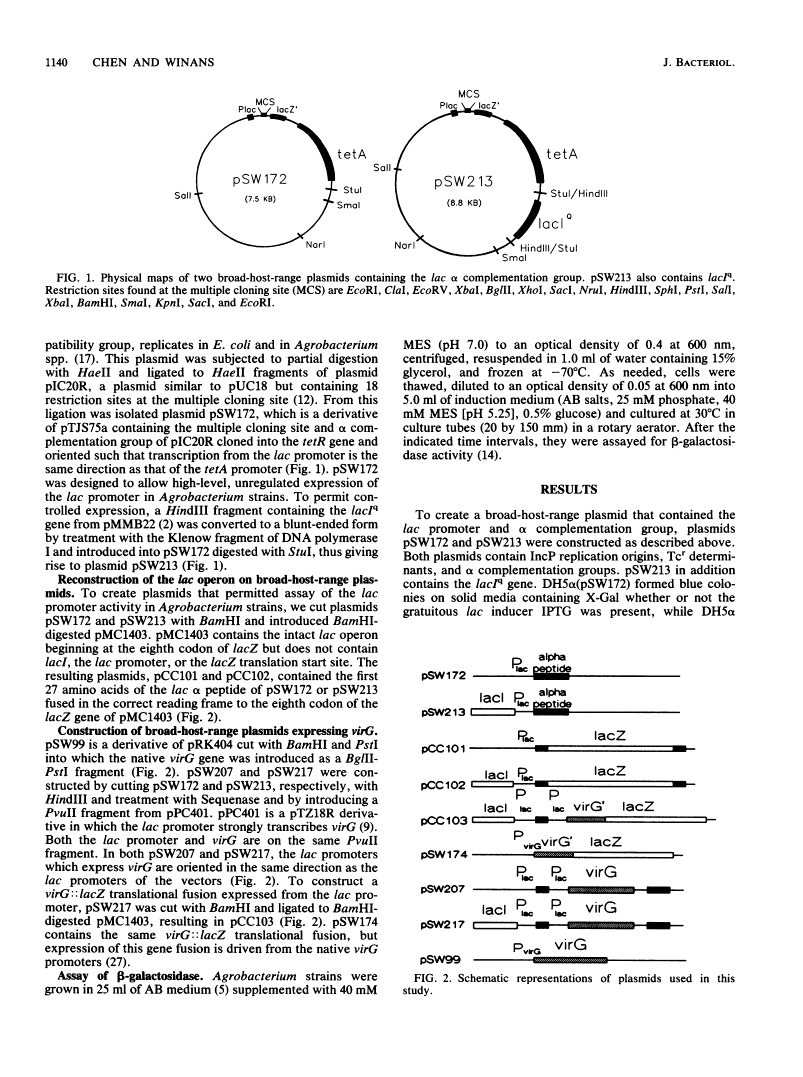
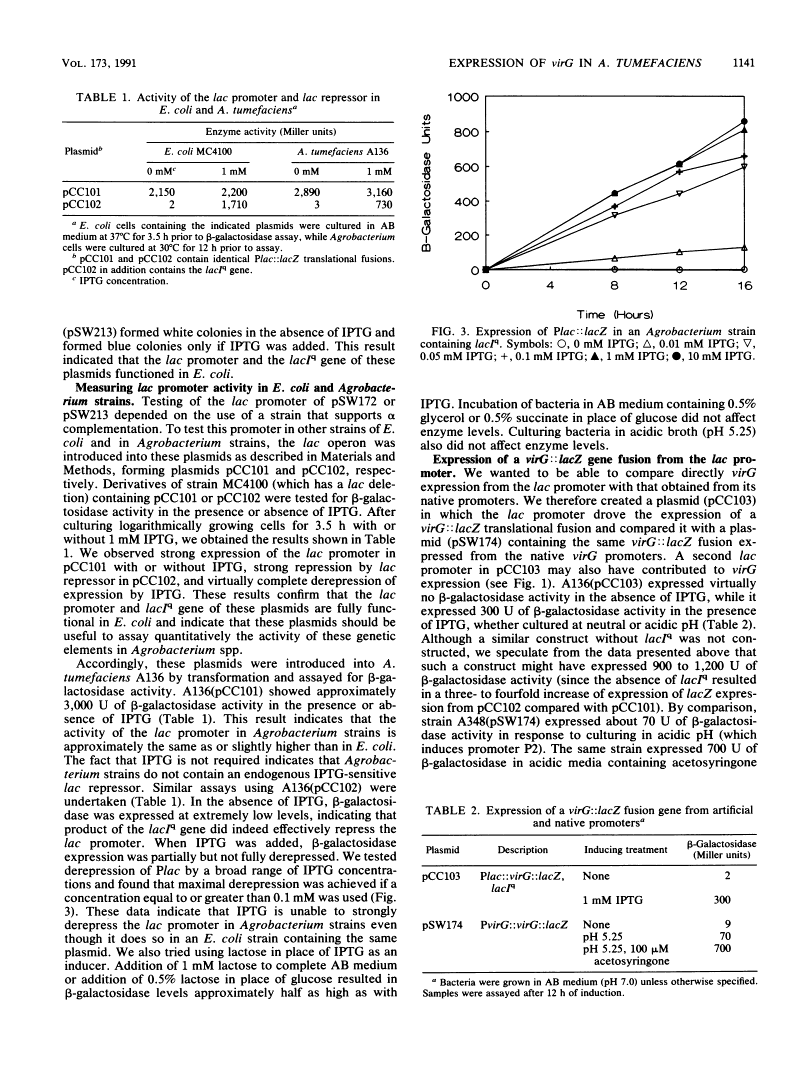
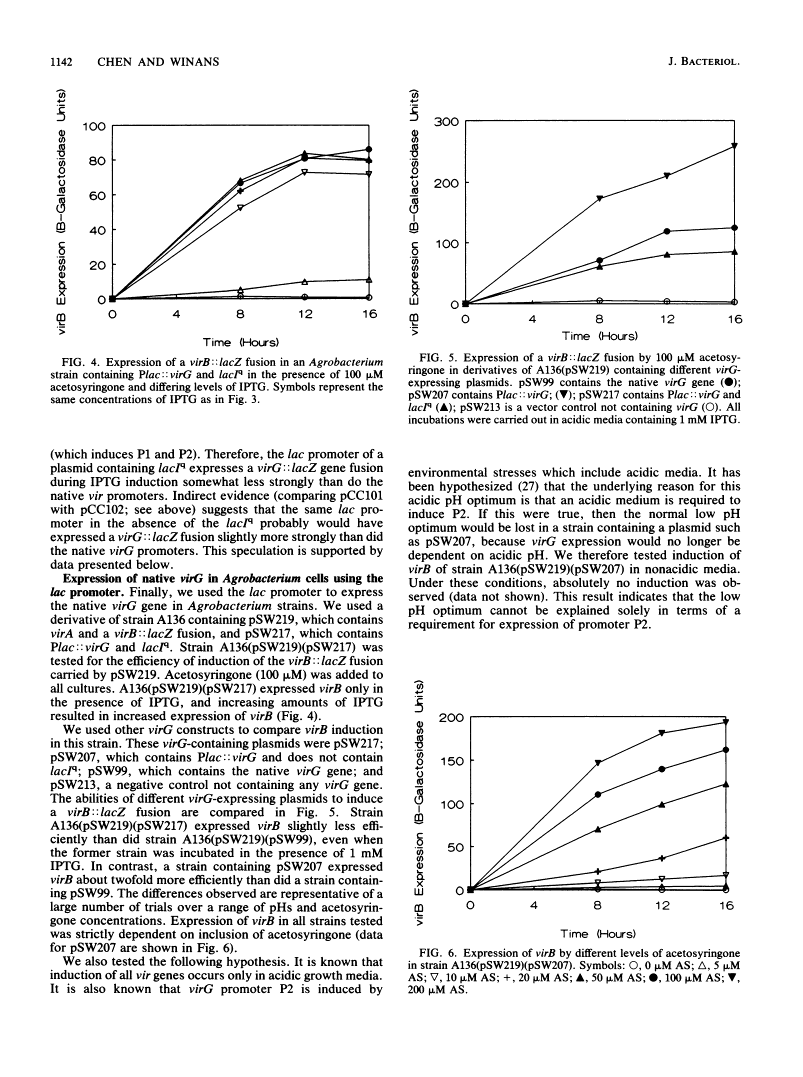
Abstract
Free full text

Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter.
Abstract
The Agrobacterium VirG protein is normally expressed from two promoters in response to multiple stimuli, including plant-released phenolics (at promoter P1) and acidic growth media (at promoter P2). To simplify the analysis of vir gene induction, we sought to create Agrobacterium strains in which virG could be expressed in a controllable fashion. To study the possibility of using the lac promoter and repressor, we constructed a plasmid containing the lac promoter fused to the lacZ structural gene. A derivative of this plasmid containing the lacIq gene was also constructed. The plasmid not containing lacIq expressed high levels of beta-galactosidase. The plasmid containing lacIq expressed beta-galactosidase at very low levels in the absence of o-nitrophenyl-beta-D-galactoside (IPTG) and at moderate levels in the presence of IPTG. We also fused the lac promoter to a virG::lacZ translational fusion and found that IPTG elevated expression of this translational fusion to moderate levels, though not to levels as high as from the stronger of the two native virG promoters. Finally, the lac promoter was used to express the native virG gene in strains containing a virB::lacZ translational fusion. virB expression in this strain depended on addition of IPTG as well as the vir gene inducer acetosyringone. In a similar strain lacking lacIq, virB expression was greater than in a strain in which virG was expressed from its native promoters. Expression of virG from the lac promoter did not alter the acidic pH optimum for vir gene induction, indicating that the previously observed requirement for acidic media was not due solely to the need to induce P2.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankenbauer RG, Nester EW. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990 Nov;172(11):6442–6446. [Europe PMC free article] [Abstract] [Google Scholar]
- Bagdasarian MM, Amann E, Lurz R, Rückert B, Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. [Abstract] [Google Scholar]
- Cangelosi GA, Ankenbauer RG, Nester EW. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6708–6712. [Europe PMC free article] [Abstract] [Google Scholar]
- Casadaban MJ, Chou J, Cohen SN. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. [Europe PMC free article] [Abstract] [Google Scholar]
- Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3672–3676. [Europe PMC free article] [Abstract] [Google Scholar]
- Close TJ, Rogowsky PM, Kado CI, Winans SC, Yanofsky MF, Nester EW. Dual control of Agrobacterium tumefaciens Ti plasmid virulence genes. J Bacteriol. 1987 Nov;169(11):5113–5118. [Europe PMC free article] [Abstract] [Google Scholar]
- Close TJ, Zaitlin D, Kado CI. Design and development of amplifiable broad-host-range cloning vectors: analysis of the vir region of Agrobacterium tumefaciens plasmid pTiC58. Plasmid. 1984 Sep;12(2):111–118. [Abstract] [Google Scholar]
- Cowing DW, Bardwell JC, Craig EA, Woolford C, Hendrix RW, Gross CA. Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci U S A. 1985 May;82(9):2679–2683. [Europe PMC free article] [Abstract] [Google Scholar]
- Jin SG, Roitsch T, Christie PJ, Nester EW. The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J Bacteriol. 1990 Feb;172(2):531–537. [Europe PMC free article] [Abstract] [Google Scholar]
- Leroux B, Yanofsky MF, Winans SC, Ward JE, Ziegler SF, Nester EW. Characterization of the virA locus of Agrobacterium tumefaciens: a transcriptional regulator and host range determinant. EMBO J. 1987 Apr;6(4):849–856. [Europe PMC free article] [Abstract] [Google Scholar]
- Marsh JL, Erfle M, Wykes EJ. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984 Dec;32(3):481–485. [Abstract] [Google Scholar]
- Melchers LS, Regensburg-Tuïnk TJ, Bourret RB, Sedee NJ, Schilperoort RA, Hooykaas PJ. Membrane topology and functional analysis of the sensory protein VirA of Agrobacterium tumefaciens. EMBO J. 1989 Jul;8(7):1919–1925. [Europe PMC free article] [Abstract] [Google Scholar]
- Rogowsky PM, Close TJ, Chimera JA, Shaw JJ, Kado CI. Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol. 1987 Nov;169(11):5101–5112. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmidhauser TJ, Helinski DR. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985 Oct;164(1):446–455. [Europe PMC free article] [Abstract] [Google Scholar]
- Sciaky D, Montoya AL, Chilton MD. Fingerprints of Agrobacterium Ti plasmids. Plasmid. 1978 Feb;1(2):238–253. [Abstract] [Google Scholar]
- Stachel SE, Nester EW. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. [Europe PMC free article] [Abstract] [Google Scholar]
- Stachel SE, Nester EW, Zambryski PC. A plant cell factor induces Agrobacterium tumefaciens vir gene expression. Proc Natl Acad Sci U S A. 1986 Jan;83(2):379–383. [Europe PMC free article] [Abstract] [Google Scholar]
- Stachel SE, Zambryski PC. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell. 1986 Aug 1;46(3):325–333. [Abstract] [Google Scholar]
- Veluthambi K, Jayaswal RK, Gelvin SB. Virulence genes A, G, and D mediate the double-stranded border cleavage of T-DNA from the Agrobacterium Ti plasmid. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1881–1885. [Europe PMC free article] [Abstract] [Google Scholar]
- Veluthambi K, Krishnan M, Gould JH, Smith RH, Gelvin SB. Opines stimulate induction of the vir genes of the Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1989 Jul;171(7):3696–3703. [Europe PMC free article] [Abstract] [Google Scholar]
- Winans SC. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J Bacteriol. 1990 May;172(5):2433–2438. [Europe PMC free article] [Abstract] [Google Scholar]
- Winans SC, Ebert PR, Stachel SE, Gordon MP, Nester EW. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. [Europe PMC free article] [Abstract] [Google Scholar]
- Winans SC, Kerstetter RA, Nester EW. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol. 1988 Sep;170(9):4047–4054. [Europe PMC free article] [Abstract] [Google Scholar]
- Yanofsky MF, Porter SG, Young C, Albright LM, Gordon MP, Nester EW. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell. 1986 Nov 7;47(3):471–477. [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.173.3.1139-1144.1991
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/173/3/1139.full.pdf
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/173/3/1139
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/173/3/1139
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jb.173.3.1139-1144.1991
Article citations
Development of constitutive and IPTG-inducible integron promoter-based expression systems for Escherichia coli and Agrobacterium tumefaciens.
3 Biotech, 13(3):91, 20 Feb 2023
Cited by: 0 articles | PMID: 36825259 | PMCID: PMC9941393
An Inducible T7 Polymerase System for High-Level Protein Expression in Diverse Gram-Negative Bacteria.
Microbiol Resour Announc, 12(2):e0111922, 16 Jan 2023
Cited by: 0 articles | PMID: 36645284 | PMCID: PMC9933640
Agrobacterium tumefaciens Type IV and Type VI Secretion Systems Reside in Detergent-Resistant Membranes.
Front Microbiol, 12:754486, 25 Nov 2021
Cited by: 3 articles | PMID: 34899640 | PMCID: PMC8656257
Inducible Expression of Agrobacterium Virulence Gene VirE2 for Stringent Regulation of T-DNA Transfer in Plant Transient Expression Systems.
Mol Plant Microbe Interact, 28(11):1247-1255, 12 Oct 2015
Cited by: 7 articles | PMID: 26292850
Mini-Tn7 vectors for stable expression of diguanylate cyclase PleD* in Gram-negative bacteria.
BMC Microbiol, 15:190, 29 Sep 2015
Cited by: 7 articles | PMID: 26415513 | PMCID: PMC4587759
Go to all (91) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media.
J Bacteriol, 172(5):2433-2438, 01 May 1990
Cited by: 83 articles | PMID: 2185220 | PMCID: PMC208880
The Agrobacterium tumefaciens vir gene transcriptional activator virG is transcriptionally induced by acid pH and other stress stimuli.
J Bacteriol, 174(4):1189-1196, 01 Feb 1992
Cited by: 34 articles | PMID: 1735712 | PMCID: PMC206411
The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes.
J Bacteriol, 172(2):531-537, 01 Feb 1990
Cited by: 73 articles | PMID: 2404941 | PMCID: PMC208474
Review: optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter.
J Ind Microbiol, 16(3):145-154, 01 Mar 1996
Cited by: 142 articles | PMID: 8652113
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (1)
Grant ID: 1 R29 GM2893-01