Abstract
Free full text

Dual Proteome-scale Networks Reveal Cell-specific Remodeling of the Human Interactome
SUMMARY
Thousands of interactions assemble proteins into modules that impart spatial and functional organization to the cellular proteome. Through affinity-purification mass spectrometry, we have created two proteome-scale, cell-line-specific interaction networks. The first, BioPlex 3.0, results from affinity purification of 10,128 human proteins – half the proteome – in 293T cells and includes 118,162 interactions among 14,586 proteins. The second results from 5,522 immunoprecipitations in HCT116 cells. These networks model the interactome whose structure encodes protein function, localization, and complex membership. Comparison across cell lines validates thousands of interactions and reveals extensive customization. While shared interactions reside in core complexes and involve essential proteins, cell-specific interactions link these complexes, ‘rewiring’ subnetworks within each cell’s interactome. Interactions covary among proteins of shared function as the proteome remodels to produce each cell’s phenotype. Viewable interactively online through BioPlexExplorer, these networks define principles of proteome organization and enable unknown protein characterization.
Abstract
Comparative analysis of large scale protein-protein interactions across two cell lines highlights context-specific interactions and proteome-scale shifts in how functional networks are arranged.
INTRODUCTION
While a cell’s genetic inheritance is fixed, its proteome adapts to external and internal cues, fostering great diversity of form and function that drives multicellular life. Myriad physical interactions assemble proteins into modules that impart spatial and functional organization and define the interactome, a network whose topology encodes each protein’s cellular environment and whose structure varies with cell state. Defining the repertoire of protein interactions and the conditions in which they occur is thus essential to understand proteome and cellular diversity.
Despite its importance, a complete map of the human interactome remains elusive due to several challenges: 1) the innumerable proteins, isoforms, and post-translational states within the proteome; 2) the biochemical properties of individual proteins; 3) variable protein expression; 4) the prevalence of transient interactions; and 5) interaction context dependence. Existing interaction profiling methods have only partially addressed these challenges. Binary methods including yeast-two-hybrid assays excel at screening large libraries, though interacting protein pairs must be detected in isolation within a foreign cellular environment (Rolland et al., 2014). Alternatively, co-fractionation detects protein interactions in native complexes, subject to limits of sensitivity, dynamic range, and resolution of fractionation (Havugimana et al., 2012; Wan et al., 2015). In contrast, affinity-purification mass spectrometry (AP-MS) enables enrichment and detection of even low-abundance proteins, though exogenous expression of tagged baits is required, and extensive sample preparation has limited scalability while precluding recovery of transient interactions (Gingras et al., 2007). Approaches that combine datasets (Drew et al., 2017, 2020) or mine literature (Oughtred et al., 2018) can compensate for limitations of individual approaches, though experimental context may be lost (Stacey et al., 2018). Thus, our view of the interactome remains static and fragmentary. While our understanding of interactome dynamics is especially limited, context-dependent interactions enable cells to adapt to variable environments and create the cellular diversity that drives tissue-specific function and disease susceptibility. Such conditional interactions combine with protein expression, localization, and post-translational modifications – the proteotype – to biochemically link genotype to phenotype.
Though no single methodology can overcome all limitations, AP-MS excels for profiling interactomes – including yeast (Gavin et al., 2002; Ho et al., 2002; Krogan et al., 2006), Drosophila (Guruharsha et al., 2011), and human (Hein et al., 2015) – due to its sensitivity and its ability to detect interactions in appropriate cellular contexts. Thus, we have established a robust AP-MS platform, generating BioPlex 1.0 and 2.0 (Huttlin et al., 2015, 2017). Here we present BioPlex 3.0, the most complete model of the human interactome to date, accompanied by a second interaction network acquired in HCT116 cells. Individually, each network encodes protein function and reveals fundamental principles of interactome organization. In tandem, these cell-specific interaction networks begin to reveal how interactomes vary with cellular state. Together, they depict shared and cell-specific modules with characteristic biological properties that often align with the unique phenotypes of each cell line. In combination, they enable biological discovery, revealing physical interactions and suggesting functions for thousands of proteins. Both networks are viewable interactively through BioPlex Explorer.
RESULTS
Interactome Profiling in Multiple Human Cell Lines
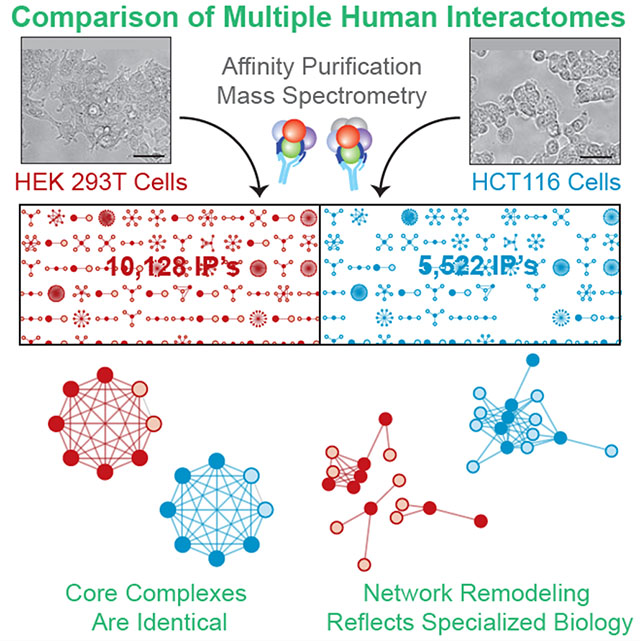
Previously we established a large-scale AP-MS platform to identify binding partners for affinity-tagged bait proteins following lentiviral expression in cultured cells. By expressing clones from the human ORFeome v. 8.1 (Yang et al., 2011) in HEK 293T cells derived from embryonic kidney tissue, we have profiled interactions for thousands of proteins to produce interaction networks of increasing scope (Figure 1A), including BioPlex 1.0 and 2.0 (Huttlin et al., 2015, 2017). These networks have been valuable for biological discovery, revealing tens of thousands of interactions that define the structural and functional organization of the interactome and afford biological context for thousands of uncharacterized proteins. Nevertheless, BioPlex incompletely models the interactome because i) only a fraction of human proteins has been profiled; and ii) although the interactome is dynamic, we have only profiled interactions in a single context.

(A) Our ongoing effort to map the human interactome has culminated in BioPlex 3.0, which builds upon two prior versions and incorporates AP-MS experiments targeting 10,128 bait proteins in 293T cells. Bars beneath each network reflect the fraction of proteins targeted as baits.
(B) We have repeated AP-MS analysis of 5,522 baits in HCT116 cells to produce a second proteome-scale interaction network. Bars beneath each network reflect the fraction of proteins targeted as baits.
(C) Our latest networks in 293T and HCT116 cells expand coverage beyond previous attempts. Y2H: yeast-two-hybrid. Circle size is proportional to interaction count.
(D) Comparison with BioGRID reveals that most BioPlex 3.0 interactions have not been previously reported. Incorporating PubMed citation counts for individual proteins suggests that much of the increased coverage comes from interactions among poorly studied proteins.
(E) Overlap among proteins in 293T and HCT116 networks.
(F) Overlap among interactions in 293T and HCT116 networks. A dashed box depicts the subset of 293T interactions matching those baits also targeted in HCT116 cells.
(G) Ternary diagram depicting the proportions of edges shared or unique to either 293T or HCT116 cells for subnetworks defined by 912 CORUM complexes. Interactions observed among proteins in each complex were extracted from the combined 293T/HCT116 network and tallied to determine numbers of edges shared or specific to each cell line. Four individual complexes are displayed as Venn diagrams; each is also represented as a point within the ternary diagram whose location reflects the relative proportions of shared and cell-specific edges. Points near the corners indicate that most edges are either shared (“Both”) or cell-specific; points near the center of the triangle indicate edges evenly distributed across shared and cell-specific categories. The ternary diagram thus summarizes Venn diagrams for 912 complexes. A box-whisker plot depicts the edge overlap across complexes.
(H) – (K) Subnetworks corresponding to four CORUM complexes highlighted in panel G.
To further enhance coverage, we have taken a bipartite approach. First, we have nearly doubled the number of baits profiled in 293T cells, completing AP-MS of all validated ORFeome v. 8.1 clones to produce BioPlex 3.0 (Figure 1A; Table S1A–B). This network, derived from 10,128 pulldowns targeting over half of human proteins, encompasses 70% of the known proteome (The UniProt Consortium, 2015). Coverage of many important protein classes is even higher, including cell fitness genes (90%) (Blomen et al., 2015; Wang et al., 2015), and kinases (85%) (kinase.com/web/current/human/). Moreover, BioPlex 3.0 contains most medically significant proteins, including 88% of cancer genes (Vogelstein et al., 2013), 65% of disease genes (Piñero et al., 2017), and 70% of drug targets (www.drugbank.ca).
Second, we have begun to repeat AP-MS of all baits in a second cell line. This effort promises large-scale validation across thousands of pulldowns and tens of thousands of interactions while producing a second proteome-scale, context-specific network as a step toward understanding interactome diversity. We have completed 5,522 pulldowns in colorectal carcinoma-derived HCT116 cells and created a second network that, while not yet as comprehensive as our 293T network, nevertheless affords a contrasting view of the interactome (Figure 1B; Table S1C–D).
Individually and in tandem, BioPlex 293T and HCT116 networks significantly increase our knowledge of human protein interactions. Compared to past efforts using yeast-two-hybrid (Luck et al., 2020; Rolland et al., 2014; Rual et al., 2005), correlation-profiling (Havugimana et al., 2012; Wan et al., 2015), AP-MS (Hein et al., 2015; Huttlin et al., 2015, 2017), and combinations thereof (Drew et al., 2017, 2020), both networks attain greater coverage, including more interactions and encompassing more of the proteome (Figure 1C).
By screening human proteins without regard to prior knowledge, we have surveyed large swaths of unexplored protein interaction space. For instance, just 9% of BioPlex edges have been previously reported and incorporated into the BioGRID database (Oughtred et al., 2018) (Figure 1D). Binning interactions according to PubMed citations for each protein reveals that those confirmed by other low- or high-throughput studies involve well-studied proteins. In contrast, BioPlex-specific interactions couple well-studied and unknown proteins alike, reflecting the sensitive, unbiased nature of our approach.
More fundamentally, creating two contrasting networks unveils context-specific interactome remodeling at proteome scale. Both networks contain similar proteins, with 67% shared (Figure 1E). Proteins detected in just one cell line are usually baits not yet targeted in the other. Though these networks share most proteins, overlap among interactions is modest, with 35,704 shared (Figure 1F, Table S1E). After removing IP’s not repeated in HCT116 cells, this represents ~50% overlap among remaining interactions.
Detecting different interactions in 293T and HCT116 cells is not unexpected. Though both cell lines grow robustly and readily express exogenous proteins, suiting them for large-scale AP-MS, they differ in sex, tissue of origin, karyotype, and driver modifications – significant variations in phenotype that can manifest in their interactomes. Nevertheless, contrasting interactions in these cells inspire several questions. To what extent do these differences reflect biological versus technical variation? What factors determine whether interactions are detected in both? How do shared and cell-specific interactions relate to the functions of their constituent proteins and to the unique phenotypes of each cell line? Finally, how do these networks enable biological discovery? Here we show that both networks faithfully model protein interactions in their respective contexts. Moreover, shared interactions often relate to essential functions while cell-specific interactions evoke unique biology of each cell line. Thus, our networks capture both core and specialized cell processes.
Internal and External Validation of the BioPlex Networks
To provide meaningful insights, our networks must faithfully model protein interactions. Since most interactions in each involve proteins lacking independent interaction data, validation has required several complementary strategies. To assess coverage of known complexes, each network was compared against CORUM (Giurgiu et al., 2018). In each network we expect subnetworks matching each CORUM complex to be highly interconnected relative to global network density (Figure S1A). Indeed, 75% of complexes detected in 293T cells are enriched, exceeding prior interaction datasets (Figure S1B). This includes prior BioPlex versions - as the network has grown, complex coverage has increased individually (Figure S1C) and collectively (Figure S1B). Likewise, the HCT116 network achieves coverage matching comparably sized BioPlex 2.0, meeting or exceeding other interaction networks.
Because CORUM complexes match just a fraction of BioPlex, we used other approaches to validate a larger share of its interactions. Since complex members at least partially co-elute via size exclusion chromatography, we correlated elution profiles from proteome co-fractionation (Heusel et al., 2019) for all interacting proteins in both networks. Strong correlations among these interacting proteins suggest that coelution confirms many interactions in both cell lines (Figure S1D). Similarly, interacting proteins must co-localize. Comparison of subcellular fractionation profiles (Orre et al., 2019) revealed that interacting proteins tend to co-purify (Figure S1E).
In addition, interactions may be confirmed within each network via reciprocal AP-MS of interacting protein pairs or repeated co-purification of protein complexes. As the fraction of baits has increased to 70% and 52% in our 293T and HCT116 networks, much of each network has become eligible for intranetwork confirmation. For reciprocal detection, both interacting proteins must be baits and must appear as preys in the relevant cell line. While technical factors sometimes prevent reciprocal detection, we observe reciprocity rates of 29% and 41% among eligible interactions in 293T and HCT116 networks (Figure S1H). Occurring far more often than null models would predict (Figure S1I), thousands of reciprocal interactions validate significant shares of each network and include associations among heterodimers (e.g. YWHAG – YWHAB; ERO1L – ERO1LB) and complex members, both direct (e.g. ARPC3 – ACTR3) and indirect (e.g. APC3 – ARPC5) (Goley and Welch, 2006), as well as pairings involving understudied proteins (e.g. ASF1B – C15orf41) (Figure S1F).
Analogously, complex co-purification may be assessed via 3-cliques: mutually interacting protein triads. Examples include SCF complexes, enolase subunits, and katanin subunits (Figure S1G). 3-cliques are also enriched in both networks (Figure S1J), reflecting modular architecture and frequent complex copurification. When both reciprocals and 3-cliques are considered, most edges (293T: 52%; HCT116: 54%) are confirmed by at least one additional IP within the same network.
While reciprocals and 3-cliques seek support for interactions among other IP’s within the same network, interactions may also be confirmed across cell lines. Remarkably, when all three are considered together, 57% of 293T interactions and 68% of HCT116 interactions are confirmed (Figure S1K).
As further validation, we repeated AP-MS of 999 baits in 293T cells (Figure S1L, Table S2A–B) and achieved a median 60% replication rate. Several replication profiles are displayed; high overlap was seen for many baits, including CASTOR1 (100%), C12orf34 (100%), and CDC20 (89%). When replication was lower, other BioPlex interactions often supported non-replicated edges. Examples include C12orf10 (25%), whose three non-replicated interactions are supported by pulldown of NOTCH2HL; similarly, though REG1B pulldown of REG1A did not replicate, the interaction was confirmed reciprocally. When these 999 293T replicates are compared with 72 HCT116 replicates (Table S2C–D) and 5,112 pulldowns in both lines (Table S2E–F), median clone-wise reproducibility rises to ~60% within the same cell line (Figure S1M), suggesting that cell context accounts for much inter-network variability.
Whether assessed through replication or inter- and intra-network confirmation, ~60% of interactions in both networks are confirmed. This compares favorably with prior AP-MS studies, particularly given the high-throughput nature of our work. Far smaller studies targeting protein families such as de-ubiquitinating enzymes (Sowa et al., 2009) or kinases (Varjosalo et al., 2013) have reported up to 70-80% replication after optimization. Since we find proteins average just 8 partners, our replication rate suggests loss of just 1-2 interactors per IP over 100-fold more baits from diverse protein families.
Another consideration is that both false positives and false negatives contribute to reproducibility, and false negatives may prevail in our dataset due to stringency of biochemical purification and statistical filtering. In fact, many non-replicated edges in Figure S1L are nonetheless confirmed by other IP’s. Because we are performing thousands of pulldowns, we must maintain a very low FDR – much lower than required of smaller studies – to avoid reporting thousands of spurious interactions. Our stringent approach accepts only ~2% of candidate interactions across all IP’s as high confidence interacting proteins. Though this high standard will cause us to miss some real interactions, it is essential for FDR control across thousands of IP’s.
To ensure accuracy of our cell line comparisons, we take three steps to focus on biological differences. First, interactions detected in one network at high stringency (top ~2%) are considered replicated if they are confirmed in the other network with relaxed stringency (top 5%). Second, rather than individual IP’s, our comparisons focus primarily on complexes, communities, and pathways: protein groups that have been immunopurified repeatedly targeting multiple baits with consistent results. Third, comparisons focus on interactions detectable in both cell lines, given the baits targeted in each.
In the following sections we survey the interaction landscape, focusing first on interactions shared between cell lines. Later, we explore cell-specific interactions that reveal extensive remodeling according to each cell’s unique physiology.
Core Protein Complexes Replicate in 293T and HCT116 Cells, Reflecting Complex Structure and Revealing Additional Complex Members
Although just 50% of interactions are shared between networks, replication is far higher within core protein complexes: among 912 drawn from the CORUM database, the median overlap is 100%, with most identical across cell lines (Figure 1G), including the exosome and COP9 signalosome (Figure 1I–J). That complexes such as these are unchanged is expected, as many are essential to cellular life. Their high conservation also complements prior observations (Gavin et al., 2006) that complexes often assemble from a central nexus of required proteins accompanied by additional peripheral proteins whose binding depends on context. In contrast, cell-specific complexes are often more specialized. For example, interactions among Fanconi Anemia complex members are largely HCT116-specific (Figure 1H), possibly reflecting differential DNA repair activity. Similarly, BCOR complex connectivity is reduced in HCT116 cells relative to 293T (Figure 1K), likely reflecting undetectable BCOR expression in the former cell line.
Detecting nearly identical interactions within several hundred well-known assemblies demonstrates the robustness of our AP-MS platform. Moreover, the specific interactions observed in these well-characterized complexes afford additional insight. To gain structural context we mapped BioPlex edges onto three-dimensional structures of complexes drawn from the Protein Data Bank (PDB) (Berman et al., 2000) (Table S3). Each structure was converted into an interaction network, assuming direct interactions among proteins separated by less than 6 Å, while inferring indirect interactions among other protein pairs. BioPlex edges were then overlaid as shown for U1 snRNP (Figure 2A). All direct interactions involving at least one bait were observed, along with many indirect interactions. Nearly all observable interactions were detected in both cell lines.
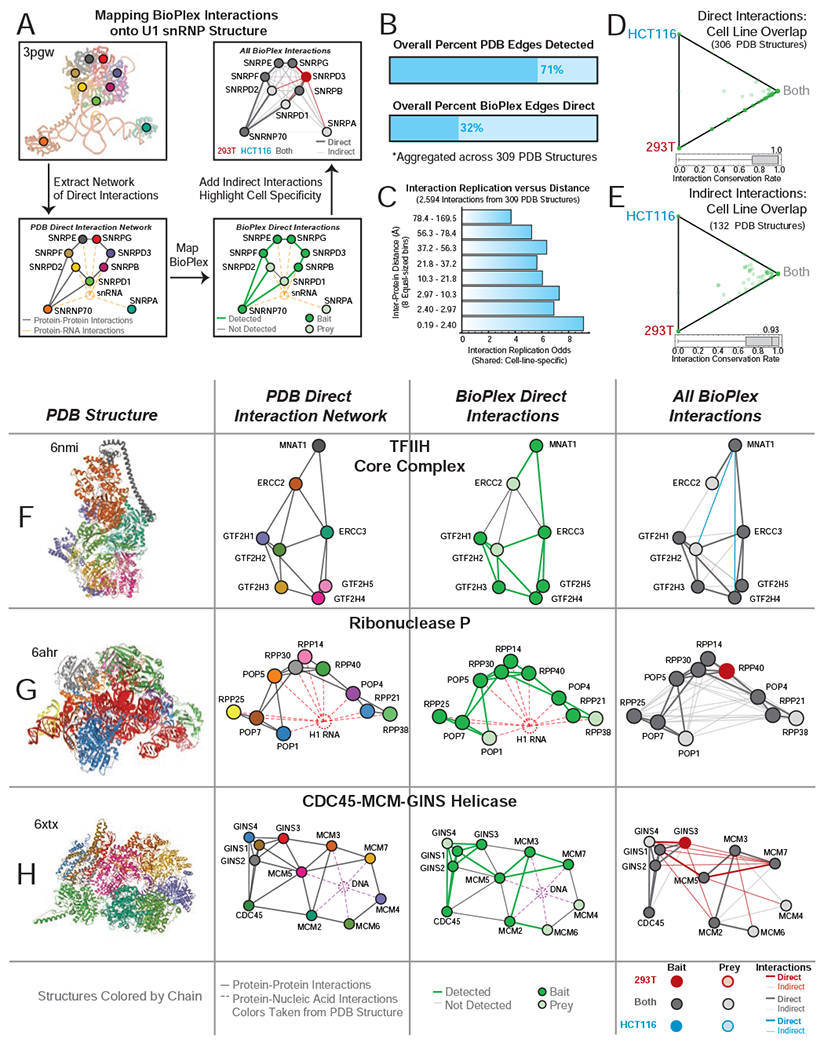
(A) Method for overlaying BioPlex interactions onto 3D structures and assessing detection and cellline specificity. See text for details.
(B) The fraction of observable interactions detected in BioPlex networks and the fraction of BioPlex interactions that match direct interactions. Results aggregated across 309 PDB structures.
(C) Relative odds that interactions are shared in 293T and HCT116 cells, versus cell-line-specific, as a function of inter-protein distance.
(D) Ternary diagram depicts sharing of direct interactions in 293T and HCT116 networks across 306 PDB structures.
(E) Ternary diagram depicts sharing of indirect interactions in 293T and HCT116 networks across 132 PDB structures with at least one indirect interaction.
(F) – (H) Selected complexes. Each structure is displayed (Column 1) along with a network visualization of all direct interactions in the structure (Column 2). BioPlex edges are then overlayed to show which direct interactions were detected (Column 3) and to show direct and indirect edges colored according to whether they were detected in 293T, HCT116, or both (Column 4).
Across 309 structures with ample coverage in BioPlex we observe 70% of all detectable edges in at least one cell line (Figure 2B). This aligns with replication and inter-/intra-network confirmation rates noted earlier (Figure S1K–M) while suggesting that false negatives may limit replication. Within these structures, 32% of BioPlex edges are direct and their propensity for detection in both networks decreases with increasing distance (Figure 2B–C). Interactions involving proteins separated by less than 2.4 Å are 9-fold more likely to be shared than cell-specific. Yet among the most distant protein pairs, shared interactions are favored only 4-fold. Thus, direct interactions are more often shared between cell lines than indirect interactions. In over half of 306 PDB structures, 100% of direct interactions are detected in both cell lines (Figure 2D); in contrast, among 132 PDB structures with indirect interactions, the median rate of indirect interaction replication is 93% (Figure 2E). This reduced propensity for indirect interactions to replicate across cell lines is both technical and biological, as interactions requiring one or more intermediate proteins are less likely to survive immunopurification and more likely to involve proteins subject to differential regulation.
Viewing intra-complex interactions in their spatial context can afford further insights. All direct interactions within Ribonuclease P are detected in both cell lines, accompanied by many indirect interactions (Figure 2G). Within the TFIIH complex, 75% of direct interactions are detected, all within both cell lines, while just a few indirect edges are cell-specific (Figure 2F). In contrast, many direct and indirect interactions within the CDC45-MCM-GINS Helicase are exclusive to 293T cells (Figure 2H). These 293T-specific edges bridge GINS-CDC45 and MCM subcomplexes detected in both cell lines. Though needed in both cell lines, the intact complex appears more readily purified from 293T cells, perhaps owing to differential abundance or stability (see below).
Since interactions within core complexes are usually consistent, we sought additional complex members that associate in both cell lines and found that the uncharacterized protein C11orf49 associates with the TTLL1 polyglutamylase complex. This complex resides in centrosomes where it polyglutamylates protein substrates, typically tubulin, modulating binding of microtubule-associated proteins (Janke et al., 2008). In both cell lines C11orf49 interacts with all complex members as well as pericentriolar marker protein PCM1 (Figure S2A). Since these interactions – detected reciprocally in both cell lines – strongly link C11orf49 to the polyglutamylase complex, we selected it for validation, demonstrating binding of endogenous C11orf49 to other polyglutamylase complex members (Figure S2B,C). Two C11orf49 isoforms localized to the pericentriolar region (Figure S2D–F), along with established polyglutamylase member LRRC49 and PCM1 – their mutual interacting partner and pericentriolar marker – in the vicinity of gamma-tubulin (Figure S2G). Since C11orf49 associates with a complex responsible for polyglutamylation, future inquiry into its role may afford key insights into the cellular function and regulation of this incompletely understood post-translational modification.
293T and HCT116 Cells Reveal Extensive Remodeling of the Human Interactome
While interactions within core complexes replicate at high rates, just 50% of observable interactions are detected in both networks. This is not unexpected, as these cell lines exhibit distinct phenotypes (Figure S3A), and quantitative proteomics (Erickson et al., 2019) revealed that 54% of proteins are differentially expressed (Figure S3B, Table S4A). While 293T-specific proteins were enriched for embryonic/nervous system development, evoking their embryonic kidney/adrenal origin (Stepanenko and Dmitrenko, 2015), proteins specific to colorectal-cancer-derived HCT116 cells were enriched for cell adhesion and cadherin binding. Marker proteins reflect the male status of HCT116 cells (EIF1AY), the potential for 293T cells to ciliate (CEP290) (Takahashi et al., 2018) and the contrasting epithelial (GRHL2, CDH1, LAMC2) and mesenchymal (TBX2, CDH2, VIM) origins of HCT116 and 293T cells (Figure S3C–F).
Cell-specific Interactions can Reflect Differential Bait and Prey Expression
Two sources of AP-MS variability are bait and prey expression. Since most proteins are differentially expressed between cell lines (Figure S3B), prey abundance will vary. Bait abundance also varies, with HCT116 cells achieving lower expression for 75% of baits (Figure S3G). These decreases in bait or prey abundance reduce median HCT116 replication rates, though variation is high (Figure S3H–I). Subnetworks surrounding 293T-specific proteins CDH2 and MYEF2 as well as HCT116-specific proteins FAM111B and RAC2 link differential expression with cell-specific interactions (Figure S3J–M).
Further insight emerges overlaying protein expression onto CORUM complexes (Figure 1H–K). Interactions within the Exosome and COP9 Signalosome are 100% replicated, and their constituent proteins express equally between cell lines (Figure S4B–C). In fact, expression variability is reduced within these complexes, reflecting that core components of essential complexes tend to express similarly (Romanov et al., 2019). In contrast, differential expression can contribute to cell specificity. Components of the HCT116-specific Fanconi Anemia Complex (Figure 1H) are expressed comparably in both cell lines (Figure S4A), while proteins BCOR and PCGF1 in the 293T-specific BCOR complex (Figure 1K) are elevated in 293T cells, likely driving cell specificity (Figure S4D). Similarly, 293T-specific CDC45-GINS-MCM helicase interactions (Figure 2H) may reflect elevated GINS1-4 and CDC45 abundance (Figure S4E). Nevertheless, while differential expression explains some interaction specificity, it does not explain most differences observed, as highlighted in Figure S4 for several additional examples described below.
This link between differential protein abundance and cell-specific interactions is partly biophysical: depending on the relevant dissociation constants, decreased bait or prey levels may reduce the likelihood of interaction while making detection more difficult, thus affecting reproducibility. Alternatively, both prey abundance and interactions may vary due to differential biology. In fact, several lines of evidence suggest that shared and cell-line-specific interactions differ in network context and biology.
Shared and Differential Interactions Reside in Contrasting Network Contexts
To assess network context, 293T and HCT116 networks were merged and edges scored for betweenness centrality (the number of ‘shortest paths’ among nodes that include each edge) and local clustering coefficient (a measure of network density). The fraction of edges shared between cell lines varied inversely with edge betweenness centrality (Figure S5A) and jointly with local clustering coefficient (Figure S5B), suggesting that shared interactions reside in dense subnetworks while cell-specific interactions bridge disparate proteins and complexes, “rewiring” connections among core modules.
To contrast the properties of proteins involved in shared versus cell-specific interactions, we next assessed how cell fitness and protein expression variability influence interaction overlap. Superimposing cell fitness data (Blomen et al., 2015; Dempster et al., 2019; Wang et al., 2015) onto our networks revealed a strong positive correlation between “essential” proteins and interaction overlap (Figure S5C). Similarly, ranking by expression variability across cancer cell lines (Nusinow et al., 2020) (Figure S5D) or human tissues (Wang et al., 2019) (Figure S5E–F) indicates that proteins with cell-specific interaction profiles are variably expressed in diverse biological contexts.
Differential Interactions Reflect Protein Evolution
Evolution is among the most potent forces shaping biological systems. To evaluate the evolutionary context of cell-specific interactions, we mapped protein evolutionary ages (Liebeskind et al., 2016) onto BioPlex (Figure S6A). Assigning each interaction the age of its youngest constituent protein split the network into subnetworks matching eight evolutionary stages (Figure S6B). While some proteins and interactions dated to the dawn of cellular life, most arose during eukaryotic and eumetazoan ages. A striking correlation was observed between cell specificity and evolutionary age with the oldest interactions overlapping 6-fold more often than their youngest counterparts (Figure S6C/D).
This relationship between shared interactions and evolutionary age also appears in individual protein interaction profiles. Among DDX31 interactors, cell-specific replication drops from 100% to 35% as younger proteins are added (Figure S6E). Similar trends are seen for HDAC1 and C15orf41 (Figure S6F–G).
Interactions within Complexes, Pathways, and Protein Families Covary According to Cellular Phenotype
While many factors contribute, biological function most strongly governs replication. Broadening our analysis of CORUM overlap to include Reactome pathways (Fabregat et al., 2017), GO categories (Ashburner et al., 2000), and DisGeNET disease associations (Piñero et al., 2017) reveals that cell specificity varies with category (Figure 3A, Table S5): those most closely associated with physical entities (e.g. CORUM, GO cellular components) overlap more than abstract functional classes (e.g. GO biological processes, disease associations).

(A) Ternary diagrams depict edge sharing across cell lines for subnetworks defined by protein functional classes. Plots are shown for CORUM complexes, Reactome pathways, GO ontologies, and DisGeNET disease associations, ordered by edge sharing among constituent protein classes. Venn diagrams match assemblies shown in panels B, C, D, F, and H.
(B) Glycolysis subnetwork (Reactome).
(C) RNA Polymerase II Transcription Initiation subnetwork (Reactome).
(D) EPH-Ephrin Signaling subnetwork (Reactome).
(E) Phosphopeptides detected for ephrin receptors and ligands expressed as baits in each cell line.
(F) “TGF-β Receptor Signaling Activates SMAD’s” subnetwork (Reactome).
(G) Expression of epithelial and mesenchymal markers regulated by Snail, ZEB, and bHLH transcription factor family members downstream of TGF-β signaling.
(H) Post-synaptic Membrane subnetwork (GO Cellular Component).
As before, entities essential to cell function are replicated in both lines. Examples include glycolysis (Figure 3B) and RNA polymerase II transcription initiation (Figure 3C) pathways. Protein expression is equivalent in both lines, and expression variation is suppressed, reflecting co-regulation (Figure S4F–G).
In contrast, cell-specific entities reveal coordinated remodeling. Among GO categories and DisGeNET disease associations, greater variation reflects cell specialization and the tissue-specific nature of disease. Among Reactome pathways, variable interactions reflect differential signaling. Some divergent interactions evoke phenotypes specific to 293T or HCT116 cells.
One example is EPH-Ephrin signaling (Figure 3D). Ephrins and ephrin receptors are cell surface proteins whose binding triggers complementary signaling in neighboring cells, contributing to cell migration, repulsion, and adhesion, especially during development (Kania and Klein, 2016). Though no consistent bias in abundance is seen (Figure S4H), their cell-specific interactions suggest increased ephrin signaling in 293T cells, which is further supported by increased phosphorylation of ephrin receptors and ligands when expressed as baits in 293T versus HCT116 cells (Figure 3E, Table S4B). This differential signaling may reflect the mesenchymal nature and fetal origins of 293T cells.
Cell-specific signaling is also reflected by SMAD2/3 which mediate TGF-β signaling by binding SMAD4 and translocating to the nucleus to modulate gene expression (Tzavlaki and Moustakas, 2020) (Figure 3F). Despite similar expression in both cell lines (Figure S4I), cell-specific interactions among SMAD2/3/4 suggest higher TGF-β signaling in 293T cells. In the nucleus these proteins act with Snail, Zeb and bHLH family transcription factors to promote the mesenchymal state (Xu et al., 2009). Since TGF-β contributes to epithelial-to-mesenchymal transition during embryogenesis, its persistence again likely reflects 293T cells’ embryonic origin and mesenchymal phenotype (Figure 3G, Figure S3C).
Finally, cell-specific interactions reflect other facets of cellular identity. Interactions among post-synaptic membrane proteins mostly favor 293T (Figure 3H). This reflects differential protein expression (p = 6e-4; data not shown) and evokes 293T cells’ neural crest derivation (Stepanenko and Dmitrenko, 2015) and partially neural phenotype, as they express many neural proteins (Figure S3A) and may be induced to form synaptic structures (Biederer and Scheiffele, 2007).
Linking Differential TP53 Signaling to Cell-Specific Interactions
Among 293T and HCT116 cells, differential TP53 signaling may be their clearest contrast. TP53 signaling governs cell cycle progression and DNA repair and its disruption can drive cancer development and viral infection (Hafner et al., 2019). While unperturbed in HCT116 (Ahmed et al., 2013), TP53 signaling has been disrupted in 293T by adenoviral transformation (Stepanenko and Dmitrenko, 2015) and SV40 Large T antigen (LgT) expression. This divergent signaling leads to cell-specific interactions and differential expression and phosphorylation of related proteins.
TP53 is a transcription factor that governs expression of CDKN1A and other cell cycle regulators, and whose abundance and activity are restricted by MDM2 and MDM4 (Figure 4A). While interactions with MDM2/4 are observed in both cell lines, interactions with ABRAXAS2 and STK11 – both involved in DNA damage response – are 293T-specific (Figure 4B). TP53 disruption results from direct binding of LgT (Topalis et al., 2013), and indeed we detect dramatic enrichment of LgT following AP-MS of TP53 in 293T cells (Figure 4C, Table S4C). When LgT binds, TP53 is stabilized and its activity blocked, increasing TP53 abundance in 293T cells while levels of TP53 targets SFN and CDKN1A (Fischer et al., 2016) fall in concert with other cell cycle regulators (Figure 4D). In response, MDM4 declines. The 293T-specific ABRAXAS2-TP53 interaction may also be compensatory, as it promotes TP53 activity by facilitating its deubiquitination and stabilization (Zhang et al., 2014).
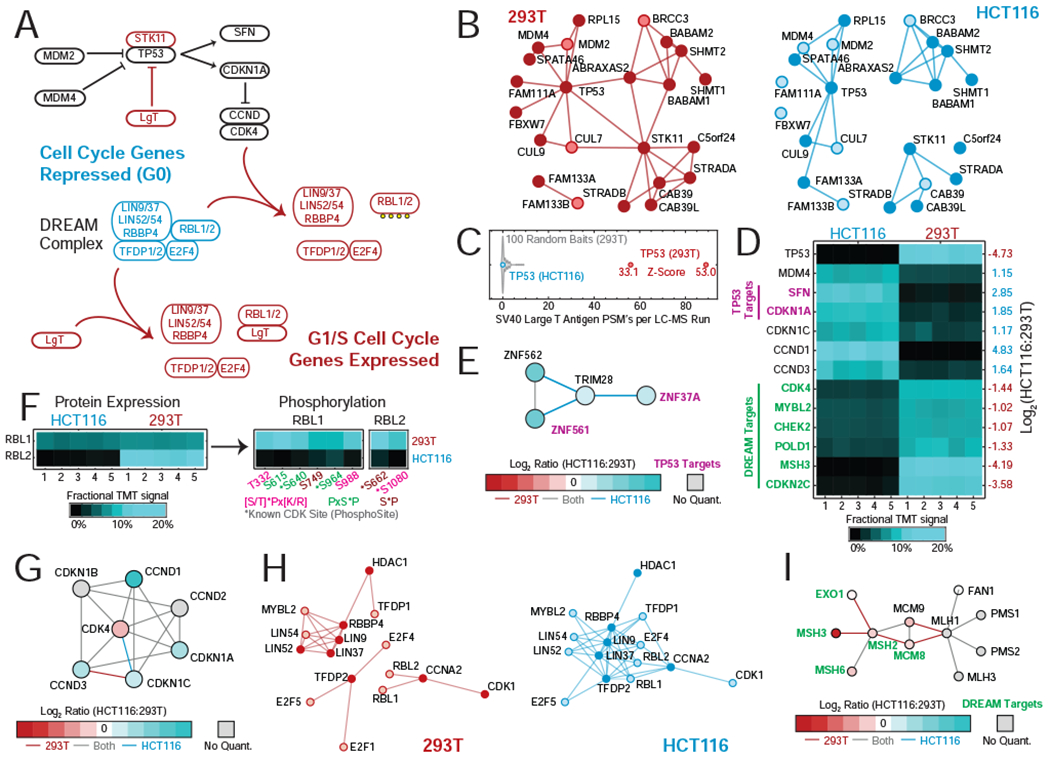
(A) Cell cycle regulation: TP53 and the DREAM Complex.
(B) Selected interactions of TP53 in 293T and HCT116 cells.
(C) Enrichment of SV40 Large T Antigen in TP53 IP’s.
(D) Expression of TP53 and related proteins in 293T and HCT116 cells.
(E) HCT116-specific interactions involving proteins whose expression is regulated by TP53.
(F) RBL1/2 abundance and phosphorylation in 293T and HCT116 cells.
(G) CDK4, Cyclin-D, and CDKN1A-C: expression and interactions in 293T and HCT116 cells.
(H) DREAM Complex interactions in 293T and HCT116 cells.
(I) 293T-specific interactions involving proteins whose expression is regulated by the DREAM complex.
Some effects of TP53 inhibition directly alter interactions: interactions of TP53 targets ZNF37A and ZNF561 (Fischer et al., 2016) with TRIM28 are detected only in HCT116 (Figure 4E), likely owing to decreased 293T expression and leading to HCT116-specific gene silencing. In contrast, interactions among CDK4, CCND1-3, and CDKN1A-C are unchanged despite decreased abundance of several components (Figure 4G). Nevertheless, 293T-specific TP53 inhibition alters CDK4 activity; this may explain increased phosphorylation of CDK4 targets RBL1/2 in 293T cells, consistent with decreased inhibition by TP53 target CDKN1A (Figure 4F, Table S4D).
RBL1/2 are themselves important cell cycle regulators whose interactions differ in 293T and HCT116 cells. Both are members of the DREAM (DP, RB-like, E2F, and MuvB) complex which represses cell cycle gene expression in quiescent cells (Figure 4A). While the DREAM complex is detected in HCT116 cells, it disintegrates in 293T (Figure 4H) following destabilization from RBL1/2 phosphorylation and RBL1/2 binding to LgT (Sadasivam and DeCaprio, 2013). In its absence, some DREAM targets (Fischer et al., 2016) are upregulated in 293T (Figure 4D), potentially contributing to cell-specific interactions among proteins responsible for mismatch repair (Figure 4I).
Data-driven Discovery of Shared and Cell-specific Network Communities
While replication within known complexes, pathways, and protein families has linked protein function to cell-specific remodeling, these analyses have included only well-studied portions of the interactome. Thus, we also used MCL clustering (Enright et al., 2002) to discover protein communities within the combined network (Table S6A–B). We then sought statistical associations among community pairs whose members were especially likely to interact (Table S6C) and quantified cell-specific overlap within and between communities (Figure 5A, TableS6D–E). Together, these communities and associations define a network (Figure 5B). Because they are defined independently of existing biological knowledge, these communities model the interactome more completely, incorporating characterized and uncharacterized proteins and complexes without bias.
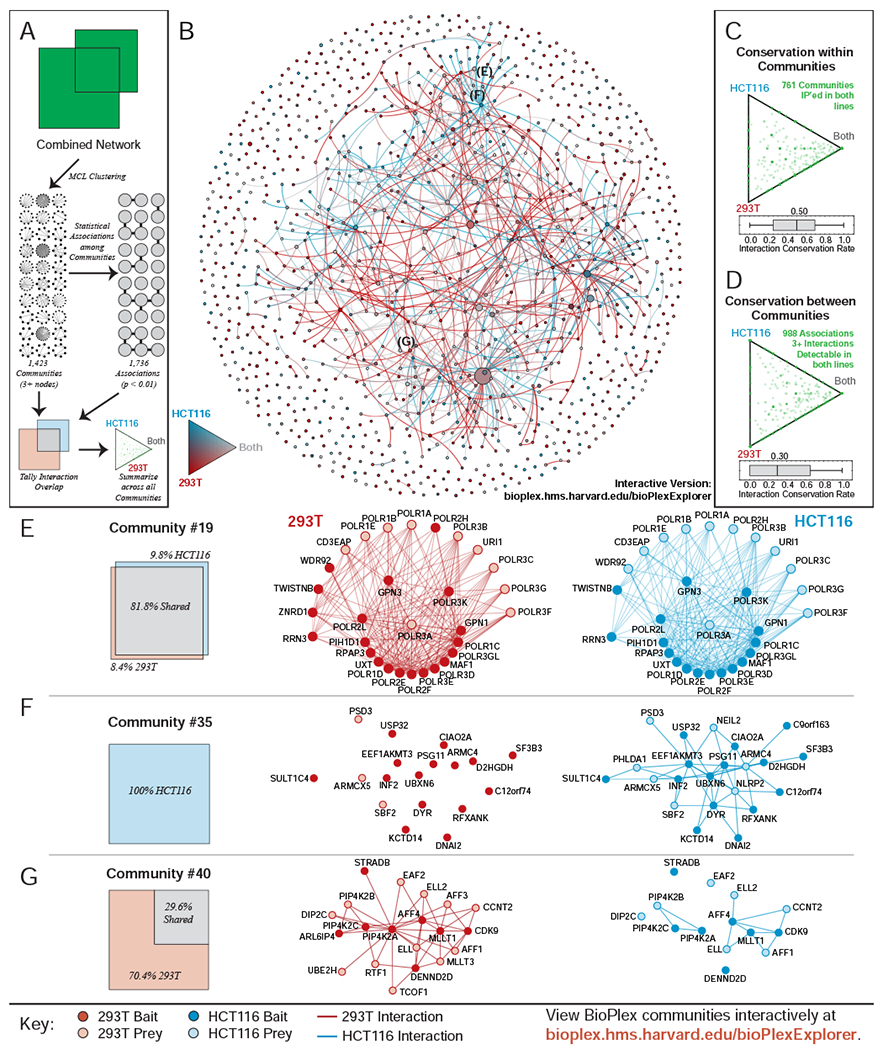
(A) 293T and HCT116 networks were combined and partitioned via MCL clustering to identify 1,423 communities with 3+ members. Interactions connecting community pairs were then tallied to identify 1,736 statistically enriched associations. Interaction overlap across cell lines was then tallied within each community and along edges that connect associated community pairs.
(B) Network of communities detected in the combined 293T/HCT116 network. Every network community with at least 3 members is represented as a node of size proportional to the number of proteins it contains. Edges connect communities that were statistically associated. Node and edge colors reflect overlap among cell lines.
(C) Ternary diagram depicting the overlap observed within each community. Only communities for which at least one member has been a bait in both cell lines are included.
(D) Ternary diagram depicting the overlap observed for edges that connect communities. Edges were only included if they were supported by 3+ edges detectable in both cell lines given the baits targeted in each.
(E) – (G) Selected Network Communities.
Overlap within communities (Figure 5C) exceeds overlap between communities (Figure 5D), implying that shared complexes interconnect in cell-specific ways. Compared to knowledge-driven protein classes (Figure 3A), overlap within and between data-driven communities is low. The median overlap within these communities trails all literature classifications – especially CORUM and GO Cellular Component categories that most directly encapsulate known complexes. This suggests that prior knowledge is biased and incomplete, focusing disproportionately on universal complexes that represent just a fraction of the interactome. Indeed, while some communities match core complexes that are equally detected in both cell lines (Figure 5E) and whose constituent proteins are equally expressed (Figure S4K), other communities are cell specific. While the role of the HCT116-specific complex in Figure 5F is uncertain, its cell-specific co-purification with many constituents implies context-dependent assembly, perhaps driven by up-regulation of some members (Figure S4L). Finally, partial edge overlap within a community can reveal cell-specific processes. In 293T cells we detect nuclear proteins bound to multiple PIP4K2 subunits; 70% of these interactions are undetectable in HCT116 cells, separating PIP4K2 subunits from nuclear proteins (Figure 5G). Since expression of the most central proteins is unchanged (Figure S4M) and PIP4K2 subunits reside in multiple compartments, these interactions suggest 293T-specific nuclear translocation.
Cell-line Specificity among Domain Associations
Although myriad interactions occur, many fit recurring motifs, as proteins bearing specific combinations of structural domains preferentially interact. Previously we demonstrated enrichment of interactions linking thousands of domain pairs within the human interactome (Huttlin et al., 2010, 2017). While these associations need not imply direct interactions between domains, they do suggest recurring structural motifs among interacting proteins. In contrast to pathways and complexes which form discrete subnetworks, interactions matching specific domain associations may distribute more broadly across the interactome; yet their structural similarity may suggest similar modes of regulation.
To understand how interactions matching domain associations covary, we mapped PFAM domains (El-Gebali et al., 2018) onto our combined network and identified domain pairs whose members interact especially often (Table S7A). We then extracted interactions linking each domain pair and calculated their overlap in both cell lines (Figure 6A, Table S7B). These domain associations define a network viewable via BioPlexExplorer. Though interactions vary, with median 40% overlap, most associations (88%) are found in both cell lines, suggesting that these interaction motifs transcend cell type.

(A) PFAM domains were mapped to proteins in the combined HCT116/293T network and domain pairs connected by unusually high edge counts identified. The overlap of edges connecting each statistically associated domain pair was then determined across cell lines. These domain associations form a network with edge colors that reflect sharing of interactions across cell lines. A ternary plot depicts sharing of edges matching each domain association across cell lines. The box-whisker plot shows the fraction of interactions shared among cell lines; a histogram highlights the fraction of domain associations that are shared or cell-line-specific.
(B) – (E) Subnetworks of PFAM domain pairs. P-values reflect enrichment of interactions among the indicated domain pair with multiple testing correction. Only edges eligible for detection in both cell lines are shown.
While most domain associations appear in both cell lines, the underlying interactions often reveal stark differences. For example, kinases partition into clusters corresponding to CDK’s, MAP kinases, and others (Figure 6B). After removing pulldowns not performed in both cell lines, few HCT116-specific edges are seen, though 42% appear only in 293T cells and suggest differential signaling. Likewise, C2H2-zinc-finger proteins often bind DNA and dimerize, frequently through SCAN or BTB domains (Figure 6C). Over 80% of these interactions are 293T-specific, though the basis for this selectivity is unclear. In contrast, interactions among Zip domain-containing proteins and 7-transmembrane-domain-containing proteins favor HCT116 cells (Figure 6D), suggesting specialized zinc transport. Finally, distinct interactions among cadherins and tyrosine kinases are observed in 293T and HCT116 cells (Figure 6E), suggesting cell surface proteome reorganization.
Interactions of Individual Proteins Reveal Cell-specific Variations on Consistent Biological Themes
Given the variation observed between cell lines, we asked whether cell-specific interactions of individual proteins share common functions, analyzing functional term enrichment among each bait’s neighbors in the combined network. Proteins matching each enriched term were categorized according to detection in one or both cell lines and each protein – function association plotted to reflect its overlap between cell lines (Figure S7A). For ECHDC2, only 4/10 interactors were observed in both cell lines; yet nearly all are mitochondrial and several link to mitochondrial organization and branched-chain amino acid metabolism. Across baits targeted in both cell lines, 90% of protein – function associations map to both (Figure S7B, Table S7C).
Though a protein’s neighbors may differ across cell lines, their function is often consistent. This is demonstrated by TRIM28, a protein known to recruit gene-silencing complexes to specific genomic regions upon binding to KRAB-domain-containing zinc-finger proteins (Ecco et al., 2017) (Figure S7C). As expected, 92% of its 157 neighbors contain KRAB domains. Only 80 TRIM28 interactors are shared; 55 are HCT116-specific and 22 were detected in 293T only. Differential expression explains only a fraction of these cell-specific interactions. That TRIM28 would bind different zinc-finger proteins in each cell line is not unexpected, as gene silencing differs in each. Similarly, we find that phosphatase inhibitor FAM122A binds distinct phosphatases (Figure S7D): while interactions with PP2A phosphatase subunits are detected in both cell lines, interactions with calcineurin are 293T-specific.
Interacting with specific sub-populations of functionally similar proteins will modulate a protein’s activity, and we can sometimes relate cell-specific interactions of individual proteins to specific phenotypes. While Kelch/BTB protein KLHL29 binds BTB proteins in both cell lines (Figure S7E), several detected specifically in 293T cells evoke known properties of the cell line. As a known regulator of neural crest specification (Werner et al., 2015), KBTBD8 reflects their neural crest lineage and retained neural qualities (Figure 3H); similarly, NUDCD3 and NPHP3 (Bergmann et al., 2008) participate in ciliary development, a process seen also in the 293T-specific proteome (Figure S3B). Likewise, we observe that the transcription factor TWIST2 interacts with cell-specific proteins that participate in DNA-templated transcription (Figure S7F). A bHLH family member, TWIST2 acts downstream of TGF-β signaling to promote a mesenchymal cell state. While TWIST2 interacts with E-box-binding proteins that activate transcription in both cell lines, we detect HCT116-specific interactions with three transcriptional repressors, confirming our previous observation (Figure 3F–G) that TGF-β signaling seems reduced in HCT116 cells compared to mesenchymal 293T cells.
Linking Physical and Functional Associations for Biological Discovery
An emerging theme is that biological function organizes interactome structure and dynamics, as proteins involved in specific processes interact preferentially and in concert to form context-specific networks. Yet because the categories considered thus far are intentionally broad, we have not yet accounted for specific functional relationships among interacting protein pairs. In one final analysis, we explore how proteins’ relative fitness effects relate to interaction replication across cell lines.
We combined BioPlex networks with genome-wide CRISPR co-essentiality profiles measured across hundreds of cancer cell lines through Project Achilles (Meyers et al., 2017). Previously, covarying fitness effects have revealed functional associations within complexes (Boyle et al., 2018; Pan et al., 2018) and identified novel complex members (Wainberg et al., 2019). We thus correlated fitness profiles for each interacting protein pair in both BioPlex networks (Figure 7A, Table S7D) and extracted subnetworks corresponding to interactions with either positive (Figure 7B) or negative (Figure 7C) correlations. While the cell specificity of negatively correlated edges matched the background distribution of edges for which fitness profiles were available, positively correlated edges were enriched for shared edges. This reflects a key structural difference: whereas the positive network includes numerous complexes, the negative correlation network contains none. This is because 3 or more proteins cannot simultaneously correlate negatively with each other. While entire complexes may correlate positively with each other – and have higher probability of conservation across cell lines – negative correlations connect specific members of complexes or span multiple complexes. Thus, positive and negative correlations reflect different functional and structural relationships.

(A) For each interacting protein pair in the combined 293T/HCT116 network, cellular fitness profiles from Project Achilles were correlated and assessed for statistical significance. Following multiple testing correction, edges with positive or negative fitness correlations were extracted and assigned as either shared or cell-specific. Only edges detectable in both cell lines are shown.
(B) – (C) BioPlex subnetworks with positive (B) or negative (C) fitness correlations. Edges connect proteins that interact in BioPlex and whose fitness profiles correlate (5% FDR).
(D) – (F) Subnetworks of panel B. Green labels summarize common biological themes in each subnetwork.
(G) – (I) Subnetworks of panel C. Green labels summarize common biological themes in each subnetwork.
Because positive correlations capture entire complexes, the positively correlated network identifies novel complex members that interact physically and share similar fitness effects (Figure 7B). We observe in both cell lines that the unknown protein C18orf21 binds physically and correlates functionally with most members of the Ribonuclease P complex highlighted previously (Figures 7C//2G).2G). In contrast, positively correlated interactions among protocadherins (Figure 7E) are mostly cell-specific, implying cell surface proteome reorganization. The positively correlated network also includes clusters of signaling proteins and CDK’s (Figure 7F). While interactions among core components (CDK1/2, cyclins, and CDK inhibitors) are found in both, interactions with regulatory proteins (PCNA, CKS1B) differ between cell lines.
In contrast, negative correlations capture antagonistic interactions among protein pairs. Examples include alternating pro- and anti-apoptotic proteins (Figure 7G); TP53 and its negative regulators MDM2 and MDM4 (Figure 7H); and mixtures of CDK’s, cyclins, and CDK inhibitors (Figure 7I). The negative fitness correlations depicted in the latter two examples encode key relationships governing TP53 signaling and cell cycle control as highlighted previously (Figure 4A).
DISCUSSION
With hundreds of cell types existing in myriad states, human cell biology is diverse and specialized. This is achieved through customization of each cell’s proteotype, fine-tuning protein expression, localization, post-translational modifications, and interactions to achieve the desired phenotype. Thus, defining interactome structure and dynamics is required to understand cellular diversity. Deploying affinity enrichment at scale to profile protein interactions has advanced these goals two-fold.
First, we have profiled interactions with singular depth and breadth. While this is mostly due to targeting over half the human proteome for AP-MS in 293T cells, repeating AP-MS in HCT116 cells has revealed additional cell-specific modules. Such context-specific interactions are important, as current knowledge favors core complexes. Maximizing coverage enhances network quality – affirming associations via repeated co-purification – and enables discovery, as BioPlex suggests function, localization, and complex membership for thousands of proteins, many uncharacterized.
Second, performing AP-MS in 293T and HCT116 cells has enabled comparison of two context-specific, proteome-scale interaction networks. Though it has been long recognized that interactions vary with context, our understanding of interactome dynamics remains rudimentary. These networks have revealed widespread reorganization. Whereas shared interactions reside in dense subnetworks and preferentially link consistently expressed, often essential proteins with long evolutionary histories, cell-specific interactions span disparate network components, coupling variably expressed proteins in context-specific configurations. Interactions among proteins of shared function covary, assembling specialized modules according to need. These variations in network structure arise via diverse mechanisms including differential protein expression, localization, and post-translational modification, and often link to specific phenotypes. Extensive proteome coverage has been essential for defining this reorganization since cell-specific interactions often reside outside core complexes. These direct observations of interactome reorganization complement reports of co-variation within protein complexes as inferred from constituent protein expression across cell lines, tissues, and individuals (Luck et al., 2020; Ori et al., 2016; Romanov et al., 2019; Ryan et al., 2017).
LIMITATIONS OF THE STUDY
Though BioPlex has increased our knowledge of human protein interactions, our understanding of the interactome remains incomplete. While we have completed AP-MS for over 10,000 human proteins, nearly as many remain untargeted. Moreover, low-affinity interactions and interactions involving certain protein classes (e.g. membrane proteins) may be challenging to detect via AP-MS. Given its relative advantages detecting weaker, more transient interactions and its compatibility with challenging proteins, large-scale proximity labeling (Go et al., 2019) is an important complement. Beyond scale and technical limitations, no two cell lines can capture the full range of cellular diversity. While systematic AP-MS will be essential for probing additional context-specific interactomes with maximum depth, parallel deployment of complementary experimental approaches like co-fractionation (Heusel et al., 2019; Rosenberger et al., 2020) will facilitate rapid interactome screening across cellular contexts.
FUTURE DIRECTIONS
Despite these limitations, this work establishes a basis for probing the interactome even more broadly. First, our AP-MS platform is limited by clone availability; as clones for remaining human genes become available, our networks can grow to encompass every amenable protein. Second, since large-scale AP-MS has proven both feasible and informative in multiple cell lines, incorporating additional cell lines will reveal interactions across diverse cellular contexts. Similarly, as seen for nutrient-sensing proteins CASTOR1/2 (Chantranupong et al., 2016) and SAMTOR (Gu et al., 2017), specific interactions afford tantalizing insights into the biology of many uncharacterized proteins that warrant hypothesis-driven study. Finally, the BioPlex networks are powerful predictive tools individually and in tandem with complementary datasets. For example, BioPlex may be combined with immunofluorescence imaging from the Human Protein Atlas (Thul et al., 2017) to model cellular structures at scales ranging from interacting protein pairs to entire organelles (Qin et al., 2020). With time we expect this work to broaden our understanding of the protein interaction landscape and to enable many complementary forms of biological discovery.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Steven Gygi.
Materials Availability
Open reading frames used in this project were taken from the Human ORFeome collection v. 8.1 which is available for purchase from Horizon Discovery (https://horizondiscovery.com/en/gene-modulation/overexpression/orfs/human-orfeome-v8-1). All clones generated for this project will be available from the DNASU plasmid repository (http://dnasu.org). Individual clones are available from the Lead Contact upon request without restriction, though requests for large numbers of clones should be directed to DNASU.
Data and Code Availability
This project has generated many types of data and code that are available for distribution via numerous venues. Items not listed here will be provided by the Lead Contact upon reasonable request.
First, both 293T and HCT116 networks are described in full in the supplementary tables included with this manuscript, as are results of all analyses described herein. These include functional and disease associations, predicted localizations, communities, PFAM domains, replicate analyses, etc. See supplementary table legends for details.
Second, both filtered and unfiltered lists of interactions in HCT116 and 293T networks are available for download on the BioPlex website at bioplex.hms.harvard.edu/downloadInteractions.php. These interactions may be accessed either as flat files or through a custom API.
Third, both 293T ( 10.18119/N9RP5D) and HCT116 ( 10.18113/N9N012) networks, as well as the intersection of these two networks ( 10.18119/N9H887), have been deposited into NDEx (https://home.ndexbio.org/index/), the repository for biological network data (Prat et al., 2015).
RAW files corresponding to both 293T and HCT116 AP-MS experiments may be accessed in multiple ways. First, if RAW files corresponding to a small number of specific baits are desired, these may be downloaded from the BioPlex website via a search interface at https://bioplex.hms.harvard.edu/downloadData.php. If larger numbers of RAW files are desired, they will be accessible via the MassIVE repository (ftp://massive.ucsd.edu). Finally, all ~30,000 RAW files corresponding to 293T and HCT116 AP-MS networks are available upon request via GLOBUS infrastructure.
CompPASS software is available as an R package (https://github.com/dnusinow/cRomppass); we also make it available for small to medium-scale AP-MS experiments online via the BioPlex website: https://bioplex.hms.harvard.edu/comppass/.
CompPASS-Plus is also available (https://github.com/HMSBioPlex/CompPASS-Plus-CLI), though we only recommend its use for very large AP-MS studies involving 1000+ IP’s.
RAW files corresponding to the RTS-MS3-TMT comparison of 293T and HCT116 proteomes have been deposited in MassIVE (ftp://massive.ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Clone Construction
Clones corresponding to human genes were obtained and prepared for AP-MS analysis as described previously (Huttlin et al., 2015). Briefly, open reading frames were taken from the human ORFeome, version 8.1 (Yang et al., 2011) and the OC collection (http://horfdb.dfci.harvard.edu) and cloned using Gateway techniques into a lentiviral recipient vector that incorporated a c-terminal FLAG-HA tag and expressed the target ORF and a puromycin resistance marker under control of a CMV promoter. All clones were sequenced to verify ORF identity.
Within version 8.1 of the ORFeome, clones were generally sorted by length and accordingly assigned to 96-well-plates. In contrast, OC collection baits were arrayed on 96-well plates in more random order. Throughout the project, clones have generally been targeted for AP-MS in batches corresponding to these 96-well plates. During initial AP-MS analysis in 293T cells, plates were run in random order, though plates corresponding to very large or small clones were avoided until late in the project. For HCT116 cells, roughly half of plates consisted of cherry-picked baits that 1) had worked in 293T cells and 2) were selected to sample evenly across the 293T interaction network; subsequently we have run remaining baits in batches according to their original ORFeome plate assignments. Baits repeated in 293T cells were also selected to sample evenly across the 293T interaction network.
Creation of Stable Cell Lines in 293T and HCT116 Cells
Cell culture and bait expression were performed as described previously (Huttlin et al., 2015). Briefly, lentivirus was produced in 293T cells following transfection of Gateway expression clones using PEI as an adjuvant. Upon subsequent lentiviral transduction of bait constructs, either 293T cells or HCT116 cells (both from ATCC) were expanded in the presence of 1 μg/mL puromycin selection (Gibco) to obtain five 10-cm dishes per cell line. Once they reached minimum 80% confluence, cells were washed with ice cold PBS phosphate-buffered saline (PBS) pH 7.2 (Gibco) and harvested via gentle scraping, followed by centrifugation to pellet cells and PBS removal. Cell pellets were stored at −80°C.
All cell lines were tested for presence of mycoplasma using the Mycoplasma Plus PCR assay kit (Agilent) and found to be free of contamination. Additionally, the identities of 293T and HCT116 cell lines were verified through GTG-banded karyotyping by the Brigham and Women’s Hospital Cytogenomics Core Laboratory.
Creation of Stable Cell Lines in U2OS Cells for Immunofluorescence Imaging
U2OS cells (American Type Culture Collection) were plated on glass coverslips (Zeiss) and transduced with lentiviral vectors expressing C-terminal Flag–HA-tagged baits. Baits included TTLL1, LRRC49, and isoforms 2 and 3 of C11orf49. At 48 h after infection, cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Immunofluorescence imaging was performed as described below.
METHOD DETAILS
Affinity-Purification Mass Spectrometry
Affinity-purification mass spectrometry experiments were performed as previously described in detail (Huttlin et al., 2015, 2017). Methods are summarized below. AP-MS experiments in each cell line were performed separately: HCT116 AP-MS experiments were not initiated until all experiments in 293T cells had been completed. Baits targeted in 293T cells are summarized in Table S1A while baits targeted in HCT116 cells are summarized in Table S1C.
Affinity Purification of Protein Complexes
After thawing, cell pellets were lysed in 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.5% (v/v) NP40 and the lysate was clarified by centrifugation and filtration. Immobilized, pre-washed mouse monoclonal anti-HA agarose resin (Sigma-Aldrich, clone HA-7) was used to immunoprecipitate affinity-tagged bait proteins and their binding partners. Beads were incubated with lysates for 4 hours at 4°C, followed by supernatant removal, four washes with lysis buffer, and two washes with PBS (pH 7.2). Elution was achieved in two steps via addition of 250ug/mL HA peptide in PBS at 37°C for 30-minute incubations with gentle shaking.
Following affinity enrichment, residual HA peptide and other non-protein material were removed through precipitation with 20% TCA followed by a wash with 10% TCA and triplicate washes with cold acetone. Pellets were then resuspended and reduced in 150 μL of 100 mM ammonium bicarbonate (Sigma-Aldrich) with 1 mM DTT (Fluka) and 200 ng of sequencing grade trypsin (Promega) was added for digestion overnight at 37°C. For HCT116, TCA precipitated pellets were resuspended in 20uL of 55mM Tris pH 8.5/10% Acetonitrile/1mM DTT, with 200ng trypsin added per sample.
5% HPLC-grade formic acid (Thermo Fisher Scientific) was subsequently added to quench digestion and samples were desalted with homemade stage tips as described previously (Rappsilber et al., 2007). After elution with 80% acetonitrile/5% formic acid, peptides were dried in a speed-vac and resuspended in 16 μL of 5% formic acid and 4% acetonitrile.
Mass Spectrometry Data Acquisition
All mass spectrometry data were acquired on first-generation Q-Exactive mass spectrometers (Thermo Fisher Scientific) equipped with Famos autosamplers (LC Packings) and Accela600 liquid chromatography (LC) pumps (Thermo Fisher Scientific). Microcapillary columns used for peptide separation were prepared in-house by packing ~0.25 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources) and ~18 cm of Accucore C18 resin (2.6 μm, 150 Å, Thermo Fisher Scientific) into a 100 μm inner diameter microcapillary column. We loaded 4 μL of sample onto the column for each analysis. Each sample was run in technical duplicate in accordance with the requirements of the CompPASS-Plus algorithm (described below).
Each LC-MS run was approximately 70 minutes long including sample loading, analytical separation, and column re-equilibration. Peptide separation occurred over 40 minutes using a gradient from 5 to 26% acetonitrile in 0.125% formic acid. The scan sequence consisted of a single MS1 spectrum followed by MS2 scans targeting up to twenty precursors. Orbitrap MS1 scans were acquired in centroid mode at 70,000 resolution spanning a range from 300-1500 Th with the automatic gain control (AGC) target set to 1.0e6 and a maximum ion injection time of 250 ms. Following higher-energy collision-induced dissociation (HCD), MS2 spectra were acquired in centroid mode at 17,500 resolution with the AGC set to 1.0e5 and a maximum ion injection time of 60 ms. The isolation window was set to 2 Th and the normalized collision energy (NCE) was 25-30. Features were targeted for MS2 analysis in order of decreasing intensity while excluding unassigned and singly charged features. Dynamic exclusion was set to automatic.
Identification of Peptides and Proteins
Upon completion of each LS-MS analysis, the resulting RAW files were converted to mzXML format using msconvert (ProteoWizard). Monoisotopic peak assignments and charge states were then verified for all features targeted for MS2 analysis and MS/MS spectra were matched with peptide sequences using the Sequest algorithm (Eng et al., 1994) along with a composite sequence database including the human Uniprot database (The UniProt Consortium, 2015), GFP (our negative control), the FLAG-HA affinity tag, and sequences of common contaminants. This Uniprot database includes both SwissProt and Trembl entries and dates to the outset of this AP-MS study in 2013. Protein sequences were listed in both forward and reversed order to facilitate false discovery rate control. During database searching, fully tryptic peptides with up to two missed cleavages were considered and MS1 and MS2 tolerances were set to 50 ppm and 0.05 Da, respectively. Variable oxidation of methionine (+15.9949) was permitted.
Following Sequest analysis, peptides and proteins identified in each LC-MS analysis were filtered in two steps using the target-decoy method (Elias and Gygi, 2007) to control both peptide- and protein-level false discovery rates. First, a linear discriminant function was trained to distinguish correct and incorrect peptide identifications using Xcorr, DCn, peptide length, charge state, mass error, missed cleavage count, and fraction of ions matched (Huttlin et al., 2010) and peptides were filtered to a 1% FDR. Next, these peptides were assembled into proteins, scored probabilistically, and filtered to a 1% protein-level FDR.
Although the filters previously described controlled the peptide- and protein-level FDR within each individual LC-MS analysis, when many datasets are combined as described in this study, the overall dataset-level FDR tends to balloon as false positives accumulate. Since our 293T and HCT116 datasets include over 20,000 and 10,000 LC-MS runs respectively, without additional filtering this problem would be especially severe. To further control the global protein FDR as AP-MS datasets were combined, entropy-based filtering was applied to remove proteins inconsistently detected across technical replicates (Huttlin et al., 2015) and protein identifications within each IP were required to be supported by multiple unique peptides when technical replicates were combined. Together these additional filters reduced the global FDR by more than 100-fold.
AP-MS Pipeline Quality Control
Several steps have been taken to ensure that sample preparation and instrument performance has been maintained at consistent levels throughout the duration of this project. First, standards were routinely run on the instrument to monitor instrument performance over time. These standards consisted of trypsin-digested yeast whole cell lysate that was diluted to approximate the complexity and sample load of a typical AP-MS sample. Second, positive and negative control IP’s (RAB11B and GFP) were analyzed with every plate. Third, every IP was analyzed in technical duplicate with replicate runs acquired in reverse order on different LC-MS systems. These replicate analyses were primarily done to aid LC carry-over removal and enhance detection of high confidence interacting proteins, though they also enabled us to continuously compare the relative performance of all instruments devoted to this project in real time, making detection of instrument problems much easier.
In addition, every LC-MS run acquired for this project was screened using an automated anomaly detection system to identify problematic runs following database searching as described below. This algorithm monitored a variety of statistics for each run such as numbers of PSM’s, unique peptides, and proteins identified, estimated maximum numbers of true positives (TPMax), missed cleavage rate, average Xcorr, numbers of MS2 spectra acquired, etc. Each run was compared with all prior runs with respect to each of these parameters and scored using a kernel-density-based probabilistic model; anomalous runs were filtered allowing 1% of runs flagged as anomalies to be false positives. This algorithm updated continually as runs were added, allowing the algorithm to adapt to changing conditions. Classifications were monitored and manually corrected as necessary to maintain accurate performance. To accommodate cell-line-specific differences, separate models were trained from runs corresponding to 293T cells and HCT116 cells.
Prior to CompPASS-Plus analysis, each IP was screened to ensure detection of the expected bait protein, thus protecting against expression problems and occasional sample mix-ups. We expected each bait to be detected at levels significantly higher than background when targeted for AP-MS enrichment. Thus, we required the expected bait to be detected at elevated levels in two technical replicates for an AP-MS experiment to be eligible for CompPASS-Plus scoring. For a small number of baits whose sequences do not directly correspond to Uniprot entries, custom searches were performed to verify bait identities. These additional searches were only used for bait verification and only searches against the standard Uniprot database were used for interaction network generation. If the expected bait was not confidently detected and could not be determined to result from a mix-up, the runs were discarded, and the IP was repeated.
Before an AP-MS experiment could be included in CompPASS-Plus analysis, two technical replicate LC-MS analyses passing all quality control checks were required. To accommodate occasional low-quality runs, we scaled our AP-MS pipeline to offer quadruplicate analysis. This provided up to two additional runs in cases where poor instrument performance compromised one of the initial runs. If two acceptable runs were not obtained after up to four injections, the bait was targeted for repeat AP-MS analysis.
Inferring Protein Interactions from LC-MS Data
Here we provide an overview of our approaches for identifying high-confidence interacting proteins from AP-MS data and creating integrated interaction networks as published previously (Huttlin et al., 2015). These procedures were performed separately for AP-MS data acquired in 293T and HCT116 cells. Additional information is provided below regarding how these networks were combined for comparative analysis. The BioPlex 3.0 network results from re-analysis of all AP-MS data in BioPlex 1.0 and 2.0, along with 4,237 additional new AP-MS datasets.
Identification of High Confidence Interacting Proteins
Identification of high confidence interacting proteins proceeded in four steps: 1) merging technical replicates; 2) CompPASS analysis; 3) Post-CompPASS filtering; and 4) CompPASS-Plus analysis. First, those AP-MS experiments that passed all quality control filters were identified; for each, their associated technical replicates were combined to produce a summary of proteins identified. Across replicates, peptides were re-assembled into proteins according to principles of parsimony and Uniprot ID’s were mapped to Entrez Gene ID’s. All subsequent analysis was performed on data assembled at the Gene ID level to address complications due to protein isoforms. For each gene product, spectral counts were averaged across replicates and entropy scores were calculated as described previously (Huttlin et al., 2015). Entropy scores (sE) were calculated according to the following equations.
Second, all AP-MS experiments were scored using CompPASS essentially as described previously (Behrends et al., 2010; Sowa et al., 2009). Merged data from all AP-MS experiments matching a single cell line were combined to create a “stats” table that indicates the number of PSM’s observed for each gene product in each AP-MS experiment. CompPASS analysis produced two scores for each protein detected in each IP: a z-score that reflects a given protein’s abundance compared to its typical background levels across all other IP’s in the dataset; and an NWD score, which takes each gene product’s abundance, detection frequency, and technical replication into account to estimate its enrichment compared to all other IP’s. NWD scores were scaled to assign values of 1.0 or higher to the top 2% of candidate interacting partners.
Third, after CompPASS analysis was complete, additional filters were applied to protein identifications in each IP to remove inconsistent or low-confidence protein identifications and avoid false positive interactions. These filters, two of which are also described above in the section titled “Identification of Peptides and Proteins” include 1) discard proteins for which only a single unique peptide sequence was observed across two technical replicates; 2) require a minimum entropy score of 0.75 (calculated using Log2) comparing spectral counts observed in two technical replicates; and 3) within each 96-well-plate, look for proteins detected with unusual frequency compared to a background defined by all other plates – if statistically significant enrichment is detected, discard observations on that plate that fall below its average. The first two of these filters help to control the global protein-level false discovery rate; furthermore, filters 2 and 3 protect against LC carry-over, especially as observed for overexpressed baits; finally, filter 3 protects miscellaneous plate-specific variations in protein detection.
Fourth, following CompPASS scoring and application of filters as described above, all remaining bait-prey associations from a single cell line were scored and classified using the supervised classifier CompPASS-Plus as described previously (Huttlin et al., 2015). To enable training, each bait-prey association was assigned one of three preliminary labels (false positive interaction, background protein, or specific interactor). All preys corresponding to decoy protein sequences were labeled as false positives; preys whose interactions were confirmed in STRING (Szklarczyk et al., 2017) or GeneMANIA (Franz et al., 2018) were labeled as true positives; all others were labeled as background. In addition, because their levels are strongly enriched by design following affinity purification, baits within each IP were also labeled as “true interactors” for modeling purposes. Moreover, because both published and unpublished versions of BioPlex data have been previously released, care was taken to ensure that the STRING and GeneMANIA data used for training did not incorporate the results of any prior BioPlex analyses. Specifically, we used archival versions of these databases that predated release of any BioPlex datasets.
Once these preliminary labels were assigned, a separate Naive Bayes classifier was trained for each 96-well plate matching either 293T or HCT116 cells, using all other 96-well-plates from that cell line for training. This plate-based, leave-one-out cross-validation scheme ensured that separate data were always used for model training and scoring. Features considered by the classifier included CompPASS NWD and Z-scores, entropy, a plate-based z-score, binned unique peptide counts, the fraction of IP’s in which a given protein was detected, the total number of PSM’s observed for a given protein across all IP’s, the ratio of observed PSMs in a given IP to the total number of PSM’s across all IPs, and the unique:total peptide ratio within the given IP. The output of this algorithm was a vector of three scores reflecting the likelihood that each interaction resulted from either an incorrect protein identification, background, or a bona fide interacting partner. CompPASS-Plus was implemented in R (RCoreTeam, 2011) using the Naïve Bayes classifier in the package e1071 (Meyer et al., 2012). Because many features did not conform to normal distributions, continuous features were discretized by splitting each into 1000 equally-sized bins for classification.
Interaction Network Assembly
Following CompPASS-Plus scoring, all AP-MS experiments corresponding to a single cell line were assembled into a network that models the human interactome. While previous versions of BioPlex excluded a small number of baits with very high numbers of interacting partners (> 100), no filter was applied limiting the numbers of interactions that could be identified in a single AP-MS experiment. In cases where the same bait protein had been targeted multiple times, often because the ORFeome contained more than one matching clone, only a single IP was retained. In these cases we preferentially kept IP’s corresponding to full-length clones and favored IP’s that returned more interacting partners. Individual interacting partners were also filtered as the network was assembled. Specifically, a few dozen remaining decoy proteins were removed, as were common contaminant proteins (e.g. keratin).
Once filtering was complete, a nonredundant list of observed bait-prey pairs was compiled accompanied by CompPASS-Plus scores for each. When pairs of baits were found to associate reciprocally, their CompPASS-Plus scores were merged as described previously (Huttlin et al., 2015) to increase the probability of interaction. Finally, this list of bait-prey pairs was filtered according to CompPASS-Plus score: interactions with an interaction score equal to or greater than 0.75 were retained to create the final interaction network. This cutoff of 0.75 corresponds to the top ~2% of candidate interactions in both 293T and HCT116 cells. Interactions observed in 293T and HCT116 networks are summarized in Table S1B and Table S1D, respectively.
Assembly of Combined and Replicated Networks
As described above, 293T and HCT116 networks were initially assembled independently through separate CompPASS and CompPASS-Plus analyses. To facilitate comparison, these networks were subsequently aligned using the following procedure:
Interactions within both 293T and HCT116 networks were merged to generate a single non-redundant list. For this purpose, each interaction was represented as a pair of linked Entrez Gene ID’s. This step effectively defines the “Combined Network” represented by the union of the Venn diagram in Figure 1F and includes every interaction among the top 2% detected in 293T cells, HCT116 cells, or both.
Each interaction in the “Combined Network” was looked up in both 293T and HCT116 datasets. By definition, each of these interactions scored in the top 2% for at least one cell line, as was required for inclusion in the original networks. If this interaction ranked among the top 2% in 293T cells and was also observed among the top 5% of candidate interactions in HCT116 cells, then it was counted as replicated in both cell lines. Similarly, if the interaction ranked among the top 2% in HCT116 cells and was also observed in the top 5% of 293T cells, it was counted as replicated. Together these define the intersection of the Venn Diagram in Figure 1F.
If the interaction ranked among the top 2% of candidate interactions in one cell line and was either undetected or failed to score among the top 5% of candidate interactions in the other cell line, then it was considered either 293T- or HCT116-specific as shown in Figure 1F.
Using a slightly relaxed threshold for judging whether edges were replicated in the opposite network was necessary due to the inherent challenges of identifying a relatively small number of bona fide interacting partners within a much larger set of background interactions. This relaxed threshold for replication enabled us to be confident that edges we call cell-line specific correspond to substantial differences in detection rather than ‘near-misses’ that were detected and fell just short of our chosen significance cutoffs.
A complete list of interactions within this combined network is provided in Table S1E; each edge is labeled to indicate whether it maps to the original 293T or HCT116 networks and whether it was classified as replicated or cell-line-specific as described above.
293T Replication Experiments
Baits used for replicate AP-MS analysis in 293T cells were selected from the set of baits that worked following first-pass analysis in 293T cells (Table S2A). Baits were selected to sample evenly across the interactome, ensuring that large complexes such as the ribosome and proteasome would not be overrepresented in the replication set and that baits falling outside of well-defined clusters would be included as well. Identical virus was used to infect a second batch of 293T cells. Cell culture, affinity purification, and mass spectrometric analysis were all performed using standard procedures as described above. These replicates were run on separate 96-well plates separated by several months to several years from the original AP-MS analysis. Replicate datasets were scored against the main 293T “stats” table just as the original IP’s were, though they were not included in the stats table for scoring other IP’s. Identical filters for unique peptides, entropy scores, etc. were applied to replicates. And replicate IP’s were scored using CompPASS-Plus models trained on the full 293T dataset using plate-based leave-one-out cross-validation as described previously. For consistency with our 293T – HCT116 comparisons, interactions identified in the original 293T IP’s were considered replicated if they were detected in the top 5% in the second IP (Table S2B).
Quantitative Proteomic Comparison of 293T and HCT116 Cells
The quantitative proteomic comparison of 293T and HCT116 proteomes presented here has been published previously (Erickson et al., 2019). Protein extraction, digestion, and TMT labeling were accomplished using the SL-TMT method (Navarrete-Perea et al., 2018). First, quintuplicate pellets of 293T and HCT116 cells were lysed in 8M urea and lysates were reduced with 5 mM TCEP and alkylated with 10 mM iodoacetamide. Proteins were then purified via methanol-chloroform precipitation, resuspended in 200 mM EPPS, pH 8.5 and digested sequentially with LysC and Trypsin. Peptides were then labeled with TMT, combined, and desalted prior to high-pH reversed-phase fractionation to produce 24 fractions. Of these, 12 non-adjacent samples were analyzed via LC-MS.
All samples were analyzed on an Orbitrap Fusion Lumos utilizing an MS3 workflow that featured a custom real-time database search method as described previously (Erickson et al., 2019). Briefly, the instrument was operated in data-dependent mode with each MS1 scan followed by multiple MS2 scans to attain a cycle time of 2 seconds. MS1 spectra were collected in the Orbitrap at 120,000 resolution allowing ion times up to 100 ms. Features with defined monoisotopic masses and charge states greater than one were targeted for collision-induced dissociation in descending order based on intensity. MS2 spectra were acquired in the ion trap using a quadrupole isolation width of 0.5 m/z; automatic gain control of 20,000; and maximum ion time of 35 ms.
Upon acquisition, each MS2 spectrum was provided via API to a custom module that performed database searches in real time. Each spectrum was searched against a database containing Uniprot human sequences plus common contaminants. When a spectrum matched a human peptide sequence with binomial score of at least 55, an MS3 scan was triggered through the API. MS3 spectra were acquired for up to 150 ms at an AGC of 50,000 utilizing a normalized collision energy of 55. Synchronous precursor selection of up to 10 fragments was employed; all selected fragments were required to account for at least 5% of base peak signal and to match a b- or y-ion corresponding to the predicted peptide.
Following acquisition, RAW files were converted to mzXML format using msconvert. Monoisotopic peak assignments were then verified for any features that were targeted for MS2 analysis. Spectra were then submitted for database searching using SEQUEST (Eng et al., 1994) along with a database of human protein sequences (Uniprot, 2014) and common contaminants in forward and reversed orientation. Fully tryptic peptides were considered for database searching assuming precursor and product ion tolerances of 50 ppm and 0.9 Th, respectively. Fixed modifications of TMT (+229.163 Da) on lysines and N-termini and carbamidomethylation (+57.021 Da) of cysteines were assumed while variable oxidation (+15.995 Da) of methionine was considered. Peptide-spectral matches were filtered to a 1% FDR using the target-decoy method in combination with linear discriminant analysis (Huttlin et al., 2010); subsequently, peptides were assembled into proteins and filtered to attain a 1% protein-level FDR and to account for peptide redundancy according to principles of parsimony. TMT quantitation of each protein was performed by summing reporter ion intensities across all matching peptide-spectral-matches with corresponding TMT data; only peptides that attained a minimum summed reporter ion signal-to-noise of at least 100 were retained for quantitation. TMT channels were subsequently normalized assuming equal protein loading. Finally, each protein’s TMT profile was scaled to sum to 100%, thus reporting the fractional TMT signal associated with each channel. The final dataset is provided in Table S4A.
C11orf49 Validation Experiments
Affinity Purification and Immunoblot analysis
293T or HCT116 cells were transduced with c-terminal FLAG-HA lentiviral constructs harboring previously characterized cDNAs of the TTLL1 polyglutamylase complex or C11orf49, a candidate complex member identified in this study. Cells were selected with puromycin as described above, expanded to five 10cm plates, and harvested via gentle scraping into PBS. Frozen cell pellets were lysed with 50mM Tris pH 7.5, 150mM NaCl, 1% NP-40, 1mM DTT), with protease inhibitors (Roche, Complete mini EDTA free) on ice. Lysates were cleared by centrifugation, and subjected to affinity purification using either HA-agarose (Sigma) or anti-FLAG magnetic beads (Sigma) for 2 hours at 4°C. Beads were washed 4x with lysis buffer, then complexes were eluted with 3xFLAG peptide or beads were boiled in 1x NuPage sample buffer (Invitrogen) prior to SDS-PAGE. Proteins were transferred to PVDF membranes and immunoblotted with the following antibodies: C11orf49 (Cat. No. 20195-1-AP, Proteintech), TPGS1 (ab184178, Abcam), and anti-FLAG (M2)-HRP (A8592, Sigma).
Confocal Microscopy
U2OS cells (American Type Culture Collection) were plated on glass coverslips (Zeiss) and transduced with lentiviral vectors expressing C-terminal Flag–HA-tagged baits. At 48 h after infection, cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Cells were washed in PBS, then blocked for 1h with 5% normal goat serum (Cell Signaling Technology) in PBS containing 0.3% Triton X-100 (Sigma). Coverslips were incubated with anti-HA antibodies (mouse monoclonal, clone HA.11, BioLegend) or anti-HA plus anti-PCM1 (#5213, Cell signaling) for 2 h at room temperature. Anti-C11orf49 (20195-1-AP, Proteintech) and anti-γ tubulin (ab11317, Abcam) were utilized to visualize C11orf49 localization in proximity to centrosomes following coverslip permeabilization in ice cold methanol for 20 minutes. Cells were washed three times with PBS, then incubated for 1 h with appropriate Alexa Fluor-conjugated secondary antibodies (ThermoFisher). Nuclei were stained with Hoechst, and cells were washed three times with PBS and mounted on slides using Prolong Gold mounting media (ThermoFisher). All images were collected with a Yokogawa CSU-X1 spinning disk confocal with Spectral Applied Research Aurora Borealis modification on a Nikon Ti-E inverted microscope equipped with a Nikon 100 × Plan Apo numerical aperture 1.4 objective lens (Nikon Imaging Center, Harvard Medical School). Both confocal and widefield images were acquired with a Hamamatsu ORCA-ER cooled CCD (charge-coupled device) camera controlled with MetaMorph 7 software (Molecular Devices). Fluorophores were excited using a Spectral Applied Research LMM-5 laser merge module with acousto-optic tuneable filter (AOTF)-controlled solid-state lasers (488 nm and 561 nm). A Lumencor SOLA fluorescence light source was used for imaging Hoechst staining. z series optical sections were collected with a step size of 0.2 μm, using the internal Nikon Ti-E focus motor, and stacked using FIJI (Image J) to construct maximum intensity projections. Image brightness and contrast were adjusted for each image equally among staining conditions using FIJI software.
Comparison of 293T and HCT116 Phosphoproteomes
Frozen cell pellets were processed using the streamlined TMT labelling protocol (Navarrete-Perea et al., 2018). Samples were lysed in 8M urea in 200mM EPPS pH 8.5 with protease (Pierce A32953) and phosphatase (Pierce A32957) inhibitors, mixed with yeast disruption beads (BioSpec 11079105z), and shaken on a bead beater (BioSpec) for 60s at 4°C three times with 60s rest in between. Lysates were cleared by centrifuging at 16,000g for 10 min in a 4°C centrifuge. Protein levels were determined by BCA assay. Samples were then reduced with 5mM TCEP, alkylated with 10mM iodoacetamide, and quenched with 5mM DTT, followed by methanol/chloroform precipitation of 1mg protein. Pellets were reconstituted in 200mM EPPS pH 8.5, digested overnight with LysC (Wako 129-02541) at 1:100 while shaking at room temperature, followed by digestion with trypsin (Pierce 90305) at 1:100 while shaking at 37°C. Samples were de-salted using a 1g SepPak Cartridge (Waters), and dried in a rotary evaporator. Peptides were reconstituted in 200uL phosphopeptide binding buffer and enriched using the Pierce High Select Fe-NTA phosphopeptide enrichment kit (A32992) according to manufacturer’s specifications, except that elutions were carried out into tubes containing 100μL 10% formic acid before rotary evaporation. After desalting using a Thermo Sola cartridge and drying overnight, enriched phosphopeptides were reconstituted in TMT labelling buffer (200mM EPPS pH 8.5, 23% anhydrous acetonitrile) and labelled with TMT10-plex reagent. 1% of each labeled sample was combined and analyzed unfractionated to ensure labeling efficiency was >97% and that the samples are mixed at a 1:1 (total amount) ratio across all conditions. Samples were quenched by adding .3% v/v hydroxylamine. After mixing, labelled peptide samples were de-salted using a 200mg SepPak Cartridge (waters) and dried. Samples were reconstituted in 0.1% TFA and fractionated using the High pH Reversed-Phase Peptide Fractionation Kit (Pierce 84868) according to manufacturer’s recommendations. Eluted fractions were dried, and reconstituted in 5% acetonitrile, 5% formic acid for LC-MS analysis.
Mass spectra were collected on Orbitrap Lumos mass spectrometer (ThermoFisher Scientific) coupled to a Proxeon EASY-nLC 1200 LC pump (ThermoFisher Scientific). Peptides were separated on a 35 cm column (100 μm i.d., Accucore, 2.6 μm, 150 Å, packed in-house) using a 180 min gradient (from 5%-30% acetonitrile with 0.1% formic acid) at 500 nl/min. Each analysis used an SPS-MS3-based TMT method (McAlister et al., 2014; Ting et al., 2011), which has been shown to reduce ion interference compared to MS2-based quantification (Paulo et al., 2016). MS1 data were collected using the Orbitrap (120,000 resolution; maximum injection time 50 ms; AGC 4e5, 400-1400 m/z). Determined charge states between 2 and 5 were required for sequencing and a 90 s dynamic exclusion window was used. MS2 scans consisted of collision-induced dissociation (CID), quadrupole ion trap analysis, automatic gain control (AGC) 2E4, NCE (normalized collision energy) 34, q-value 0.25, maximum injection time 35 ms, and isolation window of 0.7 Da using a Top10 method. MS3 scans were collected in the Orbitrap at a resolution of 50,000, NCE of 45%, maximum injection time of 100 ms, and AGC of 1.5e5.
Mass spectra were processed using a pipeline featuring the COMET search engine (Eng et al., 2013). Data were searched against forward and reversed sequences from the UniProt Human database (downloaded July 2020) with common contaminants appended, using a 20-ppm precursor ion tolerance for total protein-level analysis and 0.9 Da product ion tolerance. TMT tags on lysine residues and peptide N termini (11-plex: +229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.9949 Da), deamidation on asparagine and glutamine (+.984016 Da), and phosphorylation with neutral loss (+79.9663 Da, −97.9769 Da) were set as variable modifications. Peptide-spectrum matches (PSMs) were identified, quantified, and filtered to a 1% peptide false discovery rate (FDR) and then collapsed further to a final protein-level FDR of 1%. Sites were further localized using the AScore algorithm (Beausoleil et al., 2006) and only localized sites (AScore > 13) were retained. Phosphorylation sites and site combinations were quantified by summing reporter ion counts across all matching PSMs. Briefly, a 0.003 Da (3 millidalton) window around the theoretical m/z of each reporter ion was scanned and the maximum intensity nearest the theoretical m/z was used. Reporter ion intensities were adjusted to correct for the isotopic impurities of the different TMT reagents according to manufacturer specifications and adjusted to normalize ratios across labelling channels (column normalized). Lastly, for each site or site combination, signal-to-noise (S:N) measurements of the peptides were summed and then normalized to 100.
Identification of Phosphopeptides in AP-MS Data
To assess phosphorylation of ephrin receptors and ligands in 293T and HCT116 cells, RAW files corresponding to IP’s where these proteins were targeted as baits were selected and re-searched allowing for an expanded set of modifications. Sequest (Eng et al., 1994) searches were performed as described above (“Identification of Peptides and Proteins”), except that variable modifications included phosphorylation of Ser, Thr, and Tyr (+79.96633); oxidation of Met and Cys (+15.99491); and N-terminal acetylation (+42.01056). Peptide-spectral matches were filtered to a 1% FDR using a target-decoy approach (Elias and Gygi, 2007) coupled with linear discriminant analysis for multivariate filtering (Huttlin et al., 2010). The database used for searching included all Uniprot human protein sequences (downloaded in 2019) in forward and reverse orientation, as well as affinity tag sequences and common contaminant proteins. The numbers of unique phosphopeptide sequences observed matching each ephrin receptor or ligand as a bait were then tallied. Only ephrin receptors and ligands that have been profiled as baits in both 293T and HCT116 cell lines were considered.
Identification of SV40 Large T Antigen Peptides in AP-MS Data
To assess TP53 interactions with the SV40 Large T antigen (LgT), RAW files corresponding to technical duplicate analyses of TP53 pull-downs in 293T and HCT116 cells were re-searched using Sequest (Eng et al., 1994). Searches were performed using the standard protocol described above (“Identification of Peptides and Proteins”), except that the database was derived from Uniprot sequences downloaded in 2019 and included the SV40 Large T antigen protein. Peptides were filtered using the target-decoy approach (Elias and Gygi, 2007) coupled with linear discriminant analysis (Huttlin et al., 2010). As expected, no LgT peptides were detected in HCT116 cells, as LgT is not expressed in this cell line. To evaluate whether LgT was enriched in the TP53 pulldown, RAW files corresponding to an additional 100 randomly selected IP’s were likewise searched. The numbers of LgT peptides detected per run in these background IP’s were used to derive Z-scores for the numbers of LgT peptides observed in both technical replicates of LgT. Significant enrichment was observed in each case.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise indicated, all network analyses were performed in Mathematica 11.3 or 12.0 (Wolfram Research). All statistical tests have been adjusted for multiple testing correction using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). All figures were plotted in Mathematica and assembled in Adobe Illustrator.
BioPlex 3.0 Literature Comparison (Figure 1)
Comparison with BioGRID and PubMed
To determine the extent to which BioPlex 3.0 interactions had been reported previously in the literature, we compared our interaction network against interactions compiled by BioGRID (Oughtred et al., 2018). Human interactions reported in BioGRID version 3.5.167 (November 2018) were downloaded and filtered to remove interactions from previous published and unpublished BioPlex networks. We then searched for each BioPlex interaction in the filtered BioGRID dataset; if a match was found, we next queried whether the interaction was reported in high-throughput or low-throughput studies as recorded by BioGRID. If at least one reference pointed to a low-throughput study, that edge was counted as reported in low-throughput; alternatively, if all references pointed to high-throughput studies, the edge was counted as confirmed through high-throughput studies; finally, if no matches were found independent of previous BioPlex datasets, the edge was counted as unique to BioPlex.
PubMed publication counts associated with each human gene were obtained via FTP from NCBI in December 2018. Proteins in the BioPlex 3.0 network were then assigned to 100 bins corresponding to percentiles based on their associated citation counts. Each interaction in BioPlex was then categorized based on whether it was confirmed in low- or high-throughput or novel to BioPlex and binned according to the percentiles of its two associated proteins, producing the density plots displayed.
BioPlex Network Validation (Figure S1)
Interaction Enrichment among CORUM Complexes
CORUM (Giurgiu et al., 2018) is a manually curated database that reports core protein complexes that is frequently used as a gold standard. Each complex is reported as a list of proteins that assemble to form each complex. We employed a statistical approach to measure the extent to which each CORUM complex was reflected in the architecture of BioPlex and other human interaction networks. The basic assumption of our analysis is that the proteins within a CORUM complex will interact with each other with a frequency that significantly exceeds global connectivity of the network.
For this analysis, each human complex in CORUM’s core complex list (coreComplexes.txt; downloaded in September 2018) was mapped to the interaction network individually. To be eligible for scoring, CORUM complexes were required to have at least 3 members, one or more of which were targeted as baits in BioPlex 3.0. For each complex, the subnetwork defined by its members was extracted and the interactions within were counted. The enrichment of interactions among complex members was calculated using a one-sided binomial test assuming a background probability of interaction equal to the interaction density of the interaction network. This analysis returned p-values that were adjusted for multiple testing (Benjamini and Hochberg, 1995). A complex was considered enriched with interactions when the p-value was less than 0.01 after multiple testing correction.
In addition to BioPlex 3.0, we used this method to assess CORUM complex enrichment in the BioPlex HCT116 network and several published datasets, including BioPlex 1.0 (Huttlin et al., 2015) and BioPlex 2.0 (Huttlin et al., 2017), along with other datasets acquired via AP-MS (Hein et al., 2015), yeast-two-hybrid (Luck et al., 2020; Rolland et al., 2014; Rual et al., 2005), and correlation profiling (Havugimana et al., 2012; Wan et al., 2015). In addition, we scored two versions of HuMAP (Drew et al., 2017, 2020). HuMAP 1.0 was derived from re-analysis of data combined from several of these other studies (Hein et al., 2015; Huttlin et al., 2015; Wan et al., 2015) while HuMAP 2.0 additionally incorporates data from BioPlex 2.0 (Huttlin et al., 2017) and other datasets (Boldt et al., 2016; Gupta et al., 2015; Treiber et al., 2017; Youn et al., 2018). Importantly, to enable comparisons across datasets, identical complexes were scored in each. This means sometimes complexes were scored in a relatively small network even when its constituent proteins were not detected or when none of its member proteins had been targeted for AP-MS analysis. The CORUM coverages reported thus both reflect the scope of each network as well as the extent to which each complex was found to be highly interactive.
Randomization of the BioPlex Network
Several analyses described below use randomized interaction networks to define null distributions against which score distributions derived from BioPlex 3.0 may be compared. One important consideration in randomizing the BioPlex network is that it contains nodes of two types – baits and preys – with slightly different properties. Most significantly, baits have greater numbers of interactions on average, compared to proteins detected only as preys. This feature must thus be maintained within the randomized network. During network randomization, total numbers of edges and nodes were maintained, and vertex degree was conserved for every protein. We further ensured randomized edges still connected baits (i.e. proteins targeted for AP-MS in BioPlex 3.0) with proteins in fact detected as preys. In other words, randomized edges were not allowed to connect pairs of proteins for which neither had been targeted for affinity purification. Furthermore, randomized edges were only allowed to connect pairs of baits if at least one of them was detected as a prey as well.
Subcellular Fractionation Correlation among BioPlex Interacting Proteins
Because proteins must at least partially co-localize to interact, we used subcellular fractionation profiles (‘Subcell Bar Codes’) for human proteins taken from a previously published study (Orre et al., 2019). Within each network (293T and HCT116), every protein was matched with its subcellular fractionation profile. Then each edge was scored by calculating the Pearson correlation coefficient for the subcellular fractionation profiles of its constituent proteins. If one or both proteins could not be matched with its subcellular fractionation profile, then that interaction was skipped. In parallel, edges of an equivalent randomized network were also scored. Because separate ‘Subcell Bar Codes’ were reported for four different cell lines – A431, H322, MCF7, and HCC827 – this entire procedure was repeated four times and Pearson correlation coefficients were averaged across cell lines for each edge in the real and randomized BioPlex networks. Distributions of real and random correlation coefficients were compared using a Cramér-von Mises test.
Size-exclusion Chromatography Elution Profile Correlation among Interacting Proteins
To evaluate the tendency of interacting proteins pairs in the BioPlex to co-fractionate during size-exclusion chromatography, we mapped previously published profiles (Heusel et al., 2019) obtained from fractionation of 293T lysate to proteins in the BioPlex networks (293T and HCT116). Each interaction was scored by calculating the Pearson correlation coefficient between co-elution profiles of its two constituent proteins. If one or both proteins did not have associated co-fractionation data, that interaction was skipped. In parallel, edges of a randomized network were also scored. Distributions of correlation coefficients were compared with a Cramér-von Mises test.
Analysis of BioPlex Reciprocal Interactions
Rates of reciprocal interaction within the BioPlex 293T and HCT116 networks were determined according to the following procedure. Reciprocal interactions were tallied within the target network and 1.0 randomized versions. Counts calculated from randomized networks were used to determine a mean and standard deviation for the null distribution and convert the reciprocal count observed into a Z-score. A Z-test was subsequently performed to derive an associated p-value.
To determine the reciprocal rate observed in each BioPlex network, the total number of reciprocal interactions was compared against the number of edges that were eligible for reciprocal detection. To be eligible for reciprocal detection, an edge had to connect two proteins both of whom 1) were targeted for AP-MS as a bait; and 2) were detected as a prey in 293T cells.
Analysis of BioPlex Cliques
Cliques of three mutually interacting proteins (3-cliques) were tallied in the BioPlex networks (293T and HCT116) using Mathematica. This process was repeated in 1,000 randomized networks as well. After calculating a mean and standard deviation for the resulting null distribution, a z-score was calculated for the observed 3-clique count in each network and a Z-test was performed to determine its associated p-value.
Analysis of AP-MS Replicates within and between Cell Lines
To evaluate reproducibility of AP-MS in 293T cells, selected baits were targeted a second time for AP-MS. These replicate IP’s were performed under identical conditions and were scored against the same “stats” table used for generation of the 293T network with identical filtering to identify interacting proteins.
To determine the replication rate, each replicate IP was matched with the original pull-down of the same clone in the BioPlex 3.0 network (Table S2A). Those interactions resulting from pull-down of the target clone in the original BioPlex 3.0 network were extracted and each edge was labeled according to whether that interaction was also observed in the replicate IP. An interaction was considered replicated if it was 1) detected in the original BioPlex 3.0 IP among the top 2% of all bait-prey pairs in all 293T IP’s; and 2) was detected in the replicate IP with a score high enough to rank in the top 5% of all bait-prey pairs in all 293T IP’s. See Table S2B for a summary of interaction replication.
The same basic procedure was repeated to assess the replication rate between 293T and HCT116 cells. First, IP’s performed in both cell lines were aligned according to the specific clone used for AP-MS analysis. Then for each shared clone, interactions were extracted from the BioPlex 3.0 network that resulted from its pulldown. Each interaction was considered replicated if it was 1) detected in the original BioPlex 3.0 IP among the top 2% of all bait-prey pairs in all 293T IP’s; and 2) it was detected in the HCT116 IP with a score high enough to rank in the top 5% of all bait-prey pairs in all HCT116 IP’s.
In total, 72 replicate IP’s were also performed in HCT116 cells as part of normal AP-MS pipeline operation. Replication rates were calculated essentially as described above. One complication of this is that when the final HCT116 network was assembled and more than one IP was available for a given clone, we systematically chose to include the IP with the larger number of interacting proteins in the final network. Thus, to avoid bias when calculating the HCT116 replication rates displayed in Figure S1M, we randomly selected which replicate would be used as the basis for comparison for each clone. See Table S2.
Comparison of 293T and HCT116 Networks (Figure S3)
Quantitative Analysis of HCT116 and 293T Proteomes
Following normalization and scaling to report each protein’s TMT expression profile as a fraction of total observed signal, unpaired T-Tests were performed assuming unequal variance for each quantified protein. Differentially expressed proteins were defined based on an absolute log2 fold change greater than 0.5 coupled with a p-value smaller than 0.01 following multiple testing correction (Benjamini and Hochberg, 1995). Differentially expressed proteins were further grouped according to whether they were elevated in 293T or HCT116 cells and GO enrichment analysis was performed. GO enrichments were calculated using GO classifications downloaded from www.geneontology.org in November 2018 and used as background the set of all proteins quantified via TMT. Enrichment was calculated based on a one-sided hypergeometric test with subsequent multiple testing correction (Benjamini and Hochberg, 1995). See Table S4A.
Comparison of Bait Expression Levels
Relative bait expression levels were approximated in 293T and HCT116 cells by comparing numbers of spectral counts observed matching each bait in paired IP’s. For this comparison, 293T and HCT116 IP’s were aligned according to the precise clone used for AP-MS. A sign test was used to determine whether the median PSM difference (PSM293T – PSMHCT116) across baits is zero or not.
Quantifying Effects of Bait Expression on Interaction Replication Rates
To assess the effects of bait expression on interaction replication across 293T and HCT116 cells, IP’s were aligned according to the specific clone used for bait expression and the replication of each interaction was assessed as described above (Analysis of AP-MS Replicates within and between Cell Lines). Baits were then binned by relative expression ratio along with their associated interactions. Interactions in each bin were subsequently tallied to determine the fraction of interactions confirmed across cell lines.
Quantifying Effects of Prey Expression on Interaction Replication Rates
To assess the effects of bait expression on interaction replication across 293T and HCT116 cells, IP’s were aligned according to the clone used for bait expression. Those successfully targeted in both 293T and HCT116 cells were identified and their resulting interactions extracted from the larger networks. Replication of each interaction was assessed as described above (Analysis of AP-MS Replicates within and between Cell Lines). Preys associated with these selected interactions were then matched with their relative expression levels in 293T versus HCT116 cells as measured via TMT (see Quantitative Analysis of HCT116 and 293T Proteomes). Interactions were binned according to each prey’s expression ratio and then the rate of replication was determined within each bin.
Analysis of BioPlex Interactions in a Structural Context (Figure 2)
Converting PDB Structures to Interaction Networks
Proteins in the BioPlex networks were linked to specific PDB structures via Uniprot ID via the SIFTS project (Dana et al., 2018). PDB structures were filtered to consider only those that i) contained at least 3 human proteins; ii) contained at least 3 proteins present in the combined BioPlex network; iii) included at least two BioPlex baits; iv) included at least two proteins matching chains in the PDB structure with minimum length 25 amino acids; and v) for which at least one interaction was detected in the BioPlex network among its constituent proteins. In cases where multiple structures corresponded to the same protein complex, the most recent structure was selected and other redundant structures were set aside. Structures were drawn from the RCSB PDB website (www.rcsb.org) (Berman et al., 2000). Images of structures shown in Figure 2A, ,FF–H were produced using Mol* (Sehnal et al., 2018).
Interactions among proteins in each PDB structure were inferred by calculating the minimum distance separating atoms belonging to each protein. If the minimum distance separating atoms matching two proteins was 6 A or less, these proteins were defined to interact directly; all other pairs of proteins that occurred in the same structure were assumed to interact indirectly. Using these rules, three-dimensional structures of each protein complex were converted into network representations (Figure 2A, ,FF–H).
Mapping BioPlex Interactions onto PDB Structures
Once network representations of each structure were created, BioPlex interactions were superimposed by mapping Entrez Gene ID’s to Uniprot ID’s. When assessing the fraction of PDB interactions detected by BioPlex (Figure 2B), PDB interactions were filtered to include only those edges that would be detectable in BioPlex. To meet this requirement, at least one member of each interacting protein pair had to be targeted as a bait in at least one cell line (293T or HCT116) and both proteins had to match chains with minimum length 25 amino acids in the PDB structure. This latter condition was necessary to avoid PDB structures involving synthetic peptides and protein fragments (e.g. HLA complexes). Those BioPlex interactions that mapped to PDB structures were subsequently partitioned according to whether those interactions were classified as ‘direct’ or ‘indirect’ interactions in the PDB structure according to the definitions defined above. See Table S3.
Analysis of Interaction Replication versus Distance
The relationship between an interaction’s likelihood of detection in both cell lines and the observed distance between interacting proteins within the protein structure was examined in multiple ways. First, all 2,594 BioPlex interactions mapped to 309 PDB structures via the process described above were partitioned according to the intra-structure distance separating each interacting protein pair. These interactions were then divided into eight equally-sized bins according to distance and the fraction of edges in each bin shared between cell lines was calculated. This fraction was then expressed as the odds of replication, defined as the ratio of shared to cell-line-specific edges (Figure 2C).
Separately, PDB structures for which at least one direct interaction was identified in BioPlex were identified. Within each of these 306 PDB structures the fraction of direct edges that were 293T-specific, HCT116-specific, or detected in both cell lines was determined and plotted in a ternary diagram (Figure 2D). Likewise, the 132 PDB structures containing at least one indirect interaction detected in BioPlex were identified, and the indirect edges were subdivided according to whether they were 293T-specific, HCT116-specific, or detected equally in both cell lines (Figure 2E).
Plotting of Specific Protein Complexes
The structure of the U1 snRNP shown in Figure 2A corresponds to PDB structure 3pgw (Weber et al., 2010). Similarly, the structures of TFIIH, Ribonuclease P, and CDC45-MCM-GINS Helicase shown in Figure 2F–H correspond to PDB structures 6nmi (Greber et al., 2019), 6ahr (Wu et al., 2018), and 6xtx (Rzechorzek et al., 2020), respectively.
Analysis of Protein Expression among BioPlex Subnetworks (Figure S4)
Protein expression within complexes displayed in Figure S4 was assessed by retrieving relative protein expression data for each complex member from the TMT dataset described above (“Quantitative Proteomic Comparison of 293T and HCT116 Cells”). The average and standard deviation were calculated for all matching Log2 ratios and reported. To evaluate significance of these values, empirical p-values were determined by randomly drawing equal numbers of quantified proteins from the total set of TMT-quantified proteins and calculating their means and standard deviations. This process was repeated 10,000 times and empirical p-values were determined by calculating the fraction of random samples with mean or standard deviation at least as extreme as those observed.
Analysis of Shared and Cell-specific Interactions (Figure S5)
Quantification of Overlap among Edges Eligible for Detection in both Cell Lines
Because fewer AP-MS experiments have been completed in HCT116 cells, a significant fraction of interactions in the 293T network are unique to that network simply because the appropriate IP’s have not yet been performed in HCT116 cells. Such interactions that could only be detected in a single cell line were excluded by filtering interactions to include only those for which at least one constituent protein was targeted as a bait in both 293T and HCT116 cells. Note that the definition used here is less stringent than that described for Figure S1. In Figure S1 we required matched clones targeting the same bait protein to pull down the same prey in each cell line. Here we do not necessarily require the same protein to have been targeted as a bait in both cell lines. For example, a hypothetical interaction between two proteins A and B would be considered eligible for detection in both cell lines when protein A was a bait in 293T cells only and protein B was a bait in HCT116 cells only; moreover, this interaction could be replicated in both cell lines if we detected the directed interaction A → B in 293T cells and the complementary directed interaction B → A in HCT116 cells. This relaxed definition allowed us to retain a larger fraction of the overall interaction space for the comparative analyses presented below.
Comparison of Network Properties for Shared and Cell-line-specific Edges
To determine whether interactions shared across cell lines were likely to reside in more central network locations, edge betweenness centrality was calculated for all edges in the combined 293T/HCT116 network using Mathematica 12.0. Edges were subsequently filtered to include edges detectable in both cell lines as described above and partitioned into 10 equal bins according to their betweenness centrality. Within each bin the fraction of edges observed in both cell lines was determined. Error bars represent bootstrapped 95% confidence intervals.
Similarly, local clustering coefficients were calculated for each interaction observed in the combined 293T/HCT116 network by extracting the subnetwork bounded by the first-degree neighbors of both constituent proteins and calculating its clustering coefficient. As before, edges detectable in both cell lines were partitioned into 10 equal bins according to these clustering coefficients and within each bin the fraction of edges observed in both cell lines was determined. Error bars represent bootstrapped 95% confidence intervals.
Essentiality and Interaction Overlap Between Cell Lines
To evaluate any relationship between protein essentiality and interaction overlap between cell lines, proteins in the combined 293T/HCT116 network were labeled as either ‘essential’ or ‘not essential’ according to specific definitions described below. Interactions eligible for detection in both cell lines were then extracted from the combined network and binned according to whether 0, 1, or 2 of their constituent proteins met the indicated definition for essentiality. Finally, edges in each bin were labeled as ‘shared’ or ‘cell-line specific’ and the fraction of edges shared was calculated. Bootstrapped 95% confidence intervals were calculated as well.
This analysis was repeated three times, each time defining ‘essential’ genes according to separate datasets. First, we followed the criteria described by Wang et al. and deemed proteins with gene-based CRISPR scores < 0.1 and corrected p-value < 0.05 in KBM7 cells as ‘essential’ (Wang et al., 2015). Second, we counted as ‘essential’ all genes identified by Blomen et al. as fitness-associated in either KBM7 or HAP1 cells (Blomen et al., 2015). Third, we used ‘common essential’ genes as derived previously (Dempster et al., 2019) from Achilles data (Meyers et al., 2017; Tsherniak et al., 2017).
Interaction Overlap and Protein Expression Variability
In Figure S3, we observed that differential protein expression contributes to cell-line specificity among protein interactions. Expanding on this point, we wanted to see whether proteins involved in cell-line-specific interactions are variably expressed more generally across other biological contexts. For this purpose, we obtained datasets reporting protein expression across 378 human cancer cell lines (Nusinow et al., 2020) and gene and protein expression across 29 human tissues (Wang et al., 2019). Proteins in the cancer cell line dataset were ranked according to expression variability as measured by each protein’s standard deviation across cell lines; similarly, proteins and genes in the human tissue datasets were ranked according to relative standard deviations observed across tissues. In each case, proteins or genes were partitioned into 10 equal bins. Interactions eligible for detection in both cell lines were then extracted from the combined 293T/HCT116 network and binned according to the ranked expression variability of each constituent protein. Finally, within each bin the fraction of edges observed in both cell lines was determined.
Evolutionary Analysis of BioPlex Edges (Figure S6)
To assess the evolutionary context of shared and cell-line-specific interactions, proteins within the combined 293T/HCT116 network were matched with protein evolutionary age data as published previously (Liebeskind et al., 2016). Following the authors’ guidelines, we only accepted protein age estimates with the following parameters: entropy < 1.0, hgt_flag = false, numDBsContributing > 3, and Bimodality < 5. Proteins without evolutionary ages from this study meeting these criteria were excluded from this analysis. Each evolutionary split was assigned with an approximate date using TimeTree.org. The category Eukaryota + Bacteria was assigned the midpoint between dates associated with the emergence of Eukaryotes and Opisthokonts.
In some contexts (e.g. Figure S6D), interactions were binned according to the evolutionary ages assigned to both of their constituent proteins; however, in other situations (e.g. Figure S6A/C) it was necessary to assign a single evolutionary age to each edge. In these cases, edges were assigned the age of their younger constituent protein. Edges for which one or more proteins could not be assigned ages were excluded from this analysis.
In these analyses, edges were deemed eligible for detection in both cell lines according to the same criteria described above for Figure S5 (Quantification of Overlap among Edges Eligible for Detection in both Cell Lines).
Analysis of Functionally Defined Subnetworks (Figure 1G–K/3)
To determine the extent to which functionally defined subnetworks of the human interactome were conserved across 293T and HCT116 cell lines, we extracted subnetworks corresponding to a wide range of functionally defined categories. These include CORUM (Giurgiu et al., 2018) complexes (coreComplexes.txt; downloaded from https://mips.helmholtz-muenchen.de/corum/ in September 2018); Reactome (Fabregat et al., 2017) pathways (downloaded via www.uniprot.org in January 2019); GO Biological Process, Cellular Component, and Molecular Function (Ashburner et al., 2000) categories (www.geneontology.org; November 2018); and DisGeNET (Piñero et al., 2017) disease associations (www.disgenet.org; May 2019).
When calculating overlap of interactions within subnetworks for this analysis, interactions were only counted if at least one of their constituent proteins was targeted for AP-MS analysis in both cell types. This corrects for the fact that some interactions are not detectable in one cell line or the other simply based on the baits targeted in each. However, when subgraphs were plotted in the figures, all matching proteins and interactions were displayed including edges that could only be detected in a single cell line. Results are summarized in Table S5.
Discovery of Shared and Cell-line-specific Network Communities (Figure 4)
The combined 293T/HCT116 interaction network was partitioned into communities as described previously (Huttlin et al., 2017). Briefly, MCL clustering (Enright et al., 2002) was used to subdivide the network into communities using an inflation parameter of 2.25 and setting the ‘force-connected’ option to yes. Communities with fewer than 3 members were discarded, leaving 1,423 communities overall (Table S6A–B).
After subdividing the combined 293T/HCT116 interaction network into communities, we next sought pairs of communities whose members were found to interact with unusually high frequency, as described previously (Huttlin et al., 2017). First, the full set of interactions was trimmed to include only those interactions connecting one community to another. For each pair of communities connected by one or more edges, the numbers of edges emanating from each were determined, as were the number of edges connecting the two. Fisher’s Exact Test was then used to assess whether edges connecting the two were enriched beyond random chance, followed by multiple testing correction (Benjamini and Hochberg, 1995). A total of 1,736 statistical associations were detected between communities at a 1% FDR (Table S6C).
Having identified communities and community associations in the combined 293T/HCT116 network, we next wanted to quantify the level of overlap observed for each among cell lines. To do this, we first filtered our list of communities down to 761 that contained at least one protein which had served as an AP-MS bait in both cell lines. Within each of these we assessed the overlap as discussed below (Table S6D). Similarly, we filtered 1,736 interactions down to a subset of 988 community-community associations ensuring that at least three interactions bridging these communities were detectable in both cell lines (Table S6E).
When calculating overlap within communities or between communities, interactions were only counted if at least one of their constituent proteins was targeted for AP-MS in both cell types. This accounts for interactions detectable in only a single cell line simply due to the specific AP-MS experiments completed in each. However, Figure 5E–G include all matching proteins and interactions, including those only eligible for detection in a single cell line.
Analysis of Consistency of Function among Interactors (Figure S7)
Enrichment analysis was performed to assess consistency of function among each protein’s first-degree neighbors. For each protein in the combined 293T/HCT116 network, we extracted all first-degree neighbors. Functional categories including GO-SLIM Biological Process, Molecular Function, and Cellular Component categories (www.geneontology.org; November 2018), PFAM domains and Reactome pathways (downloaded via www.uniprot.org; January 2019) were superimposed. Enrichment of each term within a protein’s first-degree neighbors was calculated using a one-sided hypergeometric test with multiple testing correction (Benjamini and Hochberg, 1995). Those significant at an FDR less than 1% were retained.
For every functional category deemed significant after enrichment analysis, those first-degree neighbors matching the category were assessed to determine what fraction were significant to 293T cells, significant to HCT116 cells, or common to both. At least five neighboring proteins were required to match a given functional category to be included in Figure S7B. Results are provided in Table S7C.
Domain-domain Associations across Cell Lines (Figure 6)
First, the complete set of interactions in the combined 293T/HCT116 network was extracted and mined for associations among protein domains as described previously (Huttlin et al., 2015). To start, all proteins in the network were linked to PFAM domains (El-Gebali et al., 2018) as recorded in Uniprot (downloaded in January 2019). Domain pairs connected by two or more interactions were assessed for significance using Fisher’s Exact Test after accounting for 1) the number of interactions connecting both domains; 2) the numbers of interactions involving either domain individually; and 3) the number of interactions not involving either domain. After multiple correction (Benjamini and Hochberg, 1995), those domain-domain associations significant at a 1% FDR were identified (Table S7A).
It is important to note that though our domain analysis identifies pairs of PFAM domains whose parent proteins interact preferentially across the network, this does not necessarily mean that these domains themselves are responsible for the interaction. Many may simply be passengers that associate thanks to interactions mediated by contacts elsewhere in the protein sequence. Examples of this latter case include associations among zf-C2H2 and SCAN domains as well as zf-C2H2 and BTB domains. Examples of direct and indirect domain associations relating to Cullin E3 ubiquitin ligase complexes were also described in detail following analysis of BioPlex 2.0 (Huttlin et al., 2017).
Second, each statistically significant PFAM domain association was assessed for overlap between cell lines (Table S7B). For each pair of associated domains, all corresponding interactions were identified. This list of interactions was then filtered to include only interactions detectable in both cell lines, in the sense that at least one member of each interacting protein pair must be a bait in both cell types. If at least five edges remained, these were then tallied in both cell lines to determine the proportions specific to a single cell type or shared across both. These 3,249 domain-domain associations were also assembled into a network whose nodes corresponded to PFAM domains and whose edges linked statistically associated PFAM domain pairs.
Identification of Functional Associations using Achilles Data (Figure 7)
To map functional relationships onto the combined BioPlex network, gene fitness profiles defined via CRISPR knockout or RNAi knockdown across hundreds of cell lines were superimposed onto the network. Fitness profiles were originally collected through Project Achilles (Meyers et al., 2017; Tsherniak et al., 2017) and were downloaded through their public data portal in May 2019. For every pair of interacting proteins in the combined BioPlex network, fitness profiles were sought. If fitness profiles were available for both interacting proteins, their functional similarity was assessed using Spearman’s Correlation; statistical significance was assessed using the CorrelationTest function in Mathematica 12.0 with subsequent multiple testing correction (Benjamini and Hochberg, 1995). Those BioPlex interactions found to match correlated fitness profiles at a 5% FDR are listed in Table S7D and were split into groups according to whether the observed correlation was positive or negative. Edges were also labeled to indicate whether they were observed in 293T cells, HCT116 cells, or both. Subnetworks corresponding to BioPlex interactions with either positive or negative fitness correlations were extracted and plotted using Gravity Embedding.
ADDITIONAL RESOURCES
In addition to the numerous resources described previously, we have also developed a full-featured web-based viewer, BioPlexExplorer, as a companion to this paper. For best results, we recommend using this tool in Chrome, though the full range of browsers are supported, including mobile browsers. This tool, available at bioplex.hms.harvard.edu/bioPlexExplorer, provides numerous features.
An integrated search engine for locating and viewing shared and cell-specific networks centered on one or more user-selected proteins. All interactions are displayed as a fully interactive network diagram. Additional data, including evolutionary age, protein expression levels, and GO category membership may be superimposed onto the networks. These networks may be downloaded as images (png) or as Cytoscape networks (json). All interactions are also displayed in sortable tabular format and may be exported in Excel, CSV, or PDF formats. The resulting network viewer also performs GO enrichment analysis on-the-fly. Finally, these network views may be shared with colleagues and collaborators via custom URLs.
A custom ternary plot viewer that enables the user to view interactive versions of the ternary diagrams shown in Figure 1G, Figure 3A, and Figure 5C. Through a drop-down menu the user may select specific annotation sets of interest, including GO Cellular Component, Molecular Function, and Biological Process, Reactome Pathways, CORUM Complexes, DisGeNET diseases, and BioPlex communities. By zooming in on these ternary plots and clicking on specific points, the user may see cell-line-specific interaction data for thousands of complexes, communities, and functional categories. Links also enable the user to easily open a network view corresponding to each category of interest as well.
A fully interactive version of Figure 5B, which depicts the full set of communities observed in the combined BioPlex network. Statistically significant associations among BioPlex communities are displayed as well. This interface is fully searchable, enabling a user to quickly and easily locate proteins of interest; moreover, upon selecting a node, a link is available to easily view the underlying interaction data in both 293T and HCT116 networks.
A fully interactive version of the PFAM Domain Association Network depicted in Figure 6A. Nodes correspond to specific PFAM domains while edges correspond to pairs of domains that associate statistically. The color of each edge indicates the extent that each domain association is reflected in 293T and HCT116 cells individually or in common. Clicking on individual nodes reveals lists of proteins that possess each domain; in addition, a link is provided to view the interaction subnetwork defined by proteins with the indicated domain.
Fully interactive versions of Figure 7B//C,C, depicting BioPlex subnetworks that exhibit either positive or negative fitness correlations. Again, these networks are fully searchable and interactive.
Supplementary Material
1
Figure S1. Internal and External Validation of the BioPlex Networks. Related to Figure 1.
(A) – (C) BioPlex validation against CORUM complexes. To quantify whether each CORUM complex is reflected in each interaction network, the subnetwork defined by its constituent proteins was extracted from each network and the enrichment of interactions calculated relative to global network density. Focusing on enrichment, rather than interaction counts, is useful when comparing networks of variable size: smaller networks with fewer edges have lower interaction probabilities, so fewer edges are required to attain significance; in contrast, a larger network with more interactions has a higher probability of interaction and the number of interactions required for significance increases accordingly.
(A) CORUM complex coverage within the BioPlex 3.0 network. Subnetworks corresponding to four CORUM complexes are displayed. Deep red nodes were targeted as baits while light red nodes were detected as preys only. Interactions observed via AP-MS are highlighted in red.
(B) We show the fraction of CORUM complexes that were found to be significantly enriched for protein interactions (1% FDR) in nine different interaction datasets. Datasets are grouped according to the experimental methods used to create each. Note that some CORUM complexes may not be detectable in one or more AP-MS datasets because no complex members were targeted for AP-MS. No correction was applied because it would complicate comparisons across datasets, especially using different experimental approaches.
(C) TFIIH Transcription Factor Complex coverage across BioPlex networks. Dark-colored nodes were targeted in baits in each network while light-colored nodes were present only as preys. Nodes with dashed outlines were not detected in the network. Observed interactions are colored.
(D) To assess the tendency for interacting proteins to co-fractionate, size exclusion chromatography (SEC) elution profiles (Heusel et al. 2019) were correlated for all interacting proteins in BioPlex. Distributions of correlation coefficients are displayed for both real and randomized networks, as above. P-value: Cramer-von Mises test.
(E) To assess the tendency for interacting proteins to co-localize, subcellular fractionation profiles (Orre et al. 2019) were correlated for all interacting protein pairs in BioPlex. In tandem, we repeated the analysis for a randomized version of the BioPlex network. Distributions of correlation coefficients are displayed for both real and randomized networks. P-value: Cramer-von Mises test.
(F) – (J) Intra-network validation of BioPlex interactions.
(F) Representative reciprocally interacting protein pairs from BioPlex 3.0.
(G) Representative 3-cliques from BioPlex 3.0.
(H) Estimating reciprocity in the 293T and HCT116 networks. To be validated reciprocally in either network, both interacting proteins must have been targeted as baits and must be detectable as preys. The bar chart depicts the fractions of BioPlex edges either observed or eligible for reciprocal detection in each network.
(I) Assessing enrichment of reciprocal interactions in BioPlex 293T and HCT116 networks. Reciprocal interactions were counted in BioPlex 293T and HCT116 networks. The process was then repeated with 100 randomized versions of each network to derive empirical null distributions that could be used to derive Z-scores and p-values (see Methods).
(J) Assessing enrichment of 3-cliques in BioPlex 293T and HCT116 networks. 3-cliques were counted in BioPlex 293T and HCT116 networks. The process was then repeated with 100 randomized versions of each network to derive empirical null distributions that could be used to derive Z-scores and p-values (see Methods).
(K) The fraction of edges in each network that are confirmed intra-network by reciprocal interactions or 3-cliques. Interactions confirmed through replicate AP-MS in the opposite network are shown as well.
(L) To assess reproducibility, we repeated AP-MS analysis of 999 bait proteins in 293T cells. The schematic on the left highlights the fraction of BioPlex 3.0 baits repeated for validation; the box-whisker plot at the bottom displays the fraction of interactions that were confirmed upon replication for each bait. A subset of these interaction networks are displayed above the box-whisker plot in positions that approximately correspond to their observed replication rate (written in green). Replicated baits are red; replicated preys are green while preys detected in the initial IP only are light blue. Replicated edges are green while edges only detected in the initial IP are light blue. Gray nodes and dotted edges depict additional confirmatory interactions in the BioPlex 3.0 network.
2
Figure S2. C11orf49 Associates with the Tubulin Polyglutamylase Complex. Related to Figure 2.
(A) C11orf49 binds to all members of the TTLL1 polyglutamylase complex (highlighted in green) in both 293T and HCT116 cells. Interactions with the pericentriolar marker protein PCM1 are shown as well.
(B) – (C) Endogenous C11orf49 binds to members of the TTLL1 polyglutamylase complex. C-terminally FLAG-HA-tagged TTLL1 complex proteins or GFP were expressed in 293T or HCT116 cells followed by FLAG or HA purification and Western blotting with endogenous C11orf49 and TPGS1 antibodies.
(D) Localization of TTLL1 complex members and C11orf49 long and short isoforms. The longest C11orf49 isoform (isoform 2) along with a truncated isoform (isoform 3) were expressed in U2OS cells. Cells plated on high precision coverslips were fixed and immunostained with anti-HA antibodies to visualize ectopic protein localization via confocal microscopy. Max intensity projections of 2-μm z-stacks are shown; scale bar represents 20 μm.
(E) C11orf49 exhibits pericentriolar localization. U2OS cells stably expressing C11orf49 or TTLL1 complex member LRRC49 were fixed, stained with anti-HA and anti-PCM1 (pericentriolar marker) antibodies and visualized by confocal microscopy. Single planes of a z-stack are shown; scale bar represents 20 μm.
(F) Staining with PCM1 alone is shown as a control for signal bleed from the 488 channel.
(G) Cells expressing C11orf49-HA-FLAG were fixed and stained with C11orf49 and γ-tubulin endogenous antibodies. A single plane of a 2 μm z-stack is shown; scale bar represents 20 μm.
3
Figure S3. 293T and HCT116 Cells Afford Complementary Perspectives on the Human Proteome and Interactome. Related to Figure 3.
(A) 293T and HCT116 cell lines arise from different sources and exhibit distinct phenotypes as summarized.
(B) To explore how the divergent properties of 293T and HCT116 cells manifest within their proteomes, we compared biological quintuplicates of each cell line using TMT isobaric labeling coupled with Real-Time Search MS3 quantitation (Erickson et al. 2019). Protein expression profiles are summarized in the heatmap. GO enrichment analysis was applied to proteins elevated in each cell line. Enriched GO terms are summarized in the table below.
(C) – (F) TMT profiles observed for selected cell-line-specific proteins that are markers for specific phenotypes. Bar heights depict relative reporter ion intensities that have been normalized so that each TMT profile sums to 100% across all channels. P-values were calculated with T-tests and corrected for multiple testing.
(C) Expression of Y-chromosomal protein EIF1AY is specific to HCT116 cells, reflecting their male origin.
(D) 293T-specific expression of ciliary protein CEP290 reflects general increases in ciliary proteins in this cell line (Figure S3B).
(E) Epithelial marker proteins are upregulated in HCT116 cells.
(F) Mesenchymal marker proteins are upregulated in 293T cells.
(G) Histogram depicting the relative abundances of each bait expressed in 293T and HCT116 cells. Log2 ratios were calculated based on spectral counts observed for each bait in each cell line. Bait abundances are strongly reduced in HCT116 cells (p ~ 0; Sign Test).
(H) Interaction confirmation versus bait expression. Baits targeted in both cell lines were assigned to 5 bins according to relative bait expression as reflected by numbers of bait peptides in each IP. Box-whisker plots depict fractions of interactions confirmed for baits in each bin.
(I) Interaction confirmation versus prey expression. Baits targeted in both cell lines were extracted from the larger interaction networks along with their associated prey proteins. These interactions were then divided into bins based on prey expression as measured by TMT. Box-whisker plots depict fractions of interactions confirmed for preys in each bin.
(J) – (M) Interaction networks of selected cell-line-specific proteins. Node colors reflect the relative abundance of each protein in 293T and HCT116 proteomes as measured via TMT. Edges observed in both cell lines are gray while 293T-specific edges are red and HCT116 edges are blue. Numbers in parentheses after each protein are the log2 ratios of expression in HCT116 cells relative to 293T cells.
4
Figure S4. Relating Protein Expression and Cell-specific Interactions. Related to Figures 1–5.
(A) – (M) Subnetworks corresponding to specific complexes, pathways, and communities highlighted elsewhere in the paper are displayed with relative protein expression levels superimposed. Network layouts mirror those presented in other figures. Edges observed in both cell lines are gray while 293T- and HCT116-specific edges are red and blue, respectively. Relative protein expression levels were determined via TMT (Figure S3B) and are depicted by node color using a 3-color gradient ranging from red to blue, with white indicating equal expression. Proteins without quantitative data are gray.
Below each network is a plot showing where proteins in each network fall relative to all quantified proteins. The full distribution of protein expression values is depicted in gray with proteins in each community rendered in red. Outliers are labeled.
Underneath each distribution is an arrow labeling the mean expression value for all members of each complex. P-values listed for each indicate whether the observed mean differs significantly from that observed upon 10,000 random draws of equivalent protein numbers from the entire population of quantified proteins. Values are highlighted in green if the mean differs from zero at p < 0.05.
Above each distribution are brackets indicating the standard deviations among protein expression ratios for each network. Brackets are centered on the observed mean expression for each complex and span +/− one standard deviation. Dashed gray brackets depict the mean standard deviation observed across 10,000 random samples of equal size; solid brackets depict the standard deviation observed for proteins in each complex. P-values indicate whether the observed standard deviation differs significantly from that observed upon random draws of equivalent protein numbers from the entire population of quantified proteins. Values and brackets are highlighted in green if the standard deviation differs from the empirical null at p < 0.05.
(A) – (D) Protein expression data superimposed onto CORUM complexes from Figure 1H–K.
(E) Protein expression data superimposed onto the CDC45-MCM-GINS helicase complex fromc Figure 2H.
(F) – (I) Protein expression data superimposed onto Reactome pathways from Figure 3B,,CC,,DD & F.
(J) Protein expression data superimposed onto the DREAM complex from Figure 4H.
(K) – (M) Protein expression data superimposed onto BioPlex communities from Figure 5E–G.
5
Figure S5. Distinct Properties of Shared and Cell-line-specific Interactions. Related to Figures 3–6.
(A) – (B) The combined 293T/HCT116 network was filtered to include only edges eligible for detection in both cell lines (i.e. at least one protein in each interacting pair must be a bait in each cell line). These edges were then divided into deciles based on edge betweenness centrality (A) or local clustering coefficient (B) and the fraction of shared edges in each decile was calculated. Error bars: bootstrapped 95% confidence intervals.
(C) Edges detectable in both cell lines were grouped according to whether 0, 1, or 2 of their constituent proteins were classified as ‘essential’ or associated with increased cellular fitness in datasets from Wang et al. 2015, Blomen et al. 2015, or the Achilles project (Dempster et al. 2019). The edge overlap between cell lines was calculated for each class. Error bars: bootstrapped 95% confidence intervals.
(D) – (F) Edges detectable in both cell lines were binned according to the relative variability of expression observed for their constituent proteins across 378 human cancer cell lines (Nusinow et al. 2020) (D); expression patterns of proteins (E) and genes (F) across 29 human tissues were considered as well (Wang et al. 2019). The fraction of edges shared between cell lines was calculated for each bin. Bins with maximum and minimum overlap are labeled.
6
Figure S6. Differential Interactions Reflect Protein Evolution. Related to Figures 3–6.
(A) Consensus evolutionary ages from Liebeskind et al. 2016 were mapped onto the combined BioPlex network to assign ages to individual proteins; interactions were then assigned the age of their youngest constituent protein. Line plots show the fractions of BioPlex proteins and interactions that can be traced to each of 8 ages. Approximate dates for each split were taken from TimeTree.org. Only proteins whose evolutionary ages could be estimated with sufficient confidence were included in this plot (see Methods); likewise, interactions are only included if both constituent proteins could be assigned evolutionary ages. Interactions were further filtered to include only those involving at least one protein targeted for AP-MS in each cell line. In this panel and throughout the rest of the figure, a color gradient from red to blue is used to indicate the relative ages of each category. Red corresponds to the most ancient category, while blue indicates the most recent category. Evolutionary ages assigned to individual proteins and interactions may be viewed at bioplex.hms.harvard.edu/bioPlexExplorer.
(B) Simplified diagram showing phylogenetic relationships among assigned evolutionary ages.
(C) Interactions were binned according to estimated evolutionary age of their youngest constituent and filtered to include only those interactions that were detectable in both cell lines (i.e. involved at least one protein targeted as a bait in each cell line). The proportions of edges shared and unique to each cell line were then calculated within each bin. These proportions for each evolutionary age are plotted as points on a ternary diagram. The arrow highlights the progression toward higher cell-type specificity among interactions with more recent evolutionary origins.
(D) Interactions were again binned, this time accounting for the evolutionary ages of both older and younger members. Within each bin the fraction of edges shared between cell lines was calculated. This fraction is plotted on the heat map as a gradient from light to dark blue. Interaction overlap rates ranged from 6 – 86%.
(E) – (G) Heat maps depicting the evolutionary ages of interacting partners for selected BioPlex proteins. Selected proteins are labeled at the top of each plot and are depicted in the leftmost column of each heat map. Proteins identified as interactors are then displayed in decreasing order of age. Each column in the heat map is colored to indicate whether the associated interacting partner was observed in 293T cells (red), HCT116 cells (blue), or both (gray). Percentages to the right of each row indicate the overlap among interactions dating to the indicated age or before.
7
Figure S7. Interactions of Individual Proteins Reveal Cell-specific Variations on Consistent Biological Themes. Related to Figures 3–6.
(A) To evaluate consistency of biological function, individual proteins were selected from the combined 293T/HCT116 network along with their first-degree neighbors. Only proteins targeted as baits in both cell lines and found to bind at least one protein in each network were considered. For each protein, GO categories, Reactome pathways, and PFAM domains enriched among its primary neighbors were identified. As an example, the neighborhood surrounding ECHDC2 is displayed at the upper left; the target protein is black while shared nodes and edges are gray; those specific to 293T cells and HCT116 cells are red and blue, respectively. Among first-degree neighbors of ECHDC2 three terms were enriched (adjusted p < 0.01): Mitochondrion, Mitochondrial Organization, and Branched Chain Amino Acid Catabolism. These terms map to the primary neighbors as depicted in the central heat map: dark shading indicates that a prey matches the indicated classification. Within each category (i.e. row of the table) the relative proportions of edges shared or specific to a particular cell line can be tallied; each protein-function association can then be plotted within a ternary diagram as shown.
(B) Ternary diagram depicting the extent to which each association between bait and biological function is observed in both cell lines. This ternary diagram is accompanied by a histogram that shows the cell line specificity of these terms after effectively projecting the individual points onto the vertical edge of the ternary diagram. This graphically depicts the extent to which these bait – function associations are shared or cell-line-specific. To be included in this plot, at least 5 neighboring proteins had to match the indicated functional category and category had to be enriched at a 1% FDR.
(C) TRIM28 – KRAB domain functional association. Shown here are the interacting partners observed for the protein TRIM28 in both 293T and HCT116 cell lines. Nodes and edges are colored according to whether the interaction was observed in 293T cells (red), HCT116 cells (blue) or both (gray). Interactor names have been colored to indicate whether each contains a KRAB domain. The associated p-value reflects the enrichment of KRAB domains among first-degree neighbors of TRIM28. Inset histograms reflect the relative abundances of these TRIM28 interactors in 293T and HCT116 cells as measured by TMT (see Figure S2).
(D) – (F) Additional protein – function associations. Each panel depicts a protein of interest (in black) along with its first-degree neighbors, colored to indicate whether the interaction was observed in 293T cells (red), HCT116 cells (blue), or both (gray). Labels of interacting partners that match the indicated functional category or bear the indicated domain are highlighted in green. Interactions that do not involve the central proteins are rendered as thin black lines. P-values reflecting the strength of the protein-function association are displayed along with each title. Individual proteins whose functions relate to known phenotypes of 293T or HCT116 cells are labeled.
8
Table S1: BioPlex Networks, Related to Figure 1
(A) List of baits targeted for AP-MS in 293T cells
(B) List of interactions observed in 293T cells
(C) List of baits targeted for AP-MS in HCT116 cells
(D) List of interactions observed in HCT116 cells
(E) Merged list of interactions observed in either 293T cells, HCT116 cells, or both.
9
Table S2: AP-MS Replicates, Related to Figures 1 and S1.
(A) Summary of 293T replicate AP-MS experiments.
(B) Summary of interactions observed in 293T replicate experiments.
(C) Summary of HCT116 replicate AP-MS experiments.
(D) Summary of interactions observed in HCT116 replicate experiments.
(E) Summary of 293T-HCT116 replicate AP-MS experiments.
(F) Summary of interactions observed in 293T-HCT116 replicate experiments.
10
Table S3: Mapping BioPlex Interactions onto PDB Structures, Related to Figure 2.
(A) Summary of BioPlex interactions within PDB structures.
11
Table S4: Quantitative Proteomic Comparison of Cell Lines and Biological Follow-up, Related to Figures 3, ,4,4, and S3.
(A) Data from RTS-M3 TMT analysis of 293T and HCT116 proteomes in biological quintuplicate.
(B) Summary of Ephrin IP Phosphorylation Searches in 293T and HCT116 cells.
(C) SV40 Large T antigen Identifications in 293T IP’s
(D) RBL1 and RBL2 phosphorylation site abundance in 293T and HCT116 cells.
12
Table S5: Interaction Overlap among Functional Subnetworks, Related to Figure 3.
(A) Summary of edge sharing between 293T and HCT116 cells within subnetworks defined by specific DisGeNET disease associations. Only edges detectable in both cell lines are considered.
(B) Summary of edge sharing between 293T and HCT116 cells within subnetworks defined by specific GO Biological Processes. Only edges detectable in both cell lines are considered.
(C) Summary of edge sharing between 293T and HCT116 cells within subnetworks defined by specific GO Molecular Functions. Only edges detectable in both cell lines are considered.
(D) Summary of edge sharing between 293T and HCT116 cells within subnetworks defined by specific GO Cellular Components. Only edges detectable in both cell lines are considered.
(E) Summary of edge sharing between 293T and HCT116 cells within subnetworks defined by specific Reactome pathways. Only edges detectable in both cell lines are considered.
(F) Summary of edge sharing between 293T and HCT116 cells within subnetworks defined by specific CORUM complexes. Only edges detectable in both cell lines are considered.
13
Table S6: Discovery and Cell-line Specificity of BioPlex Communities, Related to Figure 5.
(A) Summary of Communities identified in the combined BioPlex 293T + HCT116 network.
(B) List of Entrez Gene ID’s and Symbols for proteins found in each community.
(C) List of community pairs whose members interact with each other unusually often, along with p-values reflecting the strength of the association between communities.
(D) Summary of the fraction of interactions within each community that are shared between cell lines. Only edges detectable in both cell lines are considered.
(E) Summary of the fraction of interactions linking each community pair that are shared between cell lines. Only edges detectable in both cell lines are considered.
14
Table S7: PFAM Domain Associations, Functional Associations, and Fitness Correlations in the Combined BioPlex Network, Related to Figures 6, ,77 and S7.
(A) A summary of all PFAM domain associations in the combined 293T/HCT116 BioPlex network.
(B) For each domain association, the fraction of edges that are specific to 293T or HCT116 cells or detected in both. Only edges detectable in both cell lines are considered.
(C) For baits targeted in both 293T and HCT116 cells, a list of enriched terms (GO, PFAM, InterPro) and the fraction of matching proteins that are unique to 293T or HCT116 cells or detected in both.
(D) List of fitness correlations calculated for those edges in the combined BioPlex network where Achilles data were available for both interacting proteins.
ACKNOWLEDGEMENTS
We thank Marc Vidal and David Hill for ORFeome 8.1 and acknowledge the Nikon Imaging Center (Harvard Medical School) for imaging support. This work was supported by the NIH (U24 HG006673 to S.P.G., J.W.H., and E.L.H.) and Biogen (S.P.G. and J.W.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
JWH is a founder and scientific advisory board member of Caraway Therapeutics and a Founding Scientific Advisor for Interline Therapeutics.
REFERENCES
- Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, and Lothe RA (2013). Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2, e71–e71. [Europe PMC free article] [Abstract] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene Ontology: tool for the unification of biology. Nat Genet 25, 25–29. [Europe PMC free article] [Abstract] [Google Scholar]
- Beausoleil SA, Villén J, Gerber SA, Rush J, and Gygi SP (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24, 1285–1292. [Abstract] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, and Harper JW (2010). Network organization of the human autophagy system. Nature 466, 68. [Europe PMC free article] [Abstract] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Royal Statistical Soc Ser B Methodol 57, 289–300. [Google Scholar]
- Bergmann C, Fliegauf M, Brüchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kränzlin B, et al. (2008). Loss of Nephrocystin-3 Function Can Cause Embryonic Lethality, Meckel-Gruber-like Syndrome, Situs Inversus, and Renal-Hepatic-Pancreatic Dysplasia. Am J Hum Genetics 82, 959–970. [Europe PMC free article] [Abstract] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, and Bourne PE (2000). The Protein Data Bank. Nucleic Acids Res 28, 235–242. [Europe PMC free article] [Abstract] [Google Scholar]
- Biederer T, and Scheiffele P (2007). Mixed-culture assays for analyzing neuronal synapse formation. Nat Protoc 2, nprot.2007.92. [Abstract] [Google Scholar]
- Blomen VA, Májek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, Diemen F.R. van, Olk N, Stukalov A, et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096. [Abstract] [Google Scholar]
- Boldt K, Reeuwijk J. van, Lu Q, Koutroumpas K, Nguyen T-MT, Texier Y, Beersum S.E.C. van, Horn N, Willer JR, Mans DA, et al. (2016). An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun 7, 11491. [Europe PMC free article] [Abstract] [Google Scholar]
- Boyle EA, Pritchard JK, and Greenleaf WJ (2018). High-resolution mapping of cancer cell networks using co-functional interactions. Mol Syst Biol 14. [Europe PMC free article] [Abstract] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, and Sabatini DM (2016). The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164. [Europe PMC free article] [Abstract] [Google Scholar]
- Dana JM, Gutmanas A, Tyagi N, Qi G, O’Donovan C, Martin M, and Velankar S (2018). SIFTS: updated Structure Integration with Function, Taxonomy and Sequences resource allows 40-fold increase in coverage of structure-based annotations for proteins. Nucleic Acids Res 47, gky1114–. [Europe PMC free article] [Abstract] [Google Scholar]
- Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, and Tsherniak A (2019). Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. Biorxiv 720243. [Google Scholar]
- Drew K, Lee C, Huizar RL, Tu F, Borgeson B, McWhite CD, Ma Y, Wallingford JB, and Marcotte EM (2017). Integration of over 9,000 mass spectrometry experiments builds a global map of human protein complexes. Mol Syst Biol 13, 932. [Europe PMC free article] [Abstract] [Google Scholar]
- Drew K, Wallingford JB, and Marcotte EM (2020). hu.MAP 2.0: Integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies. BioRxiv. [Europe PMC free article] [Abstract] [Google Scholar]
- Ecco G, Imbeault M, and Trono D (2017). KRAB zinc finger proteins. Development 144, 2719–2729. [Europe PMC free article] [Abstract] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2018). The Pfam protein families database in 2019. Nucleic Acids Res 47, gky995-. [Europe PMC free article] [Abstract] [Google Scholar]
- Elias JE, and Gygi SP (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4, nmeth1019. [Abstract] [Google Scholar]
- Eng JK, McCormack AL, and Yates JR (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectr 5, 976–989. [Abstract] [Google Scholar]
- Eng JK, Jahan TA, and Hoopmann MR (2013). Comet: An open-source MS/MS sequence database search tool. Proteomics 13, 22–24. [Abstract] [Google Scholar]
- Enright AJ, Dongen SV, and Ouzounis CA (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30, 1575–1584. [Europe PMC free article] [Abstract] [Google Scholar]
- Erickson BK, Mintseris J, Schweppe DK, Navarrete-Perea J, Erickson AR, Nusinow DP, Paulo JA, and Gygi SP (2019). Active Instrument Engagement Combined with a Real-Time Database Search for Improved Performance of Sample Multiplexing Workflows. J Proteome Res 18, 1299–1306. [Europe PMC free article] [Abstract] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. (2017). The Reactome Pathway Knowledgebase. Nucleic Acids Res 46, gkx1132-. [Europe PMC free article] [Abstract] [Google Scholar]
- Fischer M, Grossmann P, Padi M, and DeCaprio JA (2016). Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 44, 6070–6086. [Europe PMC free article] [Abstract] [Google Scholar]
- Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, and Morris Q (2018). GeneMANIA update 2018. Nucleic Acids Res 46, W60–W64. [Europe PMC free article] [Abstract] [Google Scholar]
- Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat C-M, et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147. [Abstract] [Google Scholar]
- Gavin A-C, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. (2006). Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636. [Abstract] [Google Scholar]
- Gingras A-C, Gstaiger M, Raught B, and Aebersold R (2007). Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Bio 8, 645–654. [Abstract] [Google Scholar]
- Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, and Ruepp A (2018). CORUM: the comprehensive resource of mammalian protein complexes—2019. Nucleic Acids Res 47, gky973-. [Europe PMC free article] [Abstract] [Google Scholar]
- Go CD, Knight JDR, Rajasekharan A, Rathod B, Hesketh GG, Abe KT, Youn J-Y, Samavarchi-Tehrani P, Zhang H, Zhu LY, et al. (2019). A proximity biotinylation map of a human cell. Biorxiv 796391. [Google Scholar]
- Goley ED, and Welch MD (2006). The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Bio 7, 713–726. [Abstract] [Google Scholar]
- Greber BJ, Toso DB, Fang J, and Nogales E (2019). The complete structure of the human TFIIH core complex. Elife 8, e44771. [Europe PMC free article] [Abstract] [Google Scholar]
- Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, and Sabatini DM (2017). SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818. [Europe PMC free article] [Abstract] [Google Scholar]
- Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SWT, et al. (2015). A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 163, 1484–1499. [Europe PMC free article] [Abstract] [Google Scholar]
- Guruharsha KG, Rual J-F, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. (2011). A Protein Complex Network of Drosophila melanogaster. Cell 147, 690–703. [Europe PMC free article] [Abstract] [Google Scholar]
- Hafner A, Bulyk ML, Jambhekar A, and Lahav G (2019). The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Bio 20, 199–210. [Abstract] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. (2012). A Census of Human Soluble Protein Complexes. Cell 150, 1068–1081. [Europe PMC free article] [Abstract] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. (2015). A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 163, 712–723. [Abstract] [Google Scholar]
- Heusel M, Bludau I, Rosenberger G, Hafen R, Frank M, Banaei-Esfahani A, Drogen A, Collins BC, Gstaiger M, and Aebersold R (2019). Complex-centric proteome profiling by SEC-SWATH-MS. Mol Syst Biol 15, e8438. [Europe PMC free article] [Abstract] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutilier K, et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183. [Abstract] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, and Gygi SP (2010). A Tissue-Specific Atlas of Mouse Protein Phosphorylation and Expression. Cell 143, 1174–1189. [Europe PMC free article] [Abstract] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. (2015). The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440. [Europe PMC free article] [Abstract] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505. [Europe PMC free article] [Abstract] [Google Scholar]
- Janke C, Rogowski K, and Dijk J. van (2008). Polyglutamylation: a fine-regulator of protein function? Embo Rep 9, 636–641. [Europe PMC free article] [Abstract] [Google Scholar]
- Kania A, and Klein R (2016). Mechanisms of ephrin–Eph signalling in development, physiology and disease. Nat Rev Mol Cell Bio 17, 240–256. [Abstract] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. (2006). Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643. [Abstract] [Google Scholar]
- Liebeskind BJ, McWhite CD, and Marcotte EM (2016). Towards Consensus Gene Ages. Genome Biol Evol 8, 1812–1823. [Europe PMC free article] [Abstract] [Google Scholar]
- Luck K, Kim D-K, Lambourne L, Spirohn K, Begg BE, Bian W, Brignall R, Cafarelli T, Campos-Laborie FJ, Charloteaux B, et al. (2020). A reference map of the human binary protein interactome. Nature 580, 1–7. [Europe PMC free article] [Abstract] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, and Gygi SP (2014). MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal Chem 86, 7150–7158. [Europe PMC free article] [Abstract] [Google Scholar]
- Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F, Chang C-C, and Lin C-C (2012). e1071: Misc Functions of the Department of Statistics, Probability Theory Group. R package Version 1.6–1. [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. (2017). Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784. [Europe PMC free article] [Abstract] [Google Scholar]
- Navarrete-Perea J, Yu Q, Gygi SP, and Paulo JA (2018). Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J Proteome Res 17, 2226–2236. [Europe PMC free article] [Abstract] [Google Scholar]
- Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, et al. (2020). Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402.e16. [Europe PMC free article] [Abstract] [Google Scholar]
- Ori A, Iskar M, Buczak K, Kastritis P, Parca L, Andrés-Pons A, Singer S, Bork P, and Beck M (2016). Spatiotemporal variation of mammalian protein complex stoichiometries. Genome Biol 17, 47. [Europe PMC free article] [Abstract] [Google Scholar]
- Orre LM, Vesterlund M, Pan Y, Arslan T, Zhu Y, Woodbridge AF, Frings O, Fredlund E, and Lehtiö J (2019). SubCellBarCode: Proteome-wide Mapping of Protein Localization and Relocalization. Mol Cell 73, 166–182.e7. [Abstract] [Google Scholar]
- Oughtred R, Stark C, Breitkreutz B-J, Rust J, Boucher L, Chang C, Kolas N, O’Donnell L, Leung G, McAdam R, et al. (2018). The BioGRID interaction database: 2019 update. Nucleic Acids Res 47, gky1079–. [Europe PMC free article] [Abstract] [Google Scholar]
- Pan J, Meyers RM, Michel BC, Mashtalir N, Sizemore AE, Wells JN, Cassel SH, Vazquez F, Weir BA, Hahn WC, et al. (2018). Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Syst 6, 555–568.e7. [Europe PMC free article] [Abstract] [Google Scholar]
- Paulo JA, O’Connell JD, and Gygi SP (2016). A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. J Am Soc Mass Spectr 27, 1620–1625. [Europe PMC free article] [Abstract] [Google Scholar]
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, and Furlong LI (2017). DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 45, D833–D839. [Europe PMC free article] [Abstract] [Google Scholar]
- Pratt D, Chen J, Welker D, Rivas R, Pillich R, Rynkov V, Ono K, Miello C, Hicks L, Szalma S, et al. (2015). NDEx, the Network Data Exchange. Cell Syst 1, 302–305. [Europe PMC free article] [Abstract] [Google Scholar]
- Qin Y, Winsnes CF, Huttlin EL, Zheng F, Ouyang W, Park J, Pitea A, Kreisberg JF, Gygi SP, Harper JW, et al. (2020). Mapping cell structure across scales by fusing protein images and interactions. Biorxiv 2020.06.21.163709. [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906. [Abstract] [Google Scholar]
- RCoreTeam (2011). R: A language and environment for statistical computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Rolland T, Ta§an M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. (2014). A Proteome-Scale Map of the Human Interactome Network. Cell 159, 1212–1226. [Europe PMC free article] [Abstract] [Google Scholar]
- Romanov N, Kuhn M, Aebersold R, Ori A, Beck M, and Bork P (2019). Disentangling Genetic and Environmental Effects on the Proteotypes of Individuals. Cell. [Europe PMC free article] [Abstract] [Google Scholar]
- Rosenberger G, Heusel M, Bludau I, Collins BC, Martelli C, Williams EG, Xue P, Liu Y, Aebersold R, and Califano A (2020). SECAT: Quantifying Protein Complex Dynamics across Cell States by Network-Centric Analysis of SEC-SWATH-MS Profiles. Cell Syst 11, 589–607.e8. [Europe PMC free article] [Abstract] [Google Scholar]
- Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. (2005). Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178. [Abstract] [Google Scholar]
- Ryan CJ, Kennedy S, Bajrami I, Matallanas D, and Lord CJ (2017). A Compendium of Co-regulated Protein Complexes in Breast Cancer Reveals Collateral Loss Events. Cell Syst 5, 399–409.e5. [Europe PMC free article] [Abstract] [Google Scholar]
- Rzechorzek NJ, Hardwick SW, Jatikusumo VA, Chirgadze DY, and Pellegrini L (2020). CryoEM structures of human CMG–ATPγS-DNA and CMG–AND-1 complexes. Nucleic Acids Res 48, gkaa429-. [Europe PMC free article] [Abstract] [Google Scholar]
- Sadasivam S, and DeCaprio JA (2013). The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 13, nrc3556. [Europe PMC free article] [Abstract] [Google Scholar]
- Sehnal D, Rose AS, Koca J, Burley SK, and Velankar S (2018). Mol*: towards a common library and tools for web molecular graphics. MoIVA ’18: Proceedings of the Workshop on Molecular Graphics and Visual Analysis of Molecular Data 29–33. [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, and Harper JW (2009). Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell 138, 389–403. [Europe PMC free article] [Abstract] [Google Scholar]
- Stacey RG, Skinnider MA, Chik JHL, and Foster LJ (2018). Context-specific interactions in literature-curated protein interaction databases. Bmc Genomics 19, 758. [Europe PMC free article] [Abstract] [Google Scholar]
- Stepanenko AA, and Dmitrenko VV (2015). HEK293 in cell biology and cancer research: phenotype, karyotype, tumorigenicity, and stress-induced genome-phenotype evolution. Gene 569, 182–190. [Abstract] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. (2017). The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368. [Europe PMC free article] [Abstract] [Google Scholar]
- Takahashi K, Nagai T, Chiba S, Nakayama K, and Mizuno K (2018). Glucose deprivation induces primary cilium formation through mTORC1 inactivation. J Cell Sci 131, jcs208769. [Abstract] [Google Scholar]
- The UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Res 43, D204–D212. [Europe PMC free article] [Abstract] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Blal HA, Alm T, Asplund A, Björk L, Breckels LM, et al. (2017). A subcellular map of the human proteome. Science 356, eaal3321. [Abstract] [Google Scholar]
- Ting L, Rad R, Gygi SP, and Haas W (2011). MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8, 937. [Europe PMC free article] [Abstract] [Google Scholar]
- Topalis D, Andrei G, and Snoeck R (2013). The large tumor antigen: A “Swiss Army knife” protein possessing the functions required for the polyomavirus life cycle. Antivir Res 97, 122–136. [Abstract] [Google Scholar]
- Treiber T, Treiber N, Plessmann U, Harlander S, Daiß J-L, Eichner N, Lehmann G, Schall K, Urlaub H, and Meister G (2017). A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol Cell 66, 270–284.e13. [Abstract] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a Cancer Dependency Map. Cell 170, 564–576.e16. [Europe PMC free article] [Abstract] [Google Scholar]
- Tzavlaki K, and Moustakas A (2020). TGF-β Signaling. Biomol 10, 487. [Europe PMC free article] [Abstract] [Google Scholar]
- Varjosalo M, Sacco R, Stukalov A, Drogen A. van, Planyavsky M, Hauri S, Aebersold R, Bennett KL, Colinge J, Gstaiger M, et al. (2013). Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods 10, 307. [Abstract] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, and Kinzler KW (2013). Cancer Genome Landscapes. Science 339, 1546–1558. [Europe PMC free article] [Abstract] [Google Scholar]
- Wainberg M, Kamber RA, Balsubramani A, Meyers RM, Sinnott-Armstrong N, Hornburg D, Jiang L, Chan J, Jian R, Gu M, et al. (2019). A genome-wide almanac of co-essential modules assigns function to uncharacterized genes. Biorxiv 827071. [Europe PMC free article] [Abstract] [Google Scholar]
- Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, et al. (2015). Panorama of ancient metazoan macromolecular complexes. Nature 525, 339. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang D, Eraslan B, Wieland T, Hallström B, Hopf T, Zolg DP, Zecha J, Asplund A, Li L, Meng C, et al. (2019). A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol Syst Biol 15, e8503. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, and Sabatini DM (2015). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101. [Europe PMC free article] [Abstract] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Lührmann R, and Wahl MC (2010). Functional organization of the Sm core in the crystal structure of human U1 snRNP. Embo J 29, 4172–4184. [Europe PMC free article] [Abstract] [Google Scholar]
- Werner A, Iwasaki S, McGourty CA, Medina-Ruiz S, Teerikorpi N, Fedrigo I, Ingolia NT, and Rape M (2015). Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525, 523–527. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu J, Niu S, Tan M, Huang C, Li M, Song Y, Wang Q, Chen J, Shi S, Lan P, et al. (2018). Cryo-EM Structure of the Human Ribonuclease P Holoenzyme. Cell 175, 1393–1404.e11. [Abstract] [Google Scholar]
- Xu J, Lamouille S, and Derynck R (2009). TGF-β-induced epithelial to mesenchymal transition. Cell Res 19, 156–172. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. (2011). A public genome-scale lentiviral expression library of human ORFs. Nat Methods 8, 659–661. [Europe PMC free article] [Abstract] [Google Scholar]
- Youn J-Y, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. (2018). High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell 69, 517–532.e11. [Abstract] [Google Scholar]
- Zhang J, Cao M, Dong J, Li C, Xu W, Zhan Y, Wang X, Yu M, Ge C, Ge Z, et al. (2014). ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nat Commun 5, 5059. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2021.04.011
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867421004463/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/105400524
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cell.2021.04.011
Article citations
Identification of apelin/APJ signaling dysregulation in a human iPSC-derived granulosa cell model of Turner syndrome.
Cell Death Discov, 10(1):468, 14 Nov 2024
Cited by: 0 articles | PMID: 39543104 | PMCID: PMC11564969
Epigenetic regulation of noncanonical menin targets modulates menin inhibitor response in acute myeloid leukemia.
Blood, 144(19):2018-2032, 01 Nov 2024
Cited by: 1 article | PMID: 39158067 | PMCID: PMC11561541
A Novel Single-Color FRET Sensor for Rho-Kinase Reveals Calcium-Dependent Activation of RhoA and ROCK.
Sensors (Basel), 24(21):6869, 26 Oct 2024
Cited by: 0 articles | PMID: 39517770 | PMCID: PMC11548655
Global profiling of protein complex dynamics with an experimental library of protein interaction markers.
Nat Biotechnol, 16 Oct 2024
Cited by: 0 articles | PMID: 39415059
Midnolin, a Genetic Risk Factor for Parkinson's Disease, Promotes Neurite Outgrowth Accompanied by Early Growth Response 1 Activation in PC12 Cells.
Mol Cell Biol, 44(11):516-527, 12 Sep 2024
Cited by: 0 articles | PMID: 39264361 | PMCID: PMC11529416
Go to all (347) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Data Citations (2)
- (1 citation) DOI - 10.18119/N9H887
- (1 citation) DOI - 10.18119/N9RP5D
Protein structures in PDBe (4)
-
(1 citation)
PDBe - 3pgwView structure
-
(1 citation)
PDBe - 6ahrView structure
-
(1 citation)
PDBe - 6nmiView structure
-
(1 citation)
PDBe - 6xtxView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Architecture of the human interactome defines protein communities and disease networks.
Nature, 545(7655):505-509, 17 May 2017
Cited by: 764 articles | PMID: 28514442 | PMCID: PMC5531611
Mass Spectrometry Analysis of Spatial Protein Networks by Colocalization Analysis (COLA).
Methods Mol Biol, 1636:337-352, 01 Jan 2017
Cited by: 0 articles | PMID: 28730490
The bait compatibility index: computational bait selection for interaction proteomics experiments.
J Proteome Res, 9(10):4972-4981, 01 Oct 2010
Cited by: 5 articles | PMID: 20731387
Discovery-Versus Hypothesis-Driven Detection of Protein-Protein Interactions and Complexes.
Int J Mol Sci, 22(9):4450, 24 Apr 2021
Cited by: 7 articles | PMID: 33923221 | PMCID: PMC8123138
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHGRI NIH HHS (2)
Grant ID: U24 HG006673
Grant ID: U41 HG006673
NIGMS NIH HHS (1)
Grant ID: T32 GM080189