Abstract
Free full text

Endophytic Colonization and In Planta Nitrogen Fixation by a Herbaspirillum sp. Isolated from Wild Rice Species
Abstract
Nitrogen-fixing bacteria were isolated from the stems of wild and cultivated rice on a modified Rennie medium. Based on 16S ribosomal DNA (rDNA) sequences, the diazotrophic isolates were phylogenetically close to four genera: Herbaspirillum, Ideonella, Enterobacter, and Azospirillum. Phenotypic properties and signature sequences of 16S rDNA indicated that three isolates (B65, B501, and B512) belong to the Herbaspirillum genus. To examine whether Herbaspirillum sp. strain B501 isolated from wild rice, Oryza officinalis, endophytically colonizes rice plants, the gfp gene encoding green fluorescent protein (GFP) was introduced into the bacteria. Observations by fluorescence stereomicroscopy showed that the GFP-tagged bacteria colonized shoots and seeds of aseptically grown seedlings of the original wild rice after inoculation of the seeds. Conversely, for cultivated rice Oryza sativa, no GFP fluorescence was observed for shoots and only weak signals were observed for seeds. Observations by fluorescence and electron microscopy revealed that Herbaspirillum sp. strain B501 colonized mainly intercellular spaces in the leaves of wild rice. Colony counts of surface-sterilized rice seedlings inoculated with the GFP-tagged bacteria indicated significantly more bacterial populations inside the original wild rice than in cultivated rice varieties. Moreover, after bacterial inoculation, in planta nitrogen fixation in young seedlings of wild rice, O. officinalis, was detected by the acetylene reduction and 15N2 gas incorporation assays. Therefore, we conclude that Herbaspirillum sp. strain B501 is a diazotrophic endophyte compatible with wild rice, particularly O. officinalis.
Use of nitrogen fertilizer is of great importance in rice production, as nitrogen is the major factor limiting growth under most conditions (7). Since agriculture is expected to move toward environmentally sustainable methods (31), much attention has been paid to natural methods of biological nitrogen fixation. Several diazotrophic bacteria, including Klebsiella oxytoca (11), Enterobacter cloacae (11), Alcaligenes (33), and Azospirillum (4), have been isolated from the rhizosphere of wetland rice. However, a renewed interest in endophytic diazotrophs, such as Acetobacter, Azoarcus, and Herbaspirillum, in gramineous plants has arisen because of their occurrence mainly within plant tissues and evidence for significant nitrogen fixation (18, 25). Azoarcus sp. from Kallar grass, abundantly colonize and express nif genes and nitrogenase protein inside the original host as well as in rice roots (8, 25). Acetobacter diazotrophicus, an endophytic diazotroph from sugarcane, has been found mainly associated with sugar-rich plants, such as sugarcane, sweet potato and Cameroon grass (2). In nitrogen-deficient conditions, sugarcane plants inoculated with wild-type A. diazotrophicus had a higher nitrogen content than plants inoculated with nif mutant or uninoculated plants (29). Diazotrophic Herbaspirillum seropedicae has been found in maize, sorghum, sugarcane, and other gramineous plants (3, 23). H. seropedicae strain Z67 colonized mainly subepidermal regions of rice roots (5).
Cultivated rice (Oryza sativa) originated from species of wild rice and was domesticated several thousand years ago (14, 22, 28). Wild rice species are likely to harbor unique populations of nitrogen-fixing bacteria that differ from those in extensively bred modern varieties of cultivated rice (10). Nitrogen-fixing endophytes in rice were reported to be higher in stems than in roots, indicating that rice stems probably provide a suitable niche (5). Additionally, use of rice stems for isolation of diazotrophic bacteria can minimize the possibility of contamination from soil, because the rice stems are completely covered with successive leaf sheaths before heading (14). Once endophytic diazotrophs such as Azoarcus sp. infect plants, they sometimes spread systemically and reach aerial tissues of the plant (25). Recent research has focused on the question of whether the bacteria really fix nitrogen within the plants (8, 29) and establish mutual symbiosis (16).
The aims of this study were twofold: first, to isolate nitrogen-fixing bacteria from the stems of wild and cultivated rice species, and second, to investigate endophytic colonization and in planta nitrogen fixation of a selected diazotroph in order to verify whether the bacteria function as nitrogen-fixing endophytes.
MATERIALS AND METHODS
Bacterial growth media.
For isolation of diazotrophic endophytes, we used a modified version of Rennie medium (26), Rennie medium supplemented with rice extract and malate (RMR medium). RMR medium was prepared from solutions A and B. Solution A consisted of 0.8 g of K2HPO4, 0.2 g of KH2PO4, 0.1 g of NaCl, 28 mg of Na2FeEDTA, 25 mg of Na2MoO4 · 2H2O, 100 mg of yeast extract (Difco), 3.0 g of mannitol, 5.0 g of sucrose, 0.5 ml of 60% (vol/vol) sodium lactate, 2.0 g of sodium malate, 2.0 g (for semisolid medium in test tubes) or 15 g (for agar medium for plates) of Noble agar, and 900 ml of distilled water (the final pH of solution A was adjusted to 7.0 before autoclaving). Solution B consisted of 0.2 g of MgSO4 · 7H2O, 0.06 g of CaCl2 · 2H2O, and 100 ml of distilled water. The solutions were autoclaved separately and mixed after cooling. Filter-sterilized biotin and para-aminobenzoic acid (100 μl each) were added at final concentrations of 5 and 10 μg/liter, respectively. The combined solution (approximately 1 liter) was mixed with 1.25 ml of rice shoot extract that had been extracted with ethanol and 1.25 ml of rice shoot extract that had been extracted with water as described below. RMR medium contained rice extract derived from 1 g of rice shoots in 1 liter.
To prepare rice extracts, shoots of field-grown O. sativa cv. Sasanishiki at maximum tillage stage were washed with tap water, wiped dry, and weighed. The rice shoots (200 g) were macerated in 500 ml of distilled water at 4°C or in 500 ml of 80% ethanol at room temperature using a mortar and pestle. The macerated extracts were filtered and centrifuged at 15,000 × g for 10 min to eliminate all residual substances. The fine sap was filter sterilized and kept at −20°C until use.
Acetylene reduction of bacterial culture.
The nitrogen-fixing activity of the bacterial culture was examined by acetylene reduction assay in a 27-ml test tube containing 9 ml of RMR semisolid culture. Acetylene gas was injected into the head atmosphere of the test tubes (18 ml) at a final concentration of 5% (vol/vol) and incubated for 24 h at 30°C. Ethylene concentration was assayed on a Shimadzu GC7A gas chromatograph equipped with a flame ionization detector and a Porapack R column (internal diameter, 2.2 mm; length, 2 m; Shimadzu, Kyoto, Japan).
Sources of rice and isolation of diazotrophic bacteria.
Four species of wild rice (Oryza officinalis W0012, Oryza barthii W1407, Oryza rufipogon W1989, and Oryza glandiglumis W1194) and three varieties of cultivated rice (O. sativa cv. Nipponbare, cv. Kasalath, and cv. SC41) were used. The stock seeds of wild rice were provided by the National Institute of Genetics (Mishima, Japan) and the Institute of Genetic Ecology (IGE), Tohoku University (Sendai, Japan).
O. officinalis W0012, O. barthii W1407, and O. glandiglumis W1194 were grown in Akadama soil pots in a greenhouse of the Institute of Genetic Ecology, Tohoku University. They had almost reached the heading stage when used for bacterial isolation. Stems of O. barthii W1407, O. officinalis W0012, and O. glandiglumis W1194 were separated from leaf sheaths. The stems were surface sterilized with 70% ethanol by shaking for 1 min and transferred into RMR semisolid medium. After cultivation for 1 week at 30°C, the culture was reinoculated into RMR semisolid medium in test tubes for the acetylene reduction assay.
O. rufipogon W1989, O. sativa (cv. Nipponbare, cv. Kasalath, and cv. SC41; Gemdjah Benton) were grown at the experimental station of the Institute of Genetic Ecology (Kashimadai city, Miyagi prefecture, Japan). Stem segments separated from leaf sheaths were simultaneously vigorously shaken and washed with sterile 0.01% Tween 20 for 30 min to eliminate epiphytic bacteria. The surface-washed segments were aseptically macerated in a mortar with 0.8% saline solution and quartz sand and then transferred to test tubes containing RMR semisolid medium. After 1 week of incubation at 30°C, the cultures were assayed for acetylene reduction.
The pellicles from acetylene reduction-positive tubes were streaked on RMR agar plates. Single colonies aerobically grown at 30°C on the plates were transferred separately into test tubes containing RMR semisolid medium, incubated at 30°C for 7 days, and assayed for acetylene reduction. The resultant diazotrophic bacterial culture was mixed with sterilized dimethyl sulfoxide at a final concentration of 10% (vol/vol) and stored at −80°C.
PCR amplification and sequencing of the 16S rRNA gene.
Cell lysate from each isolate for the PCR template was prepared by the method of Hiraishi et al. (13). 16S rRNA gene was amplified by PCR using universal primers pr0R (5′-AGAGTTTGATCCTGGCTCAG-3′) corresponding to positions 8 to 27 (forward primer) and 9rev (5′-AAGGAGGTGATCCAGCC-3′) corresponding to positions 1543 to 1525 (reverse primer) of the Escherichia coli numbering system (35). The PCR operating conditions were as described by Suzuki and Yamasato (32). The PCR products were visualized by electrophoresis on ethidium bromide-stained 1.0% agarose gels (SeaPlaque GTG FMC Bioproducts) and purified by using QIAquick (Qiagen) following the manufacturer's instructions. Sequencing was performed with an ABI PRISM Big Dye Terminator Cycle Sequencing Kit and a 377 DNA sequencer (Perkin-Elmer) according to the manufacturer's instructions. Six primers, f1 (5′-AGCCATGCCGCGTGTATG-3′), f2 (5′-GGGAGCAAACAGGATTAGAT-3′), f3 (5′-GAAATGTTGGGTTAAGTCCC-3′), r1 (5′-TGCAATATTCCCCACTGC-3′), r2 (5′-TTCCTCCACATCTCTACG-3′), and r3 (5′-CGCTCGTTGCGGGACTTA-3′), as well as the two PCR primers, were used to sequence both strands of the16S rRNA gene. FASTA (DDBJ, Mishima, Japan) was used to compare the resulting sequences with those identified in the DDBJ, EMBL, and GenBank DNA databases. The neighbor-joining method (27) and the Clustal W program were used to construct a phylogenetic tree.
Phenotypic characterization of diazotrophic bacteria.
Gram staining, motility, and production of pectinase and cellulase were assayed as described previously (9). Three isolates that were close to Herbaspirillum sp. on the phylogenetic tree constructed based on 16S ribosomal DNA (rDNA) sequences were further characterized. Cell shape and width were observed by phase-contrast microscopy. Catalase and oxidase assays were performed by the method of Smibert and Krieg (30). The ability of the bacteria to grow on various carbon substrates was assayed in M70 minimal medium (12) supplemented with 1% (wt/vol) of the appropriate carbon substrate.
Fluorescent labeling of diazotrophic bacteria.
To confirm the endophytic features of Herbaspirillum sp. strain B501, we incorporated DNA sequences encoding the green fluorescent protein (GFP) into the bacteria. Plasmid pUTgfpx2 (34) containing the gfp minitransposon, which expresses gfp genes downstream of a constitutive promoter, PpsbA, was transferred into strain B501 isolated from wild rice. This plasmid carried two tandem copies of gfp and kanamycin resistance genes.
Competent cells were prepared and electroporated by the methods of Unge et al. (34). Herbaspirillum sp. strain B501 was cultured in 10 ml of nutrient broth medium (Difco). Samples (1 ml) of the subculture were transferred to 1,000-ml flasks containing 200 ml of nutrient broth medium and incubated at 30°C with shaking. Cells at stationary phase were chilled on ice separately and harvested by centrifugation at 3,000 × g for 10 min at 4°C. Each pellet was then washed twice with 200 ml of cold, sterilized distilled water and resuspended in 500 μl of cold, sterile 10% glycerol–water. The cell suspension was dispensed in 100-μl aliquots and stored at −80°C until use. The competent cells (100 μl) were thawed on ice, and 200 ng of purified gfp delivery plasmid pUTgfpx2 was added to the cells and mixed quickly. The mixture was incubated on ice for 15 min and transferred to a sterile, prechilled cuvette (0.2-cm interelectrode gap) and placed in the Gene Pulser II apparatus equipped with a Pulser Controller (Bio-Rad Laboratories, Tokyo, Japan). The electroporation unit was used at the following settings: 12.5 kV/cm, 25 μF, and 200 Ω. Following the pulse, the cells were immediately diluted with 1 ml of nutrient broth medium (Difco), transferred to the sterilized tubes, and incubated at 30°C for 3 to 4 h. From each tube, 100 μl was plated onto selective nutrient agar plates containing 100 μg of kanamycin per ml. After the plates were incubated at 30°C, green fluorescence emission was examined with an Olympus SZX12 fluorescence stereomicroscope with GFP band-pass filter unit SZX-FGFPA (excitation wavelength ,460 to 490 nm; emission wavelength, 510 to 550 nm).
Rice growth media.
Nitrogen-free medium (20a) was used for colonization and nitrogen fixation studies. The medium contained the following: 0.6 mM NaH2PO4 · 2H2O, 0.3 mM K2SO4, 0.3 mM CaCl2 · 2H2O, 0.6 mM MgCl2 · 2H2O, and 0.045 mM FeEDTA as macroelements; 50 μM H3BO3, 9 μM MnSO4 · 5H2O, 0.3 μM CuSO4 · 5H2O, 0.7 μM ZnSO4 · 7H2O, and 0.1 μM Na2MoO4 · 2H2O as microelements; and 0.325% (wt/vol) agar. The pH was adjusted to 5.5. The macroelement and microelement solutions were sterilized separately and then poured into plant boxes (350 ml) (CUL-JAR300; Iwaki, Tokyo, Japan) before solidification.
Rice cultivation and inoculation with Herbaspirillum sp.
Dehulled seeds of O. officinalis W0012, O. rufipogon W2008, O. sativa cv. Sasanishiki, and O. sativa cv. Nipponbare were surface sterilized with 70% ethanol for 1 min and shaken in 10% (wt/vol) calcium hypochlorite solution (high granules; Wako Pure Chemical Co. Ltd., Osaka, Japan) for 30 min at 28°C. Seeds were then washed three times with sterilized distilled water with shaking (15 min each). To ensure the sterilization efficiency, seeds were subjected to sterility checks on RMR and nutrient agar (Difco) media.
For seed inoculation, GFP-tagged Herbaspirillum sp. strain B501 was grown on nutrient broth medium (Difco), harvested at the logarithmic phase when the turbidity (A660) reached approximately 0.9, and washed twice with sterilized distilled water. An inoculum was prepared by resuspending the pellet in sterile saline solution, and cell density was determined by direct counting with a Thoma hemocytometer. The sterilized seeds were transferred into plant boxes containing 100 ml of semisolid rice growth medium and then inoculated with GFP-tagged Herbaspirillum sp. strain B501 (105 cells/seed). The plant boxes containing the inoculated seeds were then incubated in a plant growth cabinet (LH300; NK System Co. Ltd., Osaka, Japan) under a light-dark cycle (16 h of light followed by 8 h of dark) at 25°C.
Fluorescence microscopy.
Seven-day-old seedlings of wild and cultivated rice plants that were either inoculated or not inoculated were taken from the plant box, washed lightly with sterilized distilled water, and placed separately on petri dishes. These intact seedlings were then observed and photographed using an Olympus SZX12 fluorescence stereomicroscope. Each field was photographed under three different conditions: optical light microscopy and fluorescence microscopy with a GFP filter unit (Olympus SZX-MGFP/Em510-) and GFP band-pass filter unit (Olympus SZX-MGFP/Em510-550).
To determine the localization of the inoculant bacteria in plant tissues, root and shoot segments of 7-day-old rice plants were fixed for 4 h in 4% paraformaldehyde in phosphate buffer solution (10 mM sodium phosphate buffer [pH 7.2], 150 mM NaCl) at room temperature and rinsed with the phosphate buffer. The fixed shoots and roots were cut into 80-μm-thick slices with a microslicer (DTK-1000). The specimen was observed by a confocal laser scanning fluorescence microscope (Zeiss LSM510; Carl Zeiss, Tokyo, Japan) with a laser emitting 488-nm-wavelength light and with a 514-nm barrier filter cutoff. Confocal images were reconstructed with Zeiss LSM image software.
Electron microscopy.
Seedlings (1 to 2 weeks old) inoculated with Herbaspirillum sp. strain B501 were observed with an Olympus SZX12 fluorescence stereomicroscope, and the infection zones were identified. Slices (1 to 2 mm thick) were taken from coleoptiles, leaves, and roots and fixed in 2% glutaraldehyde in 0.2 M Na-cacodylate buffer (pH 6.8) at room temperature in a vacuum for 2 h, washed four times with the same buffer, and postfixed in 1% osmium tetroxide in 0.1 M Na-cacodylate buffer (pH 6.8) for 2 h. Samples were rinsed three times with distilled water and suspended in 2% uranyl acetate for 2 h. Samples were then rinsed again with distilled water, dehydrated through a series of ethanol solutions, embedded in Spurr's low-viscosity epoxy resin, and polymerized at 60°C for 24 h. Ultrathin sections were restained with uranyl acetate and lead citrate and observed with a JEOL 100SX transmission electron microscope.
Inoculant population in surface-sterilized rice.
To estimate the population of GFP-tagged Herbaspirillum sp. inside rice tissues, 7-day-old seedlings of O. officinalis W0012, O. rufipogon W2008, and O. sativa cultivars Sasanishiki and Nipponbare inoculated with the bacteria were surface sterilized in 1% NaOCl for 30 s. The seedlings were then quickly washed with sterilized distilled water and then with saline solution. The shoots, roots, and seeds of five seedlings were excised, weighed, and macerated separately in 0.8% saline solution. The macerate was serially diluted and plated on nutrient agar plates containing 50 μg of kanamycin per ml. The green fluorescent colonies that appeared on the plates after incubation at 30°C were counted with the aid of an Olympus SZX12 fluorescence stereomicroscope.
Acetylene reduction assay of inoculated plants.
Nitrogenase activity of seedlings with and without inoculation was evaluated using the acetylene reduction assay. Seven-day-old seedlings of O. officinalis W0012 and O. sativa cv. Sasanishiki inoculated with Herbaspirillum sp. strain B501 were washed with sterilized distilled water. Each seedling was then placed in a separate 13.8- or 33.2-ml bottle sealed with rubber septa, and the bottles were injected with 10% (vol/vol) pure acetylene (17). Ethylene concentrations were measured at 8-h intervals for 24 h using a Shimadzu GC-18A gas chromatograph as described above. We drew a graph showing ethylene concentrations over time and examined the linearity of increase of ethylene concentrations. Ethylene evolution from uninoculated seedlings in the presence of 10% acetylene and inoculated seedlings in the presence of air was also assayed to discriminate acetylene reduction activity of the inoculant in planta from the background ethylene emission of rice as a plant hormone.
15N2 gas incorporation.
Ten-day-old seedlings of O. officialis W0012 inoculated with Herbaspirillum sp. strain B501 were surface sterilized in 1% NaOCl for 30 s and washed with sterilized distilled water. The seedlings were placed into 120-ml bottles sealed with butyl rubber septa. A gas phase composed of 40% (vol/vol) 15N2 (99.7 atom%), 40% (vol/vol) Ar, and 20% (vol/vol) O2 was attained in the bottle after several replacements of 80% (vol/vol) Ar and 20% (vol/vol) O2 using a vacuum line. After 24 h at 30°C in the dark, the seedlings were taken from the bottles, dried at 70°C, and powdered using a mortar and pestle. 15N concentrations were determined in duplicate using a Thermo Quest DELTA plus XL mass spectrometer (Bremen, Germany).
Nucleotide sequence accession numbers.
We deposited the sequences of diazotrophs isolated in this work under the following accession numbers to DDBJ: AB049133 (strain B501), AB049103 (strain B65), AB049104 (strain B512), AB049105 (strain B508-1), AB049106 (strain B511), AB049107 (strain B513), AB049108 (strain B509), AB049109 (strain B52), AB049110 (strain B506), AB049111 (strain B510), and AB049112 (strain B518).
RESULTS
Isolation and characterization of nitrogen-fixing bacteria from rice.
From surface-washed or sterilized stems of wild and cultivated rice plants, we obtained 11 gram-negative isolates of nitrogen-fixing bacteria grown on a modified Rennie (RMR) semisolid medium (Table (Table1;1; Fig. Fig.1).1). They consistently showed acetylene reduction activity in the medium during isolation steps. The diazotrophic isolates were closely related to Herbaspirillum, Ideonella, Enterobacter, and Azospirillum within Proteobacteria (Fig. (Fig.1).1). Isolates B65, B501, and B512 from wild rice species Oryza barthii, O. officinalis, and O. rufipogon, respectively, were closely related to Herbaspirillum spp. in β-Proteobacteria, which are known to be diazotrophic endophytes from sugarcane and other gramineous plants (3, 20, 23). Conversely, isolates B508-1, B511, and B513 from indica and javanica types of cultivated rice plants (O. sativa cultivars Kasalath and SC41, respectively) were closely related to Ideonella dechloratans (21) in β-Proteobacteria. Isolates B52, B506, B510, and B518 apparently fell into an Azospirillum cluster within α-Proteobacteria (Fig. (Fig.1).1). Isolate B509 clustered close to Enterobacter. All diazotrophic isolates in this study were motile and showed pectinase and cellulase activities except for cellulase activity of strain B511 (Table (Table1).1). These traits were found when nondiazotrophic bacteria were isolated and characterized from the stems of wild and cultivated rice varieties previously (9).
TABLE 1
Diazotrophic bacteria isolated from stems of wild rice and cultivated rice
| Strain | Close relatives based on 16S rRNAa (percent homology) | Rice genotypeb | Rice stagec | Putative endophytic featuresd
| ||
|---|---|---|---|---|---|---|
| Motility | Pectinase | Cellulase | ||||
| B65 | Herbaspirillum rubrisubalbicans (99.6) | O. barthii W1407 | Tillage | + | + | + |
| B501 | Herbaspirillum seropedicae (99.6) | O. officinalis W0012 | Tillage | + | + | + |
| B512 | Herbaspirillum seropedicae (99.8) | O. rufipogon W1989 | Tillage | + | + | + |
| B508-1 | Ideonella dechloratans (99.5) | O. sativa SC41 | MTN | + | + | + |
| B511 | Ideonella dechloratans (99.5) | O. sativa Kasalath | Tillage | + | + | − |
| B513 | Ideonella dechloratans (99.5) | O. sativa SC41 | Tillage | + | + | + |
| B509 | Enterobacter cancerogenus (99.2) | O. rufipogon W1989 | MTN | + | + | + |
| B52 | Azospirillum lipoferum (97.1) | O. glandiglumis W1194 | Tillage | + | + | + |
| B506 | Azosprillum brasilense (95.2) | O. rufipogon W1989 | MTN | + | + | + |
| B510 | Azospirillum lipoferum (96.6) | O. sativa Nipponbare | Tillage | + | + | + |
| B518 | Azospirillum lipoferum (98.1) | O. sativa Kasalath | MTN | + | + | + |
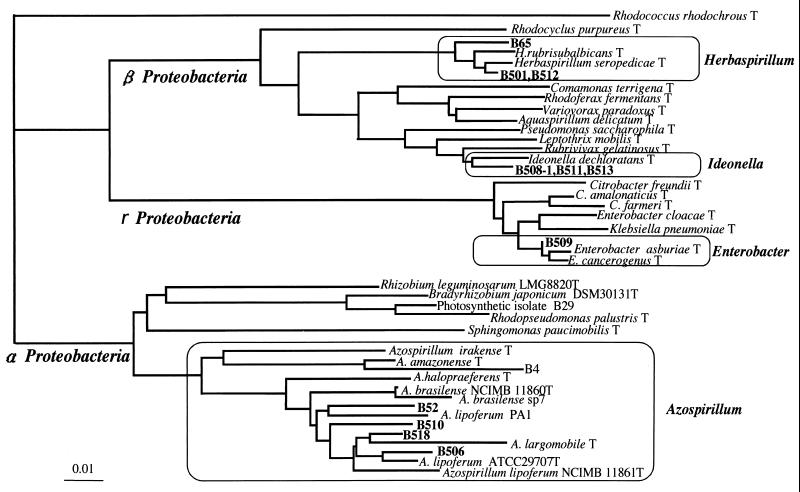
Phylogenetic tree showing relatedness of nitrogen-fixing bacteria isolated from rice and their relatives in the Proteobacteria by neighbor-joining grouping of the aligned sequences of the 16S rRNA gene. Strains B4 and B29 are nitrogen-fixing bacteria previously isolated using nutrient agar medium from surface-sterilized culms of wild rice (9). To construct the phylogenetic tree, we used the 16S rRNA sequences in the DDBJ/EMBL/GenBank database from the following bacteria (accession numbers given in parentheses): Rhodococcus rhodochrous (X79288), Rhodocyclus purpureus (M34132), Herbaspirillum rubrisubalbicans (AB021424), Herbaspirillum seropedicae (Y10146), Comamonas terrigena (AB021418), Rhodoferax fermentans (D16211), Variovorax paradoxus (D30793), Aquaspirillum delicatun (AF078756), Pseudomonas saccharophila (AB021407), Leptothrix mobilis (X97071), Rubrivivax gelatinosus (D16213), Ideonella dechloratans (X72724), Citrobacter freundii (M59291), Citrobacter amalonaticus (AF025370), Citrobacter farmeri (AF025371), Enterobacter cloacae (AJ251469), Klebsiella pneumoniae (AF130981), Enterobacter asburiae (AB004744), Enterobacter cancerogenus (Z96078), Rhizobium leguminosarum (X67227), Bradyrhizobium japonicum (X87272), Rhodopseudomonas palustris (D25312), Sphingomonas paucimobilis (U37337), Azospirillum irakense (Z29583), Azospirillum amazonense (X79735), Azospirillum halopraeferens (X79731), Azospirillum brasilense NCIMB 11860T (Z29617), Azospirillum brasilense sp7 (X79739), Azospirillum lipoferum PA1 (X79738), Azospirillum largomobile (X90759), Azospirillum lipoferum ATCC 29707T (M59061), and Azospirillum lipoferum NCIMB11861T (Z29619). Bar, 0.01 base substitution per nucleotide.
Comparison of Herbaspirillum spp. with bacterial isolates from wild rice.
Three nitrogen-fixing species of Herbaspirillum have previously been identified. H. seropedicae was first reported as a nitrogen-fixing bacterium associated with the roots of rice, maize, and sorghum (3). Subsequently, a mild plant pathogen in sugarcane was reclassified as Herbaspirillum rubrisubalbicans (1). Recently, a new species, Herbaspirillum frisingense, isolated from C4 fiber plants was proposed (20). We then examined whether diazotrophic isolates B65, B501, and B512 from wild rice could be classified as any of the known species of Herbaspirillum (Table (Table2).2).
TABLE 2
Comparison of wild-rice-derived diazotrophic bacteria B65, B501, and B512 with nitrogen-fixing Herbaspirillum spp.a
| Characteristic | Diazotrophic bacteria from wild rice
| H. seropedicae | H. rubrisubalbicans | H. frisingense | ||
|---|---|---|---|---|---|---|
| B65 | B501 | B512 | ||||
| Basic property | ||||||
 Gram staining Gram staining | − | − | − | − | − | − |
 Motility Motility | + | + | + | + | + | + |
 Cell shape Cell shape | Vibrioid | Vibrioid | Vibrioid | Vibrioid | Slightly curved rod | Slightly curved rod |
 Cell width (μm) Cell width (μm) | 0.5–0.6 | 0.5–0.6 | 0.5–0.6 | 0.6–0.7 | 0.6–0.7 | 0.5–0.7 |
 Catalase Catalase | + | + | + | + | + | + |
 Oxidase Oxidase | + | + | + | + | + | + |
| Utilization of carbon source | ||||||
 meso-Erythritol meso-Erythritol | + | + | + | − | + | − |
 N-Acetylglucosamine N-Acetylglucosamine | − | − | − | + | − | + |
 l-Rhamnose l-Rhamnose | + | + | + | + | − | − |
 meso-Inositol meso-Inositol | − | + | − | + | − | − |
 Sebacate Sebacate | + | − | − | − | − | ND |
 Adipate Adipate | − | + | + | − | − | − |
 Glucose Glucose | − | + | + | + | ND | + |
 Sucrose Sucrose | − | − | − | − | − | ND |
 Ketoglutarate Ketoglutarate | + | + | + | + | + | ND |
 Citrate Citrate | − | + | + | + | ND | ND |
| % Similarityb | ||||||
 HERB 86 (Herbaspirillum-specific) HERB 86 (Herbaspirillum-specific) | 100 | 100 | 100 | 100 | 100 | 100 |
 HERB 1432 (Herbaspirillum-specific) HERB 1432 (Herbaspirillum-specific) | 100 | 100 | 100 | 100 | 100 | 100 |
 Hsero 445 (H. seropedicae-specific) Hsero 445 (H. seropedicae-specific) | 83 | 94 | 94 | 100 | 88 | 88 |
 Hrubri 445 (H. rubrisubalbicans-specific) Hrubri 445 (H. rubrisubalbicans-specific) | 94 | 83 | 83 | 83 | 100 | 77 |
 Hfris 445 (H. frisingense-specific) Hfris 445 (H. frisingense-specific) | 77 | 94 | 94 | 88 | 77 | 100 |
Basic properties including cell shape, cell size, and production of catalase and oxidase of the tested isolates were similar to those of the known species of Herbaspirillum. Utilization of carbon sources such as meso-erythritol and N-acetylglucosamine has been used as criteria for species identification of Herbaspirillum (1, 3, 20). Isolates B65, B501, and B512 differed from H. seropedicae, H. rubrisubalbicans, and H. frisingense in the utilization of meso-erythritol, N-acetylglucosamine, l-rhamnose, and meso-inositol (Table (Table2).2). Kirchhof et al. (20) developed several diagnostic probes of a 16S rRNA-targeted oligonucleotide (18 bp) that are specific for the genus Herbaspirillum and Herbaspirillum species. Complementary sequences of the diagnostic probes were compared with DNA sequences of our 16S rRNA genes, and identity was expressed as percentage similarity in Table Table2.2. In terms of the diagnostic probe sequences, isolates B65, B501, and B512 were not identical to H. seropedicae, H. rubrisubalbicans, and H. frisingense, although the sequences of genus-specific probes, HERB 86 and HERB 1432, showed strong conservation even in the tested isolates. The results of basic properties, carbon source utilization, and the diagnostic probe sequences (Table (Table2)2) indicated that isolates B65, B501, and B512 cannot be classified as any known species of Herbaspirillum but do belong within this genus. They may be new species of Herbaspirillum, although DNA-DNA hybridization and further description are required for accurate classification. Isolates B65, B501, and B512 are thus called Herbaspirillum sp. in this work.
Endophytic colonization of Herbaspirillum sp. strain B501 from wild rice.
To examine endophytic behavior of Herbaspirillum sp. strain B501 isolated from O. officinalis W0012 in the original wild rice and cultivated rice plants, the gfp gene encoding GFP was introduced into the bacteria with a gfp minitransposon pUTgfpx2 by electroporation (34). Several transformants that showed strong GFP fluorescence were picked up, and their growth and acetylene reduction activity were compared with the parent strain Herbaspirillum sp. strain B501. Controls not exposed to electric pulse or foreign DNA did not produce any transformants. Consequently, we selected Herbaspirillum sp. strain B501gfp1, which showed the same growth curve in nutrient broth medium and acetylene reduction activity in RMR semisolid medium as the parent strain B501.
Herbaspirillum sp. strain B501gfp1 partially colonized the coleoptiles, leaves, seeds, and roots of 7-day-old seedlings of O. officinalis W0012 and was distributed in patches (Fig. (Fig.2A2A and B, lanes III). Conversely, no GFP fluorescence was observed in the seedlings of O. sativa cv. Sasanishiki, except for a weak signal in the seed (Fig. (Fig.2C2C and D, lanes III). When a filter that passes over 510-nm-wavelength light was used (lane II), chlorophylls emitted red autofluorescence in shoots. Yellow fluorescence mixed with red and green fluorescence was observed exclusively in shoots of wild rice O. officinalis (Fig. (Fig.2A2A and C, lanes II). These observations suggested that Herbaspirillum sp. strain B501gfp1 exclusively colonizes just the seedlings of wild rice O. officinalis.
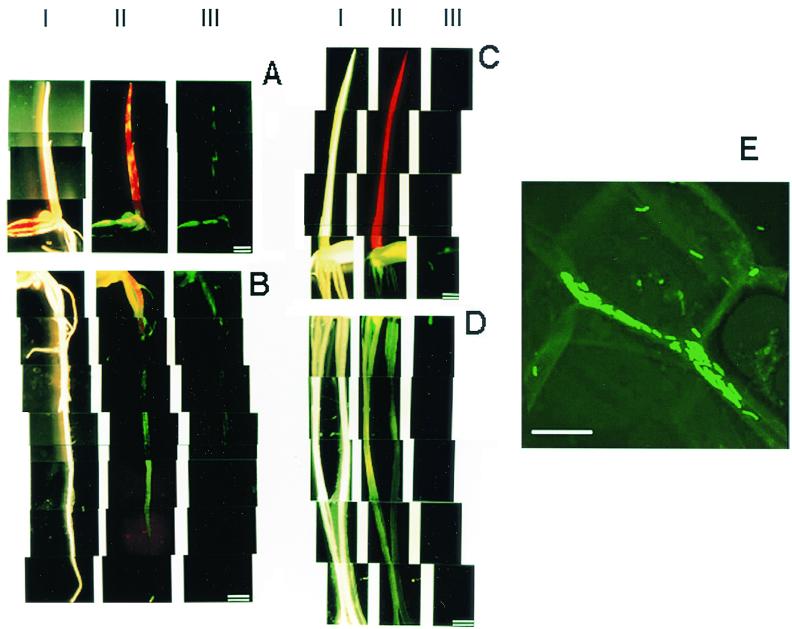
Fluorescence micrographs showing distribution of GFP-tagged Herbaspirillum sp. strain B501gfp1 in 7-day-old seedlings of wild rice (O. officinalis W0012) (A and B), cultivated rice (O. sativa cv. Sasanishiki) (C and D), and intercellular colonization of the bacterium in shoots (coleoptiles) of wild rice (O. officinalis W0012) (E) when seeds were inoculated with B501gfp1. Bars = l mm (A to D) and 10 μm (E). The seedlings were observed under conditions of light field (lanes I) and fluorescence fields with a GFP filter unit (Em510-) (lanes II) and GFP band-pass filter unit (Em510-550) (lanes III). Panel E shows a confocal image by laser scanning fluorescence microscopy.
Localization of Herbaspirillum sp. strain B501 in tissues of wild rice.
GFP-tagged cells of Herbaspirillum sp. strain B501gfp1 were apparently localized in intercellular spaces of shoot tissues of 7-day-old seedlings of O. officinalis W0012 (Fig. (Fig.22E).
Transmission electron microscopy demonstrated that Herbaspirillum sp. strain B501gfp1 colonized intercellular spaces in young tertiary leaves before the leaves expanded (Fig. (Fig.3A,3A, B, and C) and in coleoptiles that were easily seen at this stage (Fig. (Fig.3D).3D). Interestingly, the bacteria penetrated into very young tissues of quaternary leaves (Fig. (Fig.3F)3F) before leaf tissue differentiation, although they did not enter into the five-cell leaf tip (Fig. (Fig.3E).3E). No bacteria were observed in vascular tissues in shoots. Bacteria were often found outside the exodermis of roots but were rarely seen within the tissues. According to these observations, the majority of cells of Herbaspillium sp. strain B501 invaded and colonized intercellular spaces of rice shoots.
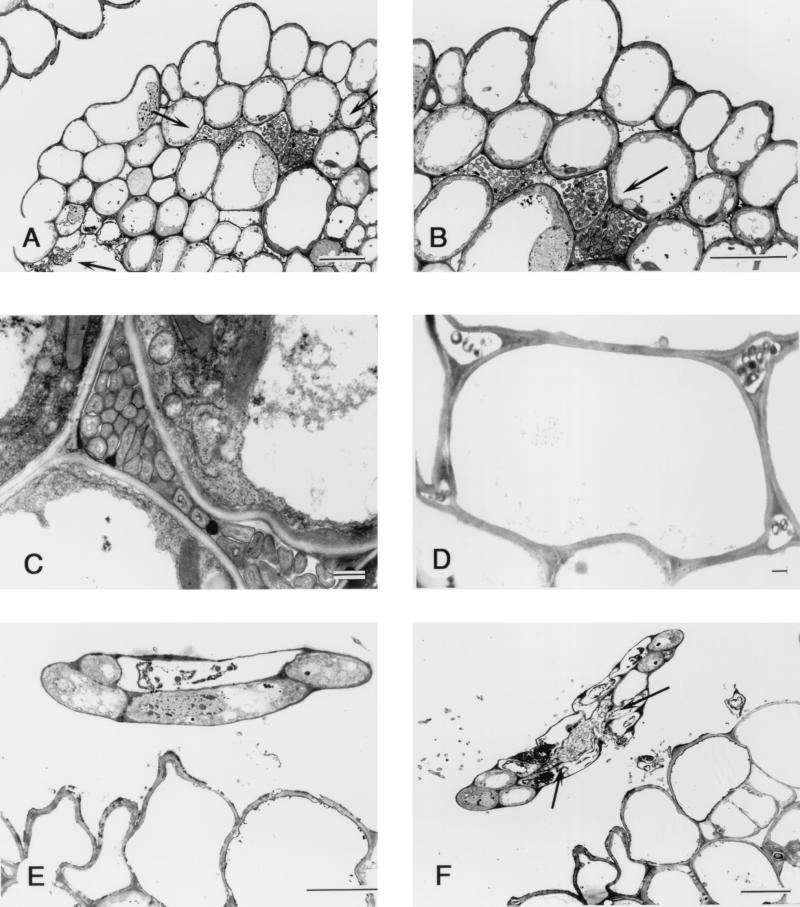
Transmission electron micrograph of transverse section through rice plant of O. officinalis W0012 seven days after seed inoculation with Herbaspirillum sp. strain B501gfp1. (A) The bacteria have invaded the shoot tissue which is a unexpanded third leaf (indicated by the two top arrows), and some bacteria were outside the leaf tissue (the lowest arrow). Bar = 10 μm. (B) High-magnification view of panel A showing bacteria colonizing the intercellular space within the rice tissue. Bar = 10 μm. (C) The bacteria colonizing the intercellular space of the third leaf of rice. Bar = 1 μm. (D) Section from rice coleoptile, showing the bacteria colonizing the intercellular space but not densely populated. Bar = 1 μm. (E) Cross section from the tip of a fourth leaf of rice shoot, showing little invasion of the undifferentiated leaf, which has only five cells. Bar = 10 μm. (F) A lower cross section from the same fourth leaf tip seen in panel E. The bacteria have entered the young fourth leaf and colonized the intercellular space (arrow). Bar = 10 μm.
Endophytic populations in surface-sterilized rice.
After 7-day-old seedlings of O. officinalis W0012, O. rufipogon W2008, O. sativa cv. Sasanishiki, and O. sativa cv. Nipponbare inoculated with the bacterium were surface sterilized, the endophytic populations of the bacterium were measured by a colony count method on nutrient agar medium containing kanamycin (Table (Table3).3). There were significantly more endophytic bacteria in the shoots of wild O. officinalis W0012 and O. rufipogon W2008 than in those of cultivated O. sativa cultivars Sasanishiki and Nipponbare (Table (Table3).3). The density of Herbaspirillum sp. strain B501 was higher in seeds of O. officinalis W0012 than in seeds of other rice varieties (Table (Table3).3). No endophytic bacteria were detected in root tissues in this study. These results are consistent with the observations from fluorescence stereomicroscopy (Fig. (Fig.2)2) and indicate that Herbaspirillum sp. strain B501 prefers wild rice and particularly prefers O. officinalis over O. sativa.
TABLE 3
Colonization of Herbaspirillum sp. strain B501 tagged with gfp and kanamycin resistance genes in wild and cultivated rice 7 days after inoculationa
| Oryza species | Cell density in surface-sterilized rice tissue (104 CFU/g [wet wt])
| ||
|---|---|---|---|
| Shoot | Root | Seed | |
| Wild rice | |||
 O. officinalis W0012 O. officinalis W0012 | 320 | <1.4 | 690 |
 O. rufipogon W2008 O. rufipogon W2008 | 190 | 0.6 | 61 |
| Cultivated rice | |||
 O. sativa cv. Sasanishiki O. sativa cv. Sasanishiki | 2.1 | <0.36 | 22 |
 O. sativa cv. Nipponbare O. sativa cv. Nipponbare | 3.6 | <0.2 | 50 |
Acetylene reduction assays of rice seedlings inoculated with Herbaspirillum sp. strain B501.
Nitrogen fixation of free-living bacteria in media is quite different from their ability to fix nitrogen in planta (8, 29). Therefore, the next question is whether Herbaspirillum sp. strain B501 has the ability to fix nitrogen in planta. In the presence of 10% acetylene, the seedlings of O. officinalis W0012 inoculated with Herbaspirillum sp. strain B501 showed significant acetylene reduction activity compared with the activity in uninoculated plants and in inoculated plants in air (controls) (Table (Table4).4). Conversely, no significant acetylene reduction activity was observed for O. sativa cv. Sasanishiki inoculated with B501 (Table (Table4).4). This result is perhaps not surprising, as the bacteria colonized O. sativa cv. Sasanishiki to a lesser degree than O. officinalis W0012 (Table (Table3).3).
TABLE 4
Acetylene reduction activity of rice seedlings inoculated with Herbaspirillum sp. strain B501gfp1a
| Rice genotype | Inoculation | Acetylene | Ethylene evolution
| |
|---|---|---|---|---|
| nmol h−1 (g [wet wt])−1 | nmol h−1 plant−1 | |||
| O. officinalis W0012 | + | + | 71.3 ± ± 22.4 22.4 | 1.60 ± ± 0.50 0.50 |
| − | + | 3.9 ± ± 1.6 1.6 | 0.07 ± ± 0.03 0.03 | |
| + | − | 1.2 ± ± 1.0 1.0 | 0.03 ± ± 0.02 0.02 | |
| 67b | 1.53 | |||
| O. sativa cv. Sasanishiki | + | + | 1.9 ± ± 0.8 0.8 | 0.18 ± ± 0.07 0.07 |
| − | + | 0.8 ± ± 0.3 0.3 | 0.08 ± ± 0.04 0.04 | |
| + | − | 1.7 ± ± 0.8 0.8 | 0.04 ± ± 0.01 0.01 | |
 1 1 | 0.1 | |||
15N2 gas incorporation.
The acetylene reduction assay is an indirect method to verify nitrogen fixation in planta of endophytes, and the possibility of ethylene emission from plants could not be completely excluded. Therefore, we designed a 15N dinitrogen incorporation experiment to follow 15N2 gas into rice plants through endophytic Herbaspirillum sp. strain B501. No incorporation of 15N was detected in O. officinalis without inoculation (Table (Table5).5). Nevertheless, significant 15N incorporation from 15N gas was detected in the rice plants with inoculation. This result clearly demonstrated that Herbaspirillum sp. strain B501 in O. officinalis fixed nitrogen within the rice plants.
TABLE 5
Incorporation of 15N2 into O. officinalis W0012 inoculated with Herbaspirillum sp. strain B501a
| Inoculation | Gas | 15N incorporationb
| |
|---|---|---|---|
| δ15N (‰) | Atom% excess | ||
| None | Air | 0.9 | <0.001 |
| None | 15N2 | 0.4 | <0.001 |
| B501 | Air | 0.8 | <0.001 |
| B501 | 15N2 | 381 | 0.135 |
DISCUSSION
Many workers have isolated diazotrophic bacteria from gramineous plants using nitrogen-free media such as semisolid NFb (3). Rennie medium was proposed as a more efficient nitrogen-free medium for isolation of nitrogen-fixing bacteria (26). Use of a modified Rennie medium in this study probably enabled us to isolate new nitrogen-fixing bacteria, such as close relatives of genera Herbaspirillum and Ideonella. In particular, Ideonella is a monospecific genus (I. dechloratans), which is capable of growing anaerobically with chlorate as an electron acceptor, although any ability to fix nitrogen is as yet unknown (21).
When the acetylene reduction activities of original rice seedlings inoculated with the corresponding diazotrophic bacteria were preliminarily assayed, the combination of bacterial isolate B501 and O. officinalis W0012 showed the highest activity (data not shown). Moreover, Herbaspirillum spp. have been found in sugarcane and other gramineous plants (3, 23) and are possibly capable of nitrogen fixation in planta (3, 16, 25). Thus, we have focused on the three isolates (B65, B501, and B512) that were closely related to Herbaspirillum spp. based on the 16S rDNA sequences. However, isolates B65, B501, and B512 did not cluster with any known species of Herbaspirillum but were determined to belong within this genus by characterization of basic properties, carbon source utilization, and the diagnostic probe sequences (Table (Table2),2), suggesting a great diversity of Herbaspirillum sp. residing in wild rice species and other gramineous plants.
Considerable progress has been made in recent years in nitrogen fixation of Acetobacter diazotrophicus (17, 29) and Azoarcus sp. (8, 15, 25) using nitrogenase antibodies and several reporter genes behind nif genes. Herbaspirillum is expected to be responsible for nitrogen fixation in sugarcane, rice, and sorghum (16, 19, 23). Except for some acetylene reduction activity in inoculated sugarcane (17) and reaction with a nitrogenase antibody in sorghum (19) however, there has been no direct evidence of nitrogen fixation in rice plants. In this study, the inoculation of wild rice, O. officinalis W0012, with Herbaspirillum sp. strain B501 apparently increased 15N concentration from 15N2 gas into the rice plants. To our knowledge, this is the first direct evidence that Herbaspirillum sp. fix nitrogen in Oryza species. Interestingly, the degree of nitrogen fixation in planta is likely to depend on rice species (Table (Table4)4) due to variable endophytic colonization of Herbaspirillum among rice species (Table (Table3).3). This suggests the presence of host specificity or preference of diazotrophic endophytes similar to those in microbe-plant mutualisms found with Rhizobium and legumes (24).
James et al. (17, 19) have reported that A. diazotrophicus, H. rubrisubalbicans, and H. seropedicae colonized xylem vessels of host plants. However, Dong et al. (7a) postulated that xylem is an unsuitable habitat for A. diazotrophicus in sugarcane from the anatomical and physiological points of view. In this work, Herbaspirillum sp. strain B501 never entered vascular tissues, apparently preferring the apoplast of shoot tissues, colonizing intercellular spaces. Conversely, microscopic observation and enumeration of surface-sterilized roots suggests that the bacterium probably colonized the root surface. Endophytic distributions and penetration of young leaf tissues (Fig. (Fig.22 and and3)3) suggest that Herbaspirillum sp. strain B501 spread in aerial parts of wild rice via apoplastic spaces. Motility and pectinase and cellulase production by Herbaspirillum sp. strain B501 might be involved in the spreading throughout shoot tissues.
The increase in 15N concentration in rice plants for 24 h (0.135 atom% excess) was low compared to the concentration of 15N2 gas used (99.7 atom%), indicating that 0.14% of total nitrogen was derived from 15N2 gas after the exposure for 24 h (Table (Table5).5). This value is similar to that reported in a combination of sugarcane and A. diazotrophicus. Sugarcane plants inoculated with A. diazotrophicus PA15 and PPe4 showed that 0.1 to 0.5% of total nitrogen was derived from 15N2 for 24-h spot exposure to 15N2 in a nitrogen-deficient condition (29). However, 15N isotope dilution studies have shown that sugarcane can fix substantial amounts of N2, up to 70% of nitrogen requirements of the plant, although the amount of fixed nitrogen is dependent on environmental conditions (6, 16). Therefore, it is possible that plant stage and environmental conditions affect the amount of nitrogen fixation by endophytes. In this work, young seedlings of the inoculated rice plants were evaluated for nitrogen fixation. Therefore, further experiments of 15N incorporation and 15N isotope dilution using mature rice plants with and without inoculation of Herbaspirillum sp. strain B501 would shed light on the real contribution of nitrogen fixation by the bacterium.
ACKNOWLEDGMENTS
This work was supported in part by grants (to K.M.) from PROBRAIN, the Ministry of Education, Science, Sports and Culture of Japan (grant 98460), Mayekawa MFG Co., and the Joint Research Program of the Institute of Genetic Ecology, Tohoku University (grant 981002). A.E. was supported by a research fellowship for young scientists from the Japan Society for the Promotion of Science, for which we are grateful.
We thank K. Yuhashi (Plant Biotechnology Institute, Ibaraki Agricultural Center) and M. Saito (National Institute of Livestock and Grassland Science) for their help with confocal laser scanning microscopy, J. K. Jansson (Stockholm University) for kindly providing plasmid pUTgfpx2, and S. Harayama (Marine Biotechnology Institute, Kamaishi Laboratories) for his continuing interest and encouragement.
REFERENCES
Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.67.11.5285-5293.2001
Read article for free, from open access legal sources, via Unpaywall:
https://aem.asm.org/content/67/11/5285.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Bioactive Compounds Produced by Endophytic Bacteria and Their Plant Hosts-An Insight into the World of Chosen Herbaceous Ruderal Plants in Central Europe.
Molecules, 29(18):4456, 19 Sep 2024
Cited by: 0 articles | PMID: 39339451 | PMCID: PMC11433698
Review Free full text in Europe PMC
Dynamic nitrogen fixation in an aerobic endophyte of Populus.
ISME J, 18(1):wrad012, 01 Jan 2024
Cited by: 0 articles | PMID: 38365250 | PMCID: PMC10833079
An extracellular polysaccharide is involved in the aluminum tolerance of Pullulanibacillus sp. CA42, a newly isolated strain from the Chinese water chestnut growing in an actual acid sulfate soil area in Vietnam.
Front Microbiol, 14:1241244, 28 Aug 2023
Cited by: 0 articles | PMID: 37700869 | PMCID: PMC10493610
Endophytic bacterial communities in wild rice (Oryza officinalis) and their plant growth-promoting effects on perennial rice.
Front Plant Sci, 14:1184489, 14 Aug 2023
Cited by: 4 articles | PMID: 37645460 | PMCID: PMC10461003
Biotization of in vitro oil palm (Elaeis guineensis Jacq.) and its plant-microbe interactions.
Front Plant Sci, 14:1150309, 18 Apr 2023
Cited by: 0 articles | PMID: 37143882 | PMCID: PMC10151813
Review Free full text in Europe PMC
Go to all (162) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 43 of 43)
- (1 citation) ENA - Z29583
- (1 citation) ENA - X67227
- (1 citation) ENA - AB004744
- (1 citation) ENA - AB021407
- (1 citation) ENA - U37337
- (1 citation) ENA - AB021424
- (1 citation) ENA - X87272
- (1 citation) ENA - D25312
- (1 citation) ENA - Y10146
- (1 citation) ENA - M59291
- (1 citation) ENA - X79735
- (1 citation) ENA - X79731
- (1 citation) ENA - D16211
- (1 citation) ENA - D16213
- (1 citation) ENA - AJ251469
- (1 citation) ENA - AB049109
- (1 citation) ENA - AB049108
- (1 citation) ENA - AB049107
- (1 citation) ENA - AB049106
- (1 citation) ENA - AB049105
- (1 citation) ENA - AB049104
- (1 citation) ENA - AF078756
- (1 citation) ENA - AB049103
- (1 citation) ENA - X90759
- (1 citation) ENA - AB021418
- (1 citation) ENA - AF130981
- (1 citation) ENA - Z96078
- (1 citation) ENA - M34132
- (1 citation) ENA - X79738
- (1 citation) ENA - X72724
- (1 citation) ENA - X79739
- (1 citation) ENA - M59061
- (1 citation) ENA - Z29619
- (1 citation) ENA - X97071
- (1 citation) ENA - X79288
- (1 citation) ENA - Z29617
- (1 citation) ENA - AB049112
- (1 citation) ENA - AB049133
- (1 citation) ENA - AB049111
- (1 citation) ENA - AB049110
- (1 citation) ENA - AF025370
- (1 citation) ENA - AF025371
- (1 citation) ENA - D30793
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants.
Int J Syst Evol Microbiol, 51(pt 1):157-168, 01 Jan 2001
Cited by: 39 articles | PMID: 11211253
Enterobacter oryzae sp. nov., a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia.
Int J Syst Evol Microbiol, 59(pt 7):1650-1655, 01 Jul 2009
Cited by: 35 articles | PMID: 19578150
Effects of the inoculation of Burkholderia vietnamensis and related endophytic diazotrophic bacteria on grain yield of rice.
Microb Ecol, 55(1):21-37, 04 Apr 2007
Cited by: 46 articles | PMID: 17406771
Expression of the nifH gene of a Herbaspirillum endophyte in wild rice species: daily rhythm during the light-dark cycle.
Appl Environ Microbiol, 71(12):8183-8190, 01 Dec 2005
Cited by: 26 articles | PMID: 16332801 | PMCID: PMC1317309





