatory transcripts in health compared to overt inflammatory disease may be worth investigating when assessing the impact of inflammatory gene changes on disease.
We observed major platelet expression differences between individuals. Sex and race accounted for some major differences. Other sources of individual variation contributed more. Our data suggest a prominent role for cis-eQTLs. Regardless of the source of variation, the propensity of a gene to vary between (or within) healthy individuals might be taken into consideration when interpreting differential disease-gene studies and designing experiments for validation. Genes with high inherent variation require larger sample sizes to reach statistical confidence. When sample sizes are small, false positives are more likely for genes with high inherent variation. Correction for known covariates might be helpful in this regard. While corrections for sex, race, and age are often considered in differential gene expression analyses, eQTLs are normally unavailable or ignored.
Stability information may also guide (along with the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE59) guidelines) the selection of reference genes for normalization controls. Within/between stable genes such as SYK, AKT1/2, GP1BA, ACTB might be good choices. On the other hand, highly within-variable genes, such as the gene TUBB1, should generally be avoided as reference genes.
As an application of our longitudinal dataset, we used repeatability as a filtering strategy to identify new platelet eQTLs. Because of the limitation of multiple testing60, even large scale gene expression association studies employ a filtering and prioritization strategy to circumvent testing thousands of genes (and even more alternative splice events) against millions of genetic loci. Common filtering strategies include hard filtering on abundance61 or total variance. However, filtering for total variance alone will also enrich for transcripts overly-influenced by technical or environmental noise. To avoid this, several studies21–23 (see introduction) have suggested using repeatability instead. Here we have used platelet longitudinal data to experimentally test this idea. We observed a significant enrichment for cis-eQTLs among the most repeatable genes that significantly improved the ability to identify eQTL genes compared to using abundance or total variation.
Although significantly enriched for eQTLs, the association with repeatability was not perfect. Cohort 1 was assayed on a different platform (RNA-seq vs microarray), was smaller (31 vs 154), and had similar, but not identical demographics to PRAX1 (race (Blacks/African Americans): 45% vs 42%, ns; sex (M): 49% vs 32%, ns; age: 42+/−11 vs 29+/−7, p < .05). Repeatability measured in the same samples used for eQTL analysis would presumably result in the best predictions. However, large longitudinal studies are often impractical because of the costs and challenges of repeated sampling. Moreover, repeatability can add a layer of confidence to eQTL results if measured in an independent cohort. For this, larger sample sizes that reflect the demographics and environment of the test cohorts will presumably fare better. Cohorts that are too small to capture genetic variation (i.e. low MAF eQTLs), or are subject to systematic environmental perturbations, will suffer from overall lower repeatability, and lack sensitivity to predict eQTLs. Additional studies are needed to determine how repeatability might be applied more generally in the analysis of additional datasets, cell types, and to handle gene-environment interactions.
We used an additional RNA-seq cohort (NL cohort) to test for unreported platelet eQTL genes among the most repeatable genes in cohort 1. Of the identified eQTLs, 22/27 (for 7/11 eQTL genes) have been reported in other tissues (The Genotype-Tissue Expression (GTEx) Project, see Online Dataset VII). Noteworthy among eQTL candidate genes is TECPR2, which was previously associated with platelet counts by GWAS62. Long non-coding RNA eQTL genes were also identified: LINC01089, MAGI2-AS3, KANSL1-AS1. Like these 3 genes, we found that non-coding RNAs are generally stable within individuals, yet more variable between individuals compared to protein-coding RNAs, suggesting more genetic diversity among non-coding RNAs. Long non-coding RNAs have gained attention for their multi-faceted ability to regulate gene expression, but are understudied in platelets. How diversity in expression of long non-coding RNAs relates to functional gene expression differences between individuals is a subject of interest for future investigation.
Repeatability was further applied to prioritize exon skipping events to find those with the greatest likelihood of identifying a biologically tractable sQTL signal. A robust association between rs6128 and SELP exon 14 skipping was identified. During the course of this work, a significant association between rs6128 splicing and exon 14 skipping was also identified in whole blood samples63, further strengthening the findings.
To establish causality, we performed transfection experiments in HEK 293 cells, which advantageously do not express endogenous SELP. The results strongly implicate rs6128 as causal for differential SELP exon 14 splicing in platelets, but do not rule out potential contributing effects of the endogenous promoter, or of additional associated or linked variants. The confirmation of a platelet observation in an unrelated cell line suggests rs6128 may affect SELP splicing in multiple tissues such as endothelial cells—another major producer of P-selectin.
Surface P-selectin mediates leukocyte interactions and inflammation, is involved in atherogenesis, and plays a role in tumor metastasis. Soluble P-selectin is a functionally and clinically relevant platelet protein in the circulation associated with a variety of diseases47,64. While a major source of soluble P-selectin is related to activation induced shedding, a significant amount is heritable. Associations between rs6128 and soluble P-selectin protein levels in plasma have been reported50,52. The observed effects of rs6128 on exon 14 SELP splicing and soluble versus surface P-selectin localization establishes a link between these observations. They may explain previous clinical studies that have associated rs6128 with plasma P-selectin and diabetic retinopathy in African Americans50. Finally, the finding that SELP is differentially spliced according to race may be therapeutically relevant in light of promising clinical trials that effectively used P-selectin blockade to treat pain crisis in Sickle Cell Anemia, a disease that predominantly affects individuals of African descent47.
Supplementary Material
315215_DataSets
315215 Uncut Gel Blots
315215 Online Supplement
ACKNOWLEDGEMENTS
We thank Brad Demarest for insights into statistics. We also thank Diana Lim for preparation of the figures, critical comments, and consultation regarding effective display of the images.
SOURCES OF FUNDING
This work was supported by HL066277 and HL112311 (ASW), AG040631, HL126547, and HL092161 (MTR), HL118049 (DV), HL144957, and GM103806 (JWR). HS was supported by a Post-Doctoral fellowship (0625098Y), a Beginning-Grant-in-Aid (09BG1A2250381) from the American Heart Association Western States Affiliate, and a Lichtenberg-Professorship from the Volkswagen Foundation. This investigation was supported by the University of Utah Population Health Research (PHR) Foundation, with funding in part from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant 5UL1TR001067–05 (formerly 8UL1TR000105 and UL1RR025764). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This material is the result of work supported with resources and the use of facilities at the George E. Wahlen VA Medical Center, Salt Lake City, Utah. The sponsor had no role in the design or preparation of paper. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal (V8) on 10/01/19.
Nonstandard Abbreviations and Acronyms:
| QTL | quantitative trait loci |
| eQTL | expression QTL |
| sQTL | splice QTL |
| GWAS | genome wide association study |
| ASE | allele specific expression |
| SNP | single nucleotide polymorphism |
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1161/circresaha.119.315215
Read article for free, from open access legal sources, via Unpaywall:
https://www.ahajournals.org/doi/pdf/10.1161/CIRCRESAHA.119.315215
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1161/circresaha.119.315215
Article citations
Whole genome sequencing based analysis of inflammation biomarkers in the Trans-Omics for Precision Medicine (TOPMed) consortium.
Hum Mol Genet, 33(16):1429-1441, 01 Aug 2024
Cited by: 0 articles | PMID: 38747556
A Platelet Reactivity ExpreSsion Score derived from patients with peripheral artery disease predicts cardiovascular risk.
Nat Commun, 15(1):6902, 20 Aug 2024
Cited by: 1 article | PMID: 39164233 | PMCID: PMC11336089
Prospective, international, multisite comparison of platelet isolation techniques for genome-wide transcriptomics: communication from the SSC of the ISTH.
J Thromb Haemost, 22(10):2922-2934, 03 Jul 2024
Cited by: 0 articles | PMID: 38969303
Mitofusin-2 Regulates Platelet Mitochondria and Function.
Circ Res, 134(2):143-161, 29 Dec 2023
Cited by: 4 articles | PMID: 38156445
Heterogeneity of platelets and their responses.
Res Pract Thromb Haemost, 8(2):102356, 01 Feb 2024
Cited by: 0 articles | PMID: 38666061 | PMCID: PMC11043642
Go to all (33) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject (2)
- (1 citation) BioProject - PRJNA531691
- (1 citation) BioProject - PRJNA353588
Ensembl Genome Browser
- (1 citation) Ensembl - ENST00000263686
SNPs (2)
- (3 citations) dbSNP - rs6128
- (1 citation) dbSNP - rs1168663
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genetic Effects on Transcriptome Profiles in Colon Epithelium Provide Functional Insights for Genetic Risk Loci.
Cell Mol Gastroenterol Hepatol, 12(1):181-197, 16 Feb 2021
Cited by: 18 articles | PMID: 33601062 | PMCID: PMC8102177
Regional Variation of Splicing QTLs in Human Brain.
Am J Hum Genet, 107(2):196-210, 25 Jun 2020
Cited by: 23 articles | PMID: 32589925 | PMCID: PMC7413857
Alu insertion variants alter mRNA splicing.
Nucleic Acids Res, 47(1):421-431, 01 Jan 2019
Cited by: 46 articles | PMID: 30418605 | PMCID: PMC6326789
Multiplexed primer extension sequencing: A targeted RNA-seq method that enables high-precision quantitation of mRNA splicing isoforms and rare pre-mRNA splicing intermediates.
Methods, 176:34-45, 21 May 2019
Cited by: 5 articles | PMID: 31121301 | PMCID: PMC6868320
Review Free full text in Europe PMC
Funding
Funders who supported this work.
CSRD VA (1)
Grant ID: I01 CX001696
NCATS NIH HHS (3)
Grant ID: UL1 TR001067
Grant ID: UL1 TR000105
Grant ID: UL1 TR002538
NCRR NIH HHS (1)
Grant ID: UL1 RR025764
NHLBI NIH HHS (10)
Grant ID: K23 HL092161
Grant ID: R01 HL066277
Grant ID: R35 HL145237
Grant ID: U54 HL112311
Grant ID: R01 HL102482
Grant ID: R01 HL141424
Grant ID: R01 HL144957
Grant ID: R01 HL118049
Grant ID: R01 HL126547
Grant ID: R01 HL142804
NIA NIH HHS (2)
Grant ID: R03 AG040631
Grant ID: K01 AG059892
NIGMS NIH HHS (1)
Grant ID: K01 GM103806