Abstract
Free full text

The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding
Abstract
Nucleosome remodelling complexes CHRAC and ACF contribute to chromatin dynamics by converting chemical energy into sliding of histone octamers on DNA. Their shared ATPase subunit ISWI binds DNA at the sites of entry into the nucleosome. A prevalent model assumes that DNA distortions catalysed by ISWI are converted into relocation of DNA relative to a histone octamer. HMGB1, one of the most abundant nuclear non-histone proteins, binds with preference to distorted DNA. We have now found that transient interaction of HMGB1 with nucleosomal linker DNA overlapping ISWI-binding sites enhances the ability of ACF to bind nucleosomal DNA and accelerates the sliding activity of limiting concentrations of remodelling factor. By contrast, an HMGB1 mutant with increased binding affinity was inhibitory. These observations are consistent with a role for HMGB1 as a DNA chaperone facilitating the rate-limiting DNA distortion during nucleosome remodelling.
Introduction
Chromatin is intrinsically involved in the regulation of all nuclear processes, most prominently transcription, replication, repair and recombination (Flanagan and Peterson, 1999; Muchardt and Yaniv, 1999; Kwon et al., 2000; de La Serna et al., 2001; Fyodorov and Kadonaga, 2001). DNA and histones within nucleosomes are held together by a large number of weak interactions, which render the nucleosome a rather stable particle (Luger et al., 1997). In vitro, these interactions can be overcome by non-physiological conditions, such as elevated temperature or ionic strength, which induce the sliding of histone octamers on DNA (Beard, 1978; Pennings et al., 1991; Guschin and Wolffe, 1999; Widom, 1999). In a more physiological environment, nucleosomes can be moved by enzymes called nucleosome remodelling factors. These multi-subunit complexes contain dedicated ATPases that are able to convert the energy stored in ATP into displacement of nucleosomal DNA relative to the histone octamer (Workman and Kingston, 1998; Aalfs and Kingston, 2000; Peterson, 2000; Narlikar et al., 2002). Members of the three most prominent subfamilies of remodelling ATPases, represented by yeast SWI2/SNF2, Drosophila and Xenopus Mi-2 and Drosophila ISWI, are all able to alter histone–DNA interactions such that nucleosomes can move (for reviews, see Vignali et al., 2000; Längst and Becker, 2001b; Becker and Hörz, 2002; Peterson, 2002a).
Nucleosome sliding presumably involves the detachment of a DNA segment from the histone octamer surface, possibly as a local loop or bulge, or as otherwise distorted DNA, at sites where the DNA enters its path around the particle (Becker and Hörz, 2002). Propagation of this detached segment over the surface of the histone octamer would lead to displacement of nucleosomal DNA with respect to hallmarks of the histone octamer surface. Models of this kind are attractive because they explain how a nucleosome can be mobilized even though, at any given time during the process, only a limited number of histone–DNA contacts are disrupted. Nucleosome remod elling enzymes may catalyse the initial detachment of a DNA segment, possibly by some kind of distortion, and may endow its ‘propagation’ through the nucleosome with directionality.
The ATPase ISWI serves as one paradigm for analysis of the mechanism of nucleosome remodelling (Längst et al., 1999; Längst and Becker, 2001a). ISWI can interact with linker DNA next to a nucleosome and induce the movement of the histone octamer towards it (effectively ‘pushing’ DNA over the octamer surface; Längst and Becker, 2001a). Its ability to modify DNA structure is nicely illustrated by the fact that it is able to introduce superhelical torsion into linear DNA if nucleosomes are present (Havas et al., 2000). We recently found that ISWI can facilitate nucleosome sliding even on DNA containing single-stranded DNA breaks (‘nicks’), which indicates that if nucleosome mobility involves propagation of a segment of detached DNA, the integrity of the phosphodiester backbone is not required for it (Längst and Becker, 2001a). Interestingly, we observed that nicks at strategic positions in the DNA, at the nucleosomal edge where ISWI binds, facilitate nucleosome sliding. We speculated that nicking at this position might increase the flexibility of DNA and might therefore facilitate the kind of DNA distortion necessary for initiating the nucleosome sliding process (Längst and Becker, 2001a).
The current manuscript elaborates on this idea. If the rate-limiting step in nucleosome sliding is the distortion of linker DNA, e.g. the formation of a small local loop or bulge, proteins that can generate and/or stabilize such distortions might facilitate nucleosome sliding. HMGB1 is the archetype of proteins that bend and distort DNA, and, because it has been shown to interact with linker DNA at sites overlapping ISWI-binding sites, is a good candidate to help ‘lubricate’ nucleosome sliding.
HMGB1 is present in all mammalian nuclei at high concentration: each nucleus contains more than one million HMGB1 molecules, statistically about one molecule per every 10–20 nucleosomes. HMGB1 binds to the minor groove of the DNA, causing a local bending and untwisting of the double helix; on the other side, it has binding preference for altered DNA structures, like bends, bulges and four-way junctions (for reviews, see Bianchi and Beltrame, 1998; Travers, 2000; Thomas and Travers, 2001). Although HMGB1 binds naked DNA with low affinity and no sequence specificity, it interacts structure specifically with DNA at the entry into a nucleosome (Schroter and Bode, 1982; Nightingale et al., 1996; An et al., 1998). Although HMGB1 and histone H1 both interact with nucleosomal DNA, their mode of binding is very different (Thomas and Travers, 2001). Nevertheless, HMG-D, the Drosophila HMGB1 counterpart, is able to replace histone H1 from nucleosomal DNA in vitro (Ner et al., 2001), although the reverse replacement is seen during the course of Drosophila embryonic development (Ner and Travers, 1994). The observation of relatively unfolded chromatin domains characterized by depletion of H1 and enrichment of HMGB1/2 to levels stoichiometric with nucleosomes (Jackson et al., 1979) supports the idea that HMGB1 might substitute for H1 under certain circumstances in vivo, leading to drastically different chromatin structure. The interaction of HMGB1 with chromatin in vivo is highly dynamic: the protein has a very high rate of diffusion in the nucleus, which indicates that the binding to sites in vivo is very transient (Scaffadi et al., 2002).
In this work, we show that HMGB1 is indeed able to enhance the activity of native CHRAC (Varga-Weisz, 1995) and recombinant ACF, a nucleosome remodelling factor consisting of ISWI and ACF1 (Ito et al., 1999; Eberharter et al., 2001), apparently by increasing the affinity of ACF for its nucleosomal substrate. The results are compatible with a model in which HMGB1 distorts DNA at its entry into the nucleosome and interacts with it in a highly dynamic manner thus effectively ‘presenting’ the DNA bend to ACF. Our results are particularly relevant in light of the recent discussion about the mechanisms governing nucleosome sliding (Havas et al., 2000; Längst and Becker, 2001b; Narlikar et al., 2001; Becker and Hörz, 2002; Peterson, 2002b). They are consistent with models assuming that DNA distortions are rate-limiting steps of nucleosome sliding, and identify HMGB1 as a potential regulator of ATP-dependent nucleosome remodelling processes.
Results
Interaction of HMGB1 with mononucleosomes
Several proteins, among them histone H1 and HMGB1, that are able to interact with bent or otherwise distorted DNA (Lilley, 1992; Varga-Weisz et al., 1994) are also known to bind DNA at the entry points into the nucleosome. Several investigators have already documented the interaction of HMGB1 with nucleosomes (Schroter and Bode, 1982; Nightingale et al., 1996; An et al., 1998). Recombinant HMGB1 also interacts efficiently with the mononucleosomes that are used as substrates in nucleosome sliding assays (Figure 1). Electrophoretic mobility shift assays (EMSA) document the binding of HMGB1 to a nucleosome assembled centrally on a 248 bp DNA fragment derived from the ribosomal RNA promoter. The titration of HMGB1 yields a single retarded band, confirming the existence of one preferred binding site (Figure 1A, lanes 4–6). HMGB1 also supershifts a nucleosome positioned at the end of the same DNA fragment (Figure 1A, lanes 10–12). This interaction does not lead to disruption of the nucleosome: excess unlabelled DNA added to an EMSA reaction before gel loading effectively competes HMGB1 off the labelled nucleosome, which is released intact (Figure 1B, lanes 6–9).
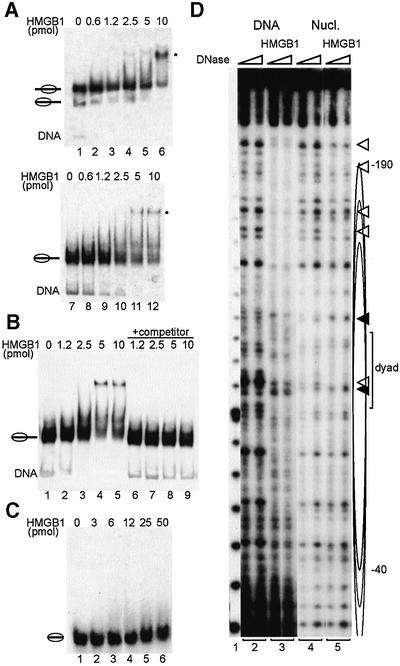
Fig. 1. Specific interaction of HMGB1 with nucleosomes exhibiting protruding DNA. (A) Increasing amounts of full-length HMGB1 (as indicated) were incubated with nucleosomes (60 fmol) positioned at the centre (lanes 1–6) or at the end (lanes 7–12) of the 248 bp rDNA fragment. The complexes were resolved by electrophoresis on a native polyacrylamide gel. An autoradiograph of the gel is shown. The major nucleosome–HMGB1 complex is marked by an asterisk. Nucleosomes are schematized by ellipses with protruding DNA. (B) HMGB1 binding to the nucleosome does not disrupt the histone octamer. HMGB1 binding was analysed as described in (A) without (lanes 2–5) and with the addition of competitor DNA (lanes 6–9) prior to gel loading. (C) Nucleosomes (60 fmol) assembled on a 146 bp DNA fragment were incubated with increasing amounts of HMGB1 as indicated and analysed by electromobility shift assay as above. (D) DNA (lanes 2 and 3) or purified nucleosomes (lanes 4 and 5) were incubated with HMGB1. DNase I-treated free DNA, nucleosomes and nucleosome–HMGB1 complexes were resolved by PAGE, the DNA isolated from the corresponding bands and analysed on sequencing gels. The cluster of central nucleosome positions is indicated by ellipses, and the area of perturbation of histone–DNA interaction upon HMGB1 binding is marked by white (DNase protection) or black triangles (DNase hypersensitivity). A 10 bp DNA ladder was used as size marker (lane 1).
In agreement with earlier results (Schroter and Bode, 1982; Nightingale et al., 1996; An et al., 1998), stable interaction of HMGB1 requires the presence of some linker DNA protruding from the nucleosomal core. It is unable to associate with a nucleosome core particle, consisting of 146 bp DNA wrapped tightly around a histone octamer, even at very high concentrations (Figure 1C). Thus, the interaction of HMGB1 with nucleosomes is reminiscent of the properties of the remodelling ATPase ISWI: ISWI cannot bind to a nucleosome core, but requires linker DNA on at least one side of the nucleosome particle (Brehm et al., 2000; Längst and Becker, 2001a).
We attempted to document the preferred interaction of HMGB1 by footprinting (Figure 1D). Centrally positioned nucleosomes were isolated, incubated with HMGB1 and partially digested with DNase I. HMGB1–nucleosome complexes were identified on native polyacrylamide gels, excised and the purified DNA was analysed on sequencing gels. While HMGB1 protects free DNA non-specifically from DNase I digestion (Figure 1D, lane 3), the protection of nucleosomal DNA was modulated (Figure 1D, lane 5). Since the ‘central nucleosome’ actually corresponds to a series of nucleosomes with different translation positions between position –20 and –190 on the DNA, the footprint naturally lacks some precision. However, obvious protection of DNase I digestion on the nucleosomal fragment is seen in the area corresponding to one end of the nucleosomes (the –190 position), but not at the other end (the –40 position). This interaction is reminiscent of earlier studies involving model nucleosomes, which described asymmetrical interactions (Nightingale et al., 1996; An et al., 1998). Enhancements of DNase I cleavage are also seen close to the nucleosomal pseudodyad, which suggest a close interaction of HMGB1 with the nucleosome rather than a detached interaction with linker DNA.
The fact that HMGB1 and ISWI approach nucleosomes with overlapping binding specificity raises questions about the consequences of the presence of HMGB1 in ISWI-induced nucleosome sliding.
HMGB1 affects the sliding activity of CHRAC and ACF
In order to test for an effect of HMGB1 on ISWI- and ACF-induced nucleosome mobility, we employed an established assay that allows visualization of single nucleosome movement, catalysed by ATP-dependent nucleosome-remodelling factors (Längst et al., 1999; Brehm et al., 2000; Clapier et al., 2001; Eberharter et al., 2001). The wrapping of short DNA fragments around histone octamers frequently leads to multiple translational positions that can be separated electrophoretically. In our case, nucleosomes occupy prominent positions on a 248 bp fragment derived from the rDNA promoter: a series of internal positions, which are not separated by the gel system, are dictated by the anisotropic flexibility of the DNA sequence, and in addition, nucleosomes can abut the fragment ends. Isolated nucleosomes positioned either at the centre or at the periphery of the DNA fragment serve as a substrate for remodelling machines. We previously showed that the remodelling ATPase ISWI is able to move nucleosomes from internal positions to fragment ends (Längst et al., 1999; Längst and Becker, 2001a). Association of ACF1 with ISWI reconstitutes the heteromeric ACF (Ito et al., 1999). The presence of two small histone fold subunits distinguishes CHRAC from ACF (Corona et al., 2000). ACF1 not only increases the efficiency of ISWI-dependent remodelling by an order of magnitude, it also leads to a qualitative change in nucleosome sliding: in contrast to isolated ISWI, ACF and CHRAC can catalyse the movement of nucleosomes from fragment ends to more central positions (Längst et al., 1999; Eberharter et al., 2001). While these simple model reactions on short linear DNA fragments are unlikely to reflect all physiological constraints of nucleosome movements in nuclei, they provide an excellent opportunity to study fundamental parameters that affect nucleosome mobility.
The first evidence for an effect of HMGB1 on nucleosome sliding was obtained in reactions catalysed by native CHRAC (Figure 2A). Stopping the reactions at different time points with competitor DNA, followed by native gel electrophoresis, measured the kinetics of nucleosome sliding as a function of fixed amounts of HMGB1. We found that nucleosomes moved significantly faster in the presence of HMGB1 (Figure 2A, compare lanes 2 and 3 with 6 and 7), but the overall level of nucleosome mobilization at the end of an extended incubation did not change (Figure 2A, compare lane 5 with 9). We next explored whether HMGB1 would also affect the kinetics of nucleosome sliding by the recombinant two-subunit ACF (Eberharter et al., 2001). In the presence of 2 pmol of HMGB1, a clear increase of nucleosome sliding, particularly at the early time points, was detectable (Figure 2B, lanes 2–4 and 8–10). A similar stimulation was observed when the HMGB1 concentrations were increased 5-fold, but raising the concentrations of remodelling factor abolished the effect (data not shown). In order to better document the kinetic effect of HMGB1 on nucleosome sliding, we quantified the degree of nucleosome sliding throughout the sliding reaction and determined the degree of stimulation by HMGB1 for each time point. The graph in Figure 2C displays the average data derived from four independent experiments. It documents a kinetic effect of HMGB1 on nucleosome sliding, which is mainly observable at the early times.

Fig. 2. HMGB1 stimulates CHRAC- and ACF-mediated nucleosome remodelling. (A) Nucleosomes positioned at the end of the DNA fragment (lane 1) were incubated with 5 fmol of native CHRAC and ATP, in the absence (lanes 2–5) or presence of 2 pmol of HMGB1 (lanes 6–9). The reactions were incubated at 16°C and stopped at the indicated time points by the addition of competitor DNA. Nucleosome positions were subsequently analysed by electrophoresis. Nucleosomes are schematized by ellipses with protruding DNA. (B) Nucleosome remodelling assay, as described in (A), using 5 fmol of recombinant ACF in the absence (lanes 1–6) or presence of 2 pmol of HMGB1 (lanes 7–12). (C) Quantification of the HMGB1 effect on nucleosome remodelling. The ratio of nucleosomes moved in the presence of HMGB1 versus nucleosomes moved in the absence of HMGB1 was calculated for four independent experiments and displayed in a graph. (D) Nucleosomes positioned at the centre of the DNA fragment were incubated with 30 fmol of ISWI and ATP, in the absence (lanes 1–5) or presence of 2 pmol of HMGB1 (lanes 6–10) and assayed as described in (A).
In order to establish whether ISWI by itself would also profit from the presence of HMGB1, we added the factor to reactions in which ISWI mobilized a centrally positioned histone octamer to slide to a fragment end (Figure 2D). Surprisingly, ≥2 pmol amounts (data not shown) of HMGB1 did not affect the kinetics of ISWI-dependent nucleosome sliding, suggesting a critical contribution of the ACF1 moiety for the functional synergism with HMGB1.
HMGB1 promotes ACF binding to the nucleosome
The stimulatory effect could be due to interactions of HMGB1 with ACF, with the nucleosomal linker DNA or both. We have used mouse HMGB1 in the context of a nucleosome remodelling machinery from Drosophila to minimize the probability of direct interactions, and, accordingly, we were so far unable to detect any significant interactions between HMGB1 and ACF in biochemical pull-down experiments (data not shown). Therefore we next analysed the effect of HMGB1 on the interaction of the remodelling factors with the nucleosomes. Binding of ACF to the nucleosome can be visualized by EMSA as a single prominent band if no competitor DNA is added prior to gel loading (Figure 3A, lanes 4–6). As before, HMGB1 shifts the nucleosome into a position close to, but distinct from, the ACF–nucleosome complex (Figure 3A, lanes 13 and 14). Importantly, the presence of 0.6 pmol of HMGB1 does not shift the nucleosomal band significantly (Figure 3A, lane 10). However, if this amount of HMGB1 was included in a nucleosome-binding reaction with ACF, 4-fold lower concentrations of ACF were required to shift the nucleosome (Figure 3A, compare lanes 1–6 with 15–20). At higher, saturating concentrations of ACF, the degree of nucleosome binding was unaffected by HMGB1, in close correspondence with the nucleosome sliding profiles. Under these conditions, no evidence for the existence of a ternary complex of ACF, HMGB1 and the nucleosome was detected using specific antibodies in ‘supershift’ strategies (data not shown). Thus, HMGB1 may function as a DNA chaperone that facilitates ACF binding to the nucleosome at suboptimal conditions without being a stable component of the complex.
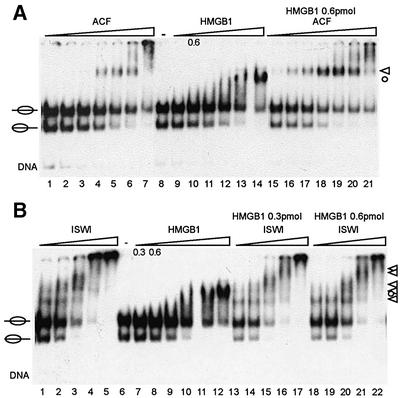
Fig. 3. HMGB1 promotes the formation of a ACF–nucleosome complex. (A) Nucleosomes preferentially positioned at the centre of the DNA fragment were incubated with increasing amounts of ACF (lanes 1–7; 15, 30, 60, 120, 240, 480 and 960 fmol), HMGB1 (lanes 9–14; 0.3, 0.6, 1.2, 2.4, 4.8 and 9.6 pmol) and increasing amounts of ACF in the presence of 0.6 pmol HMGB1 (lanes 15–21). Complexes were analysed in EMSAs, as before. Nucleosome–ACF complexes are marked by a triangle and nucleosome–HMGB1 complexes are indicated by a circle. (B) Positioned nucleosomes were incubated with increasing amounts of ISWI (lanes 1–5; 60, 120, 240, 480 and 960 fmol), HMGB1 (lanes 7–12, 0.3, 0.6, 1.2, 2.4, 4.8 and 9.6 pmol) and ISWI with fixed amounts of HMGB1 (lanes 13–17, 0.3 pmol HMGB1; lanes 18–22, 0.6 pmol HMGB1). Nucleosome– ISWI complexes are marked by triangles and the nucleosome– HMGB1 complex is indicated by a circle.
A similar experiment, in which we assayed for an effect of HMGB1 on the ISWI-nucleosome interaction, revealed no stimulation (Figure 3B). We conclude that the potential of HMGB1 to facilitate nucleosome sliding correlates with its ability to lead to enhanced interaction of the remodelling machine with the nucleosomal substrate. The data identify the ACF1 moiety of ACF as crucial for this effect.
An HMGB1 mutant that binds nucleosomes with high affinity inhibits nucleosome sliding
Previously, several investigators observed that the C-terminal acidic tail of HMGB1 controls the DNA-binding properties of the HMG boxes (Stros et al., 1994; Lee and Thomas, 2000; Muller et al., 2001). In agreement with these studies, we also observed that deletion of the C-terminal domain increased the activity of the HMG boxes in protein–protein interaction and transactivation assays (T.Bonaldi and M.E.Bianchi, unpublished observations). Therefore, we wished to test a C-terminally deleted HMGB1 (HMGB1ΔC) in nucleosome sliding assays. Considering the absolute amounts of input protein, HMGB1ΔC interacted ~100-fold better with free DNA than full-length HMGB1 (Figure 4A). Similarly, HMGB1ΔC interacted two orders of magnitude better than HMGB1 with either peripheral or central nucleosomes (Figure 4B, compare with Figure 1A). Higher concentrations of HMGB1ΔC yielded several bands, which, in light of the pattern obtained by interaction of HMGB1ΔC to free DNA (Figure 4A), are due, presumably, to interaction of several molecules with the free DNA next to the nucleosome.
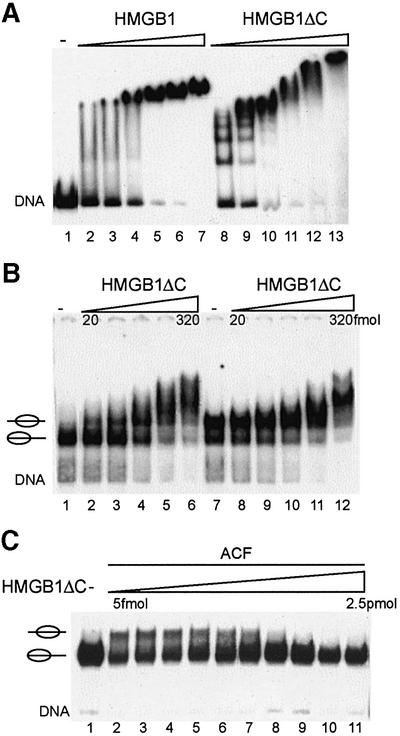
Fig. 4. HMGB1ΔC inhibits ACF-mediated nucleosome remodelling. (A) Comparison of HMGB1 and HMGB1ΔC binding to the 248bp rDNA fragment. The DNA fragment was incubated with increasing amounts of HMGB1 (lanes 2–7; 1.5, 2.3, 3.4, 5, 7.6 and 11 pmol) and HMGB1ΔC (lanes 8–13; 50, 75, 112, 169, 253 and 380 fmol). HMG–DNA complexes were separated on native polyacrylamide gels. (B) HMGB1ΔC interacts with nucleosomes positioned at the border or the centre of the rDNA fragment. Positioned nucleosomes were incubated with increasing amounts of HMGB1ΔC (20, 40, 80, 160 and 320 fmol). (C) ACF-mediated nucleosome remodelling is inhibited by the addition of HMGB1ΔC. Nucleosomes positioned at the border of the DNA fragment were incubated with ACF, ATP and increasing amounts of HMGB1ΔC (lanes 2–11; 5, 10, 20, 40, 80, 160, 320, 640, 1240 and 2560 fmol). Remodelling reactions were stopped by the addition of competitor DNA and nucleosome positions were analysed on a 4.5% polyacrylamide gel.
Surprisingly, when HMGB1ΔC was titrated into the nucleosome mobility assay, it inhibited the nucleosome sliding catalysed by a fixed amount of ACF (Figure 4C) rather than facilitating it. Nucleosome movement decreased as a function of HMGB1ΔC, and complete inhibition was achieved upon addition of 80–160 fmol. This amount of HMGB1ΔC is similar to the amount required to yield the first complex with the nucleosome in the EMSA, which presumably reflects the interaction of a single HMGB1ΔC molecule with its preferred binding site (Figure 4B, lane 4). This result suggests that the tight binding of HMGB1ΔC with the nucleosome prevents efficient nucleosome mobilization by ACF.
Discussion
Nucleosome remodelling complexes containing the ATPase ISWI are likely to contribute significantly to the flexibility of chromatin, enabling dynamic structural transitions throughout the lifetime of a cell. Among the Drosophila ISWI complexes, only the nucleosome remodelling factor (NURF) seems to be involved in targeted transcriptional regulation (Mizuguchi et al., 1997, 2001; Xiao et al., 2001). By contrast, the features of the related ACF and CHRAC complexes suggest more global roles in the assembly and maintenance of dynamic chromatin (Längst and Becker, 2001b). The experiments described above indicate that HMGB1, one of the most abundant non-histone proteins in nuclei, is able to increase the efficiency of ACF-dependent nucleosome sliding in a defined model system. This observation may point to an important, novel function of HMGB1, but at the same time reveals unexpected insight into the mechanism underlying catalysed nucleosome sliding.
HMGB1, a hallmark of dynamic chromatin
HMGB1 diffuses throughout the nucleus with the highest rates, indicating a rather dynamic association with chromatin (Scaffadi et al., 2002). Since HMGB1 binds nucleosomes at sites overlapping those recognized by the linker histone H1, HMGB1-like proteins can compete with histone H1 for nucleosomal-binding sites (Nightingale et al., 1996; Ner et al., 2001). Association of histone H1 prevents spontaneous nucleosome sliding at elevated salt and temperature (Pennings et al., 1994) and inhibits the remodelling action of the SWI/SNF complex on mononucleosome substrates (Hill and Imbalzano, 2000) and nucleosomal arrays (Horn et al., 2002). Whether histone H1 affects nucleosome sliding or other ATP-dependent functions of ISWI has not been determined in a defined system yet (but see Varga-Weisz et al., 1995). Given that the interaction of H1 with linker DNA (Widom, 1998) is likely to interfere with ISWI binding, some kind of inhibition is expected. Therefore, HMGB1 may facilitate nucleosome remodelling in two ways: (i) by ‘unlocking’ the nucleosome through displacing linker histones, and (ii) by stimulating the activity of ACF/CHRAC as shown here and discussed below.
Early experiments indicated that a fraction of chromatin exists containing reduced levels of histone H1 and corresponding enrichment of HMGB1/2 to stoichiometric levels (with respect to nucleosomes) (Jackson et al., 1979). Our findings suggest that chromatin of this type should be characterized by high levels of nucleosome mobility. A further example for chromatin with unusual dynamic properties is found in the preblastula nuclei of Drosophila embryos, which lack histone H1 but contain large amounts of the Drosophila HMGB1-like protein, HMG-D, which also interacts with the nucleosomal linker DNA (Ner et al., 2001). We imagine the resulting chromatin to be extraordinarily dynamic, consistent with the high replication and chromatin assembly rates observed before cellularization (Ner and Travers, 1994).
HMGB1 and nucleosome sliding: mechanistic considerations
Prevalent models assume that the critical step in nucleosome mobilization is the distortion of a DNA segment at the edge of the nucleosome by the remodelling ATPase, and the introduction of a segment of ‘bulged’ DNA on the surface of the histone octamer. Propagation of this ‘bulge’ over the histone surface eventually would lead to displacement of the DNA relative to hallmarks on the octamer surface (Havas et al., 2000; Längst and Becker, 2001b; Narlikar et al., 2001; for review, see Becker and Hörz, 2002). In the context of this model, HMGB1 could, in principle, facilitate at least two steps: the initial distortion of DNA or the propagation of the distortion over the nucleosomal surface (Figure 5). Several arguments lead us to favour the first mechanism. (i) The band-shift data shown in Figure 3 demonstrate an improved binding of ACF to nucleosomal DNA in the presence of HMGB1. The magnitude of this effect correlates with the kinetic acceleration of nucleosome sliding. (ii) Conceivably, the initial distortion of a relatively short segment of DNA must be rate-limiting the overall reaction, while the propagation of the ‘bulge’, once formed, is presumed to require relatively little further energy input (Längst and Becker, 2001a; Becker and Hörz, 2002). The ‘HMG-effect’ is particularly prominent at early times during the course of the experiment, pointing to an effect on initiation. (iii) Consistent with these ideas, we recently observed that single-stranded nicks in DNA close to the entry into the nucleosome—the site of ISWI interaction—facilitated ATP-dependent nucleosome sliding (Längst and Becker, 2001a). We hypothesized that the presence of these nicks might enhance the flexibility of DNA at this site, such that it could be more easily bent or otherwise distorted. (iv) HMGB1 affected nucleosome sliding by ACF and CHRAC, but not by recombinant ISWI alone. Previous results already indicated profound differences in the initiation of nucleosome mobilization by these factors (Eberharter et al., 2001). Apparently, ACF and ISWI approach the nucleosomal substrate somehow differently, since ISWI only triggers the sliding of nucleosomes to fragment ends whereas ACF is able to move nucleosomes from ends to more central positions on DNA. At the same time, ACF mobilizes nucleosomes by an order of magnitude more efficiently than ISWI alone, which may be due to the ability of ACF1 to contribute to substrate recognition.

Fig. 5. Model showing the stimulatory effect of HMGB1 on ACF-mediated nucleosome remodelling. The wrapping of DNA (black line) around a histone octamer (grey sphere) is shown sideways. See text for details.
In summary, we hypothesize that HMGB1 acts as a DNA chaperone (Crothers, 1993; Travers et al., 1994) to flexibilize or distort DNA (or to stabilize an intrinsic bend) at the nucleosomal edge (Figure 5). This flexibilization and/or distortion might then enhance the ability of ACF to interact with this site. Since HMGB1 does not form ternary complexes with ACF and the nucleosome under these conditions, we speculate that ACF1 might enable the remodelling factor to recognize DNA structure within the nucleosomal linker and hence sensitize it for pre-bent DNA. Remarkably, ACF1-like molecules appear to be obligatory subunits of ISWI-containing remodelling ATPases (Längst and Becker, 2001b; Xiao et al., 2001), and therefore seem to be tightly involved in the remodelling process itself. Although a direct allosteric effect on ACF cannot be excluded in light of the fact that HMGB1 is able to interact with several regulators of chromatin structure (Dintilhac and Bernues, 2001), this appears not to be the basis of our current findings since we have been unable to detect a direct interaction of HMGB1 with ISWI and ACF in the absence of nucleosomal DNA.
The observation that catalytic concentrations of HMGB1 accelerate the rate of nucleosome sliding, but not a mutant HMGB1 with 100-fold increased DNA binding, highlights the requirement for a dynamic association of HMGB1 with nucleosomes. Due to its higher affinity for the nucleosomal linker DNA, HMGB1ΔC might resist displacement by ACF and hence continue to ‘lock’ the nucleosome in analogy to histone H1. In agreement with previous findings (Lee and Thomas, 2000; Muller et al., 2001), the acidic C-terminus of HMGB1 is responsible for its dynamic interaction with nucleosomal DNA. Differential calorimetry experiments, both in the presence (Muller et al., 2001) or the absence of DNA (Ramstein et al., 1999), indicated that the acidic tail binds to one of the HMG boxes of HMGB1 (most likely box A). By dynamic competition with DNA this intramolecular binding might render the HMG box–DNA interaction reversible and transient. The DNA bulge released from box A could then be recognized by ACF.
Widespread roles for HMGB1-like DNA chaperones in facilitating protein–DNA interactions
Our conclusion on a potential role of HMGB1 to promote ACF–nucleosome interactions through DNA distortion extends a wealth of prior relevant observations on the function of HMG box proteins. HMGB1/2 molecules are known to facilitate the DNA binding of various regulatory factors, such as HOX proteins, steroid hormone receptors, p53 and the site-specific recombinase RAG1 (Boonyaratanakornkit et al., 1998; Aidinis et al., 1999; Mitsouras et al., 2002; for a review of the older literature, see Bianchi and Beltrame, 1998). In yeast, the HMGB1-related Nhp6 proteins facilitate the interaction of the heterodimeric Spt16/Pob3 proteins with nucleosomes (Brewster et al., 2001; Formosa et al., 2001). Together, these three latter factors are functionally homologous to the mammalian FACT complex, which remodels chrom atin to facilitate transcription elongation (Orphanides et al., 1999). The BAP111 subunit of the Drosophila BRM complex, and the SSRP1 subunit of human FACT, which are important for nucleosome remodelling during transcription initiation and elongation, respectively, contain HMG boxes (Orphanides et al., 1999; Papoulas et al., 2001). The HMG domain of the BAF57 subunit of the SWI/SNF-like complexes in mice appears to be required for nucleosome remodelling in vivo (Chi et al., 2002). HMG domains may thus be more generally involved in the recognition and modification of a nucleosomal substrate by nucleosome remodelling factors.
Materials and methods
Nucleic acids
Mononucleosomes were reconstituted on a 248 bp murine ribosomal DNA fragment representing promoter sequences between –232 and +16 relative to the transcription start site (+1; Längst et al., 1999). This DNA fragment was synthesized by PCR and body labelled by inclusion of [α32P]dCTP in the PCR reaction.
Proteins
Histones were extracted from Drosophila embryos and purified by chromatography on a hydroxylapatite column as described (Simon and Felsenfeld, 1979). ISWI, ACF and CHRAC were prepared according to Eberharter et al. (2001).
HMGB1 and HMGB1ΔC protein expression and purification was described in Muller et al. (2001); HMGB1ΔC was identified there as the HMGB1 AB truncation.
Nucleosome assembly and purification
Nucleosomes were assembled using purified histones and polyglutamic acid (PGA; Sigma P4886) according to Stein et al. (1979). Histones and PGA were mixed in a 2:1 ratio in 0.15 M NaCl and incubated for 1 h at room temperature. Precipitates were removed by centrifugation at room temperature (RT) for 4 min at 11 kr.p.m. (Eppendorf, 5415R), the supernatant (HP-mix) was stored at –20°C. Different ratios of DNA and HP-mix were incubated for 3 h at 37°C and analysed by electromobility shift to reveal optimal conditions for nucleosome assembly. Translational positions of the nucleosomes were separated by electrophoresis on 4.5% polyacrylamide gels in 0.4× TBE and isolated as described previously (Längst et al., 1999).
Nucleosome mobility assay
Sixty femtomoles of nucleosomes with defined translational positions were incubated with ISWI (30 fmol), ACF (5 fmol), CHRAC (5 fmol), HMGB1 and HMGB1ΔC for 0–90 min at 16°C. Reaction mixes contained EX70 buffer (70 mM KCl, 10 mM Tris, 1 mM EDTA, 1.5 mM MgCl2 and 10% glycerol) supplemented with 1 mM ATP, 200 ng/µl chicken egg albumin (CEA; Sigma) and 1 mM dithiotheritol (DTT) in a final volume of 10 µl. Reactions were stopped by the addition of 300 ng competitor DNA and separated on 4.5% polyacrylamide gels in 0.4× TBE. Gels were scanned with a phosphoimager (Fuji) and the individual bands were quantified with Aida software.
Electromobility shift assays
Sixty femtomoles of purified nucleosomes were incubated with increasing amounts of ISWI, ACF, HMGB1 and HMGB1ΔC in EX80 buffer containing 200 ng/µl CEA and 1 mM DTT. Reactions were incubated for 10 min at RT and then separated on 4.5% polyacrylamide gels containing 0.4× TBE. ACF-/HMGB1–nucleosome complexes were supershifted with the specific HMGB1 (Pharmingen) and anti-Flag (Sigma) antibodies.
Nucleosome footprinting
Purified nucleosomes localized at the centre of the 248 bp rDNA fragment, labelled at position +16, were used for footprinting assays (Längst and Becker, 2001a). Optimal HMGB1–nucleosome ratios were estimated in ESMAs. The electromobility shift reactions were scaled up 50-fold and treated with increasing amounts of DNase I. Reactions were stopped by the addition of EDTA at a final concentration of 5 mM and directly loaded on preparative electromobility shift gels. DNase I-treated nucleosomes and HMGB1–nucleosome complexes were isolated from the gel, treated with proteinase K and analysed on 6% sequencing gels.
Acknowledgements
This work was supported by Network Grant ERBFMRXCT98–191 from the European Commission, by a grant to M.E.B. from Associazione Italiana Ricerca sul Cancro and through SFB 594, funded by Deutsche Forschungsgemeinschaft.
References
- Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [Abstract] [Google Scholar]
- Aidinis V., Bonaldi,T., Beltrame,M., Santagata,S., Bianchi,M.E. and Spanopoulou,E. (1999) The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol., 19, 6532–6542. [Europe PMC free article] [Abstract] [Google Scholar]
- An W., van Holde,K. and Zlatanova,J. (1998) The non-histone chromatin protein HMG1 protects linker DNA on the side opposite to that protected by linker histones. J. Biol. Chem., 273, 26289–26291. [Abstract] [Google Scholar]
- Beard P. (1978) Mobility of histones on the chromosome of simian virus 40. Cell, 15, 955–967. [Abstract] [Google Scholar]
- Becker P.B. and Hörz,W. (2002) ATP-dependent nucleosome remodelling. Annu. Rev. Biochem., 71, 247–273. [Abstract] [Google Scholar]
- Bianchi M.E. and Beltrame,M. (1998) Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet., 63, 1573–1577. [Europe PMC free article] [Abstract] [Google Scholar]
- Boonyaratanakornkit V. et al. (1998) High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol., 18, 4471–4487. [Europe PMC free article] [Abstract] [Google Scholar]
- Brehm A., Längst,G., Kehle,J., Clapier,C.R., Imhof,A., Eberharter,A., Muller,J. and Becker,P.B. (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J., 19, 4332–4341. [Europe PMC free article] [Abstract] [Google Scholar]
- Brewster N.K., Johnston,G.C. and Singer,R.A. (2001) A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol., 21, 3491–3502. [Abstract] [Google Scholar]
- Chi T.H., Wan,M., Zhao,K., Taniuchi,I., Chen,L., Littman,D.R. and Crabtree,G.R. (2002) Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature, 418, 195–199. [Abstract] [Google Scholar]
- Clapier C.R., Lan gst,G., Corona,D.F., Becker,P.B. and Nightingale,K.P. (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol., 21, 875–883. [Europe PMC free article] [Abstract] [Google Scholar]
- Corona D.F., Eberharter,A., Budde,A., Deuring,R., Ferrari,S., Varga-Weisz,P., Wilm,M., Tamkun,J. and Becker,P.B. (2000) Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC). EMBO J., 19, 3049–3059. [Europe PMC free article] [Abstract] [Google Scholar]
- Crothers D.M. (1993) Architectural elements in nucleoprotein complexes. Curr. Biol., 3, 675. [Abstract] [Google Scholar]
- de La Serna I.L., Carlson,K.A. and Imbalzano,A.N. (2001) Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet., 27, 187–190. [Abstract] [Google Scholar]
- Dintilhac A. and Bernues,J. (2001) HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J. Biol. Chem., 277, 7021–7028. [Abstract] [Google Scholar]
- Eberharter A., Ferrari,S., Längst,G., Straub,T., Imhof,A., Varga-Weisz,P., Wilm,M. and Becker,P.B. (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J., 20, 3781–3788. [Europe PMC free article] [Abstract] [Google Scholar]
- Flanagan J.F. and Peterson,C.L. (1999) A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res., 27, 2022–2028. [Europe PMC free article] [Abstract] [Google Scholar]
- Formosa T., Eriksson,P., Wittmeyer,J., Ginn,J., Yu,Y. and Stillman,D.J. (2001) Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J., 20, 3506–3517. [Europe PMC free article] [Abstract] [Google Scholar]
- Fyodorov D.V. and Kadonaga,J.T. (2001) The many faces of chromatin remodeling: SWItching beyond transcription. Cell, 106, 523–525. [Abstract] [Google Scholar]
- Guschin D. and Wolffe,A.P. (1999) SWItched-on mobility. Curr. Biol., 9, R742–R746. [Abstract] [Google Scholar]
- Havas K., Flaus,A., Phelan,M., Kingston,R., Wade,P.A., Lilley,D.M. and Owen-Hughes,T. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133–1142. [Abstract] [Google Scholar]
- Hill D.A. and Imbalzano,A.N. (2000) Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry, 39, 11649–11656. [Abstract] [Google Scholar]
- Horn P.J., Carruthers,L.M., Logie,C., Hill,D.A., Solomon,M.J., Wade,P.A., Imbalzano,A.N., Hansen,J.C. and Peterson,C.L. (2002) Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol., 9, 263–267. [Abstract] [Google Scholar]
- Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [Europe PMC free article] [Abstract] [Google Scholar]
- Jackson J.B., Pollock,J.M.,Jr and Rill,R.L. (1979) Chromatin fractionation procedure that yields nucleosomes containing near-stoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry, 18, 3739–3748. [Abstract] [Google Scholar]
- Kwon J., Morshead,K.B., Guyon,J.R., Kingston,R.E. and Oettinger,M.A. (2000) Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell, 6, 1037–1048. [Abstract] [Google Scholar]
- Längst G. and Becker,P.B. (2001a) ISWI induces nucleosome sliding on nicked DNA. Mol. Cell, 8, 1085–1092. [Abstract] [Google Scholar]
- Längst G. and Becker,P.B. (2001b) Nucleosome mobilization and positioning by ISWI-containing chromatin- remodeling factors. J. Cell Sci., 114, 2561–2568. [Abstract] [Google Scholar]
- Längst G., Bonte,E.J., Corona,D.F. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [Abstract] [Google Scholar]
- Lee K.B. and Thomas,J.O. (2000) The effect of the acidic tail on the DNA-binding properties of the HMG1,2 class of proteins: insights from tail switching and tail removal. J. Mol. Biol., 304, 135–149. [Abstract] [Google Scholar]
- Lilley D.M. (1992) DNA–protein interactions. HMG has DNA wrapped up. Nature, 357, 282–283. [Abstract] [Google Scholar]
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 389, 251–260. [Abstract] [Google Scholar]
- Mitsouras K., Wong,B., Arayata,C., Johnson,R.C. and Carey,M. (2002) The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol. Cell. Biol., 22, 4390–4401. [Europe PMC free article] [Abstract] [Google Scholar]
- Mizuguchi G., Tsukiyama,T., Wisniewski,J. and Wu,C. (1997) Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell, 1, 141–150. [Abstract] [Google Scholar]
- Mizuguchi G., Vassilev,A., Tsukiyama,T., Nakatani,Y. and Wu,C. (2001) ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J. Biol. Chem., 276, 14773–14783. [Abstract] [Google Scholar]
- Muchardt C. and Yaniv,M. (1999) ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol., 293, 187–198. [Abstract] [Google Scholar]
- Muller S., Bianchi,M.E. and Knapp,S. (2001) Thermodynamics of HMGB1 interaction with duplex DNA. Biochemistry, 40, 10254–10261. [Abstract] [Google Scholar]
- Narlikar G.J., Phelan,M.L. and Kingston,R.E. (2001) Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell, 8, 1219–1230. [Abstract] [Google Scholar]
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [Abstract] [Google Scholar]
- Ner S.S. and Travers,A.A. (1994) HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J., 13, 1817–1822. [Europe PMC free article] [Abstract] [Google Scholar]
- Ner S.S., Blank,T., Perez-Paralle,M.L., Grigliatti,T.A., Becker,P.B. and Travers,A.A. (2001) HMG-D and histone H1 interplay during chromatin assembly and early embryogenesis. J. Biol. Chem., 276, 37569–37576. [Abstract] [Google Scholar]
- Nightingale K., Dimitrov,S., Reeves,R. and Wolffe,A.P. (1996) Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J., 15, 548–561. [Europe PMC free article] [Abstract] [Google Scholar]
- Orphanides G., Wu,W.H., Lane,W.S., Hampsey,M. and Reinberg,D. (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature, 400, 284–288. [Abstract] [Google Scholar]
- Papoulas O., Daubresse,G., Armstrong,J.A., Jin,J., Scott,M.P. and Tamkun,J.W. (2001) The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl Acad. Sci. USA, 98, 5728–5733. [Europe PMC free article] [Abstract] [Google Scholar]
- Pennings S., Meersseman,G. and Bradbury,E.M. (1991) Mobility of positioned nucleosomes on 5 S rDNA. J. Mol. Biol., 220, 101–110. [Abstract] [Google Scholar]
- Pennings S., Meersseman,G. and Bradbury,E.M. (1994) Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl Acad. Sci. USA, 91, 10275–10279. [Europe PMC free article] [Abstract] [Google Scholar]
- Peterson C.L. (2000) ATP-dependent chromatin remodeling: going mobile. FEBS Lett., 476, 68–72. [Abstract] [Google Scholar]
- Peterson C.L. (2002a) Chromatin remodeling enzymes: taming the machines. Third in review series on chromatin dynamics. EMBO rep., 3, 319–322. [Europe PMC free article] [Abstract] [Google Scholar]
- Peterson C.L. (2002b) Chromatin remodeling: nucleosomes bulging at the seams. Curr. Biol., 12, R245–R247. [Abstract] [Google Scholar]
- Ramstein J., Locker,D., Bianchi,M.E. and Leng,M. (1999) Domain–domain interactions in high mobility group 1 protein (HMG1). Eur. J. Biochem., 260, 692–700. [Abstract] [Google Scholar]
- Scaffadi P., Misteli,T. and Bianchi,M.E. (2002). Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature, 418, 191–195. [Abstract] [Google Scholar]
- Schroter H. and Bode,J. (1982) The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur. J. Biochem., 127, 429–436. [Abstract] [Google Scholar]
- Simon R.H. and Felsenfeld,G. (1979) A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res., 6, 689–696. [Europe PMC free article] [Abstract] [Google Scholar]
- Stein A., Whitlock,J.P.,Jr and Bina,M. (1979) Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc. Natl Acad. Sci. USA, 76, 5000–5004. [Europe PMC free article] [Abstract] [Google Scholar]
- Stros M., Stokrova,J. and Thomas,J.O. (1994) DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res., 22, 1044–1051. [Europe PMC free article] [Abstract] [Google Scholar]
- Thomas J.O. and Travers,A.A. (2001) HMG1 and 2 and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci., 26, 167–174. [Abstract] [Google Scholar]
- Travers A. (2000) Recognition of distorted DNA structures by HMG domains. Curr. Opin. Struct. Biol., 10, 102–109. [Abstract] [Google Scholar]
- Travers A.A., Ner,S.S. and Churchill,M.E. (1994) DNA chaperones: a solution to a persistence problem? Cell, 77, 167–169. [Abstract] [Google Scholar]
- Varga-Weisz P., van Holde,K. and Zlatanova,J. (1994) Competition between linker histones and HMG1 for binding to four-way junction DNA: implications for transcription. Biochem. Biophys. Res. Commun., 203, 1904–1911. [Abstract] [Google Scholar]
- Varga-Weisz P.D., Blank,T.A. and Becker,P.B. (1995) Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J., 14, 2209–2216. [Europe PMC free article] [Abstract] [Google Scholar]
- Vignali M., Hassan,A.H., Neely,K.E. and Workman,J.L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [Europe PMC free article] [Abstract] [Google Scholar]
- Widom J. (1998) Chromatin structure: linking structure to function with histone H1. Curr. Biol., 8, R788–R791. [Abstract] [Google Scholar]
- Widom J. (1999) Equilibrium and dynamic nucleosome stability. Methods Mol. Biol., 119, 61–77. [Abstract] [Google Scholar]
- Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [Abstract] [Google Scholar]
- Xiao H., Sandaltzopoulos,R., Wang,H.M., Hamiche,A., Ranallo,R., Lee,K.M., Fu,D. and Wu,C. (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell, 8, 531–543. [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1093/emboj/cdf692
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc139112?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/emboj/cdf692
Article citations
Hmo1: A versatile member of the high mobility group box family of chromosomal architecture proteins.
World J Biol Chem, 15(1):97938, 01 Aug 2024
Cited by: 0 articles | PMID: 39156122 | PMCID: PMC11325855
Review Free full text in Europe PMC
Single-molecule study reveals Hmo1, not Hho1, promotes chromatin assembly in budding yeast.
mBio, 14(4):e0099323, 11 Jul 2023
Cited by: 2 articles | PMID: 37432033 | PMCID: PMC10470511
Negatively charged, intrinsically disordered regions can accelerate target search by DNA-binding proteins.
Nucleic Acids Res, 51(10):4701-4712, 01 Jun 2023
Cited by: 16 articles | PMID: 36774964 | PMCID: PMC10250230
Reappraisal of oxidized HMGB1 as a mediator and biomarker.
Future Sci OA, 8(10):FSO828, 01 Dec 2022
Cited by: 1 article | PMID: 36874369 | PMCID: PMC9979160
Review Free full text in Europe PMC
The high mobility group protein HMG20A cooperates with the histone reader PHF14 to modulate TGFβ and Hippo pathways.
Nucleic Acids Res, 50(17):9838-9857, 01 Sep 2022
Cited by: 4 articles | PMID: 36124662 | PMCID: PMC9508832
Go to all (142) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer.
Cell, 97(7):843-852, 01 Jun 1999
Cited by: 233 articles | PMID: 10399913
Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling.
EMBO J, 20(14):3781-3788, 01 Jul 2001
Cited by: 94 articles | PMID: 11447119 | PMCID: PMC125259
The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF.
Mol Cell, 13(2):265-277, 01 Jan 2004
Cited by: 47 articles | PMID: 14759371
H1 and HMGB1: modulators of chromatin structure.
Biochem Soc Trans, 40(2):341-346, 01 Apr 2012
Cited by: 68 articles | PMID: 22435809
Review




