Abstract
Free full text

A Positively Selected fur-R88H Mutation Enhances Helicobacter pylori Fitness in a High-Salt Environment and Alters Fur-Dependent Regulation of Gene Expression
ABSTRACT
Both Helicobacter pylori infection and a high-salt diet are risk factors for gastric cancer. We previously showed that a mutation in fur (encoding the ferric uptake regulator variant Fur-R88H) was positively selected in H. pylori strains isolated from experimentally infected Mongolian gerbils receiving a high-salt diet. In the present study, we report that continuous H. pylori growth in high-salt conditions in vitro also leads to positive selection of the fur-R88H mutation. Competition experiments with strains containing wild-type fur or fur-R88H, each labeled with unique nucleotide barcodes, showed that the fur-R88H mutation enhances H. pylori fitness under high-salt conditions but reduces H. pylori fitness under routine culture conditions. The fitness advantage of the fur-R88H mutant under high-salt conditions was abrogated by the addition of supplemental iron. To test the hypothesis that the fur-R88H mutation alters the regulatory properties of Fur, we compared the transcriptional profiles of strains containing wild-type fur or fur-R88H. Increased transcript levels of fecA2, which encodes a predicted TonB-dependent outer membrane transporter, were detected in the fur-R88H variant compared to those in the strain containing wild-type fur under both high-salt and routine conditions. Competition experiments showed that fecA2 contributes to H. pylori fitness under both high-salt and routine conditions. These results provide new insights into mechanisms by which the fur-R88H mutation confers a selective advantage to H. pylori in high-salt environments.
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that colonizes the stomach in about 50% of the world's population (1,–3). A gastric mucosal inflammatory response (gastritis) develops in individuals who are colonized with H. pylori. While most individuals colonized with H. pylori remain asymptomatic, the presence of this organism is a risk factor for the development of peptic ulcer disease or gastric cancer (2, 3).
The clinical outcome of H. pylori infection is determined by multiple factors, including genetic characteristics of H. pylori strains and genetic characteristics of human hosts (4). Environmental factors such as diet can also impact the risk of gastric disease. For instance, the consumption of a diet high in salt content is associated with an increased risk of gastric cancer (5,–8). The association between high dietary salt intake and increased gastric cancer risk has been observed in both humans and a Mongolian gerbil model of H. pylori infection (9, 10).
To understand how a high-salt diet contributes to adverse disease outcome, we and others have investigated the effects of various sodium chloride concentrations on H. pylori in vitro (11,–17). When H. pylori grown under high-salt conditions is compared with bacteria grown under routine conditions, numerous genes are differentially expressed (11, 13, 15, 17). Similarly, there are differences in the steady-state levels of various proteins (15, 16). For example, the levels of several proteins that cause alterations in host cells, including CagA, VacA, and HopQ, are higher in H. pylori cultured under high-salt conditions than in H. pylori cultured under routine conditions (13, 15,–17).
A high-salt diet also shapes the evolution of H. pylori within the stomach (14, 18). For example, in two previous studies of H. pylori-infected Mongolian gerbils, we detected a mutation in fur (encoding the ferric uptake regulator variant Fur-R88H) more commonly in H. pylori strains isolated from Mongolian gerbils fed a high-salt diet than in strains isolated from gerbils fed a regular-salt diet (14, 18). The fur-R88H polymorphism was also positively selected in Mongolian gerbils fed an iron-deficient diet (19).
Fur is a transcriptional regulator that modulates the expression of genes involved in iron uptake and utilization. Previous studies have shown that H. pylori Fur can act as either a repressor or activator of specific genes and can regulate gene expression in both iron-bound and apo forms (20,–28). Amino acids H42, E90, H97, H99, and E110 are important for the binding of iron by H. pylori Fur and constitute an iron binding site (29, 30). Mutagenesis of these residues leads to a conformational change in the DNA-binding domain (29), and mutations to Fur amino acids R87 and Y89 affect the regulation of the Fur target amiE (30). Given the proximity of the R88H mutation to these amino acid residues (E90, R87, and Y89), we hypothesized that the fur-R88H mutation influences the capacity of Fur to regulate H. pylori gene expression.
In the present study, we undertook experiments to further evaluate how the fur-R88H mutation influences H. pylori fitness and systematically investigate effects of the fur-R88H mutation on H. pylori gene expression. We show that the fur-R88H mutation enhances H. pylori fitness in vitro under high-salt conditions but reduces H. pylori fitness under routine culture conditions. The fitness advantage of the fur-R88H mutant in high-salt conditions is abrogated by the addition of supplemental iron. We show that the transcriptional profile of a strain containing wild-type (WT) fur differs from the transcriptional profile of a strain containing fur-R88H under both routine and high-salt conditions. One of the transcripts with increased levels in the fur-R88H mutant compared to the WT strain is fecA2, a Fur-regulated gene that is annotated as an ortholog of Escherichia coli FecA (31, 32), a TonB-dependent outer membrane protein. E. coli FecA is known to transport ferric citrate (33,–35). We show that fecA2 contributes to H. pylori fitness under both high-salt and routine conditions. These results provide new insights into mechanisms by which the fur-R88H mutation confers a selective advantage to H. pylori in high-salt conditions.
RESULTS
H. pylori evolution in response to high-salt conditions in vitro.
In a previous study (14), we showed that a single nucleotide polymorphism (SNP) in fur (encoding Fur-R88H) was positively selected in H. pylori strains isolated from experimentally infected gerbils fed a high-salt diet. To directly investigate the effects of high-salt conditions on the selection of fur-R88H and other SNPs, we serially passaged H. pylori strain 7.13 in vitro for 4 months on sulfite-free Brucella agar supplemented with fetal bovine serum (BB-FBS agar), containing either 0.5% added NaCl (routine salt concentration) or 0.9% added NaCl (high salt). The H. pylori populations were analyzed by whole-genome sequencing at 4-, 6-, and 16-week time points. Analyses of the whole-genome sequences allowed us to identify and compare nucleotide variations that emerged in the H. pylori populations cultured under the two conditions. In comparison to the parental strain, the passaged populations contained numerous nucleotide substitution mutations, as well as insertions and deletions, many of which were detected in only a small proportion of the sequence reads. We focused on SNPs that were detected in >80% of sequence reads from the passaged strains and either absent or uncommon (<15% of sequence reads) in the parental strain.
As shown in Fig. 1A, H. pylori passaged for 16 weeks on routine medium (0.5% NaCl) contained nonsynonymous SNPs in genes encoding the flagellar biosynthesis protein FlhA (A205V) (36, 37), galactose epimerase GalE (H339Q) (38), and the immunomodulatory autotransporter protein (ImaA, originally designated VacA5 [39, 40]; W2537* premature termination), as well as a synonymous SNP in niuB (originally designated ceuE3), which encodes a periplasmic nickel-binding protein (41,–43). H. pylori passaged under high-salt conditions contained four unique SNPs different from those detected in the bacteria cultured under routine conditions (Fig. 1B). Specifically, SNPs were detected in the coding region of fur (R88H), galE (R200C), and the catalase gene katA (G125D) (Fig. 1B). A SNP in the 5′ untranslated region (UTR) of hopD (encoding an outer membrane protein) (44) was also detected (Fig. 1B). These SNPs became dominant in the population at different time points during the passage of H. pylori
in vitro. The fur-R88H SNP was detected in 100% of sequence reads at the earliest time point sampled (4
weeks on routine medium (0.5% NaCl) contained nonsynonymous SNPs in genes encoding the flagellar biosynthesis protein FlhA (A205V) (36, 37), galactose epimerase GalE (H339Q) (38), and the immunomodulatory autotransporter protein (ImaA, originally designated VacA5 [39, 40]; W2537* premature termination), as well as a synonymous SNP in niuB (originally designated ceuE3), which encodes a periplasmic nickel-binding protein (41,–43). H. pylori passaged under high-salt conditions contained four unique SNPs different from those detected in the bacteria cultured under routine conditions (Fig. 1B). Specifically, SNPs were detected in the coding region of fur (R88H), galE (R200C), and the catalase gene katA (G125D) (Fig. 1B). A SNP in the 5′ untranslated region (UTR) of hopD (encoding an outer membrane protein) (44) was also detected (Fig. 1B). These SNPs became dominant in the population at different time points during the passage of H. pylori
in vitro. The fur-R88H SNP was detected in 100% of sequence reads at the earliest time point sampled (4 weeks).
weeks).
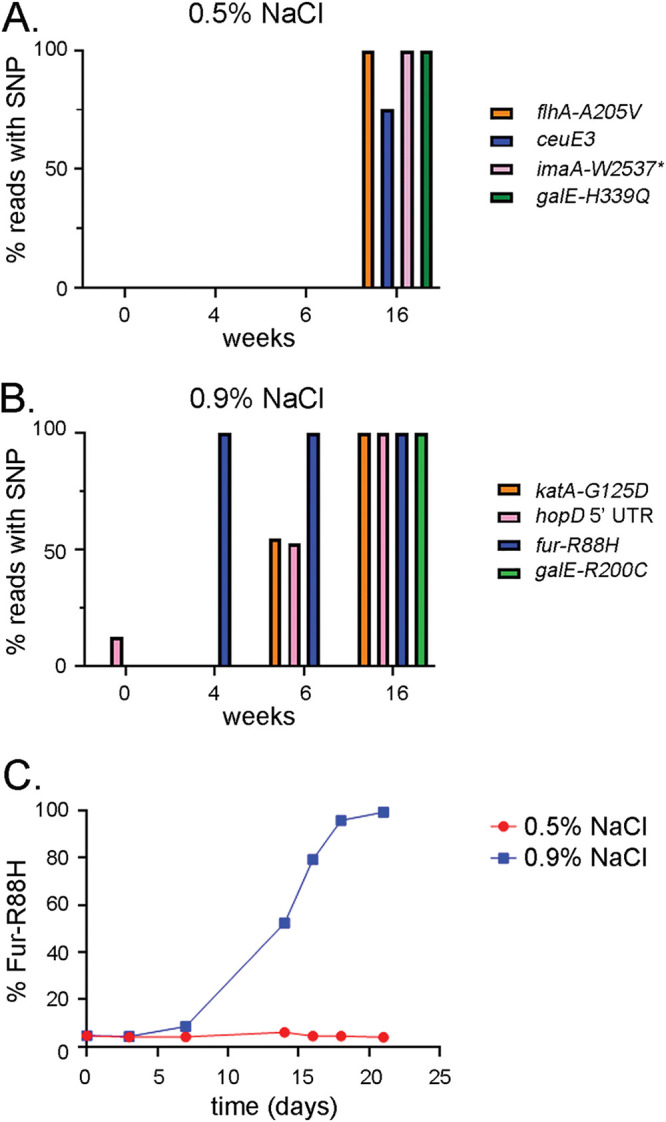
Identification of positively selected SNPs in H. pylori strains grown under routine or high-salt conditions. H. pylori strain 7.13 was cultured continuously for 4 months on medium containing either 0.5% or 0.9% added NaCl. At the indicated time points, bacteria were harvested, genomic DNA was isolated, and whole-genome DNA sequencing analysis was performed. (A and B) The figure illustrates SNPs detected in greater than 75% of sequence reads from H. pylori passaged for 4
months on medium containing either 0.5% or 0.9% added NaCl. At the indicated time points, bacteria were harvested, genomic DNA was isolated, and whole-genome DNA sequencing analysis was performed. (A and B) The figure illustrates SNPs detected in greater than 75% of sequence reads from H. pylori passaged for 4 months on medium containing 0.5% (A) or 0.9% (B) added NaCl but detected at low levels (<15% of sequence reads) in the parental strain. The proportions of sequence reads containing the indicated SNPs at the indicated time points are shown. Nonsynonymous SNPs in genes encoding the flagellar biosynthesis protein FlhA (A205V), the immunomodulatory autotransporter protein ImaA (VacA5) (W2537*), and galactose epimerase GalE (H339Q) and a synonymous SNP in the coding region of niuB (originally designated ceuE3) were identified after 4
months on medium containing 0.5% (A) or 0.9% (B) added NaCl but detected at low levels (<15% of sequence reads) in the parental strain. The proportions of sequence reads containing the indicated SNPs at the indicated time points are shown. Nonsynonymous SNPs in genes encoding the flagellar biosynthesis protein FlhA (A205V), the immunomodulatory autotransporter protein ImaA (VacA5) (W2537*), and galactose epimerase GalE (H339Q) and a synonymous SNP in the coding region of niuB (originally designated ceuE3) were identified after 4 months of passaging on medium containing 0.5% added NaCl. Nonsynonymous SNPs were identified in genes encoding catalase (G125D), Fur (R88H), and GalE (R200C) and a SNP in the 5′ UTR of hopD following 4
months of passaging on medium containing 0.5% added NaCl. Nonsynonymous SNPs were identified in genes encoding catalase (G125D), Fur (R88H), and GalE (R200C) and a SNP in the 5′ UTR of hopD following 4 months of passaging on medium containing 0.9% added NaCl. (C) H. pylori strain 7.13 was cultured for 21
months of passaging on medium containing 0.9% added NaCl. (C) H. pylori strain 7.13 was cultured for 21 days on BB-FBS agar plates containing either 0.5% or 0.9% added NaCl. Genomic DNA samples isolated from bacteria at the indicated time points were analyzed by qPCR methods to quantify the proportional abundance of fur alleles containing the fur-R88H mutation (see Materials and Methods).
days on BB-FBS agar plates containing either 0.5% or 0.9% added NaCl. Genomic DNA samples isolated from bacteria at the indicated time points were analyzed by qPCR methods to quantify the proportional abundance of fur alleles containing the fur-R88H mutation (see Materials and Methods).
To further define the kinetics of the process through which the fur-R88H mutation becomes predominant in the population, we repeated the experiment, analyzed earlier time points, and utilized a quantitative PCR (qPCR)-based approach to quantify the proportion of strains containing wild-type fur or fur-R88H in the bacterial population (45). As shown in Fig. 1C, the fur-R88H mutation was present at low levels (~3 to 4% fur-R88H and 96% WT fur) in the starting population (t =
= 0). In the bacterial population passaged on high-salt medium, the fur-R88H mutation was detected in almost 100% of the population at the 18-day time point. In contrast, there was little or no increase in the levels of fur-R88H in bacteria passaged on routine medium. Collectively, these results indicate that H. pylori variants containing the fur-R88H SNP are positively selected under high-salt conditions in vitro.
0). In the bacterial population passaged on high-salt medium, the fur-R88H mutation was detected in almost 100% of the population at the 18-day time point. In contrast, there was little or no increase in the levels of fur-R88H in bacteria passaged on routine medium. Collectively, these results indicate that H. pylori variants containing the fur-R88H SNP are positively selected under high-salt conditions in vitro.
Relative fitness of strains containing wild-type fur or fur-R88H under various conditions.
To compare the relative fitness of strains containing wild-type fur or fur-R88H, we introduced unique nucleotide barcodes into H. pylori 7.13 strains that were previously engineered to contain either wild-type fur or fur-R88H (14) (Fig. 2A and andB).B). Strain JL1 (containing wild-type fur) was used as a control strain, and strain JL2 contains the fur-R88H mutation. Growth curve analysis of these strains individually revealed that they grew at similar rates in medium containing 0.5% added NaCl (Fig. 2C). The maximum optical densities and growth rates of both strains were lower in medium containing 1.0% added NaCl than those in medium containing 0.5% added NaCl, but there were no substantial differences in growth characteristics of the two strains (Fig. 2C). To determine the effects of NaCl on H. pylori survival, cultures of JL1 (WT fur) and JL2 (fur-R88H) grown for 15 h in medium containing either 0.5% NaCl or 1.0% NaCl were normalized based on optical density, and the normalized cultures were plated on BB-FBS agar plates containing 0.5% NaCl. As shown in Fig. 2D, the number of CFU from broth cultures grown under high-salt conditions was lower than the number of CFU cultured from the same strains grown under regular salt conditions. These results suggest that exposure to high-salt conditions reduces H. pylori survival. CFU counts were slightly reduced for strain JL2 (fur-R88H) grown in 0.5% NaCl compared to those for JL1 (WT fur) grown in 0.5% NaCl, but there was no significant difference in CFU counts when JL1 and JL2 grown under high-salt conditions (1.0% NaCl) were compared. We speculate that small differences in growth and/or survival of the two strains could account for fitness advantages over longer periods of time.
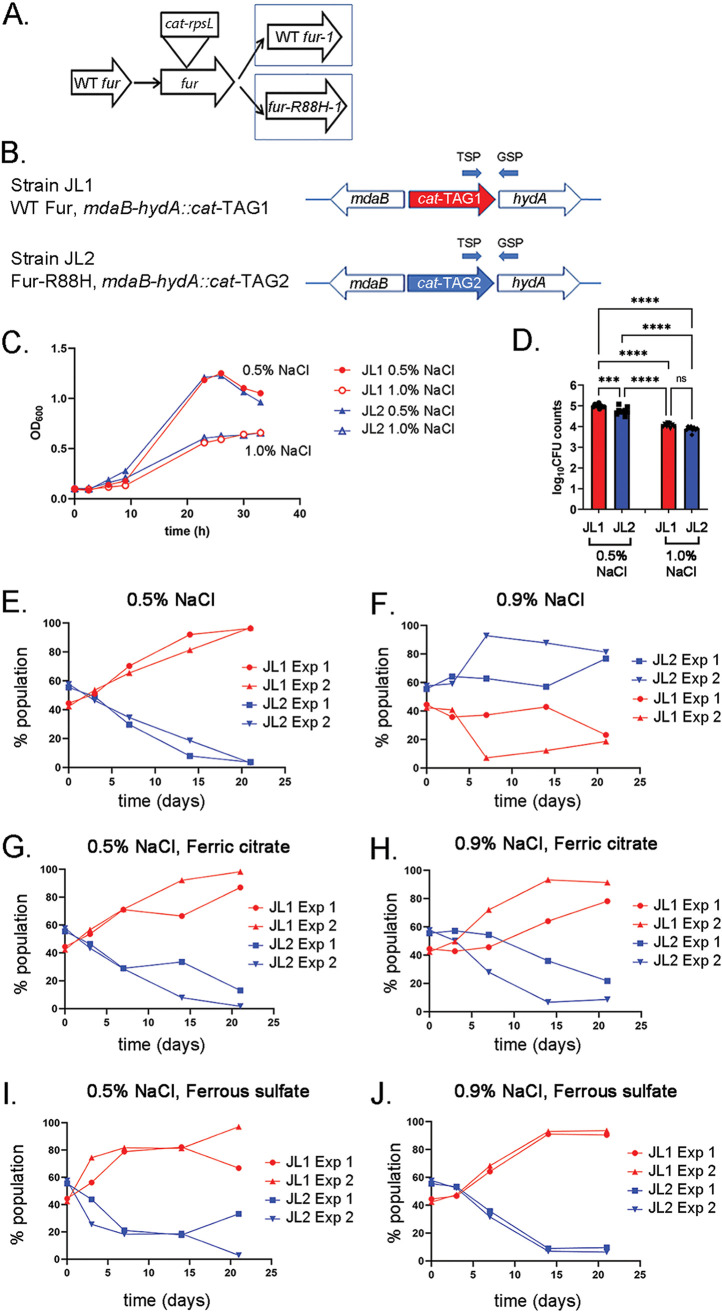
Fitness of H. pylori strains containing WT fur or fur-R88H under various conditions. (A) H. pylori strains containing either WT fur (i.e., 7.13 WT Fur-1) or fur-R88H (i.e., 7.13 Fur-R88H-1) were generated previously (14). (B) Strain JL1 is derived from 7.13 WT Fur-1 and contains a cat-TAG1 cassette (chloramphenicol acetyltransferase gene and unique DNA barcode TAG1) inserted between mdaB and hydA genes (designated HP0630 and HP0631, respectively, in H. pylori strain 26695, and HPB8_829 and HPB8_830, respectively, in H. pylori strain B8). Strain JL2 is derived from 7.13 Fur-R88H-1 and contains a cat-TAG2 cassette inserted in the mdaB-hydA intergenic region. (C) Strains JL1 and JL2 were grown individually in BB-FBS broth containing either 0.5% or 1.0% added NaCl. OD600 values were measured at the indicated time points. (D) Strains JL1 and JL2 were grown for 15 h in BB-FBS broth containing either 0.5% added NaCl or 1.0% added NaCl. Cultures grown in 0.5% added NaCl reached an OD600 of ~0.5, while cultures grown in 1.0% added NaCl reached an OD600 of ~0.25. All cultures were normalized to an OD600 of 0.2, and serial dilutions of these normalized cultures were plated on BB-FBS agar plates containing 0.5% added NaCl. The numbers of CFU for strain JL1 (red bars) and JL2 (blue bars) were enumerated 6 days after plating. (***, P = 0.0001; ****, P < 0.0001, one-way analysis of variance [ANOVA] with Tukey’s multiple-comparison test). (E to J) Strains JL1 and JL2 were mixed 1:1 and cultured on medium containing either 0.5% (E) or 0.9% (F) added NaCl. Competition experiments were also done using medium supplemented with 100
days after plating. (***, P = 0.0001; ****, P < 0.0001, one-way analysis of variance [ANOVA] with Tukey’s multiple-comparison test). (E to J) Strains JL1 and JL2 were mixed 1:1 and cultured on medium containing either 0.5% (E) or 0.9% (F) added NaCl. Competition experiments were also done using medium supplemented with 100 μM ferric citrate (G and H) or 100
μM ferric citrate (G and H) or 100 μM ferrous sulfate (I and J). The bacteria from the plates were subjected to continuous passage (once every 3.5
μM ferrous sulfate (I and J). The bacteria from the plates were subjected to continuous passage (once every 3.5 days) for 21
days) for 21 days. At the indicated time points, bacteria were harvested and genomic DNA was isolated. The proportion of each strain in the bacterial population was then quantified by qPCR as described in Materials and Methods. The results from two independent experiments are shown. qPCR analysis was performed in triplicate on samples from each time point, and mean values are reported. qPCR primer pairs involving a TSP and a GSP are illustrated in panel B, and the sequences are listed in Table S3 in the supplemental material.
days. At the indicated time points, bacteria were harvested and genomic DNA was isolated. The proportion of each strain in the bacterial population was then quantified by qPCR as described in Materials and Methods. The results from two independent experiments are shown. qPCR analysis was performed in triplicate on samples from each time point, and mean values are reported. qPCR primer pairs involving a TSP and a GSP are illustrated in panel B, and the sequences are listed in Table S3 in the supplemental material.
To further evaluate the relative fitness of strains JL1 (WT fur) and JL2 (fur-R88H), H. pylori mixtures containing equal proportions of the two barcoded strains were serially cultured for 21 days on BB-FBS agar plates containing either 0.5% or 0.9% added NaCl (see details in Materials and Methods regarding the use of 0.9% added NaCl for experiments with plates instead of 1.0% NaCl). We also cultured the bacteria on these media supplemented with ferric citrate (100
days on BB-FBS agar plates containing either 0.5% or 0.9% added NaCl (see details in Materials and Methods regarding the use of 0.9% added NaCl for experiments with plates instead of 1.0% NaCl). We also cultured the bacteria on these media supplemented with ferric citrate (100 μM) or ferrous sulfate (100
μM) or ferrous sulfate (100 μM). We then analyzed changes in the proportional abundance of strains within the mixed populations, using a qPCR method (45). As shown in Fig. 2E, the strain containing WT fur (JL1) comprised the major component of the population following 21
μM). We then analyzed changes in the proportional abundance of strains within the mixed populations, using a qPCR method (45). As shown in Fig. 2E, the strain containing WT fur (JL1) comprised the major component of the population following 21 days of growth on medium containing 0.5% added NaCl. The addition of either ferric citrate or ferrous sulfate to the medium did not alter the results (Fig. 2G and andI).I). In contrast, the strain containing fur-R88H (JL2) comprised the major component of the population after 21
days of growth on medium containing 0.5% added NaCl. The addition of either ferric citrate or ferrous sulfate to the medium did not alter the results (Fig. 2G and andI).I). In contrast, the strain containing fur-R88H (JL2) comprised the major component of the population after 21 days of growth on medium containing 0.9% added NaCl (Fig. 2F). Interestingly, the addition of either ferric citrate or ferrous sulfate to the medium containing 0.9% NaCl resulted in a change in the relative fitness of the strains. Specifically, the strain containing WT fur (JL1) comprised the major component of the population after 21
days of growth on medium containing 0.9% added NaCl (Fig. 2F). Interestingly, the addition of either ferric citrate or ferrous sulfate to the medium containing 0.9% NaCl resulted in a change in the relative fitness of the strains. Specifically, the strain containing WT fur (JL1) comprised the major component of the population after 21 days of growth on medium containing 0.9% NaCl, supplemented with ferric citrate or ferrous sulfate (Fig. 2H and andJ).J). These results indicate that both salt concentration and iron concentration influence the relative fitness of strains containing WT fur or fur-R88H.
days of growth on medium containing 0.9% NaCl, supplemented with ferric citrate or ferrous sulfate (Fig. 2H and andJ).J). These results indicate that both salt concentration and iron concentration influence the relative fitness of strains containing WT fur or fur-R88H.
Transcriptional profiling of the Fur-R88H mutant.
To test the hypothesis that the fur-R88H mutation impacts Fur-mediated gene transcription, we compared the transcriptional profiles of H. pylori strains containing WT fur or fur-R88H (Fig. 3A). Each strain was grown for 15 h in medium supplemented with either 0.5% NaCl or 1.0% NaCl (Fig. 3A). Growth under 1.0% NaCl conditions resulted in a lower optical density (mean optical density at 600 nm [OD600] = 0.45 ±
± 0.09) than growth under 0.5% NaCl conditions (mean OD600 = 0.89
0.09) than growth under 0.5% NaCl conditions (mean OD600 = 0.89 ±
± 0.04). RNA was isolated and gene expression was analyzed by transcriptome sequencing (RNA-seq), as described in Materials and Methods. Differences in the patterns of gene expression were first analyzed by unsupervised principal-component analysis (Fig. 3B). The most striking variation among samples was linked to the culture conditions (growth under routine salt or high-salt conditions). Differences between the two strains were detectable for cultures grown under routine conditions, but there was very little variance between the two strains grown under high-salt conditions (Fig. 3B).
0.04). RNA was isolated and gene expression was analyzed by transcriptome sequencing (RNA-seq), as described in Materials and Methods. Differences in the patterns of gene expression were first analyzed by unsupervised principal-component analysis (Fig. 3B). The most striking variation among samples was linked to the culture conditions (growth under routine salt or high-salt conditions). Differences between the two strains were detectable for cultures grown under routine conditions, but there was very little variance between the two strains grown under high-salt conditions (Fig. 3B).
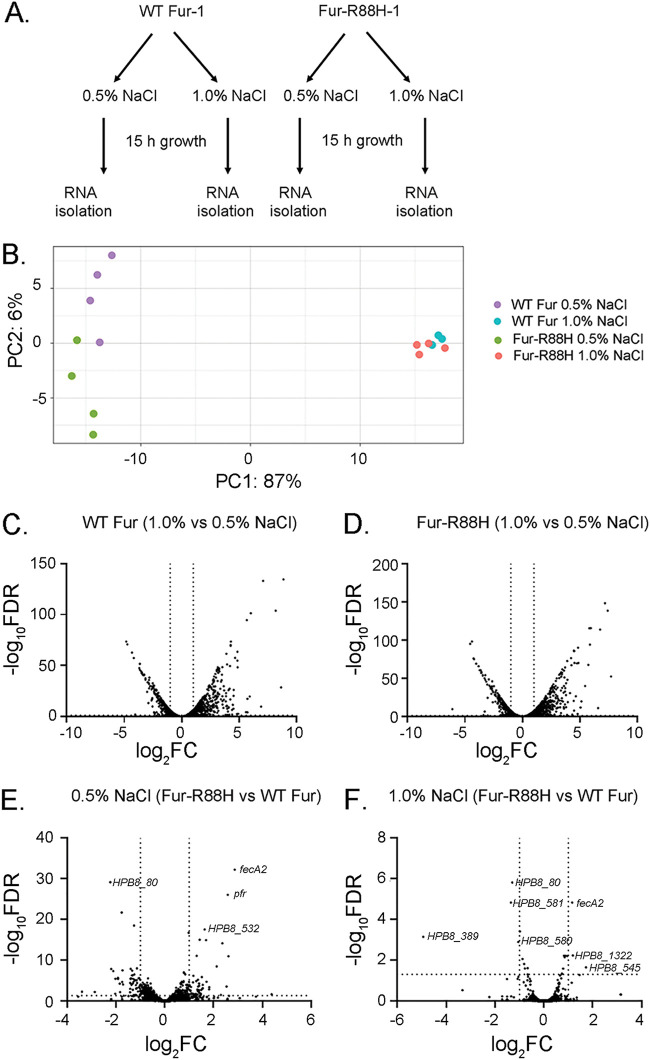
Transcriptional profiling of H. pylori strains. (A) Schematic illustrating the methods used for transcriptional analysis of H. pylori strains. H. pylori strains (7.13 WT Fur-1 and 7.13 Fur-R88H-1) (Fig. 2A) were each grown in medium containing either 0.5% or 1.0% added NaCl for 15 h. Bacteria then were harvested and processed for RNA isolation and RNA-seq analysis, as described in Materials and Methods. (B) Principal-component analysis was used to assess variation in gene expression patterns. (C and D) For each strain, the transcript levels of genes in bacteria grown under high-salt conditions were compared to the corresponding transcript levels in bacteria grown under routine conditions. (E and F) The transcript levels of each gene in the fur-R88H mutant were compared to the corresponding transcript levels in the H. pylori strain containing WT fur under routine conditions (E) or high-salt conditions (F). The y axes in panels C to F show the statistical significance of differences in transcript abundance (negative log10 FDR values; a higher value indicates greater significance). The x axes show the magnitude of differences (log2 fold change [FC] values). Vertical dotted lines correspond to 2.0-fold changes. The dotted horizontal line indicates an FDR of 0.05, with points above the line having an FDR value of <0.05 and points below the line having an FDR value of >0.05.
of 0.05, with points above the line having an FDR value of <0.05 and points below the line having an FDR value of >0.05.
To identify genes that were differentially expressed, we calculated transcript abundance ratios for each gene based on the RNA-seq data (described in Materials and Methods). In an initial analysis, we compared the transcript levels of bacteria grown in high-salt conditions with those of the same bacterial strain grown in routine conditions (i.e., transcript abundance under high-salt conditions divided by transcript abundance under routine conditions). We designated differentially expressed genes as those with transcript abundance ratios of >2 or <0.5 and false discovery rates (FDR) of <0.05. As expected based on the principal-component analysis, we detected numerous genes that were differentially expressed when an individual strain cultured under routine conditions was compared with the same strain cultured under high-salt conditions. Similar numbers of genes were upregulated or downregulated in response to high-salt conditions in the strain containing WT fur and the strain containing fur-R88H (Fig. 3C and andD).D). As shown in Fig. S1A in the supplemental material, a total of 336 genes were upregulated by high-salt conditions in both the fur-R88H mutant and the strain containing WT fur, and a total of 222 genes (Fig. S1B) were downregulated by high-salt conditions in both the fur-R88H mutant and the strain containing WT fur. The relatively large number of genes differentially expressed in response to high-salt conditions in the present study compared to a previous study (13) is likely attributable to differences in the length of time that the bacteria were exposed to high-salt conditions (15 h in the present study and 6 h in a previous study).
We next compared gene expression in the fur-R88H mutant with gene expression in the strain containing WT fur. In an analysis of bacterial growth under routine (0.5% added NaCl) conditions, transcript levels of 57 genes were increased and 53 were decreased in the fur-R88H mutant compared to the strain containing WT fur (Fig. 3E; Table S1). The majority of the differentially expressed genes encoded hypothetical proteins. Importantly, several of the genes differentially expressed in strains containing WT fur or fur-R88H, each grown under routine conditions (Table S1), are known to be regulated by Fur and are known or predicted to have roles in iron homeostasis. For example, transcript levels of genes encoding FecA- and FrpB-like proteins (FrpB1 and FecA2) (21, 22, 46) and iron storage proteins (ferritin [Pfr]) (21, 23) were increased in the fur-R88H mutant compared to those of the strain containing WT Fur. Transcript levels of other Fur-regulated genes, including those encoding ferrodoxin (Fer), OorA, and OorB (2-oxoglutarate:acceptor oxidoreductase) (47), were decreased in the fur-R88H mutant. Consistent with the principal-component analysis (Fig. 3B), only a few genes were differentially expressed when the fur-R88H and WT fur-containing strains grown under high-salt (1.0% NaCl) conditions were compared (Fig. 3F; Table S2). Only one gene known to be regulated by Fur (fecA2, predicted to encode a TonB-dependent outer membrane receptor) (33,–35) exhibited increased expression in the fur-R88H mutant strain compared to the strain containing WT fur in both routine and high-salt conditions (Fig. 3E and andF;F; Tables S1 and S2 and Fig. S2).
We evaluated expression of fecA2 using quantitative reverse transcription-PCR (RT-qPCR) in strains containing WT fur or fur-R88H, using RNA isolated from independent cultures grown under the same conditions as in the RNA-seq experiments. Consistent with the RNA-seq data, the RT-qPCR analyses showed upregulated expression of fecA2 in the fur-R88H mutant compared to that in the strain containing WT fur, each grown under high-salt conditions (Fig. 4A). In contrast, the RT-qPCR analyses revealed little or no difference in transcript levels of the other genes considered to be differentially expressed in the two strains grown under high-salt conditions, based on the RNA-seq experiments (Table S2) or the control gene gyrB (Fig. 4A). Multiple factors may account for the poor concordance between the RNA-seq and RT-qPCR results for these genes, including low levels of expression for several genes and adverse effects of high-salt conditions on bacterial growth and viability. Since the fecA2 results were validated by RT-qPCR and fecA2 is known to be regulated by Fur (46), we focused further studies on this gene.
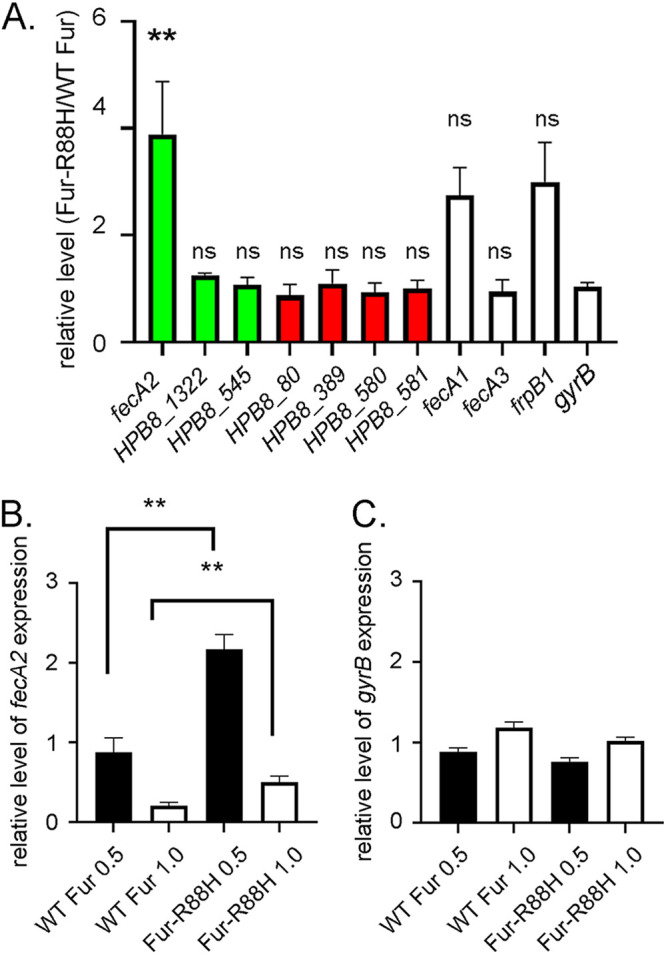
RT-qPCR analysis of differential gene expression. (A) H. pylori strains harboring either WT fur or fur-R88H were cultured for 15 h under high-salt (1.0% added NaCl) conditions. Transcript levels in the fur-R88H variant were then compared to those found in the strain containing WT fur, using RT-qPCR. The y axis shows the relative level of each transcript in the strain containing fur-R88H compared to the strain containing WT fur, each grown under high-salt conditions. Green bars indicate genes with increased expression in the fur-R88H-containing strain compared to the strain containing WT fur, and red bars indicate genes with decreased expression in fur-R88H- versus WT fur-containing strains, based on the RNA-seq analysis. Genes that did not meet the threshold of being transcriptionally different between fur-R88H- and WT fur-containing strains in RNA-seq experiments are indicated by open bars. Results represent an analysis of RNA isolated from H. pylori in at least five independent experiments. **, P value < 0.01 (one-way ANOVA with Tukey’s multiple-comparison test); ns, nonsignificant. Statistical significance is indicated for each gene compared to gyrB. (B and C) RT-qPCR analysis of fecA2 and gyrB expression in H. pylori strains containing either WT fur or fur-R88H (grown in medium containing either 0.5% or 1.0% added NaCl). Transcript levels of fecA2 and gyrB were compared to the levels present in the strain containing WT fur grown under routine conditions (0.5% added NaCl). Transcript levels for strains grown under high-salt conditions (1.0% added NaCl) and compared to the strain containing WT fur grown under routine conditions are indicated by white bars. Transcript levels for strains grown under 0.5% added NaCl and compared to the strain containing WT fur grown under routine conditions are indicated by black bars. Results represent an analysis of RNA isolated from eight independent experiments. **, P value <
< 0.01 (Mann-Whitney test).
0.01 (Mann-Whitney test).
The RT-qPCR analyses revealed higher fecA2 transcript levels in the fur-R88H-containing strain than in the strain containing WT fur when the cultures were grown in medium containing either 0.5% NaCl (2.46-fold increase in fur-R88H mutant compared to WT fur) or 1.0% added NaCl (2.42-fold increase) (Fig. 4B). The analyses also revealed decreased fecA2 expression in both WT fur and fur-R88H-containing strains under high-salt conditions (1.0% added NaCl) compared to that under routine conditions (0.5% added NaCl) (Fig. 4B). The latter observation is consistent with the salt-dependent decrease in fecA2 expression reported previously (13).
We also used RT-qPCR to analyze the expression of several additional fecA- or frpB-like genes (i.e., fecA1, fecA3, and frpB1) that showed nonsignificant trends for differential expression when fur-R88H- and WT fur-containing strains were compared in the RNA-seq experiments. H. pylori frpB1 is reportedly capable of binding to hemoglobin (48), and fecA3 has a role in nickel acquisition (49,–51). Both fecA1 and frpB1 are reported to be regulated by Fur (20,–22, 24, 46, 52), while regulation of fecA3 is reported to be regulated by NikR (46, 49). Increased expression of fecA1 and frpB1 was observed in the fur-R88H-containing strain compared to the WT fur-containing strain (Fig. 4A), but the increase was not statistically significant. Little to no difference in expression of fecA3 was observed between fur-R88H- and WT fur-containing strains (Fig. 4A).
Role of fecA2 in H. pylori fitness.
Based on the observation that fecA2 transcript levels were higher in the strain containing fur-R88H than in the strain containing WT fur, we next investigated the role of fecA2 in H. pylori fitness. We generated H. pylori strains that were tagged as shown in Fig. 5A. Strain JL1 (containing WT fur) was used as a control strain, strain JL2 contains fur-R88H, strain JL3A contains WT fur and a ΔfecA2 mutation, and strain JL4 contains fur-R88H and a ΔfecA2 mutation. There were no discernible differences in growth among the four strains when cultured individually in medium containing 0.5% NaCl. Similarly, there were no differences in growth when the strains were cultured in medium containing 1.0% added NaCl (data not shown). Equal proportions of JL1 (WT fur) and JL3A (WT fur, ΔfecA2), or JL2 (fur-R88H) and JL4 (fur-R88H, ΔfecA2), were mixed and serially cultured for 21 days on medium containing either 0.5% or 0.9% added NaCl. The composition of the bacterial population was analyzed at various time points using qPCR techniques (described in Materials and Methods). As shown in Fig. 5B and andC,C, strain JL1 (containing WT fur) comprised the major component of the bacterial population following continuous passage on medium containing 0.5% or 0.9% added NaCl, with strain JL3A (containing the ΔfecA2 mutation) exhibiting a marked defect in the ability to compete with JL1 under both conditions. Similarly, strain JL4 (fur-R88H, ΔfecA2) exhibited a fitness defect compared to its parental strain JL2 (fur-R88H) when passaged under both conditions (Fig. 5D and andE).E). These results suggest that fecA2 is important for H. pylori fitness under all the conditions tested (medium containing either 0.5% or 0.9% added NaCl and strains containing either WT fur or fur-R88H).
days on medium containing either 0.5% or 0.9% added NaCl. The composition of the bacterial population was analyzed at various time points using qPCR techniques (described in Materials and Methods). As shown in Fig. 5B and andC,C, strain JL1 (containing WT fur) comprised the major component of the bacterial population following continuous passage on medium containing 0.5% or 0.9% added NaCl, with strain JL3A (containing the ΔfecA2 mutation) exhibiting a marked defect in the ability to compete with JL1 under both conditions. Similarly, strain JL4 (fur-R88H, ΔfecA2) exhibited a fitness defect compared to its parental strain JL2 (fur-R88H) when passaged under both conditions (Fig. 5D and andE).E). These results suggest that fecA2 is important for H. pylori fitness under all the conditions tested (medium containing either 0.5% or 0.9% added NaCl and strains containing either WT fur or fur-R88H).
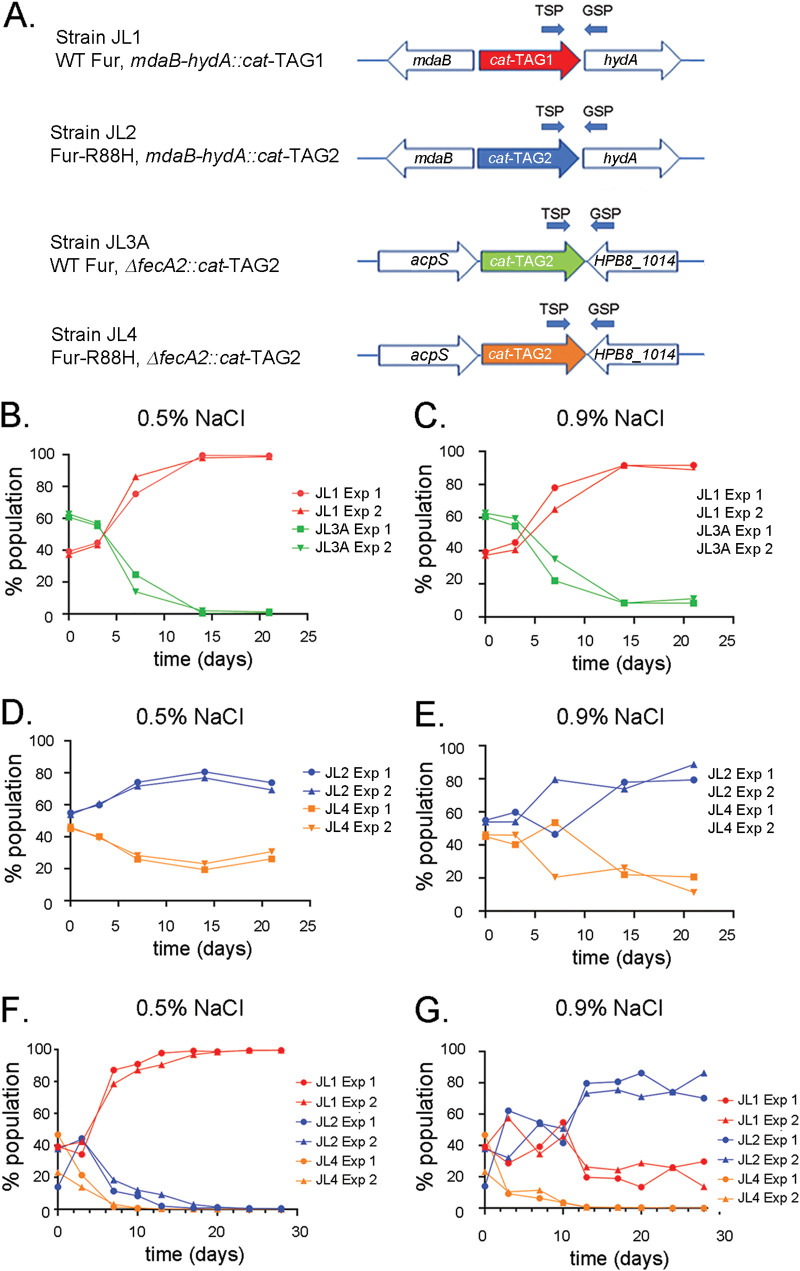
fecA2 expression is important for H. pylori fitness. (A) Depiction of strains used for this analysis. Strain JL1 contains WT fur, and strain JL2 contains fur-R88H (see Fig. 2 for further details about DNA barcodes in these strains). Strain JL3A contains WT fur and a cat-TAG2 sequence inserted into the deleted fecA2 locus. Strain JL4 contains fur-R88H and a cat-TAG2 sequence inserted into the deleted fecA2 locus. (B to G) The indicated strains were mixed 1:1 (B to E), or strains JL1, JL2, and JL4 were mixed 1:1:1 (F and G). All mixtures were serially passaged (once every 3.5 days) on medium containing either 0.5% or 0.9% added NaCl. At the indicated time points, bacteria were harvested and genomic DNA was isolated. The proportion of each strain in the bacterial population was then quantified by qPCR as described in Materials and Methods. The primers for the qPCR analyses (see Table S4 in the supplemental material) included a TSP and a GSP. The results are from two independent experiments. qPCR analysis for each time point was performed in triplicate, and the mean value is reported.
days) on medium containing either 0.5% or 0.9% added NaCl. At the indicated time points, bacteria were harvested and genomic DNA was isolated. The proportion of each strain in the bacterial population was then quantified by qPCR as described in Materials and Methods. The primers for the qPCR analyses (see Table S4 in the supplemental material) included a TSP and a GSP. The results are from two independent experiments. qPCR analysis for each time point was performed in triplicate, and the mean value is reported.
We also compared the fitness of JL1 (WT fur), JL2 (fur-R88H), and JL4 (fur-R88H, ΔfecA2) strains in a mixed population comprising equal proportions of the three strains. As shown in Fig. 5F, strain JL1 (containing WT fur) comprised the major component of the bacterial population following continuous passage of the bacterial mixture on medium containing 0.5% added NaCl. In contrast, strain JL2 (containing fur-R88H) exhibited enhanced fitness and comprised the major component (80% at day 21) of the bacterial population passaged on medium containing 0.9% added NaCl (Fig. 5G). This result is consistent with the finding that fur-R88H-containing strains are positively selected under high-salt conditions, compared to strains containing WT Fur (Fig. 2F). Consistent with the results observed in Fig. 5B to toE,E, the fur-R88H (ΔfecA2) mutant (JL4) exhibited a marked defect in the ability to compete with JL1 or JL2 under both routine (0.5% added NaCl) (Fig. 5F) and high-salt (0.9% added NaCl) (Fig. 5G) conditions.
To further examine the role of fecA2 in H. pylori fitness, we compared the fitness of fecA2 mutants containing either WT fur (i.e., JL3B containing cat-TAG1) or fur-R88H (JL4 containing cat-TAG2). As shown in Fig. S3, strain JL4 (fur-R88H, ΔfecA2) was more fit than strain JL3B (WT fur, ΔfecA2) under either routine salt conditions (0.5% added NaCl [Fig. S3B]) or high-salt conditions (0.9% added NaCl [Fig. S3C]). The latter result suggests that additional factors besides fecA2 contribute to the differential fitness of H. pylori strains containing WT fur or fur-R88H.
As previous results (Fig. 2) demonstrated that the population dynamics of a mixture of WT fur- and fur-R88H-containing strains was altered by the addition of ferric citrate or ferrous sulfate, we examined whether the addition of supplemental iron could restore the fitness of a fecA2 mutant. Consistent with earlier results (Fig. 5B and andC),C), the fitness of strain JL3A (WT fur, ΔfecA2) was reduced compared to that of strain JL1 (WT fur) in competition assays conducted under both routine and high-salt conditions (Fig. S4B and D). The addition of ferric citrate did not increase the fitness of JL3A (WT fur, ΔfecA2) compared to that of JL1 (WT fur) in either routine or high-salt conditions (Fig. S4C and E). Similarly, the fitness of strain JL4 (fur-R88H, ΔfecA2) was reduced compared to that of strain JL2 (fur-R88H), and the addition of iron did not increase the fitness of JL4 (fur-R88H, ΔfecA2) (data not shown). These results suggest that ferric citrate supplementation does not reverse the fitness defect of H. pylori fecA2 mutants.
To further evaluate a role of fecA2 in H. pylori fitness, we generated a complemented fecA2 mutant strain. The strain was generated by inserting an intact copy of the fecA2 gene into the intergenic region between genes HPB8_1388 (neuB) and HPB8_1389 (efp) in the H. pylori ΔfecA2 strain JL5, yielding strain JL7 (Fig. 6A). Strain JL5 contains fur-R88H and an aacC4-rpsL insertion (described in Materials and Methods) (53) that facilitates mutagenesis of the region between the neuB and efp loci. In parallel, a control transformation of the ΔfecA2 strain JL5 was performed to restore the original DNA sequence in the intergenic region between neuB and efp (i.e., JL6) (Fig. 6A). Three different pairwise competition assays were then performed. Strain JL2 (fur-R88H-containing strain) was mixed 1:1 with either the fecA2-complemented strain JL7 or the ΔfecA2 strains (JL4 or JL6), and the mixtures were passaged on medium containing 0.9% added NaCl for 28 days. As shown in Fig. 6, JL2 was the dominant strain in the competition assays between JL2 and ΔfecA2 strain JL4 (Fig. 6B) or JL6 (Fig. 6C). In contrast, the complemented fecA2 mutant strain JL7 did not exhibit a fitness defect when in competition with strain JL2 (Fig. 6D). This result corroborates the finding that fecA2 contributes to H. pylori fitness.
days. As shown in Fig. 6, JL2 was the dominant strain in the competition assays between JL2 and ΔfecA2 strain JL4 (Fig. 6B) or JL6 (Fig. 6C). In contrast, the complemented fecA2 mutant strain JL7 did not exhibit a fitness defect when in competition with strain JL2 (Fig. 6D). This result corroborates the finding that fecA2 contributes to H. pylori fitness.

Complementation restores the fitness of a fecA2 mutant. (A) Relevant characteristics of strains used in this analysis. JL2 is a strain containing fur-R88H and a cat-TAG2 insertion in the intergenic region between mdaB and hydA. Strain JL4 contains a cat-TAG2 insertion in the deleted fecA2 locus. JL5 contains an aacC4-rpsL cassette inserted in the intergenic region between genes HPB8_1388 (neuB) and HPB8_1389 (efp) of strain JL4. Transformation of strain JL5 with a plasmid that restored the genetic sequence between neuB and efp resulted in strain JL6, while transformation of JL5 with a plasmid that introduced an intact copy of fecA2 into the neuB and efp intergenic loci resulted in strain JL7. (B to D) Competition assays were performed by mixing the indicated strains in a 1:1 proportion and serially culturing the bacterial mixtures on medium containing 0.9% added NaCl for 28 days. At the indicated time points, H. pylori bacteria were harvested and genomic DNA was extracted. The relative proportions of each bacterial strain in the population were quantified using qPCR as described in Materials and Methods. The results are from three independent experiments. Means and standard errors of the mean are reported.
days. At the indicated time points, H. pylori bacteria were harvested and genomic DNA was extracted. The relative proportions of each bacterial strain in the population were quantified using qPCR as described in Materials and Methods. The results are from three independent experiments. Means and standard errors of the mean are reported.
DISCUSSION
In the present study, we show that the fur-R88H mutation confers a selective advantage to H. pylori under high-salt conditions in vitro. Similarly, we showed in previous studies that H. pylori strains containing fur-R88H were positively selected in experimentally infected Mongolian gerbils receiving a high-salt diet (14, 18). We compared the transcriptomes of H. pylori strains containing wild-type fur or fur-R88H, each grown under routine or high-salt conditions, and we detected numerous differences between the two strains, including differential transcript abundances of known Fur-regulated genes. These results provide strong evidence that the fur-R88H mutation alters Fur regulatory properties.
Our analysis of H. pylori evolution during serial culture in vitro under either routine or high-salt conditions showed that the fur-R88H polymorphism becomes predominant relatively quickly under high-salt conditions (Fig. 1). The same result was obtained using both whole-genome sequencing methods and analysis by qPCR methods. The latter analysis provided evidence that the fur-R88H polymorphism was present in low abundance in the 7.13 parental population. The fur-R88H mutation in strains selected in high-salt environments consistently resulted from a CGC-to-CAC transition (14, 18), which provides additional evidence that the mutation was present in the parental population (instead of repeatedly arising as new mutations). The presence of fur-R88H at low levels in the parental 7.13 population helps to explain why the fur-R88H mutation has been repeatedly selected in response to high-salt conditions, whereas other mutations in fur have not been commonly identified (14, 18). In contrast to the fur-R88H polymorphism, which became predominant relatively quickly under high-salt conditions, SNPs in other genes became predominant after longer time periods (16 weeks of culture in high-salt medium). The temporal sequence of mutation emergence suggests that the fur-R88H allele and the other positively selected SNPs were not all present together in individual bacteria in the initial H. pylori population. While the fur-R88H mutation was positively selected in high-salt conditions both in vitro (in the current study) and in vivo (in previous studies) (14), there was relatively little overlap in the other SNPs that were positively selected under high-salt conditions in vivo or in vitro. Notably, the fur-R88H mutation has been identified in multiple H. pylori strain backgrounds besides the strain analyzed in the present study. For example, in one study, fur-R88H was detected in 46 (14%) of 339 clinical H. pylori isolates (19).
weeks of culture in high-salt medium). The temporal sequence of mutation emergence suggests that the fur-R88H allele and the other positively selected SNPs were not all present together in individual bacteria in the initial H. pylori population. While the fur-R88H mutation was positively selected in high-salt conditions both in vitro (in the current study) and in vivo (in previous studies) (14), there was relatively little overlap in the other SNPs that were positively selected under high-salt conditions in vivo or in vitro. Notably, the fur-R88H mutation has been identified in multiple H. pylori strain backgrounds besides the strain analyzed in the present study. For example, in one study, fur-R88H was detected in 46 (14%) of 339 clinical H. pylori isolates (19).
Competition experiments showed that strains containing WT fur were more fit than strains containing fur-R88H under routine culture conditions. In contrast, strains containing fur-R88H were more fit than the WT fur-containing strains under high-salt conditions. Interestingly, the supplementation of high-salt medium with iron resulted in a change in the relative fitness of the WT fur-containing strain compared to the fur-R88H-containing strain. This suggests that the fur-R88H SNP modulates H. pylori physiology and fitness in different ways, depending on the environmental conditions.
Fur is an important transcriptional regulator and has been shown to be important for H. pylori colonization of Mongolian gerbils (54, 55). Many different physiological functions have been attributed to Fur in H. pylori (24, 54, 56). For instance, Fur has an important role in iron homeostasis and H. pylori resistance to oxidative stress (20, 24, 56), and a fur deletion mutant exhibited reduced survival under high-salt conditions (12). As Fur is a transcriptional regulator, we investigated the possibility that Fur-mediated transcriptional regulation might be different in H. pylori strains containing WT fur and H. pylori strains containing fur-R88H. Importantly, not only is the R88H mutation located close to the Fur amino acid residue E90 that is important for binding of the Fur iron cofactor (29), but the R88H SNP is also located between 2 amino acid residues (R87 and Y89) that are important for Fur-mediated regulation of amiE (28). Protein modeling with AlphaFold does not indicate any obvious structural difference between WT Fur and Fur-R88H (data not shown). However, given the proximity of the R88H mutation to residues important for binding of the Fur iron cofactor (29, 30), the R88H mutation could potentially influence the iron-binding properties of Fur.
About 110 genes were differentially expressed when strains containing WT fur or fur-R88H were compared, each grown under routine culture conditions. A much smaller number of genes were differentially expressed when these strains cultured under high-salt conditions were compared. The reason for the difference in the number of genes that are differentially expressed between the two strains depending on the culture conditions (0.5% or 0.9% added NaCl) is unclear. We speculate that the differences in the transcriptional profiles of the two strains may be blunted under high-salt conditions because of the effects of high-salt conditions on bacterial growth and bacterial viability, so that only the most pronounced differences are detected under high-salt conditions. In addition, we speculate that the actions of multiple regulatory systems, including systems independent of Fur and iron, may contribute to the observed patterns of gene expression under high-salt conditions.
We focused on fecA2 (encoding an outer membrane protein whose transcription is repressed when the fecA2 promoter is bound by Fur [21, 24, 46]) for several reasons: the RNA-seq experiments showed a highly significant difference in fecA2 transcript levels in WT fur- and fur-R88H-containing strains (Fig. 3E and andF;F; see Tables S1 and S2 in the supplemental material), fecA2 was the only gene differentially expressed in WT fur- and fur-R88H-containing strains under both routine and high-salt conditions, and this difference was validated by RT-qPCR. fecA2 is predicted to encode a TonB-dependent outer membrane transporter (34, 35), but its exact function in H. pylori has not been experimentally determined. We explored a possible role of fecA2 in H. pylori fitness under high-salt conditions and found that the fitness of a fur-R88H-containing strain with a fecA2 deletion was markedly reduced compared to that of a fur-R88H-containing strain with an intact fecA2 gene. Complementation of the fecA2 mutant restored the ability of the bacteria to grow under high-salt conditions. While these data suggest that the increased expression of fecA2 in fur-R88H-containing strains compared to that in WT fur-containing strains under high-salt conditions may allow H. pylori to tolerate a high-salt environment, it may be oversimplistic to attribute the increased fitness of the fur-R88H mutant under high-salt conditions solely to the alteration in fecA2 expression. For example, we found that a fecA2 mutant in a fur-R88H-containing strain has increased fitness compared to a fecA2 mutant in a WT fur-containing strain (Fig. S3), which suggests that other genes besides fecA2 are relevant. Further experiments are needed to understand the function of FecA2, its potential role of metal acquisition, and its relationship to other FecA proteins and FrpB proteins.
Fur-R88H mutant strains are positively selected under both high-salt and low-iron conditions (14, 18, 19). This suggests that these two different conditions might have related effects on bacterial physiology. A review of previous literature suggests that multiple H. pylori genes altered in expression under high-salt conditions are also altered in expression under low-iron conditions, and many of these are regulated by Fur. For example, the expression of ferritin (pfr), superoxide dismutase (sodB), and hydrogenase (hydA and hydB) is decreased in response to both high-salt and low-iron conditions (23, 24, 57). Conversely, the effects of high-salt and low-iron conditions on the expression of other genes are not the same. For example, fecA2 is repressed by high salt, and fecA2 is upregulated in response to low-iron conditions (24, 46). Similarly, frpB1, fecA1, and amiE (24, 48, 58) are repressed by high salt, whereas low-iron conditions stimulate expression of these genes (24, 46, 48, 58). High salt reduces expression of fur, whereas expression of fur is increased in low-iron conditions (59). We speculate that multiple regulatory systems may be differentially activated under these two conditions. Interestingly, previous studies suggested that iron bioavailability in Bacillus may be modulated by NaCl concentration (60, 61). Further studies are required to further evaluate the effects of NaCl concentration on iron acquisition and iron homeostasis in H. pylori.
The effects of the fur-R88H mutation on H. pylori gene expression observed in this study help to explain mechanisms by which this polymorphism impacts H. pylori fitness under high-salt conditions. A previous study showed that the fur-R88H mutation was also positively selected in H. pylori-infected Mongolian gerbils that were fed an iron-depleted diet and during H. pylori growth under low-iron conditions in vitro (19). The presence of the fur-R88H mutation has been associated with increased H. pylori survival under oxidative stress conditions (14) and increased ability of the bacteria to survive in cocultures with neutrophils (18). In future studies, it will be important to ascertain if the fur-R88H mutation modulates H. pylori gene expression in the same way under all these conditions, leading to increased fitness, or if effects of the fur-R88H mutation on gene expression vary among these conditions. Interestingly, a previous study reported that H. pylori strains containing the fur-R88H allele were more commonly isolated from patients with premalignant lesions than from patients with nonatrophic gastritis (19). This suggests that the effects of the fur-R88H mutation may extend beyond impacts on H. pylori fitness and might directly contribute to H. pylori-induced gastric disease.
In conclusion, we show that the fur-R88H SNP confers a fitness advantage to H. pylori under high-salt conditions and a fitness disadvantage under routine conditions, and we show that the fitness advantage conferred by the fur-R88H SNP is abrogated in the presence of excess iron. We demonstrate that there are differences in the transcriptional profiles of strains containing WT fur or fur-R88H, indicating that the fur-R88H mutation modulates Fur regulatory activity. In future studies, it will be important to further evaluate mechanisms underlying the differences in the regulatory activities of WT Fur and Fur-R88H, elucidate mechanisms by which FecA2 contributes to H. pylori fitness, and evaluate interrelated effects of environmental salt and iron concentrations on H. pylori physiology.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Table S3 in the supplemental material lists the plasmids and H. pylori strains used in this study. For routine growth, H. pylori strains were passaged on Trypticase soy agar plates containing 5% sheep blood or on sulfite-free Brucella (BB) agar plates containing 5% fetal bovine serum (FBS) in room air supplemented with 5% CO2 at 37°C. When necessary, the BB-FBS agar plates were supplemented with chloramphenicol (5 μg/mL), gentamicin (10
μg/mL), gentamicin (10 μg/mL), or streptomycin (25
μg/mL), or streptomycin (25 μg/mL). Escherichia coli strains were grown on Luria-Bertani medium. When necessary, the E. coli culture medium was supplemented with ampicillin (50
μg/mL). Escherichia coli strains were grown on Luria-Bertani medium. When necessary, the E. coli culture medium was supplemented with ampicillin (50 μg/mL), chloramphenicol (25
μg/mL), chloramphenicol (25 μg/mL), gentamicin (10
μg/mL), gentamicin (10 μg/mL), or streptomycin (25
μg/mL), or streptomycin (25 μg/mL). H. pylori liquid cultures were grown in sulfite-free Brucella broth supplemented with 5% FBS (Gibco) (BB-FBS). The broth medium used for routine H. pylori culture contains 0.5% added sodium chloride. To generate high-salt conditions, the broth was supplemented with 1.0% added NaCl. To generate high-salt conditions for bacterial growth on plates, the medium was supplemented with 0.9% added NaCl, because bacterial growth was inhibited by higher concentrations of NaCl.
μg/mL). H. pylori liquid cultures were grown in sulfite-free Brucella broth supplemented with 5% FBS (Gibco) (BB-FBS). The broth medium used for routine H. pylori culture contains 0.5% added sodium chloride. To generate high-salt conditions, the broth was supplemented with 1.0% added NaCl. To generate high-salt conditions for bacterial growth on plates, the medium was supplemented with 0.9% added NaCl, because bacterial growth was inhibited by higher concentrations of NaCl.
Generation of barcoded H. pylori strains.
We previously reported the use of nucleotide barcodes to monitor population dynamics in a pool of H. pylori strains (45). Plasmids used for the generation of barcoded mutant strains contained unique 21-nucleotide barcodes linked to a chloramphenicol acetyltransferase gene as a selectable marker (i.e., cat-TAG), flanked by 500 bp of sequence on either side of the targeted insertion site. These plasmids are unable to replicate in H. pylori. Following transformation, H. pylori bacteria were plated on BB-FBS agar plates containing chloramphenicol (5
bp of sequence on either side of the targeted insertion site. These plasmids are unable to replicate in H. pylori. Following transformation, H. pylori bacteria were plated on BB-FBS agar plates containing chloramphenicol (5 μg/mL). Chloramphenicol-resistant transformants were validated by sequencing PCR amplicons generated using primers flanking the predicted barcoded cassette insertion site (about 600
μg/mL). Chloramphenicol-resistant transformants were validated by sequencing PCR amplicons generated using primers flanking the predicted barcoded cassette insertion site (about 600 bp upstream and 600
bp upstream and 600 bp downstream of the insertion site).
bp downstream of the insertion site).
Plasmids p630cat-TAG1 and p630cat-TAG2 were previously used to introduce cat-TAGs into the intergenic region between genes HP0630 (mdaB) and HP0631 (hydA) in H. pylori strain 26695 (45). The intergenic region between HP0630 and HP0631 is not expected to affect bacterial fitness (45). In the present study, the plasmid p630cat-TAG1 was used to introduce cat-TAG1 into the intergenic region between mdaB (HPB8_829) and hydA (HPB8_830) of H. pylori strain 7.13 WT Fur-1. Similarly, plasmid p630cat-TAG2 was used to introduce cat-TAG2 into the intergenic loci between mdaB and hydA of strain 7.13 Fur-R88H-1. Plasmid pΔfecA2cat-TAG2 was used to delete the fecA2 gene, with cat-TAG2 sequences inserted into the mutated locus. To generate the pΔfecA2cat-TAG2 plasmid, pΔfecA2 was synthesized (GenScript) to contain KpnI and EcoRV restriction sites engineered between 500 bp of DNA sequence upstream and downstream of the deleted fecA2 gene. The cat-TAG2 cassette was PCR amplified from p630cat-TAG2 and inserted into the KpnI/EcoRV-digested pΔfecA2 plasmid.
bp of DNA sequence upstream and downstream of the deleted fecA2 gene. The cat-TAG2 cassette was PCR amplified from p630cat-TAG2 and inserted into the KpnI/EcoRV-digested pΔfecA2 plasmid.
Generation of a complemented fecA2 mutant strain.
To complement an H. pylori
fecA2 mutant, a counterselection method using an aacC4-rpsL cassette was employed as previously described (53). This cassette confers resistance to gentamicin mediated by the aminoglycoside-3-acetyltransferase IV (aacC4) gene and susceptibility to streptomycin mediated by the intact rpsL gene from H. pylori 26695. Strain JL4, containing a ΔfecA2 mutation, was first transformed with a nonreplicating plasmid containing a mutated H. pylori
rpsL gene (A-to-G mutation at nucleotide 128 in rpsL) (62). This mutation results in an amino acid substitution [Lys (K) to Arg (R)] at position 43 of RpsL that confers streptomycin resistance. The streptomycin-resistant rpsL-K43R ΔfecA2 mutant strain was next transformed with p177::aacC4-rpsL, a nonreplicating plasmid that allows the insertion of an aacC4-rpsL cassette (conferring gentamicin resistance) into the intergenic region between genes HPB8_1388 (neuB) and HPB8_1390 (efp). These correspond to the HP0177-HP0178 locus in H. pylori 26695. The accC4-rpsL cassette was obtained by digesting plasmid pADgent-rpsL (53) with KpnI/ApaI, and the 1.5-kb aacC4-rpsL cassette was inserted into the p177 plasmid that contains the cloned intergenic region between neuB and efp. After ligation with the aacC4-rpsL cassette, the resultant plasmid (p177::aacC4-rpsL), which is unable to replicate in H. pylori, was transformed into the rpsL-K43R mutant ΔfecA2
H. pylori strain, and single colonies resistant to gentamicin (10 μg/mL) but sensitive to streptomycin (25
μg/mL) but sensitive to streptomycin (25 μg/mL) were selected. Introduction of the aacC4-rpsL cassette into the neuB-efp intergenic region was confirmed by PCR amplification and DNA sequencing, and strain JL4 (rpsL-K43R, ΔfecA2, neuB-efp::aacC4-rpsL) was chosen for transformation with pfecA2, a plasmid that contains the fecA2 gene flanked by neuB and efp sequences for homologous recombination. Streptomycin-resistant transformants were screened by PCR and confirmed by sequencing of the amplicons. Expression of fecA2 in the resultant clone was confirmed by real-time RT-qPCR.
μg/mL) were selected. Introduction of the aacC4-rpsL cassette into the neuB-efp intergenic region was confirmed by PCR amplification and DNA sequencing, and strain JL4 (rpsL-K43R, ΔfecA2, neuB-efp::aacC4-rpsL) was chosen for transformation with pfecA2, a plasmid that contains the fecA2 gene flanked by neuB and efp sequences for homologous recombination. Streptomycin-resistant transformants were screened by PCR and confirmed by sequencing of the amplicons. Expression of fecA2 in the resultant clone was confirmed by real-time RT-qPCR.
H. pylori genome sequence analysis.
H. pylori strain 7.13 was passaged on BB-FBS agar plates containing either 0.5% (routine) or 0.9% (high salt) added NaCl, with passages to fresh plates performed every 2 days. At various time points, H. pylori was harvested and DNA samples were prepared using a Wizard genomic purification kit (Promega). Methods similar to those previously used for H. pylori genome sequencing were next employed (14, 18, 53). Briefly, H. pylori DNA samples were subjected individually to enzymatic fragmentation using the NEBNext double-stranded DNA (dsDNA) fragmentase kit (NEB) according to the manufacturer’s instructions, with an average fragment length of 600
days. At various time points, H. pylori was harvested and DNA samples were prepared using a Wizard genomic purification kit (Promega). Methods similar to those previously used for H. pylori genome sequencing were next employed (14, 18, 53). Briefly, H. pylori DNA samples were subjected individually to enzymatic fragmentation using the NEBNext double-stranded DNA (dsDNA) fragmentase kit (NEB) according to the manufacturer’s instructions, with an average fragment length of 600 bp (range of 400 to 1,000
bp (range of 400 to 1,000 bp). Libraries of DNA were prepared from purified fragmented DNA samples using the JetSeq library prep kit with unique indexes per the manufacturer’s protocol (Meridian Biosciences, Inc., Cincinnati, OH, USA). Quantification of these library preps was performed with the JetSeq library quantification kit (Meridian Biosciences, Inc., Cincinnati, OH, USA), and the libraries were sequenced on a MiSeq sequencer using the 600V3 kit (Illumina, Inc., San Diego, CA, USA). For the analysis, raw reads were quality trimmed and aligned to the reference sequence (H. pylori strain B8 [63] [GenBank accession no. NC_014256.1], a closely related strain whose complete genome sequence is available) by using CLCbio Genomics workbench version 11.
bp). Libraries of DNA were prepared from purified fragmented DNA samples using the JetSeq library prep kit with unique indexes per the manufacturer’s protocol (Meridian Biosciences, Inc., Cincinnati, OH, USA). Quantification of these library preps was performed with the JetSeq library quantification kit (Meridian Biosciences, Inc., Cincinnati, OH, USA), and the libraries were sequenced on a MiSeq sequencer using the 600V3 kit (Illumina, Inc., San Diego, CA, USA). For the analysis, raw reads were quality trimmed and aligned to the reference sequence (H. pylori strain B8 [63] [GenBank accession no. NC_014256.1], a closely related strain whose complete genome sequence is available) by using CLCbio Genomics workbench version 11.
RNA-seq analysis.
The strains used for RNA-seq analysis were H. pylori 7.13 strains that contained either WT Fur (7.13 WT Fur-1) or Fur-R88H (7.13 Fur-R88H-1) (Fig. 3A) (14). H. pylori cultures were grown for 15 h in BB-FBS medium containing either 0.5% or 1.0% added NaCl. Cultures were pelleted by centrifugation, and bacterial pellets were resuspended in RNAlater (Ambion) for 40 min. The cell suspensions were centrifuged at 3,500
min. The cell suspensions were centrifuged at 3,500 ×
× g, supernatants were decanted, and the pellets were stored at −70°C. A total of 15 independent samples were analyzed; 8 RNA preparations were from 7.13 Fur-R88H-1 grown in medium containing either 0.5% NaCl (4 independent samples) or 1.0% NaCl (4 independent samples), and 7 RNA preparations were from 7.13 WT Fur-1 grown in medium containing 0.5% NaCl (4 independent samples) or 1.0% NaCl (3 independent samples). Total RNA was isolated as previously described (13, 15, 64) using TRIzol reagent, according to the manufacturer’s instructions. All samples were subjected to DNase digestion (Turbo DNA-free kit; Ambion) to remove contaminating DNA, followed by a cleanup step using RNeasy columns (Qiagen). Each RNA sample was eluted in 100
g, supernatants were decanted, and the pellets were stored at −70°C. A total of 15 independent samples were analyzed; 8 RNA preparations were from 7.13 Fur-R88H-1 grown in medium containing either 0.5% NaCl (4 independent samples) or 1.0% NaCl (4 independent samples), and 7 RNA preparations were from 7.13 WT Fur-1 grown in medium containing 0.5% NaCl (4 independent samples) or 1.0% NaCl (3 independent samples). Total RNA was isolated as previously described (13, 15, 64) using TRIzol reagent, according to the manufacturer’s instructions. All samples were subjected to DNase digestion (Turbo DNA-free kit; Ambion) to remove contaminating DNA, followed by a cleanup step using RNeasy columns (Qiagen). Each RNA sample was eluted in 100 μL of water. RNA quality was assessed using a 2100 Bioanalyzer (Agilent). At least 200
μL of water. RNA quality was assessed using a 2100 Bioanalyzer (Agilent). At least 200 ng of the DNase-treated total RNA (RNA integrity number greater than 8) was used to generate rRNA-depleted/mRNA-enriched libraries using TruSeq Ribo-Zero bacterial RNA kits (Illumina). Library quality was assessed using a 2100 Bioanalyzer (Agilent) and KAPA library quantification kits (KAPA Biosystems). Libraries were sequenced on a NovaSeq 6000 system with paired reads of 150-bp length, according to the manufacturer’s protocol. Bcl2fastq2 conversion software (Illumina) was used to generate demultiplexed Fastq files. The number of sequence reads for each sample ranged from 24 to 33 million.
ng of the DNase-treated total RNA (RNA integrity number greater than 8) was used to generate rRNA-depleted/mRNA-enriched libraries using TruSeq Ribo-Zero bacterial RNA kits (Illumina). Library quality was assessed using a 2100 Bioanalyzer (Agilent) and KAPA library quantification kits (KAPA Biosystems). Libraries were sequenced on a NovaSeq 6000 system with paired reads of 150-bp length, according to the manufacturer’s protocol. Bcl2fastq2 conversion software (Illumina) was used to generate demultiplexed Fastq files. The number of sequence reads for each sample ranged from 24 to 33 million.
RNA-seq data were trimmed to remove all bases below a quality of Q3, and adapter sequences were removed using FastQ quality control software (FaQCs). Kallisto pseudocounting was applied to all annotated genes in the H. pylori B8 genome (63) (GenBank accession no. NC_014256.1). Estimated abundance counts from Kallisto outputs were used for all analysis steps. Transcripts associated with a total of 1,589 H. pylori genes were identified by RNA-seq. Data from individual samples were normalized within EdgeR and analyzed using the generalized linear model (GLM).
Fold change values were calculated by comparing the means of normalized pseudocounts for samples from each group (calculated by edgeR). A false-discovery rate (FDR), corresponding to a Benjamini-Hochberg adjusted P value, was calculated using the p.adjust program within edgeR. Differentially expressed genes were defined as those exhibiting an FDR value of <0.05, as well as a >2.0 or <0.5-fold difference in transcript abundance in the above comparisons.
RT-qPCR analyses of gene expression.
Overnight broth cultures of H. pylori strains were subcultured into fresh medium and grown for 15 h. Total RNA was isolated as described for RNA-seq analysis and analyzed as previously described (13, 14, 64). cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) was performed with 100 ng of purified RNA. As controls, parallel first-strand cDNA synthesis reactions without reverse transcriptase were performed. The cDNA and control preparations were diluted 1:20 and used in real-time PCRs. Real-time PCR was performed with an ABI real-time PCR machine (StepOnePlus) with SYBR green as the fluorochrome. Transcript abundance was assessed by the ΔΔCT method (CT, cycle threshold), with each transcript signal normalized to the abundance of the 16S rRNA internal control. The normalized transcript signal for each biological sample was then divided by similarly normalized values from control samples to obtain a relative expression ratio. The primers used for real-time analysis are listed in Table S4. As noted above, the results of RNA-seq experiments involving strain 7.13 were annotated based on the use of the genome of H. pylori B8, a strain closely related to strain 7.13 (63). A BLAST search revealed that frpB genes in the published genome of H. pylori strain B8 were designated differently from those of H. pylori strain 26695. For example, the gene designated frpB1 in H. pylori strain 26695 was designated frpB7 in H. pylori strain B8. As frpB1 is the commonly used name for this gene (48), we refer to HPB8_frpB7 as frpB1 in the present study.
ng of purified RNA. As controls, parallel first-strand cDNA synthesis reactions without reverse transcriptase were performed. The cDNA and control preparations were diluted 1:20 and used in real-time PCRs. Real-time PCR was performed with an ABI real-time PCR machine (StepOnePlus) with SYBR green as the fluorochrome. Transcript abundance was assessed by the ΔΔCT method (CT, cycle threshold), with each transcript signal normalized to the abundance of the 16S rRNA internal control. The normalized transcript signal for each biological sample was then divided by similarly normalized values from control samples to obtain a relative expression ratio. The primers used for real-time analysis are listed in Table S4. As noted above, the results of RNA-seq experiments involving strain 7.13 were annotated based on the use of the genome of H. pylori B8, a strain closely related to strain 7.13 (63). A BLAST search revealed that frpB genes in the published genome of H. pylori strain B8 were designated differently from those of H. pylori strain 26695. For example, the gene designated frpB1 in H. pylori strain 26695 was designated frpB7 in H. pylori strain B8. As frpB1 is the commonly used name for this gene (48), we refer to HPB8_frpB7 as frpB1 in the present study.
Detection of Fur-R88H allele.
A qPCR SNP genotyping method (45) was used to quantify the relative abundances of the wild-type fur allele and the fur-R88H allele in a population of H. pylori. Primers specific for fur-R88H and WT fur were designed by Applied Biosystems and used in qPCR TaqMan-based assays. The quantitative PCR (qPCR) TaqMan SNP genotyping assay, designed by Applied Biosystems, includes two differentially labeled, allele-specific TaqMan MGB probes specific for fur-R88H (FAM [6-carboxyfluorescein]/NFQ-labeled 5′-CGGTCGGCACTATG-3′) and WT fur (VIC/NFQ-labeled 5′-CGGTCGGCGCTATG-3′) and a PCR primer pair (5′-AGAAAAAGAAAATTTTATCTGTGTTTTAGAGACTTCAAA-3′ and 5′-CATGGTGTTCTTTAGCCGCAATTT-3′) that specifically amplifies the fur allele. Standard curves for alleles encoding either fur-R88H or WT fur were generated using 3-fold dilutions (up to a 729-fold dilution) of purified DNA, starting with 40 ng/well. The relative concentrations of alleles encoding either WT Fur or Fur-R88H in a population were then determined by comparison with the appropriate standard curve for each DNA target.
ng/well. The relative concentrations of alleles encoding either WT Fur or Fur-R88H in a population were then determined by comparison with the appropriate standard curve for each DNA target.
Growth curve analysis.
The H. pylori barcoded mutant strains described above were grown overnight in BB-FBS medium in room air supplemented with 5% CO2 at 37°C. All cultures were pelleted, resuspended in BB-FBS, and subsequently inoculated into fresh BB-FBS medium to a starting OD600 of 0.1 in medium containing either 0.5% or 1.0% added NaCl and allowed to grow for 36 h. At various time points, OD600 values were measured.
Competition assays.
Barcoded mutant strains were grown on blood agar plates for 1 day and then were resuspended in 5
day and then were resuspended in 5 mL of fresh BB-FBS. OD600 measurements of the suspensions were used to quantify bacterial concentrations. Bacterial mixtures were generated for competition experiments using equal concentrations of each bacterial strain. The bacterial mixtures were serially passaged for 28
mL of fresh BB-FBS. OD600 measurements of the suspensions were used to quantify bacterial concentrations. Bacterial mixtures were generated for competition experiments using equal concentrations of each bacterial strain. The bacterial mixtures were serially passaged for 28 days on BB-FBS agar plates containing either 0.5% or 0.9% added NaCl, with passaging of bacterial cultures onto fresh plates performed twice per week. To test the effects of supplemental iron, ferrous sulfate or ferric citrate (Sigma) was added (final concentration of 100
days on BB-FBS agar plates containing either 0.5% or 0.9% added NaCl, with passaging of bacterial cultures onto fresh plates performed twice per week. To test the effects of supplemental iron, ferrous sulfate or ferric citrate (Sigma) was added (final concentration of 100 μM) to the medium. At various time points, genomic DNA was extracted (Promega Wizard genomic DNA kit) and real-time qPCR analysis of DNA samples was performed with SYBR green fluorophore (iQ SYBR green supermix; Bio-Rad) on an ABI StepOnePlus machine as described previously (45) Primer sets used in qPCR analyses of competition experiments included a barcode tag-specific primer (TSP) and a gene-specific primer (GSP) (Table S4). The locations of these primers are illustrated in Fig. 2, ,4,4, and and5.5. A standard curve of each DNA target was generated using 3-fold dilutions starting at 40
μM) to the medium. At various time points, genomic DNA was extracted (Promega Wizard genomic DNA kit) and real-time qPCR analysis of DNA samples was performed with SYBR green fluorophore (iQ SYBR green supermix; Bio-Rad) on an ABI StepOnePlus machine as described previously (45) Primer sets used in qPCR analyses of competition experiments included a barcode tag-specific primer (TSP) and a gene-specific primer (GSP) (Table S4). The locations of these primers are illustrated in Fig. 2, ,4,4, and and5.5. A standard curve of each DNA target was generated using 3-fold dilutions starting at 40 ng/well (up to a 729-fold dilution) and used to calculate the abundance of each DNA target in the bacterial population.
ng/well (up to a 729-fold dilution) and used to calculate the abundance of each DNA target in the bacterial population.
Data availability.
Genome sequence data were deposited in NCBI (Bioproject ID PRJNA882804). RNA-seq data were deposited in the GEO database under accession number GSE214216.
ACKNOWLEDGMENTS
The work described in this paper was supported by the NIH (grant no. CA116087, AI039657, and AI118932) and the Department of Veterans Affairs (grant no. I01 BX004447). RNA-seq experiments were supported by the Vanderbilt-Ingram Cancer Center (grant no. P30 CA068485).
Footnotes
Supplemental material is available online only.
Supplemental file 1
Fig. S1 to S4 and Tables S1 to S4. Download iai.00420-22-s0001.pdf, PDF file, 0.8 MB
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.00420-22
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9933627
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/141149994
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/iai.00420-22
Article citations
High salt condition alters LPS synthesis and induces the emergence of drug resistance mutations in <i>Helicobacter pylori</i>.
Antimicrob Agents Chemother, 68(10):e0058724, 06 Sep 2024
Cited by: 0 articles | PMID: 39240098
Essential role of Helicobacter pylori apolipoprotein N-acyltransferase (Lnt) in stomach colonization.
Infect Immun, 91(12):e0036923, 08 Nov 2023
Cited by: 1 article | PMID: 37937999 | PMCID: PMC10715074
High-Salt Diet Exacerbates H. pylori Infection and Increases Gastric Cancer Risks.
J Pers Med, 13(9):1325, 28 Aug 2023
Cited by: 3 articles | PMID: 37763093 | PMCID: PMC10533117
Review Free full text in Europe PMC
Clinical Pathogenesis, Molecular Mechanisms of Gastric Cancer Development.
Curr Top Microbiol Immunol, 444:25-52, 01 Jan 2023
Cited by: 1 article | PMID: 38231214 | PMCID: PMC10924282
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (1 citation) BioProject - PRJNA882804
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE214216
RefSeq - NCBI Reference Sequence Database
- (2 citations) RefSeq - NC_014256.1
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Helicobacter pylori adaptation in vivo in response to a high-salt diet.
Infect Immun, 83(12):4871-4883, 05 Oct 2015
Cited by: 16 articles | PMID: 26438795 | PMCID: PMC4645402
The role of the Ferric Uptake Regulator (Fur) in regulation of Helicobacter pylori iron uptake.
Helicobacter, 7(4):237-244, 01 Aug 2002
Cited by: 69 articles | PMID: 12165031
Proteomic analysis of a ferric uptake regulator mutant of Helicobacter pylori: regulation of Helicobacter pylori gene expression by ferric uptake regulator and iron.
Proteomics, 4(7):2014-2027, 01 Jul 2004
Cited by: 30 articles | PMID: 15221763
The ferric uptake regulator of Helicobacter pylori: a critical player in the battle for iron and colonization of the stomach.
Future Microbiol, 8(6):725-738, 01 Jun 2013
Cited by: 32 articles | PMID: 23701330 | PMCID: PMC3852439
Review Free full text in Europe PMC
Funding
Funders who supported this work.
BLRD VA (1)
Grant ID: I01 BX004447
HHS | National Institutes of Health (3)
Grant ID: CA116087
Grant ID: AI039657
Grant ID: AI118932
NCI NIH HHS (2)
Grant ID: P30 CA068485
Grant ID: P01 CA116087
NIAID NIH HHS (2)
Grant ID: R01 AI039657
Grant ID: R01 AI118932
U.S. Department of Veterans Affairs (1)
Grant ID: BX004447
 a
,
b
,
c
,
d
,
e
a
,
b
,
c
,
d
,
e




