Abstract
Free full text

Proteomic analysis of zygote and ookinete stages of the avian malaria parasite Plasmodium gallinaceum delineates the homologous proteomes of the lethal human malaria parasite Plasmodium falciparum
Abstract
Delineation of the complement of proteins comprising the zygote and ookinete, the early developmental stages of Plasmodium within the mosquito midgut, is fundamental to understand initial molecular parasite-vector interactions. The published proteome of Plasmodium falciparum does not include analysis of the zygote/ookinete stages, nor does that of P. berghei include the zygote stage or secreted proteins. P. gallinaceum zygote, ookinete, and ookinete-secreted/released protein samples were prepared and subjected to Multidimensional protein identification technology (MudPIT). Peptides of P. gallinaceum zygote, ookinete, and ookinete-secreted proteins were identified by MS/MS, mapped to ORFs (>50 amino acids) in the extent P. gallinaceum whole genome sequence, and then matched to homologous ORFs in P. falciparum. A total of 966 P. falciparum ORFs encoding orthologous proteins were identified; just over 40% of these predicted proteins were found to be hypothetical. A majority of putative proteins with predicted secretory signal peptides or transmembrane domains were hypothetical proteins. This analysis provides a more comprehensive view of the hitherto unknown proteome of the early mosquito midgut stages of P. falciparum. The results underpin more robust study of Plasmodium-mosquito midgut interactions, fundamental to the development of novel strategies of blocking malaria transmission.
1 Introduction
Malaria is the most important parasitic disease affecting humans, causing more than 100 million clinical cases and 1.1 million deaths each year worldwide primarily due to Plasmodium falciparum [1, 2]. The Plasmodium life cycle consists of alternating infections of the intermediate host, a vertebrate, and the definitive host, the mosquito. Plasmodium must accomplish a complex developmental program within the mosquito [3], yet the molecular mechanisms by which the Plasmodium ookinete invades the mosquito host is only understood in simple outline [4]. Upon biting a human carrier, gametocyte stages of the parasites are taken up along with blood and enter the mosquito gut, where they transform into male and female gametes that fertilize to form zygotes. Within 8-10 h, zygotes transform into the invasive ookinete, whose combined gliding motility [5] and constitutive secretion through its apical apparatus [4, 6] allow the parasite to penetrate the chitin-containing peritrophic matrix [7-11] and midgut epithelium [12] at which point it transforms into the oocyst stage on the midgut wall.
Within the mosquito midgut, the transition from gametes to zygotes and then to ookinetes is a genetic bottleneck in the lifecycle of Plasmodium, as the number of parasites drops by several orders of magnitude [13]. During this time, the parasite remains exposed to human antibody (taken up along with the gametocyte), as well as mosquito proteases and immune molecules, and reactive oxygen, all of which can reduce parasite infectivity for the mosquito [14]. Therefore, zygote and ookinete surface proteins which may be involved in resisting mosquito attack on the parasite are potential targets for antibody-mediated interventions (transmission-blocking vaccine) to interrupt the mosquito stages of the parasite. Monoclonal and polyclonal antibodies raised against a limited number of zygote and ookinete proteins have been reported as potential immunological targets for blocking/reducing malaria transmission in experimental systems [15]. A more complete understanding of the zygote and ookinete proteome will facilitate the development of new tools to prevent malaria transmission.
Proteomic and transcriptomic analyses of all the stages of P. falciparum, except zygote, and ookinete, have been published [16, 17]. Limitations to these analyses include the lack of proteomic and expression profiling data for P. falciparum zygote and ookinete stages, although proteomic and expression profiling data for rodent malaria parasite for all stages other than zygotes and ookinete-secreted protein have been published [18]. We hypothesized that understanding the proteome of zygote and ookinete stages from Plasmodium model species will provide insight into mechanisms of P. falciparum-mosquito interaction given the close affinity of the species [19-21].
Plasmodium gallinaceum, an avian malaria parasite for which zygotes, ookinetes, and secreted/released proteins in chemically defined, serum-free medium are readily obtainable, is perhaps the most relevant animal model of the P. falciparum sexual stages. Based on circumsporozoite protein, chitinase, cytochrome b, and ribosomal RNA genes sequence analysis of the bird malaria parasite, P. gallinaceum has been suggested to be evolutionarily closer to P. falciparum [19-23] than are the rodent malaria parasites. P. gallinaceum zygotes and ookinetes can be produced in vitro using serum-free media, which has not been possible with P. falciparum and has not been done with the rodent malaria parasites. Here, we present a proteomic analysis of P. gallinaceum zygote, ookinete, and ookinete-secreted/released proteins using Multidimensional protein identification technology (MudPIT). Thousands of proteins from P. gallinaceum were identified by MS/MS and mapped to translated genomic ORFs. These translated ORFs were then matched to homologous ORFs in P. falciparum. This analysis provides a new view of the hitherto unknown proteome of the early mosquito midgut stages of P. falciparum.
2 Materials and methods
2.1 In vitro culture of P. gallinaceum mosquito stages (zygote, ookinete, and supernatant)
The 8A strain of P. gallinaceum was used to infect 4-6 wk-old White Leghorn chickens. A gametocyte line of P. gallinaceum was maintained by subpassage through chicken and mosquito. Purified zygotes were extensively washed with PBS then ookinetes were cultured in vitro using serum-free M199 media for 24 h [24] and six biological replicates were prepared. Supernatants from the six ookinete cultures were pooled and protein was precipitated by standard acetone method. Zygote pellets were also obtained from six different cultures in a similar way to the preparation of the ookinete culture but parasites were harvested 2-3 h after suspending in M199 media.
2.2 Sample preparation and MudPIT analysis
Purified zygote and ookinetes were used for MudPIT analysis using trypsin and proteinase K separately. For trypsin analysis, 1.15 × 107 cells (zygotes) and 1.6 × 106 cells (ookinetes) were used. For proteinase K analysis, 1.01 × 107 cells (zygotes) and 1.5 × 106 cells (ookinetes) were used. For whole cell fractionation and trypsin-based digestion, cell pellets were resuspended in 100 μL of 100 mM phosphate buffer pH 6.7, 5 mM MgCl2, 250 mM sucrose, 5 mM CaCl2. One microgram of trypsin (sequencing grade, Promega) was added and then incubated at 37°C for 30 min, followed by two-fold dilution with 100 μL of 100 mM phosphate buffer pH 6.7, then addition of 200 μL of 100 mM phosphate buffer pH 6.7, 5 mM MgCl2, 250 mM sucrose, 2 mM CaCl2. The mixture was centrifuged at 18 000 × g 30 min at 4°C, and the supernatant (extracellular, E) was obtained. The pellet was then resuspended in 100 μL of 10 mM Tris-HCl pH 8.5 and incubated for 1 h on ice to lyse cells. The sample was then centrifuged at 18 000 rcf for 30 min at 4°C, and the supernatant was transferred (soluble lysate, S). The pellet was resuspended in 100 μL of 0.1 M Na2CO3 pH 11.5, incubated on ice for 1 h, centrifuged at 18 000 rcf for 30 min at 4°C, and the supernatant was transferred (salt-eluted membrane fraction, SP). The pellet (membrane fraction, P) was dried via speed-vac.
S and SP fractions were subjected to protease digestion. Solid urea was added to 8 M, and TCEP was added to a final concentration of 5 mM, and incubated at 27°C for 30 min. The mixture was brought to 10 mM iodoacetamide (20 mM for P samples) and incubated at 27°C for 30 min in the dark. For SP, the pH was adjusted to 8.5 with 1 M Tris-HCl pH 8.5 and 1 μg of endoproteinase Lys-C (sequencing grade, Roche) was added to ookinete and zygote samples, respectively, and incubated overnight at 37°C, with shaking. Samples were diluted to 2 M urea with 100 mM Tris-HCl pH 8.5, CaCl2 was added to 2 mM, 0.5 μg of trypsin was added, and incubated 37°C overnight, with shaking. Digestions were stopped by adding formic acid to 5%, and then stored at -80°C until analysis.
P fractions were subjected to cyanogen bromide-treatment followed by protease digestion. The P fraction was resuspended in 100 μL of 90% formic acid (this and all subsequent steps performed in a hood), 500 mg/mL CNBr, and incubated overnight in the dark. NH4OH (30%) was then added dropwise until the pH was ~8.5 (300 μL). Tris-HCl pH 8.5 was added to a final concentration of 0.1 M. Solid urea was added to 8 M, TCEP was added to a final concentration of 5 mM, and incubated at room temperature (RT) for 30 min. The mixture was brought to 20 mM iodoacetamide and incubated at RT for 30 min in the dark. Lys-C (1 μg) was added and incubated overnight at 37°C. Samples were diluted to 2 M urea with 100 mM Tris-HCl pH 8.5. CaCl2 was added to 2 mM, 0.5 μg of trypsin was added and incubated at 37°C overnight with shaking. Digestions were stopped by adding formic acid to 5%, and concentrated on a 4 mg SPEC-PT-C18 (Varian), then washed and eluted as directed by the manufacturer. Eluates were dried down via speed-vac, resuspended in 20 μL of MudPIT-buffer A (see below), and stored at -80°C until analysis.
Each digest was loaded onto a three-phase MudPIT column [25] as previously described [26]. Briefly, digests were loaded onto a multidimensional 250 μm id fused-silica column encountering 3.5 cm of C18 resin (5 μm, 110A, Aqua, Phenomenex) followed by 3.5 cm of SCX resin (5 μm, 110A, Partishere, Whatman) via a high pressure loading device. After loading, a 100 μm fused-silica analytical column with a 5 μm laser-pulled tip containing 10.5 cm on C18 resin was attached adjacent to the SCX-side of the multidimensional column via a union (Upchurch). This three-phase MudPIT column was then attached to an HP1100 HPLC (Agilent) and coupled to an LCQ Deca IT mass spectrometer (Thermo Electron). Online LC/LC-MS/MS was then carried out using salt steps, RP gradients, instrument settings, and data-dependent collection of MS/MS spectra as previously described [25, 26]. For all digests of ookinete supernatants, 6-step MudPIT runs were performed; for trypsin and proteinase K digests of subcellular fractions, 12-step MudPIT runs were performed. Collected MS/MS spectra were searched against the database described below using the algorithm SEQUEST [26].
A P. gallinaceum ORF database was created using current available genome databases (12/31/06 for contigs and 1/3/2007 for reads downloaded from ftp://ftp.sanger.ac.uk/pub/pathogens/Plasmodium/gallinaceum/). The nucleotide sequences of both reads and contigs were translated in all six reading frames, and a FASTA format database was then created for those entries containing at least 50 amino acids stretches using an in-house Perl script (J. Johnson, unpublished). This database was then combined with all chicken (Gallus gallus) protein entries (downloaded from ftp://ftp.ensembl.org/pub/current_gallus_gallus/data/fasta/pep/), to which was manually added a list of common contaminants (e.g., human keratin and trypsin), and the protein sequences for P. gallinaceum chitinase (PgCHT1 and PgCHT2) and all of the above sequences were reversed. This database was then used for searching MS/MS spectra. The program DTA select [27] was used to filter peptide identifications, requiring that all identifications pass standard XCorr (1.8, 2.5, 3.5 for 11, 12, and 13 charged peptides, respectively) and deltaCN (0.08) score thresholds [28]. By concatenating a reversed database, we estimated the false positive rate of the searches based on how many hits were obtained to reversed sequence. Additionally, identifications against chicken and contaminant entries were removed from the final list.
2.3 Homologous protein identification and annotation
P. gallinaceum ORFs identified from the above search were then subjected to a BLAST (BLASTP) alignment of protein sequences based on ORFs found in the most current version P. falciparum database (PlasmoDB v5.4 downloaded from http://www.plasmodb.org/download/release-5.3/Pfalciparum/). This was done en masse using an in-house Perl script (J. Johnson and Tao Xu, unpublished). Hits better than 0.00001E are reported as being homologs of P. falciparum. TMHMM 2.0 and SignalP 3.0 (http://www.cbs.dtu.dk) was used to determine transmembrane (TM) and signal peptide (SP) sequences, respectively, within identified proteins. GO annotations, Class ID, and class code for each protein were obtained from PlasmoDB (PlasmoDB.org).
3 Results and discussion
To identify proteins specifically expressed during Plasmodium developmental stages in the mosquito midgut, we cultured zygote and ookinete stages of P. gallinaceum in vitro using chemically defined media. In Plasmodium berghei, in vitro culture of zygotes and ookinete are carried out in conditioned media containing FCS [29], which is one of the major limitations to analyze the secretome of this parasite species. P. gallinaceum zygotes and ookinetes were cultured in chemically defined and serum-free medium, which allowed us to analyze ookinete-secreted proteins. Protein samples were subjected to proteomic analysis by MudPIT where protein mixtures are subjected to proteolytic digestion followed by multidimensional LC/LC-MS/MS. Isolated zygote and ookinete cells were subjected to biochemical fractionation into soluble versus insoluble fractions followed by proteolytic digestion by trypsin and proteinase-K, respectively. This approach yields more protein identifications by MudPIT compared to using only one digestion strategy.
The proteome of zygote, ookinete, and ookinete culture supernatant was analyzed resulting in 19 869 partial ORFs (>50 amino acids) identified in the P. gallinaceum genome and with some redundancy. The complete genome of P. gallinaceum has been sequenced to 3X coverage but not been assembled. Because the P. gallinaceum genome has not been annotated, orthologs were searched in the most recently annotated P. falciparum complete genome as reflected in PlasmoDB 5.4 (reflecting the October 2007 update). P. gallinaceum is evolutionarily closer to P. falciparum [23] than to other Plasmodium species. Therefore P. falciparum orthologs present in zygote and ookinete stages of P. gallinaceum will further improve the understanding of P. falciparum host parasite interaction and more precisely guide novel intervention approaches.
3.1 Total proteome of zygote, ookinete, and secretome
From the P. gallinaceum partial ORFs identified in the analysis of zygote, ookinete, and ookinete-secreted protein (ookinete culture supernatant), a total of 966 orthologous proteins of P. falciparum were identified. These predicted proteins account for 18.3% of the total predicted proteins in the P. falciparum genome. Among these identified 966 P. falciparum orthologs, 460 (47.6%) proteins had more than two spec count (spec count ranges from 2-397); another 506 proteins had only one spectrum counts. The details of identified proteins with sequence and spectrum counts presented as the Supporting Information. (Table S1).
The P. falciparum orthologs were classified according to their molecular function (class code 1-13) (Table 1). Forty percent of the orthologous proteins were hypothetical proteins matching to P. falciparum (36% hypothetical and 4% conserved hypothetical). Other important classes of proteins, belonging to cell surface (3%), signal transduction (3%), and protein fate (10%) accounts for 3-10% the total orthologs identified in P. falciparum. These classes of proteins are necessary for invasion, defense, and cell communication, which are critical for the successful transmission of the parasite.
Table 1
Functional orthologs of P. gallinaceum zygote, ookinete, and ookinete-secreted proteins identified in the annotated P. falciparum genome as determined by MudPIT analysis
| Class code | Molecular function | P. falciparum | % |
|---|---|---|---|
| 1 | Cell cycle (DNA processing) | 51 | 5.3 |
| 2 | Cell rescue defense (virulence) | 6 | 0.6 |
| 3 | Cellular communication (signal transduction) | 29 | 3.0 |
| 4 | Cellular transport (transport mechanism) | 55 | 5.7 |
| 5 | Metabolism (energy) | 109 | 11.2 |
| 6 | Protein fate | 102 | 10.6 |
| 7 | Protein synthesis | 104 | 10.7 |
| 8 | Transcription | 49 | 5.1 |
| 9 | Transport facilitation | 17 | 1.8 |
| 10 | Conserved hypothetical | 40 | 4.1 |
| 11 | Hypothetical | 352 | 36.4 |
| 12 | Cell surface (apical organelles) | 29 | 3.0 |
| 13 | Unclassified | 23 | 2.3 |
| Total numbers | 966 |
Proteins with predicted secretory SP and TM domains are of particular importance due to their secretory fate or presence on the cell surface [23]. Few proteins (Chitinase, Pfs230, CTRP, WARP, and Pfs25) have been studied in detail in zygote and ookinete stages, some of which are characterized as malaria transmission-blocking vaccine candidates [4]. We have analyzed the presence of SP/TM domain(s) containing proteins in different functional classes of P. falciparum orthologs (Fig. 1). Surprisingly, a majority of SP and TM containing proteins are hypothetical, which suggests that a substantial number of hitherto uncharacterized proteins may have particularly interesting roles in parasite-vector interactions.
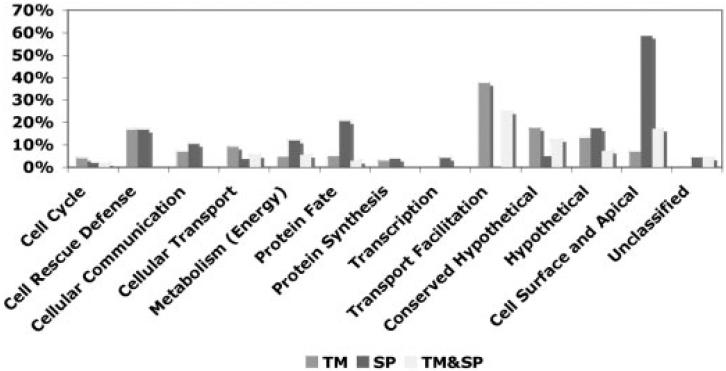
The bar diagram shows the percentage of P. falciparum orthologous proteins with secretory/cell surface associated domains among the different functional class of proteins. The P. falciparum orthologs were identified in the zygote, ookinete, and ookinete secretomes of P. gallinaceum.
In P. falciparum about 5268 predicted proteins are present in the genome, 65% of which are hypothetical or uncharacterized [30]. Earlier using similar MudPITapproaches, 2415 proteins from sprorozoites, merozoites, trophozoites, and gametocytes were described. This number was later increased to 2904 by adding proteins of rings, schizont, and gamete stages [31]. Le Roch et al. [13] reported that at least 4557 genes (88%) were transcribed in one or more stages of the life cycle of P. falciparum, but this analysis did not include analysis of proteins from gametocytes, zygotes, or ookinetes. In P. berghei, 1836 predicted proteins were detected with high confidence from asexual and ookinete stages of the parasite [18]. However, because the P. falciparum zygote and ookinete stages have not obtainable in sufficient number in vitro for proteomic analysis these stages have remained a “black box” in the life cycle of the human parasite.
3.2 Comparative proteome of zygote, ookinete, and secretome
In the present study, using proteins of P. gallinaceum zygote, ookinete, and culture supernatant, we identified 381-749 predicted orthologs shared by P. gallinaceum and P. falciparum in the zygote, ookinete, and ookinete-secreted protein proteomes (Fig. 2). These observations further demonstrated the close evolutionary affinity of P. falciparum and P. gallinaceum at the mosquito stages of both malaria parasites. The close evolutionary affinity of P. falciparum and P. gallinaceum allows for a more complete estimate of P. falciparum zygote and ookinete proteome. The identified zygote, ookinete, secreted proteome corresponded to 7-14.2% of the annotated protein databases from P. falciparum genome. Zygotes, ookinete, and ookinete-secreted proteins share only 16.9% of the identified P. falciparum orthologs. As reported earlier with P. falciparum, asexual blood stages, gametocytes, and sporozoites (but not zygotes and ookinetes) share 6% total predicted proteins [16], which indicates that each stage is highly specialized in its proteome. In zygotes, 418 P. falciparum orthologs were identified, of which 68 (16%) were unique to this stage. In ookinetes, 749 P. falciparum orthologs were identified, of which 343 (46%) proteins were unique to this stage. Zygotes and ookinete shared 338 (45-80%) proteins. In earlier published reports, 1147 proteins were detected in P. falciparum gametocytes, where 33% proteins were unique to the stage [16]. In P. falciparum sporozoites, of 1049 proteins identified, almost half (49%) were unique to the stage [16]. These findings, including our results, suggest that ookinete and sporozoites express a large number of unique proteins inside mosquito hosts. In the P. gallinaceum ookinete secretome, 381 P. falciparum orthologous proteins were detected, of which a remarkable 36% were not found in either zygote or in ookinete lysate.
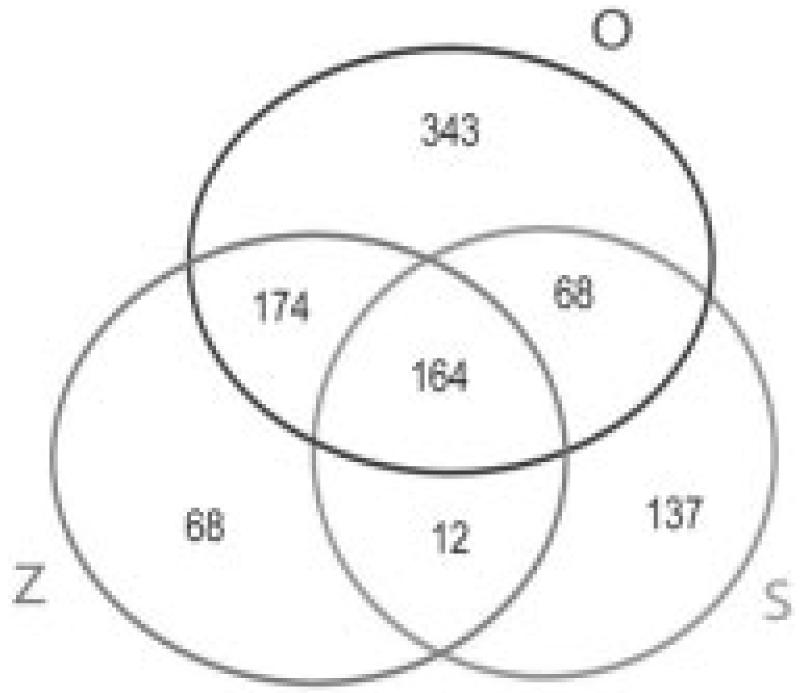
Venn Diagram showing the unique and shared orthologs of P. falciparum found in zygotes (Z), ookinetes (O), and ookinete-secreted proteins (S) of P. gallinaceum by MudPIT analysis.
The number of unique proteins in the ookinete was higher than either zygote or ookinete secretome, hence it may be particularly interesting to investigate their role in host-parasite interactions. These results indicate that, like the asexual blood stages, gametocytes, and sporozoites that the midgut stages also have a largely unique proteome, likely related to the stage-specific functions including immune evasion, coping with the mosquito gut environment, and invasion of the peritrophic matrix.
3.3 Functional specific protein expression in zygote, ookinete, and secretome
Micronemes are the only secretory organelles that have been found in ookinete stages of Plasmodium. Some micronemal proteins such as Chitinase, have well defined biological functions while others such CTRP, SOAP, and WARP are thought to play critical roles in host invasion, motility, and host cell recognition (Fig. 3). Micronemal proteins are targets of disrupting malaria transmission from vertebrate host to mosquito [4]. Only a small number of secretory and cell-surface associated proteins of ookinete stages of P. falciparum have been identified and functionally characterized [4]. These data indicate that analysis of proteins of P. gallinaceum zygote and ookinete stages is a powerful method to provisionally identify zygote- and ookinete-expressed proteins in P. falciparum.

Pie chart showing distribution of different functional classes of P. falciparum ortholog proteins in zygote, ookinete, and ookinete-secreted proteins of P. gallinaceum. (A, zygote; B, ookinete; C, secretome).
Many cell surface proteins (class 12) are candidate targets for transmission-blocking vaccines [13] and we have identified 29 P. falciparum orthologs within this class. Twenty-two of these 29 P. falciparum orthologs have a secretory signal (SP) indicative of a secreted or membrane associated protein. Known transmission-blocking targets such as Pfs230 (PFB0405w) were found to have very high spectra counts in zygote, ookinete, and ookinete supernatant. Other known transmission-blocking targets are antigen precursor 48/45 (PF13_0247), Pf47 (PF13_0248), and Pfs25 (PF10_0303) were also detected higher spectra count in ookinete lyaste. CTRP (PFC0640w) and Chitinase (PF11_0224) were detected abundantly only in the ookinete and secretome but not in the zygote stages (Table 2). In the ookinete lysate, ten proteins were found to be unique to this group including CSP (PF11_0224, Spec count 19), exported protein 2 (PF14_0678, Spec count 5), and MAEBL protein (PF11_0486) known for molecular apical receptor binding function. Surprisingly, we have also detected rhoptry-associated protein (PF14_0102) in ookinete (spec count 82) and zygote lysate (spec count 12), with no hit in supernatant. The other known blood stage antigen MSP-1 (PFI1475w) was also detected in ookinetes. Both of these proteins have already been detected in the proteome of gametocyte and sporozoite stages (PlasmoDB.org).
Table 2
Transmission-blocking antigens of P. falciparum identified in the P. gallinaceum zygote, ookinete, and ookinete secretome
| Locus | Protein description | Zygote sequence count (spec count) | Ookinete sequence count (spec count) | Ookinete secretome sequence count (spec count) |
|---|---|---|---|---|
| PFB0405W | Transmission-blocking target antigen s230 precursor | 153(345) | 123(302) | 190(604) |
| PF13_0247 | Transmission-blocking Target antigen precursor Pfs45/48 | 53(194) | 26(81) | 34 (86) |
| PF13_0248 | Pfs47 | 53(176) | 47 (240) | 17(58) |
| PF13_0011 | Gamete antigen 27/25 | 1(2) | 1(1) | 0 |
| PFC0640w | CTRP | 0 | 54 (87) | 19(27) |
| PF10_0303 | Pfs25 | 12(20) | 34(127) | 1(1) |
| PFL2510w | Chitinase | 0 | 102(193) | 28(102) |
| PF08_0136B | Von willebrand a domain-related | 0 | 39 (79) | 14(40) |
Proteins involved in signal transduction and cell communication (class 3) are crucial throughout the parasite life cycle. In this class we identified 29 Pf orthologs proteins expressed in all stages. For the calmodulin-domain protein kinase (PF07_0072,), Ser/Thr protein kinase (MAL13P1.278) more spectra were found in the ookinete supernatant, than in the ookinete and zygote lysate. The kinases found only in the supernatants were exported Ser/Thr Kinase (PFC0105w), calcium-dependent protein kinase (PF14_0227), and PK4 protein kinase (PFF1370w). Interestingly, calmodulin-like protein (PF14_0181) was only found in the ookinete lysate and IRP-like protein (PF13_0229) was found to have higher spectra counts in the ookinete than in the zygote, and was absent in the ookinete supernatant. In ookinete and zygote stages, 14-3-3 protein and RabGDI binding proteins were found to be highly overexpressed. The parasite-specific signaling pathway is an attractive focus for the identification of novel transmission-blocking targets and understanding the functions of zygote and ookinete expressed signaling protein may be very useful in the development of this therapeutic research area.
Several anti-Plasmodium genes are expressed in midgut tissue and by hemocytes [32] of mosquitoes, which are able to attack midgut-stage Plasmodia and are found to extensively overlap between P. falciparum and P. berghei infection in similar hosts. In our findings, only four P. falciparum orthologs belong to cell rescue and defense (class 2) are found in ookinete and zygote with spectra counts ranging from 1 to 27 hits. Thioredoxin peroxidase (PFL0725w) are found only in the ookinete and zygote stages, whereas Cg4 protein is highly expressed (SPEC count>15) in all the three stages.
The ookinete is a specialized cell that has gliding motility and rigidity of its apical end to allow it to escape the blood meal and invade the mosquito midgut epithelial cell layer. The structure of the parasite therefore requires high actin and tubulin expression. Actin, tubulin, and myosin are highly expressed in both zygotes and ookinetes. However, α-tubulin II (PFD 1050w, SPEC count 92) and myosin A tail domain protein (PFL2225w, SPEC count 111) are abundantly and relatively overexpressed in the ookinete stage, consistent with their putative roles in ookinete gliding motility.
4 Concluding remarks
In summary, high-throughput analysis of peptides in P. gallinaceum zygote, ookinete, and secreted proteins has permitted the identification of 966 predicted orthologous proteins in the P. falciparum genome that are likely present in the zygote and ookinete stages. Importantly, we were able to separately identified ookinete-secreted and zygote proteins of P. gallinaceum, which was not previously reported for the rodent malaria parasite P. berghei. As direct proteome analysis of these stages in P. falciparum is not yet possible, using the evolutionarily close species of malaria parasite, the avian-infecting parasite P. gallinaceum, has allowed the delineation of a comprehensive list of zygote, ookinete, and ookinete-secretome proteins that have orthologs in P. falciparum. The identified orthologous proteins with predicted cell surface-localizing motifs or signals for secretion will provide new targets of blocking malaria transmission and permit comprehensive experimental approaches to study mechanisms of parasite-vector interactions.
Acknowledgments
This project was supported by U.S. Public Health Service grants R21AI053781 and R01AI45999 to J. M. V. from the National Institute of Health. The genome sequencing of P. gallinaceum was carried out by the Pathogen Sequencing Unit, Wellcome Trust Sanger Institute, supported by the Wellcome Trust, which provided permission to analyze this dataset at the whole genome level. This work was additionally supported by NIH grant P41 RR11823-10 to J. L. Y. and an NRSA fellowship 32AI062061 to G. C. We thank Dr. Fengwu Li and Karen Chin for their help and support in the laboratory. We especially thank Dr. Laurence Florens for her enormous support in initiating this project.
Abbreviations
| MudPIT | multidimensional protein identification technology |
| SP | signal peptide |
| TM | transmembrane |
Footnotes
6 Note added in proof Gene expression analysis for P. falciparum zygotes and ookinetes has recently been published (Zhou, et al., Evidence-based annotation of the malaria parasite's genome using comparative expression profiling. PLoS One 2008, 3, e1570).
References
Full text links
Read article at publisher's site: https://doi.org/10.1002/pmic.200700727
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2637033?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1002/pmic.200700727
Article citations
Decrypting the complexity of the human malaria parasite biology through systems biology approaches.
Front Syst Biol, 2:940321, 16 Sep 2022
Cited by: 0 articles | PMID: 37200864 | PMCID: PMC10191146
Evolutionary Insights into the Microneme-Secreted, Chitinase-Containing High-Molecular-Weight Protein Complexes Involved in Plasmodium Invasion of the Mosquito Midgut.
Infect Immun, 90(1):e0031421, 04 Oct 2021
Cited by: 4 articles | PMID: 34606368 | PMCID: PMC8788677
Review Free full text in Europe PMC
A Hetero-Multimeric Chitinase-Containing Plasmodium falciparum and Plasmodium gallinaceum Ookinete-Secreted Protein Complex Involved in Mosquito Midgut Invasion.
Front Cell Infect Microbiol, 10:615343, 08 Jan 2021
Cited by: 4 articles | PMID: 33489941 | PMCID: PMC7821095
Transcriptome analysis based detection of Plasmodium falciparum development in Anopheles stephensi mosquitoes.
Sci Rep, 8(1):11568, 01 Aug 2018
Cited by: 5 articles | PMID: 30068910 | PMCID: PMC6070505
Plasmodium Parasites Viewed through Proteomics.
Trends Parasitol, 34(11):945-960, 23 Aug 2018
Cited by: 14 articles | PMID: 30146456 | PMCID: PMC6204299
Review Free full text in Europe PMC
Go to all (30) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Plasmodium falciparum ookinete expression of plasmepsin VII and plasmepsin X.
Malar J, 15:111, 24 Feb 2016
Cited by: 19 articles | PMID: 26911483 | PMCID: PMC4765185
A Hetero-Multimeric Chitinase-Containing Plasmodium falciparum and Plasmodium gallinaceum Ookinete-Secreted Protein Complex Involved in Mosquito Midgut Invasion.
Front Cell Infect Microbiol, 10:615343, 08 Jan 2021
Cited by: 4 articles | PMID: 33489941 | PMCID: PMC7821095
Chitinases of the avian malaria parasite Plasmodium gallinaceum, a class of enzymes necessary for parasite invasion of the mosquito midgut.
J Biol Chem, 275(14):10331-10341, 01 Apr 2000
Cited by: 68 articles | PMID: 10744721
The development of malaria parasites in the mosquito midgut.
Cell Microbiol, 18(7):905-918, 24 May 2016
Cited by: 95 articles | PMID: 27111866 | PMCID: PMC5089571
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCRR NIH HHS (2)
Grant ID: P41 RR011823
Grant ID: P41 RR11823-10
NIAID NIH HHS (11)
Grant ID: R01 AI045999
Grant ID: R21 AI053781-03
Grant ID: R01 AI045999-06A2
Grant ID: R21AI053781
Grant ID: F32 AI062061
Grant ID: R21 AI053781
Grant ID: R21 AI053781-01
Grant ID: R01 AI045999-04
Grant ID: R01 AI045999-05
Grant ID: R01AI45999
Grant ID: R21 AI053781-02
 1
1



