Abstract
Free full text

Remodeling of Yeast Genome Expression in Response to Environmental Changes
Abstract
We used genome-wide expression analysis to explore how gene expression in Saccharomyces cerevisiae is remodeled in response to various changes in extracellular environment, including changes in temperature, oxidation, nutrients, pH, and osmolarity. The results demonstrate that more than half of the genome is involved in various responses to environmental change and identify the global set of genes induced and repressed by each condition. These data implicate a substantial number of previously uncharacterized genes in these responses and reveal a signature common to environmental responses that involves ~10% of yeast genes. The results of expression analysis with MSN2/MSN4 mutants support the model that the Msn2/Msn4 activators induce the common response to environmental change. These results provide a global description of the transcriptional response to environmental change and extend our understanding of the role of activators in effecting this response.
INTRODUCTION
The ability to respond rapidly to fluctuations in temperature, nutrients, and other environmental changes is important for competitive fitness and cell survival. Understanding the response of cells to environmental changes is of interest because it can provide clues to the molecular apparatuses that enable cells to adapt to new environments and the molecular mechanisms that have evolved to regulate the remodeling of gene expression that occurs in new environments.
Significant clues to the mechanisms involved in adaptation to new environments have come from studies of the genes that are expressed in response to specific stresses. For example, cells exposed to elevated temperatures induce transcription of genes encoding heat shock proteins (Craig, 1992). The heat shock proteins are a family of approximately a dozen proteins that are evolutionarily conserved. Studies of heat shock proteins led to the realization that many function as molecular chaperones (Ellis, 1999). Molecular chaperones are critical regulators of protein structure and function and have roles in almost every cellular process. Some molecular chaperones may even facilitate evolutionary processes (Rutherford and Lindquist, 1998). The importance of molecular chaperones suggests that it will be valuable to identify and further study the complete set of stress-inducible genes. If the number of stress-responsive genes is substantial, their identification could make a significant contribution to functional annotation of an important set of previously uncharacterized genes.
Cells must coordinate adjustments in genome expression to accommodate changes in their environment. Despite our lack of knowledge about the complete set of genes involved in these changes, investigators have identified transcriptional activators and repressors that likely contribute to coordinate remodeling of genome expression. For example, the yeast heat shock transcriptional activator Hsf1 and the canonical sequence it binds have been identified (Parker and Topol, 1984; Wu, 1985; Kingston et al., 1987; Sorger and Pelham, 1987). In the absence of heat shock, Hsf1 is inactive; the molecular chaperone Hsp90 is thought to contribute to this inactivation by binding and sequestering the activator (Ali et al., 1998; Duina et al., 1998; Zou et al., 1998; Bharadwaj et al., 1999). Another set of activators, Msn2 and Msn4, act in concert to induce expression of genes under almost any stress condition. Msn2 and Msn4, normally resident in the cytoplasm, are transported into the nucleus during stress, where they bind to stress response elements (STREs) in promoters (Estruch and Carlson, 1993; Marchler et al., 1993; Martinez-Pastor et al., 1996; Schmitt and McEntee, 1996; Görner et al., 1998). The complete set of genes induced by various environmental changes has not been established; therefore, it is not yet clear that these activators are responsible for coordinate induction of these genes.
Here we describe the temporal expression profiles of yeast cells exposed to seven environmental changes. These transcriptional responses demonstrate that a much larger fraction of the genome is involved in responses to environmental changes than previously appreciated, identify the global set of genes induced and repressed by new conditions, and reveal a signature common to each of the environmental responses. Furthermore, expression profiles of strains deleted for Msn2/Msn4 reveal the contributions of these activators to coordinate regulation of environmental responses.
MATERIALS AND METHODS
Strains
American Type Culture Collection (ATCC)-201388, Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 (S288c).
Z985 (1097), Mata ade2-1 can1–100 GAL+ his3-11,15 leu2-3,112 psi+ trp1-1 ura3 msn2Δ::HIS3 msn4Δ::TRP1 (W303).
wt-P82a, Mata ade2-1 can1-100 leu2-3112 trp1-1 ura3-1 hsc82Δ::LEU2 hsp82Δ::LEU2 his3::HSP82 (W303).
Heat Shock
An overnight culture of strain wt-P82a was grown in YPD (1% yeast extract/2% peptone/2% glucose) and used to inoculate two (4 l) flasks containing 1500 ml of YPD. These were grown to OD600 = 0.5 (~1.5 × 107 cell/ml) at 25°C. The temperature shift to 37°C was carried out by the addition of an equal volume of YPD prewarmed to 49°C. Cells were harvested by centrifugation 15, 30, 45, 60, and 120 min after the temperature shift and from duplicate cultures immediately before the temperature shift (time 0 min).
The strain wt-P82a was used in this study because it is isogenic to many strains that are used by this and other laboratories to investigate the heat shock responses. Although this strain harbors a null mutation in the HSC82 gene, it is functionally wild type, because it constitutively expresses the HSP82 gene. The HSP82 and HSC82 gene products are ~97% identical and are functionally equivalent (Borkovich et al., 1989).
Acid
An overnight culture of strain ATCC-201388 was grown in YPD and used to inoculate two (2 l) flasks containing 200 ml of YPD. These were grown to OD600 = 0.5 (~1.5 × 107 cell/ml) at 30°C. To a final concentration of 0.05 M, 20 ml of succinic acid (0.5 M, titrated to pH 3.0 with Tris base) was added. This brought the pH from 6.0 to 4.0. The media remained at pH 4.0 throughout the experiment. Cells were harvested by centrifugation 10, 20, 40, 60, 80, and 100 min after the addition of acid and from duplicate cultures immediately before the addition of acid (time 0 min). The pH of each sample was measured using a pH meter from Orion Research Inc. (Beverly, MA).
Z985 (1097) was grown under the same conditions and cells were harvested 0, 10, and 20 min after the addition of acid.
Alkali
The experiment was carried out as described for acid except that 20 ml of Tris-HCl (1 M, pH 8.25) was added to a final concentration of 0.1 M. This brought the pH from 6.0 to 7.9. The media remained at pH 7.9 throughout the experiment.
Hydrogen Peroxide
An overnight culture of strain ATCC-201388 was grown in YPD and used to inoculate two (2 l) flasks containing 200 ml of YPD. These were grown to OD600 = 0.7 (~2.1 × 107 cell/ml) at 30°C. Hydrogen peroxide was added to a final concentration of 0.4 mM. Cells were harvested by centrifugation 10, 20, 40, 60, and 100 min after the addition of hydrogen peroxide and from duplicate cultures immediately before the addition of hydrogen peroxide (time 0 min).
Salt
An overnight culture of strain ATCC-201388 was grown in YPD and used to inoculate seven (2 l) flasks containing 200 ml of YPD. These were grown to OD600 = 0.6–0.8 (~1.8–2.4 × 107 cell/ml) at 30°C. Sodium chloride was added to a final concentration of 1.0 M. Cells were harvested by centrifugation 15, 30, 45, 60, 90, and 120 min after the addition of sodium chloride and from duplicate cultures immediately before the addition of sodium chloride (time 0 min).
Sorbitol
The experiment was carried out as described for salt except that sorbitol was added to a concentration of 1.5 M.
RNA Preparation, Probe Preparation, and DNA Chip Hybridization
mRNA isolation, cDNA preparation, biotin-labeled cRNA generation, and DNA microarray analysis were performed as described previously (Holstege et al., 1998), and detailed protocols can be found at the author's web site: http://web.wi.mit.edu/young/expression/. Briefly, total RNA was first extracted from frozen cell pellets by a hot phenol method (Schmitt et al., 1990). Cells were thawed on ice and then mixed with 3 ml of acid phenol/chloroform (5:1; prewarmed for 10 min at 65°C) and 3 ml of TES (10 mM Tris, pH 7.5/10 mM EDTA/0.5% SDS) per 200 OD600 U. The mixture was incubated at 65°C for 1 h with frequent vortexing, and the aqueous phase was separated from the organic phase by centrifugation. The aqueous layer was extracted once more with an equal volume of phenol/chloroform (5:1) and then with chloroform-isoamyl alcohol before ethanol precipitation. The mRNA was purified from the total RNA with an oligo(dT) selection step (Oligotex; QIAGEN, Chatsworth, CA). Then, double-stranded cDNA was made from mRNA using a modified oligo(dT) primer with a 5′-T7 RNA polymerase promoter sequence (GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24) and the Superscript Choice System for cDNA synthesis (Life Technologies, Gaithersburg, MD). Double-stranded cDNA was purified by phenol-chloroform extraction and ethanol precipitation. In vitro transcription was performed with T7 RNA polymerase (T7 Megascript kit; Ambion, Austin, TX) and 0.75–0.95 μg of double-stranded cDNA template in the presence of a mixture of unlabeled ATP, CTP, GTP, and UTP and biotin-labeled CTP and UTP (biotin-11-CTP, biotin-16-UTP from ENZO Diagnostics, Farmingdale, NY). cRNA was purified on an affinity resin (RNeasy; QIAGEN). The amount of labeled cRNA was determined by measuring absorbance at 260 nm and using the convention that 1 OD at 260 nm corresponds to 40 μg/ml RNA.
For heat shock, acid, alkali, and hydrogen peroxide experiments, Affymetrix (Santa Clara, CA) YE6100 gene chips were used. Ten micrograms of cRNA samples were heated in the hybridization solution (1.0 M NaCl/10 mM Tris-HCl, pH 7.6/0.005% Triton X-100/0.1 mg/ml unlabeled, sonicated herring sperm DNA; Promega, Madison, WI) to 95°C for 5 min followed by 40°C for 7 min, before being placed in a hybridization cartridge. Hybridizations were carried out at 40°C for 16 h with mixing on a rotisserie at 60 rpm. After hybridization, the solutions were removed, and the arrays were washed in a fluidics station. First, the arrays are rinsed with 6× SSPE-T (0.9 M NaCl/60 mM NaH2PO4/6 mM EDTA/0.005% Triton X-100 adjusted to pH 7.6), incubated with 6× SSPE-T for 1 h at 50°C, and then washed with 0.5× SSPE-T at 50°C for 15 min. After the arrays were washed, the hybridized RNA was fluorescently labeled by incubating with 2 μg/ml streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) and 1 mg/ml acetylated bovine serum albumin (Sigma, St. Louis, MO) in 6× SSPE-T at 40°C for 10 min. Unbound streptavidin-phycoerythrin was removed by rinsing at room temperature before scanning. The arrays were read at a resolution of 7.5 μm using a GeneChip Scanner (Affymetrix).
For salt and sorbitol experiments, the Affymetrix S98 yeast genome chips were used because of discontinuation of YE6100 GeneChip by the manufacturer. The S98 gene chips are different from the YE6100 GeneChip series in that they contain more probes and can detect expression of the whole genome with just one chip, instead of four chips. The total amount of cRNA hybridized to the chip is the same as to the low density YE6100 chips, i.e. between 10 and 12 μg. The protocol is also the same as that for YE6100 except that biotin-labeled antibody against streptavidin was used after the last staining step to increase the signal.
Data Acquisition and Processing
The output files from the scanner were downloaded as text files and then loaded into a custom-built database (ChipDB) for further analysis. The classification of genes into functional groups was as described in the Yeast Proteome Database (http://www.proteome.com).
Clustering Analysis of Gene Expression Profiles in All Conditions (Figure (Figure11)
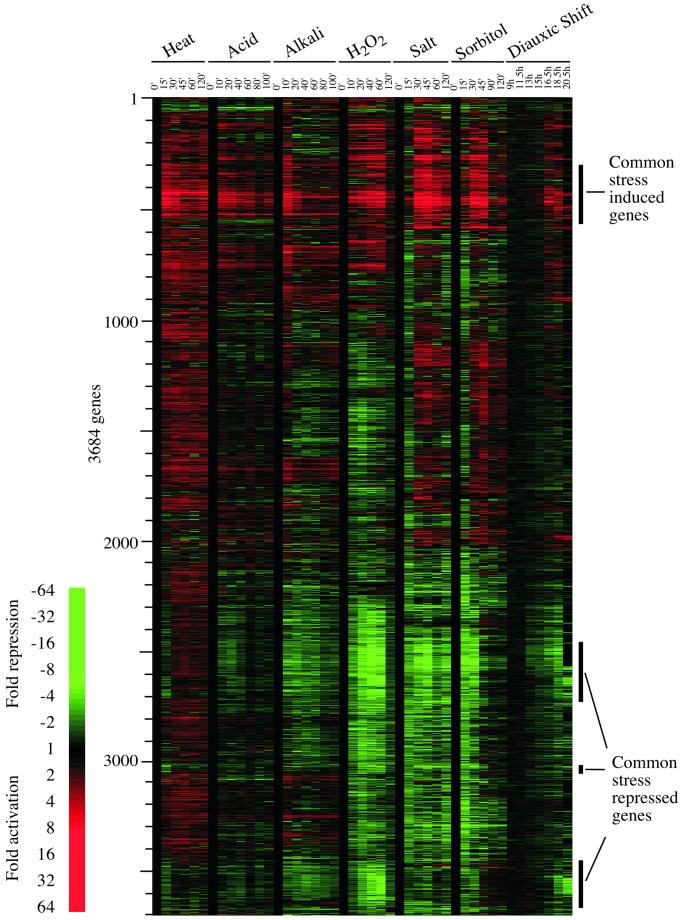
Time-course expression profiles for cells exposed to changes in environment. The expression profiles of 3684 genes whose transcript levels changed by threefold in at least one of the time courses are represented. See MATERIALS AND METHODS for further details of data analysis. Each horizontal strip represents a single gene. The fold change is represented by a color (see color bar). The genes that are induced or repressed in most of the responses to environmental changes are indicated.
The expression profiles corresponding to individual time points were scaled to a common reference profile based on fluorescent intensities of five DNA controls that were added after the preparation of total RNA. Specifically, a scaling ratio between the sample chip and the reference chip was calculated by using the following formulas:

where n is the total number of controls, a is the reference sample and aI is the sample to be normalized. The rescaled value for sample i was then calculated as:

where ai is the sample to be normalized, and ai′ is the rescaled value.
The normalized data were floored at 20 and downloaded from ChipDB as a text file. The SAS software package was used to calculate the fold changes for each gene as follows:

A gene's fold change value was considered to be reliable and was used for analysis if the fluorescence intensity value was scored as “present” (P) according to the GeneChip software (Affymetrix) in at least one of the time points after the treatment or in both of the t0 time points.
Genes whose expression changed more than threefold (fc ≥ 3) in at least one treatment and whose rescaled fluorescent intensities differed from that of the untreated controls by more than 100 U (‖αti − αt0‖ ≥ 100) were selected. Genes that could not be reliably detected in more than 30% of the time points for a given time course were removed from the resulting data set. Genes that passed these selection criteria were considered to be the set of genes whose expression is changed in response to environmental changes (Figure (Figure1).1). The log-transformed expression values [ln(fc)] for the selected genes (a total of 3864 genes) were then exported to a text file for further analysis.
The Cluster program (Eisen et al., 1998) was used to cluster the above genes into a hierarchical tree. The clustering is based on a nonparametric correlation matrix (Kendall's Tau similarity metric) calculated for every pair of genes.
Identification of Common Environmental Response (CER) Genes (Figures (Figures22 and and33)
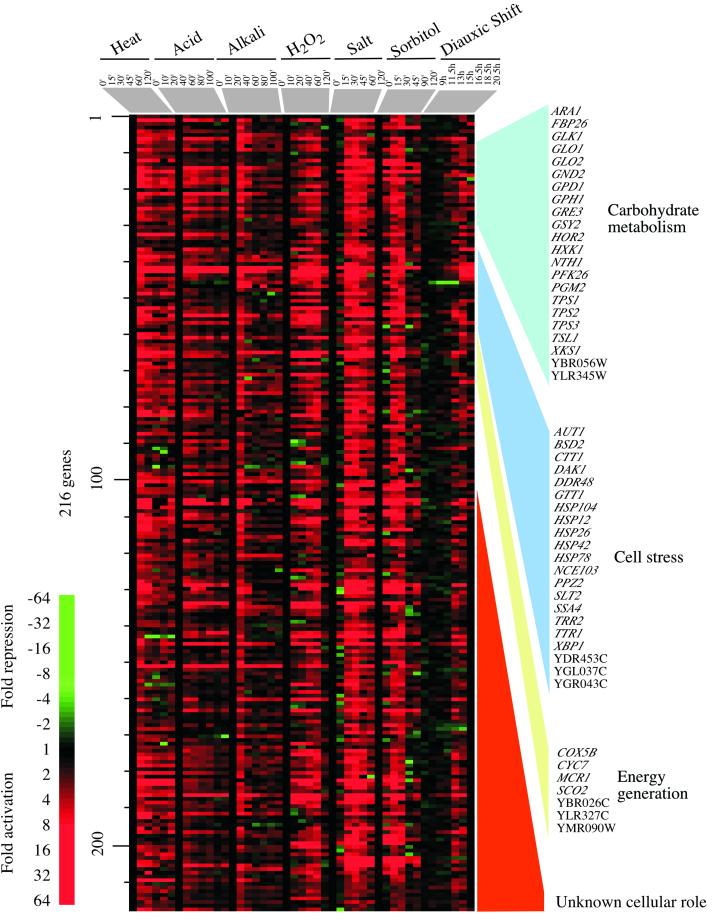
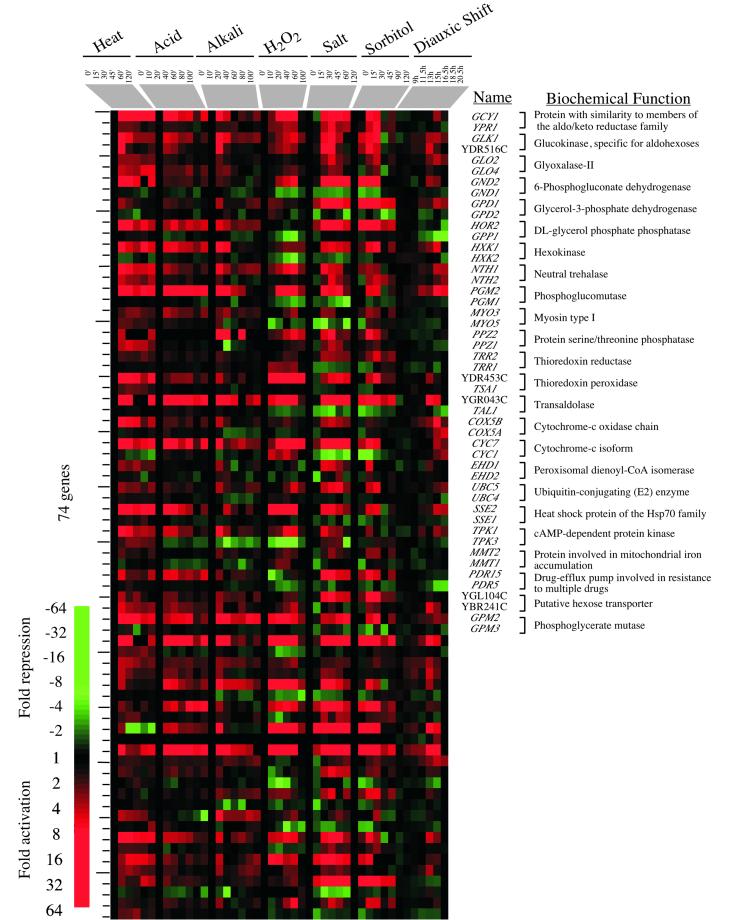
A CER: gene activation. One of the clusters from the hierarchical tree in Figure Figure1,1, containing genes whose expression is induced in most of the environmental responses, is represented. These data were sorted so that genes with similar cellular functions, as listed in the Yeast Proteome Database (http://www.proteome.com), are listed together. Details are as described in the legend to Figure Figure11.

A CER: transient repression of the translation apparatus. Three clusters from the hierarchical tree in Figure Figure1, 1, containing genes whose expression was repressed in most of the environmental responses, are represented. These clusters were combined, and the genes were sorted according to their cellular roles. Details are as described in the legend to Figure Figure11.
The CER genes were identified from the hierarchical tree of the clustering output as genes whose expression was induced, or repressed, in all conditions by visual inspection. From this list, the genes that changed at least twofold in five or more time courses were selected as CER genes.
Identification of Specific Responses to Each Environmental Change (Figure (Figure55)
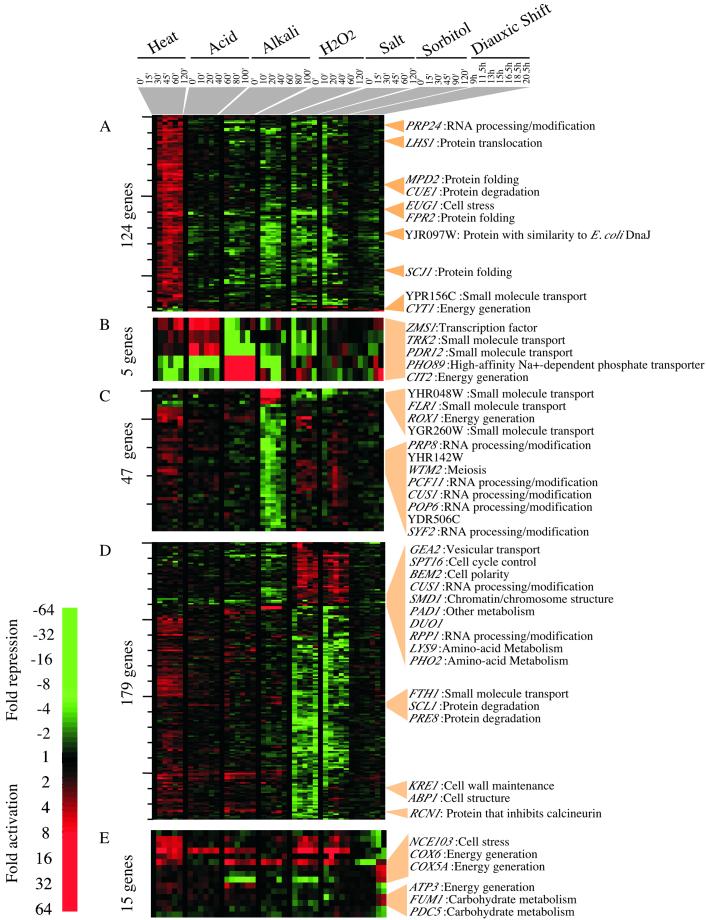
Environmental response-specific gene expression. Environmental response-specific genes were selected based on the criteria described in MATERIALS AND METHODS. The genes displayed are a subset of the environmental response-specific genes. (A) Genes whose expression is uniquely remodeled in response to heat. (B) Genes whose expression is diametrically regulated in response to acid and alkali. (C) Genes whose expression is uniquely remodeled in response to hydrogen peroxide. (D) Genes that respond similarly to salt and sorbitol. (E) Genes whose expression is specific to the diauxic shift. Some of the response-specific genes are listed on the right.
The environment-specific response genes were identified as genes whose expression was induced, or repressed, at least threefold in response to a specific change in environment but were not included in the CER.
To identify genes whose expression is specifically reset in response to the shift to 37°C, the genes whose expression was induced, or repressed, by more than threefold in response to the temperature shift, were selected (fc = 3 or 0.33 for heat shock time course). To confer specificity, a second criterion required that genes induced in response to heat were not induced or repressed by more than two-fold in response to any other environment change (0.5< fc < 2 for other time courses). Linear correlation coefficients (LCCs) were calculated for the log-transformed fold change [ln(fc)] of each gene against a synthetic reference pattern. The reference pattern required that gene expression was high at each time point after the temperature shift and low before the temperature shift and in response to other environmental changes (heat 0 min = 0, heat 15 min = 1, heat 30 min = 1, heat 45 min = 1, heat 60 min = 1, heat 120 min = 1, acid 0 min = 0, acid 10 min = 0, etc; 0 represents an arbitrary low value and 1 represents an arbitrary high value). Genes whose profiles gave an LCC 0.60 were defined as genes whose expression remodeled in response to a change in temperature. This strategy was used to identify genes reset in response to other environmental changes, except that the reference pattern was modified accordingly. For example, to find genes whose expression is modified in response to hyperoxia, the reference pattern required a high value after the addition of hydrogen peroxide and a low value for other time courses. For the salt-sorbitol response, the reference pattern demanded a high value after the addition of salt or sorbitol and low before the addition of salt or sorbitol or in response to other environmental changes. For the diauxic shift (DeRisi et al., 1997), the only criterion was that the expression was induced, or repressed, more than threefold during diauxic shift but less than twofold in other environmental changes.
To identify genes whose expression levels were reset in a reciprocal manner in response to acid and to alkali, the transcription profile of the PDR12 gene was used as reference pattern against which LCCs were calculated. The genes selected are those for which the LCC 0.60 and whose expression level induced, or repressed, by more than threefold in acid or alkali and less than twofold in response to any other environmental changes (as described for the selection of heat-specific genes).
Identification of Msn2/Msn4-dependent CER Genes (Figure (Figure66)

Induction of most acid-induced genes depends on Msn2/Msn4. The genes whose expression changed by at least threefold in response to acid are shown for a wild-type strain and a strain deleted for MSN2 and MSN4. Msn2/Msn4-dependent genes are shown with a bar. Details are as described in the legend to Figure Figure11.
The genes induced in the CER were examined under acid conditions in wild-type yeast and in msn2msn4 strains. Among the 216 CER acid-induced genes, the transcript levels of 193 genes could be reliably measured in the msn2msn4 mutants. From these genes, 147 genes whose expression is induced at least twofold after addition of acid (compared with wild-type cells) were used for analysis. The expression values of these genes in wild-type and msn2msn4 strains were clustered using the Cluster program (Eisen et al., 1998). Genes that are dependent on Msn2/Msn4 for their induction were identified as those with a signature showing a high level of induction in wild type and no induction in the msn2/msn4 strain.
RESULTS
We identified environmental conditions that have been frequently selected for study by other investigators. Saccharomyces cerevisiae cells in logarithmic phase growth were exposed to various environmental changes and the transcriptional response was monitored using high-density oligonucleotide arrays. These changes involved heat (a shift from 25 to 37°C), acid (pH 6.0 to 4.0), alkali (pH 6.0 to 7.9), hyperoxia (0.0 to 0.4 mM H202), salt (addition of NaCl to 1.0 M), and osmotic stress (addition of sorbitol to 1.5 M). These environmental changes were of limited severity; therefore, in each case yeast cells continued to grow and divide. For each of the conditions cells were grown in YPD and subjected to the new environment when cultures reached OD600 0.5–0.8. Labeled “target” RNA was prepared from cultures harvested immediately before and at various times after the change in environment and hybridized to Affymetrix GeneChips, as described previously (Holstege et al., 1998). Additional detailed information and interactive databases supporting this study can be found on the World Wide Web at web.wi.mit.edu/young/environment/. Data on the transcriptional response to nutrient depletion at the diauxic shift were taken from DeRisi et al. (1997).
The clustered results shown in Figure Figure11 reveal several interesting features of the response yeast cells undergo to various environmental changes. A remarkable fraction of the genome is subjected to expression remodeling during these responses. Of the 5594 genes whose expression could be scored in these time courses, expression of 66% (3684) is altered significantly when the data are analyzed as described in MATERIALS AND METHODS. It is clear from much previous work that cells have evolved responses that enhance cell survival and fitness in the dynamic environments in which they live, but the extent of the genome involved in these responses in yeast is impressive (Mager and De Kruijff, 1995). The involvement of many genes with unknown cellular roles in these environmental changes implicates these genes in specific responses, and these results should therefore contribute to further functional annotation of the genome. Given the broad scope of expression remodeling that occurs when cells encounter new environments, which is a frequent occurrence outside of the laboratory, the term “stress response” seems inadequate to describe these events. To avoid confusing the large-scale effects observed in this study with previously described stress responses, we will tend to refer to the broader effects on gene expression as environmental responses rather than stress responses.
It is also evident from the results shown in Figure Figure11 that the kinetics of global change, and whether the change is transient or constitutive, varies with the environmental change. For example, cells exposed to a shift in temperature activate expression of heat-responsive genes within 15 min of the temperature shift, whereas cells exposed to an increase in salinity take longer to respond. In most responses, there were genes whose expression levels changed transiently and others whose levels remained altered through the entire time course. We infer that the products of the genes that exhibited transient increases are involved in facilitating the transition to the new environment. The genes whose expression levels change to a new level and remain so altered likely encode products that have a continuous role in the cell under the new conditions.
We note that there are a substantial number of genes whose expression patterns are common to most of the environmental changes. We henceforth refer to these as CER genes.
CER
Genes Induced
The response that is common to most of the environmental changes examined here involves 499 genes, of which 216 are induced and 283 are repressed (the criteria used for analysis are described in MATERIALS AND METHODS). The CER thus involves ~10% of the genome; that such a large fraction of the genome is remodeled under a wide range of environmental changes attests to the importance of these genes in cellular adaptation to the environment. The expression data for the induced CER genes are displayed in Figure Figure2.2. The genes induced in the CER include those with functions in carbohydrate metabolism, cell stress, and the generation of energy.
Many of the genes induced are involved in glycolysis, an increase in which could provide energy needed for the functions of ATP-dependent molecular chaperones and other machinery involved in the response to cellular stress. Genes encoding all the subunits of trehalose synthetase (TPS1, TPS2, TPS3, TSL1) were also activated. This might be anticipated from previous studies (Jelinsky and Samson, 1999; Rep et al., 2000), because trehalose is thought to protect cellular components from the detrimental effects of stress by providing energy for the renaturation of cellular structures and possibly by protecting cells and membranes from denaturation.
The “cell stress” genes in the CER include classical heat shock genes (HSP12, HSP26, HSP42, HSP78, HSP104, SSA4, SSE2), many of which encode molecular chaperones that facilitate protein folding or maintenance of a particular protein conformation. Genes whose products are involved in protein degradation (PHB2, RPN5, UBC5, UBC8, YPS6) were also induced, consistent with the notion that damaged or partially denatured proteins need to be degraded to prevent the accumulation of protein aggregates.
Other genes induced in the CER include those involved in defenses against oxidizing agents (Moradas-Ferreira et al., 1996). These function in the degradation of reactive species such as hydrogen peroxide that can potentially damage proteins and nucleic acids (CTT1). Ion homeostasis genes are also represented in the CER: these are involved in sequestration or metal transport (BSD2) or in thioredoxin or glutathione regulation (TTR1, YDR435C, YCL035C). These genes are important for maintaining the reducing environment within the cell. Many of the CER genes were thought to be specifically induced in response to changes in the tonicity of the environment; however, our data suggest that many changes in the environment also result in increased membrane permeability and thus induce systems involved in ion transport.
The energy generation genes include those required for respiration, some of which have consensus binding sites for the HAP2,3,4 complex. It has been observed previously that these genes are induced upon nutrient limitation at the diauxic shift (DeRisi et al., 1997). More than one-half of the induced CER genes do not have known cellular roles, and these can now be annotated as being involved in the CER.
A subset of the CER genes was previously observed to be involved in a variety of stress responses and has been termed “general stress response” genes (Moskvina et al., 1998; Treger et al., 1998). These ~60–190 genes have STRE consensus sequences in their promoter regions; the Msn2 and Msn4 transcription factors bind to these elements and activate transcription of these genes under stress conditions (Martinez-Pastor et al., 1996; Schmitt and McEntee, 1996; Boy-Marcotte et al., 1998; Görner et al., 1998). The relationship between the CER genes and the previously identified general stress genes will be discussed further below.
Genes Repressed
The expression data for the repressed set of genes in the CER are displayed in Figure Figure3.3. The 283 repressed genes are dominated by genes associated with translation and protein synthesis. Many of these have previously been observed to be repressed in response to specific stresses (Kim and Warner, 1983; DeRisi et al., 1997; Lashkari et al., 1997; Eisen et al., 1998; Jelinsky and Samson, 1999; Rep et al., 2000). The repressed set includes genes for cytoplasmic ribosomal proteins, polymerase I, II, and III transcription, tRNA synthetases, proteins required for processing rRNAs, and a subset of translation initiation factors. The genes repressed in all environmental changes include 106 genes of unknown function.
The transient but significant reduction in transcripts for the translation apparatus is consistent with the previously noted transient translational arrest that occurs on heat shock, because cellular resources are redirected toward synthesis of stress proteins (Miller et al., 1979, 1982). The loss and then reestablishment of transcript levels for genes encoding the translation apparatus and its regulators are remarkably coordinated for each environmental change, although the dynamics of each response are different, as observed for the induced set of CER genes. For example, genes are significantly repressed for ~60 min after treatment with hydrogen peroxide but are only transiently repressed in the heat and sorbitol time courses (Figure (Figure33).
Differential Expression of Isozymes
Previous reports have suggested that isozymes and other members of multigene families can be differentially expressed under specific conditions (Rep et al., 2000). To determine whether this is the case for the CER genes under conditions explored in this study, we examined the 405 pairs of homologous genes identified by Wolfe and Shields (1997). Of the 316 pairs of genes for which expression data were obtained, 79 pairs of genes contain at least one member of the CER. In 37 of these pairs, one gene of the pair is CER induced and expression of the other member of the pair is not (Figure (Figure4).4). Many of these enzymes are involved in glycolysis (GLK1, GLO2, GND2, HXK1, PGM2) or energy generation (COX5A, CYC7). The observation that cells differentially express these particular isozymes during changes in environment suggests that one member of these pairs plays a particularly important role in the adaptation to new environments.
Environment-specific Responses
Although the CER genes exhibit expression changes in response to all the conditions tested, there is a substantial number of genes whose expression is altered in response to a specific change in environment (Table 1). The CER accounts for 18–38% of the total population of genes whose expression is altered in response to an environmental change. The responses to specific changes in environment, such as salt concentration, involve the remodeling of up to one-third of the genome.
Heat
Genes whose expression is induced and remains elevated after a shift to higher temperature are shown in Figure Figure5A.5A. Many of these induced genes have functions in protein folding and transport, including EUG1 and LHS1, which are regulated by the unfolded protein response (Craven et al., 1996; Chapman et al., 1998; Travers et al., 2000). The induction of protein folding genes is consistent with the need to contend with the widespread protein denaturation that occurs on heat stress. It has been proposed that retention of proteins in the endoplasmic reticulum and the refolding of heat-denatured glycoproteins is also part of the cellular stress response. Lhs1 is one of the proteins required for the return to secretion competence of heat-denatured proteins in the endoplasmic reticulum (Saris and Makarow, 1998). Although the cellular response to heat stress has been extensively studied, 50% of the 854 heat-responsive genes do not have defined functions.
Acid and Alkali
PDR12 is one of the genes that is regulated in a pH-specific manner (Figure (Figure5B).5B). PDR12 encodes an ATP-dependent membrane transporter that is highly induced at low pH but is repressed at high pH. Pdr12 was first identified as a protein induced by sorbic acid and may function to export carboxylate anions out of cells (Piper et al., 1998; Holyoak et al., 1999). The transcription profile of PDR12 was used as a template to identify genes whose expression is regulated in a reciprocal manner in acid and in alkali. We were interested in identifying those genes whose expression is either reset and maintained at a higher level or reset and maintained at a lower level during the environmental change. We identified four genes by these criteria (Figure (Figure5B).5B). Two of these genes, ZMS1 and TRK2, are activated by acid treatment and repressed by alkali treatment. They encode a zinc-finger family transcription factor and a potassium transporter (Ko et al., 1990), respectively. Another two, CIT2 and PHO89, are repressed by acid and activated by alkali. They encode a peroxisomal citrate synthase and sodium phosphate symporter. The Trk2 potassium transporter is responsible for a K+ current at low pH, and its activity is low at neutral or high pH (Bihler et al., 1999). By contrast, the Pho89 sodium phosphate symporter catalyzes sodium-dependent phosphate uptake, and its activity is high at alkaline pH (Martinez and Persson, 1998). Thus, for Trk2 and Pho89, both differential transcriptional regulation and protein activity contribute to adaptation of yeast to changes in the pH environment.
Hydrogen Peroxide
The response to hyperoxia involves about one-third of the genome and differs from the other stress responses in that the maximal effects on gene expression occur slightly later than in the other stress time courses (Figure (Figure1).1). Despite this, much of the transcriptome has returned to prestress levels by 2 h after the addition of hydrogen peroxide. As expected, ROX1, which encodes a transcriptional repressor of hypoxic genes, was among the hyperoxia-specific genes (Figure (Figure5C; 5C; Deckert et al., 1995a). ROX1 was induced and then repressed, consistent with its autoregulatory activity (Deckert et al., 1995b).
High Salt and High Osmolarity
The high-salt response is likely to be a composite response to both the osmotic and ionic changes caused by the addition of sodium chloride, whereas the sorbitol response is expected to be specific for osmotic changes. However, the sets of genes involved in the responses to high salt and to sorbitol are remarkably similar to each other (Figure (Figure5D),5D), suggesting that most of the changes in gene expression are in response to the change in osmotic conditions. There are some genes whose expression is reset in response to a change in the ion concentration. For example, the levels of RCN1 (YKL159C) decrease 8-fold in salt and only 1.4-fold in sorbitol over the time course. Rcn1 is a known inhibitor of calcineurin, which functions to stimulate the transcription of the gene encoding the primary sodium transporter, ENA1. It is notable that many of the genes known to be induced in high-saline conditions, including ENA1, are induced by multiple stresses, suggesting that ion homeostasis is a critical response for most environmental changes.
Diauxic Shift
There are only a few genes whose expression changes at least threefold in response to nutrient limitation at the diauxic shift but whose expression changes less than twofold in response to other environmental changes. These include genes required for respiration (e.g., COX5A and COX6), indicative of a shift from fermentative to respiratory growth, as observed previously (DeRisi et al., 1997).
Requirement for the Msn2/Msn4 Activators
Because the transcriptional activators Msn2 and Msn4 are thought to be involved in induction of genes common to many stress responses, we tested whether Msn2/Msn4 are required for activation of the induced set of CER genes. We selected the acid response for this experiment because the CER contributes to a large percentage of the acid response. The response of wild type and msn2msn4-deleted strains is compared to acid in Figure Figure6.6. Of the 193 genes induced in the CER whose expression could be measured in the msn2msn4 strain, 147 were induced more than twofold in acid. Msn2/Msn4 appear to be required for the induced expression of 136 (93%) of these genes. It is also notable that these activators appear to have a function in overcoming transcriptional repression, because most acid-induced genes are repressed upon treatment with acid in the msn2msn4 strain.
DISCUSSION
Genome-wide expression analysis was used to explore how gene expression is remodeled in response to changes in extracellular environment, including changes in temperature, oxidation, nutrients, pH, and osmolarity. We found that approximately two-thirds of the genome is involved in the response to environmental changes. The inclusion of a large fraction of genes in environmental responses reveals the importance of expression remodeling in adapting to environmental changes and implicates a substantial set of genes with previously uncharacterized cellular roles in these responses. The results of this study identify a common set of genes that are induced and repressed in most of these responses, and these we call CER genes. We found that Msn2/Msn4 are involved in the activation of nearly all of the genes that are induced in the CER.
Approximately 66% of yeast genes are involved in responding to the changes in environment that we have examined here when using stringent analysis criteria. Only genes whose expression changed more than threefold in at least one treatment and whose rescaled fluorescent intensities differed from that of the untreated controls by 100 U were selected. In addition, those genes that could not be reliably detected in 30% of the time points were discarded. Given the large number of genes involved in these responses, it is striking that only ~1000 (17%) yeast genes are thought to be essential for viability under standard laboratory conditions (Winzeler et al., 1999). The large difference in these numbers emphasizes the fact that life has evolved under conditions in which the environment is continually changing, in contrast to those in the laboratory.
CER
Our results indicate that ~10% of yeast genes are induced or repressed in common when yeast cells respond to a wide variety of environmental changes. It seems likely that this CER is due to a common effect on cells when exposed to almost any change that affects permeability of the cell wall or integrity of protein structure. The permeability of the cell wall increases in response to heat, spheroplasting, and ethanol, and the consequences of increased permeability may contribute to a common response (Adams and Gross, 1991; Carratùet al., 1996). Diverse changes in environmental conditions may generally induce molecular chaperones because of their effects on the structural integrity of proteins, and this, in turn, may require substantial increases in energy production because many molecular chaperones use the energy of ATP hydrolysis (Lindquist, 1992).
The identification of a CER helps to explain the phenomena of tolerance and cross-protection, in which pretreatment of cells with a mild environmental change provides protection against a more severe change of either the same or a different nature (Lewis et al., 1995; Park et al., 1997). Not all changes in conditions provide cross-protection, e.g., ethanol treatment does not result in increased tolerance to heat, although the converse is true (Piper, 1995). This suggests that some of the environmental change-specific genes play important roles in the response to individual changes in conditions or that there may be a graded response, depending on the extent to which the cell is challenged.
Regulation of Responses to Environment
We have shown that Msn2/Msn4 are required for the activation of 90% of the CER induced genes, at least in response to acid treatment. Msn2/Msn4 have been shown to bind to an upstream STRE (Görner et al., 1998). Of the 216 CER-induced genes, 47 have a perfect STRE consensus within 600 base pairs (bp) of the transcription start site. Msn2/Msn4 may also bind upstream of genes with other consensus sites or they may bind in concert with other, perhaps environmental change-specific, factors.
We note that, among the 283 CER repressed genes, 133 have a GATGAG site within 600 bp of the transcription start site. This is significant because only 4% of all yeast genes have this sequence within 600 bp of the ATG. In addition, 114 have an AAA(A/T)TTTCA within 600 bp of the transcription start site, whereas only 5% of all yeast genes have this sequence.
Msn2/Msn4 shuttle between the nucleus and the cytoplasm, by a variety of mechanisms that are thought to involve the protein kinase A, Hog1, and TOR kinase signal transduction pathways, depending on the environmental conditions (Schmitt and McEntee, 1996; Beck and Hall, 1999; Rep et al., 1999). The protein kinase A and TOR kinase pathways are also involved in the response to nutritional signals, consistent with our observation that nearly 40% of the genes affected by nutrient limitation at the diauxic shift are also CER genes. These pathways appear to play roles in regulation of both the CER-induced and -repressed genes, via a variety of mechanisms (Klein and Struhl, 1994; Neuman-Silberg et al., 1995; Boy-Marcotte et al., 1998; Görner et al., 1998; Smith et al., 1998; Beck and Hall, 1999).
The regulation of some of the CER-repressed genes has also been reported to involve the protein kinase C pathway (Nierras and Warner, 1999). Our data show that many of the genes that function to restore membrane integrity and to deal with the consequences of membrane damage are part of the CER, consistent with the involvement of the protein kinase C pathway, which is thought to respond to changes in membrane integrity.
The set of CER-repressed genes include a large number of genes that encode components of the translation apparatus, including cytoplasmic ribosomal protein genes. The 137 cytoplasmic ribosomal protein genes are coordinately regulated, and their transcripts have relatively short half-lives (Li et al., 1999). Transcription of ribosomal protein genes can account for up to one-half of the RNA polymerase II-mediated transcription initiation events in the cell (Warner, 1999). A transient reduction in the synthesis of ribosomal protein mRNAs would permit energy and other resources to be diverted toward the synthesis and use of molecular chaperones, along with other mechanisms involved in surviving a change in the environment (Jelinsky and Samson, 1999).
The results described here implicate a substantial set of genes with previously uncharacterized cellular roles in the response to environmental change. These include 118 of the 216 CER-induced genes and 106 of the 283 CER-repressed genes. These genes can now be defined as environmental change responsive genes.
Exercising the Genome
The time course expression profiles described here involve changes in a substantial fraction of the genes in the yeast genome. Studies of the yeast cell cycle revealed that 500–800 genes change expression levels significantly during cell cycle progression (Cho et al., 1998; Spellman et al., 1998). Environmental change also “exercises” a large fraction of the genome, in many cases a larger portion than seen with the cell cycle. The availability of genome-wide expression data involving significant portions of the genome should prove to be valuable for efforts to map the regulatory circuits of yeast cells and should serve as a useful foundation for future efforts to increase our understanding of the molecular mechanisms involved in adaptation to environmental change. The large number of genes involved and the temporal changes that occur in these environmental responses should also provide a rich source of information for computational modeling of regulatory networks.
ACKNOWLEDGMENTS
Strain Z985 (1097) was the kind gift of Helmut Ruis. We were alerted to the possibility that isozymes are differentially expressed during environmental changes in public talks by members of the laboratory of Dr. Patrick Brown. This work was supported by funding from the Bristol-Meyers Squibb Company, Affymetrix, Inc., and Millenium Pharmaceuticals, Inc., and grants from the National Institutes of Health (R.A.Y.), the Helen Hay Whitney Foundation (B.R.), and Howard Hughes Medical Institute (E.G.J.).
REFERENCES
- Adams CC, Gross DS. The yeast heat shock response is induced by conversion of cells to spheroplasts and by potent transcriptional inhibitors. J Bacteriol. 1991;173:7429–7435. [Europe PMC free article] [Abstract] [Google Scholar]
- Ali A, Bharadwaj S, O'Carroll R, Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol. 1998;18:4949–4960. [Europe PMC free article] [Abstract] [Google Scholar]
- Beck T, Hall MN. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. [Abstract] [Google Scholar]
- Bharadwaj S, Ali A, Ovsenek N. Multiple components of the HSP90 chaperone complex function in regulation of the heat shock factor 1 in vivo. Mol Cell Biol. 1999;19:8033–8041. [Europe PMC free article] [Abstract] [Google Scholar]
- Bihler H, Gaber RF, Slayman CL, Bertl A. The presumed potassium carrier Trk2p in Saccharomyces cerevisiae determines an H+-dependent, K+- independent current. FEBS Lett. 1999;447:115–120. [Abstract] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. [Europe PMC free article] [Abstract] [Google Scholar]
- Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bacteriol. 1998;180:1044–1052. [Europe PMC free article] [Abstract] [Google Scholar]
- Carratù L, Franceschelli S, Pardini CL, Kobayashi GS, Horvath I, Vigh L, Maresca B. Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc Natl Acad Sci USA. 1996;93:3870–3875. [Europe PMC free article] [Abstract] [Google Scholar]
- Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Dev Biol. 1998;14:459–485. [Abstract] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. [Abstract] [Google Scholar]
- Craig E. The heat shock response of Saccharomyces cerevisiae. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 501–537. [Google Scholar]
- Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [Europe PMC free article] [Abstract] [Google Scholar]
- Deckert J, Perini R, Balasubramanian B, Zitomer RS. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics. 1995a;139:1149–1158. [Europe PMC free article] [Abstract] [Google Scholar]
- Deckert J, Torres AMR, Simon JT, Zitomer RS. Mutational analysis of Rox1, a DNA-binding repressor of hypoxic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1995b;15:6109–6117. [Europe PMC free article] [Abstract] [Google Scholar]
- DeRisi JL, Iyer V, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. [Abstract] [Google Scholar]
- Duina AA, Kalton HM, Gaber RF. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem. 1998;273:18974–18978. [Abstract] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression paptterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. [Europe PMC free article] [Abstract] [Google Scholar]
- Ellis RJ. Molecular chaperones: pathways and networks. Curr Biol. 1999;9:R137–R139. [Abstract] [Google Scholar]
- Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. [Europe PMC free article] [Abstract] [Google Scholar]
- Görner W, Durchschlag E, Martinex-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. [Europe PMC free article] [Abstract] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TS, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. [Abstract] [Google Scholar]
- Holyoak CD, Bracey D, Piper PW, Küchler K, Coote PJ. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J Bacteriol. 1999;181:4644–4652. [Europe PMC free article] [Abstract] [Google Scholar]
- Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. [Europe PMC free article] [Abstract] [Google Scholar]
- Kim CH, Warner JR. Mild temperature shock alters the. transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983;3:457–465. [Europe PMC free article] [Abstract] [Google Scholar]
- Kingston RE, Schuetz TJ, Larin Z. Heat-inducible human factor that binds to a human hsp70 promoter. Mol Cell Biol. 1987;7:1530–1534. [Europe PMC free article] [Abstract] [Google Scholar]
- Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. [Europe PMC free article] [Abstract] [Google Scholar]
- Ko CH, Buckley AM, Gaber RF. TRK2 is required for low affinity K+ transport in Saccharomyces cerevisiae. Genetics. 1990;125:305–312. [Europe PMC free article] [Abstract] [Google Scholar]
- Lashkari DA, DeRisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, Brown PO, Davis RW. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. [Europe PMC free article] [Abstract] [Google Scholar]
- Lewis JG, Learmonth RP, Watson K. Induction of heat, freezing and salt tolerance by heat and salt shock in Saccharomyces cerevisiae. Microbiology. 1995;141:687–694. [Abstract] [Google Scholar]
- Li B, Nierras CR, Warner JR. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol. 1999;19:5393–5404. [Europe PMC free article] [Abstract] [Google Scholar]
- Lindquist S. Heat-shock proteins and stress tolerance in microorganisms. Curr Opin Genet Dev. 1992;2:748–755. [Abstract] [Google Scholar]
- Mager WH, De Kruijff AJ. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. [Europe PMC free article] [Abstract] [Google Scholar]
- Marchler G, Schüller C, Adams G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. [Europe PMC free article] [Abstract] [Google Scholar]
- Martinez P, Persson BL. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:628–638. [Abstract] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [Europe PMC free article] [Abstract] [Google Scholar]
- Miller M, Xuong N-H, Geiduschek EP. Quantitative analysis of the heat shock response of Saccharomyces cerevisiae. J Bacteriol. 1982;151:311–327. [Europe PMC free article] [Abstract] [Google Scholar]
- Miller MJ, Xuong N, Geidushek EP. A response of protein synthesis to temperature shift in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:5222–5225. [Europe PMC free article] [Abstract] [Google Scholar]
- Moradas-Ferreira P, Costa V, Piper P, Mager W. The molecular defenses against reactive oxygen species in yeast. Mol Microbiol. 1996;19:651–658. [Abstract] [Google Scholar]
- Moskvina E, Schüller C, Maurer CTC, Mager WH, Ruis H. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast. 1998;14:1041–1050. [Abstract] [Google Scholar]
- Neuman-Silberg FS, Bhattacharya S, Broach J. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol Cell Biol. 1995;15:3187–3196. [Europe PMC free article] [Abstract] [Google Scholar]
- Nierras CR, Warner JR. Protein kinase C enables the regulatory circuitry that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:13235–13241. [Abstract] [Google Scholar]
- Park J-I, Grant CM, Attfield PV, Dawes IW. The freeze-thaw stress response of the yeast Saccharomyces cerevisiae is growth phase specific and is controlled by nutritional state via the RAS-cyclic AMP signal transduction pathway. Appl Environ Microbiol. 1997;63:3818–3824. [Europe PMC free article] [Abstract] [Google Scholar]
- Parker CS, Topol J. A Drosophila RNA polymerase II transcription factor specific for the heat-shock gene binds to the regulatory site of an hsp70 gene. Cell. 1984;37:273–283. [Abstract] [Google Scholar]
- Piper P, Mahé Y, Thompson S, Pandjaitan R, Holyoak C, Egner R, Mühlbauer M, Coote P, Kuchler K. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998;17:4257–4265. [Europe PMC free article] [Abstract] [Google Scholar]
- Piper PW. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol Lett. 1995;134:121–127. [Abstract] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. J Biol Chem. 2000;275:8290–8300. [Abstract] [Google Scholar]
- Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. [Europe PMC free article] [Abstract] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–310. [Abstract] [Google Scholar]
- Saris N, Makarow M. Transient ER retention as stress response: conformational repair of heat-damaged proteins to secretion-competent structures. J Cell Sci. 1998;111:1575–1582. [Abstract] [Google Scholar]
- Schmitt AP, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;13:3556–3564. [Europe PMC free article] [Abstract] [Google Scholar]
- Sorger PK, Pelham HRB. Purification and characterization of a heat shock element binding protein from yeast. EMBO J. 1987;6:3035–3041. [Europe PMC free article] [Abstract] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen M, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. [Europe PMC free article] [Abstract] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. [Abstract] [Google Scholar]
- Treger JM, Schmitt AP, Simon JR, McEntee K. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharamyces cerevisiae. J Biol Chem. 1998;273:26875–26879. [Abstract] [Google Scholar]
- Warner J. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. [Abstract] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittman M, Pai C, Reibischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahacleo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmerman K, Philippsen P, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. [Abstract] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. [Abstract] [Google Scholar]
- Wu C. An exonuclease protection assay reveals heat-shock element and TATA-box binding proteins in crude nuclear extracts. Nature. 1985;317:84–87. [Abstract] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. [Abstract] [Google Scholar]
Articles from Molecular Biology of the Cell are provided here courtesy of American Society for Cell Biology
Full text links
Read article at publisher's site: https://doi.org/10.1091/mbc.12.2.323
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc30946?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Genomic analysis and mechanisms exploration of a stress tolerance and high-yield pullulan producing strain.
Front Genet, 15:1469600, 20 Sep 2024
Cited by: 0 articles | PMID: 39371418 | PMCID: PMC11449735
Calcineurin promotes adaptation to chronic stress through two distinct mechanisms.
Mol Biol Cell, 35(10):ar123, 31 Jul 2024
Cited by: 1 article | PMID: 39083354
Shared and redundant proteins coordinate signal cross-talk between MAPK pathways in yeast.
Mol Biol Cell, 35(10):ar126, 31 Jul 2024
Cited by: 0 articles | PMID: 39083355
Natural variation in yeast reveals multiple paths for acquiring higher stress resistance.
BMC Biol, 22(1):149, 04 Jul 2024
Cited by: 0 articles | PMID: 38965504 | PMCID: PMC11225312
Gene Expression Analysis of Yeast Strains with a Nonsense Mutation in the eRF3-Coding Gene Highlights Possible Mechanisms of Adaptation.
Int J Mol Sci, 25(12):6308, 07 Jun 2024
Cited by: 0 articles | PMID: 38928012
Go to all (828) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth.
Mol Microbiol, 35(2):397-406, 01 Jan 2000
Cited by: 56 articles | PMID: 10652100
[Alternative ways of stress regulation in cells of Saccharomyces cerevisiae: transcriptional activators Msn2 and Msn4].
Tsitologiia, 51(3):271-278, 01 Jan 2009
Cited by: 1 article | PMID: 19435282
Review
Genomic expression programs in the response of yeast cells to environmental changes.
Mol Biol Cell, 11(12):4241-4257, 01 Dec 2000
Cited by: 2887 articles | PMID: 11102521 | PMCID: PMC15070
The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress.
Mol Cell, 9(3):563-573, 01 Mar 2002
Cited by: 106 articles | PMID: 11931764