Abstract
Free full text

Gliomas display a microRNA expression profile reminiscent of neural precursor cells
Associated Data
Abstract
Gliomas express many genes that play a role in neural precursor cells (NPCs), but no direct comparison between glioma and stem cell (SC) gene expression profiles has been performed. To investigate the similarities and differences between gliomas and SCs, we compared the microRNA (miRNA) expression signatures of glial tumors, embryonic SCs (ESCs), NPCs, and normal adult brains from both human and mouse tissues. We demonstrated that both human gliomas (regardless of their grade) and methylcholanthrene-induced mouse glioma shared an miRNA expression profile that is reminiscent of NPCs. About half of the miRNAs expressed in the shared profile clustered in seven genomic regions susceptible to genetic/epigenetic alterations in various cancers. These clusters comprised the miR17 family, mir183-182, and the SC-specific clusters mir367-302 and mir371-373, which are upregulated in gliomas, ESCs, and NPCs. The bipartite cluster of 7 + 46 miRNAs on chromosome 14q32.31, which might represent the largest tumor suppressor miRNA cluster, was downregulated in the shared expression profile. This study provides the first evidence for association between these clusters and gliomas. Despite the broad similarity in the miRNA expression profiles, 15 miRNAs showed disparate expression between SC and gliomas. Ten miRNAs belong to the 2 SC-specific clusters and the remaining (mir135b, mir141, mir205, mir200C, and mir301a) have been previously shown to associate with malignancies. Our finding showed that all gliomas displayed NPC-like miRNA signatures, which may have implications for studies of glioma origins. Furthermore, careful study of the 15 miRNAs that differ in expression between SCs and gliomas, particularly those 5 that are not SC-specific, may enhance our understanding of gliomagenesis.
Many studies have investigated the protein-coding genes and gene products that participate in the self-renewal and differentiation of stem cells (SCs) and cancer SCs. Recently, it has been recognized that a class of endogenous, small, nonprotein coding single-stranded RNA molecules, termed microRNA (miRNA), plays a crucial role in the posttranscriptional regulation of gene expression. It has also been demonstrated that human miRNA genes are frequently located at the fragile genomic regions involved in cancer.1 Emerging evidence suggests that many miRNAs may be oncogenes and/or tumor suppressors. In addition, recent studies indicate that miRNAs also play essential roles in SC self-renewal and differentiation.2 Some miRNAs are specifically expressed in SCs and control both self-renewal and differentiation through negative regulation of the expression of certain key genes. Despite the obvious similarities in the self-renewal capacity of SCs and cancer cells, it is not yet known how miRNA profiles of cancer cells compare to the miRNA profiles of nontransformed normal SCs.
Brain tumors of glial origin contain heterogeneous neoplastic cell populations that phenotypically resemble undifferentiated or immature glial cells, rather than a fully differentiated lineage. This correlates with the observed biological characteristics of glial tumors, including the ability to grow as neurospheres in culture, self-renewal, migration, and invasion. Moreover, it is in agreement with the current view of cancer as an aberrant organ3 containing a SC-like population that drives tumor growth.4 Understanding the growth and differentiation pathways responsible for glioma SC proliferation and survival will aid in the discovery of novel therapeutic targets. As many pathways may participate in regulating glioma–stem cell growth, we chose to study the miRNA expression profiles to attempt to identify the specific growth and signaling pathways involved. This study compares the miRNA profiles of glial tumors, embryonic SCs (ESCs), neural precursor cells (NPCs), and normal adult brain tissues of both human and mouse origin.
Materials and Methods
Samples
We used an miRNA array to compare the expression profiles of 180 mature mRNAs (miRNAs) in ESCs, NPCs, gliomas, and normal brains. Our initial panel contained 15 RNA samples derived from glial tumors of various grades, ESCs, NPCs, and normal adult brains from both humans and mice.
For human miRNA testing, fresh frozen tumors were acquired from the brain tumor bank of Hadassah Hebrew-University Medical Centre, in accordance with the regulations and approved procedures of the committee of human research. The initial human panel included a commercial mix of normal brain (Ambion, Austin, TX), an astrocytoma (A), an oligoastrocytoma, an oligodendroglioma (O), two glioblastoma multiforme (GBM), cell lines of an anaplastic astrocytoma (U87MG), and of a GBM (A172), a human ESC (hESC) line (HES-2)5 and hESC-derived neural precursors cells (hNPCs).6
The mouse panel included brains from adult C57BL/6 mice, a mouse glioma cell line (GL261), and one sample of each of the following: NPCs from a 14-day-old embryo (meNPCs), neonatal NPCs (mnNPCs), oligodendrocyte progenitor cells (mOPCs), and astrocytes (mAs), all isolated from brains of 1-day-old C57BL/6 mice. Each sample of precursor or progenitor cells contained pooled RNA extracted from different cell preparations. There were 3 to 7 biological replicates per cell line or tissue type and 3 technical replicates for each sample.
Later, we extended the panel by exploring and quantifying the expression of 5 additional miRNAs (a total of 13 miRNAs located in the Dlk1-Dio3 miRNAs cluster). For this evaluation, we used 12 human astrocytic tumors and 3 Os that do not contain a 1p/19q loss of heterozygosity (LOH).
We also analyzed DNA samples from 40 human tumors (20 Os and 20 glioblastomas) previously extracted for routine genetic analysis. DNA was extracted from paraffin-embedded sections of the tumors used for histological diagnosis. The purpose of this analysis was to evaluate whether chromosomal deletion was associated with downregulation of the Dlk1-Dio3 miRNA cluster analyzed in the extended panel. All patients signed an informed consent form prior to DNA extraction and a neuropathologist (Y.F.) reviewed all tumor samples for verification of histological diagnosis and tumor grade.
RNA and DNA Extractions
Total RNA was isolated from snap frozen gliomas or cell cultures using the TRI Reagent® (Sigma-Aldrich). Each RNA sample used for the analysis was composed of a pool of numerous extractions (3 to 7), each obtained from individual tumor tissue, different cell preparations, or normal mouse brains. The RNA isolated from adult brain was a commercial mix pooled from multiple donors and several brain regions (Ambion Inc.).
Tumor DNA was extracted from paraffin-embedded tissue using the Paraffix kit (Syntezza Bioscience, Jerusalem, Israel), according to the manufacturer's instructions.
miRNA Quantification
A total of 180 miRNA genes were quantified in the miRNA array using the TaqMan® miRNA Assays Panel Early Access Kit (Applied Biosystems), according to the manufacturer's instructions. Briefly, the assay included two steps: RT and PCR. Each RT reaction was incubated for 30 minutes at 16°C and then at 42°C. Real-time PCR was performed on an ABI 7000 sequence detection system in triplicate for each sample. The fold-change normalization was based on cycle threshold (CT) changes of the mean calculated values obtained by subtracting individual CTs for each tissue from a median CT (ΔCT). For the individual miRNA quantification, the fold change was normalized to the RNU6B transcript (ΔCT). Following normalization, the fold change of each miRNA was calculated between the analyzed tissue (tumor, cell line or SC) and normal brain reference (ΔΔCT).
LOH Analysis
For the microsatellite PCR-based LOH analysis, primer pairs for the microsatellite loci D14S65 at 14q32.2 and D14s292 at 14q32.31 were labelled with NED and FAM, respectively (Applied Biosystems). PCR was performed on each sample in a final volume of 25 µl, containing the two primer pairs (3 pmoles of each primer), 25 ng of DNA, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 0.2 mM deoxyribonucleoside triphosphates, 2.5 mM MgCl2, and 0.6 unit of AmpliTaq Gold DNA polymerase (Applied Biosystems). PCR cycling conditions were 95°C for 9 minutes once, 42 cycles at 94°C for 45 seconds, 55°C for 45 seconds, and 72°C for 60 seconds, with a final elongation step of 45 minutes at 60°C. Amplified PCR products were electrophoresed in denaturing 5% polyacrylamide gels on an ABI Prism 310 automated DNA sequencer (Applied Biosystems). Data analysis was performed with GeneScan Analysis software version 3.1 (Applied Biosystems).
Data Analysis and Visualization
Hierarchical clustering was performed on those miRNAs that displayed deviated expression (defined as either less than 0.5-fold or more than 2-fold of adult reference brain). MATLAB Version 7.5.0.342 (R2007b) was used to generate the clustergram.
To identify significant differences in the expression levels of miRNAs between SCs and gliomas, a standard t-test was performed on all miRNA data using Spotfire DecisionSite 9.1.1. A P value threshold of .05 was used for subsequent interpretation.
To characterize the trends exhibited by these data and explore correlations between samples, a principal components analysis (PCA) was carried out. Using PCA we first analyzed all raw data, then separately 92% of the data that excluded the extreme values, and finally the remaining 8% of the data that were excluded from the previous analysis. This created 2 miRNAs data sets. The first set included all miRNA except the 15 miRNAs that showed differential expression between gliomas and SCs (total of 165 miRNAs). The second set included only the 15 miRNAs that displayed significant differential expression between SCs and gliomas. The visualization of the PCA was performed with the Partek Genomics Suite 6.4.
Results
To explore a possible association between the miRNA profiles of gliomas and SCs, we compared the expression profiles of 180 mature miRNAs in ESCs, NPCs, gliomas, and normal brains. The initial analysis included 15 samples derived from glial tumors of various grades, ESCs, NPCs, and normal adult brains from both humans and mice. The results of this analysis are shown in the Supplementary Material, Table S1 and are detailed below. To test the validity of these results we analyzed the partially available raw data of Silber et al.7, a study that used the same miRNA array, and compared it with our results. Although Silber et al. used a different normalization method and different analysis software; the findings obtained from the analysis of Sibler's data set were similar to the results obtained from our data set (Supplementary Material, Fig. S1). Consequently, our conclusions remained unchanged and the comparison emphasized the validity of our results.
Gliomas Display an miRNA Expression Profile That Is Reminiscent of the Profile From SCs
As described above, miRNA expression was quantified by RT real-time PCR, using the expression profile of the normal adult brains as a reference for all other tissues, separately for each species. It is interesting to note that we observed 80% identity between the miRNA expression signature of human gliomas and the miRNA expression profile of methylcholanthrene-induced mouse glioma GL-261. In the human glioma panel, 71 of the 180 miRNAs showed a distinct expression pattern (defined as either less than 0.5-fold or more than 2-fold compared with adult brain). The expression pattern of these 71 miRNAs was remarkably reminiscent of the expression pattern observed in ESCs and NPCs (Fig. 1A, Table 1).
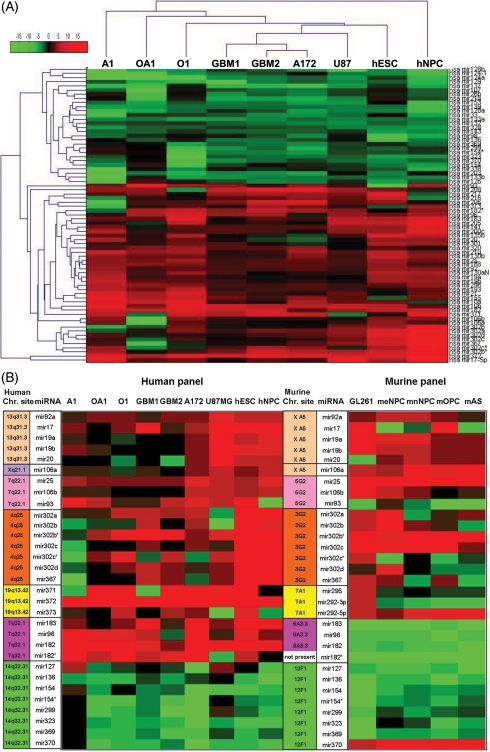
Heat maps displaying the expression of the miRNAs that showed distinct expression in gliomas, ESCs and NPCs, when compared with normal adult brains. (A) Hierarchical clustering of the 71 miRNAs showed distinct expression (less than 0.5-fold and more than 2-fold) in human gliomas, hESCs, and NPCs, when compared with normal brains. The clustergram was generated using MATLAB Version 7.5.0.342 (R2007b). The expression values ranged between + 20 log2 to −20 log2. (B) Heat map of the clustered miRNAs that showed significantly different expression when compared with normal adult brains. The chromosomal location of the miRNAs is specified in the right row of each species panel: human (right) and murine (left) panels. This figure also demonstrates the similarity in miRNA expression between human and murine gliomas. The expression values ranged between + 10 log2 to −10 log.
Table 1.
Summary of upregulated and downregulated miRNAs in SCs and gliomas
| Upregulated miRNA | Downregulated miRNA | ||||||
|---|---|---|---|---|---|---|---|
| miRNA | hESC | hNPC | Gliomas (average) | miRNA | hESC | hNPC | Gliomas (average) |
| mir106a | 4.55 | 15.33 | 4.32 | mir33 | 0.03 | 0.92 | 0.16 |
| mir106b | 10.87 | 13.57 | 5.68 | mir98 | 0.02 | 0.001 | 0.5 |
| mir10a | 0.28 | 9.47 | 30.53 | mir107 | 0.19 | 2.08 | 0.28 |
| mir10b | 4.89 | 65.21 | 107.6 | mir124-1 | 0.002 | 0.07 | 0.25 |
| mir17 | 252,108.25 | 48.57 | 40627.83 | mir124a | 0.01 | 0.34 | 0.06 |
| mir19a | 19.78 | 23.1 | 10.35 | mir126 | 0.22 | 0.03 | 0.27 |
| mir19b | 23.11 | 23.87 | 17.21 | mir128a | 0.01 | 0.08 | 0.14 |
| mir20 | 19.41 | 18.66 | 7.08 | mir128b | 0.002 | 0.06 | 0.11 |
| mir21 | 7.7 | 0.81 | 9.77 | mir129 | 0.01 | 0.18 | 0.08 |
| mir25 | 8.86 | 23.04 | 9.8 | mir132 | 0.08 | 0.16 | 0.19 |
| mir92 | 12.93 | 17.91 | 6.97 | mir133a | 0.11 | 0.11 | 0.08 |
| mir93 | 48.59 | ND | 13.52 | mir133b | 0.07 | 0.1 | 0.38 |
| mir96 | 23.23 | 8.6 | 19.04 | mir136 | 0.1 | 0.004 | 0.44 |
| mir130aN | 41.55 | 44.57 | 9.41 | mir137 | 0.02 | 0.4 | 0.1 |
| mir130b | 14.93 | 83.4 | 8.04 | mir139 | 0.01 | 0.12 | 0.09 |
| mir135b | 65.69 | 469.9 | 2.5 | mir143 | 0.07 | 0.04 | 0.23 |
| mir141 | 551.68 | 21.2 | 6.33 | mir153 | 0.004 | 1.04 | 0.09 |
| mir155 | 9.36 | 0.18 | 33.51 | mir154 | 0.04 | 0.09 | 0.44 |
| mir182 | 156.25 | 97.14 | 195.16 | mir154* | 0.35 | 0.23 | 0.29 |
| mir182* | 14.47 | 1.21 | 49.62 | mir203 | 1.07 | 0.08 | 0.2 |
| mir183 | 467.46 | 48.57 | 90.75 | mir218 | 0.25 | 2.5 | 0.29 |
| mir188 | 4.04 | 17.11 | 8.52 | mir26b | 0.18 | 0.57 | 0.38 |
| mir193 | 6.76 | 1.76 | 20.84 | mir299 | 0.47 | 0.17 | 0.25 |
| mir200c | 854.35 | 24.68 | 6.17 | mir323 | 0.05 | 0.03 | 0.27 |
| mir205 | 1680.51 | 106.9 | 18.8 | mir328 | 0.04 | 0.07 | 0.19 |
| mir206 | 1.38 | 15.71 | 4.99 | mir338 | 0.02 | 0.21 | 0.34 |
| mir208 | 0.48 | 11.42 | 34.31 | mir369 | 0.18 | 0.55 | 0.32 |
| mir210 | 11.99 | 54.8 | 57.02 | mir370 | 0.17 | 0.07 | 0.27 |
| mir216 | 0.07 | 127.5 | 5.93 | ||||
| mir217 | 0.21 | 944.3 | 4.32 | ||||
| mir296 | 18.02 | 5.87 | 13.37 | ||||
| mir301a | 22.26 | 39.29 | 3.8 | ||||
| mir302a | 1 002 856.52 | 356.3 | 9.84 | ||||
| mir302b | 599 617.84 | 22.96 | 3.56 | ||||
| mir302b* | 10 769.65 | 192.2 | 56.14 | ||||
| mir302c | 187 779.99 | 141 | 2.92 | ||||
| mir302c* | 36 704.87 | 34.85 | 10.5 | ||||
| mir302d | 408 134.12 | 482.7 | 4.85 | ||||
| mir320 | 1.67 | 4.9 | 10.42 | ||||
| mir367 | 814 572.61 | 211.7 | 4.98 | ||||
| mir371 | 1 508.28 | 1.77 | 104.64 | ||||
| mir372 | 263 909.50 | 630.4 | 1,797.98 | ||||
| mir373 | 28.58 | 0.2 | 7.1 | ||||
Abbreviations: SC, stem cell; hESCs, human embryonic stem cells; hNPC, human neural precursor cells. miRNAs that showed disparate expression (t-test, P ≤ .05) in SCs when compared with gliomas are marked in bold.
Clustering of the miRNAs That Constitute the Shared Profile of SCs and Gliomas
About half of the 71 miRNAs exhibiting similar expression profiles in gliomas, hESCs, and NPCs clustered in seven genomic regions: (1–3) the miR17 family, containing the miRNA clusters miR17-92, miR106b-25, and miR106a-363 and encoded by three gene clusters on chromosomes 13q31.3, 7q22.1, and Xq21.1, respectively; (4) mir183-182 on chromosome 7q32.1; (5) mir367-302 on chromosome 4q25; (6) mir371-373 on chromosome 19q13.42; and (7) the miRNA cluster in the Dlk1-Dio3 genomic region at 14q32.31 (Fig. 1B). Five of these clusters were recently linked to SCs (Table 2).8
Table 2.
The miRNAs clusters showed distinct expression in gliomas, with similarities to expression profiles of hESCs and NPCs, their involvement in cancer development and their relationship to SC signatures.
| miRNA cluster | Gliomas and SC in current study | Gliomas in prior studies | Nonglial tumors | SC in previous studies | Related genomic aberrations in cancer |
|---|---|---|---|---|---|
| miR17 family clusters: miR17-92, miR106b-25, and miR106a-363 on chromosomes 13q31.3, 7q22.1 and Xq21.1, respectively. | Upregulated: mir17-92, mir106b-25 mir106a | No reports | Upregulated: mir17-92 in haematopoietic malignancies and solid tumors (breast, colon, lung, pancreas, prostate, stomach40,41), and mir17-92 and mir106b-25 in medulloblastoma42,43 | Upregulated: mir17-92 and mir106a in hESCs8 | Amplification of chromosomal region 13q31.3, in lymphomas39 |
| mir183-182 cluster on chromosome 7q32.1 | Upregulated: human gliomas and hESCs | No reports | Upregulated: colorectal cancer23 | Upregulated: mir183 in murine ESCs and embryoid bodies13 | Frequent structural abnormalities of chromosome 7 in splenic lymphoma with villous lymphocytes,44 leukaemia and lymphoma. 7q32.1 translocations and deletions45–47 |
| Downregulated: murine gliomas and mNPCs | |||||
| mir367-302 on chromosome 4q25; mir371-373 on chromosome 19q13.42 | Upregulated | No reports | Upregulated: mir373 in retinoblastoma, breast and testicular germ cell cancers,31,48 mir302 in leukaemia cell lines49 | Upregulated in both hESCs8,28 and murine ESCs2,11,13,29,30 | Amplification of 0.89 Mb at chromosome band 19q13.42 covering the miRNA cluster in a pediatric embryonal tumor with abundant neutropil and true rosettes |
| Downregulated: mir372 in medulloblastoma42,43 | |||||
| The large Dlk1-Dio3 7 + 46 bipartite miRNA cluster on chromosome 14q32.31 | Downregulated | No reports | Downregulated: epithelial ovarian cancer27 | Downregulated in SC8 | Frequently altered in both haematopoietic and solid tumors (more than 160 articles summarized in Supplementary Material, Table S2) |
Abbreviations: SC, stem cell; hESCs, human embryonic stem cells; NPC, neural precursor cells. These miRNAs are presented according to their genomic region.
The miRNA Cluster on Chromosome 14q32.31 is Downregulated in All of the Investigated Gliomas
The large 7 + 46 bipartite Dlk1-Dio3 miRNA cluster on chromosome 14q32.31 was of special interest. This cluster was downregulated in the initial panel in all gliomas, ESCs, and NPCs. This suggests that this cluster might represent the largest tumor-suppressor miRNA cluster. For this reason, we further explored this region and quantified the expression of 13 miRNAs located along the Dlk1-Dio3 miRNA cluster. For this evaluation, we used 12 astrocytic tumors and 3 Os that did not contain 1p/19q LOH. All of the miRNAs that were tested appeared to be downregulated in hESCs, hNPCs, and in all of the gliomas (Fig. 3A). Because the miRNAs from this large miRNA cluster are expressed only from the maternally inherited allele,9 deletion of the active allele may result in complete silencing of these miRNAs. To examine whether a chromosomal deletion was the likely reason for the downregulation of these miRNAs, LOH analysis was performed on paraffin-embedded samples of other gliomas, specifically 20 Os and 20 GBMs. D14S65 at 14q32.2 and D14s252 at 14q32.31 were used as markers for this analysis. Only 5% (1/20) of the Os and 35% (7/20) of the GBMs showed LOH of 14q in this region (data not shown).
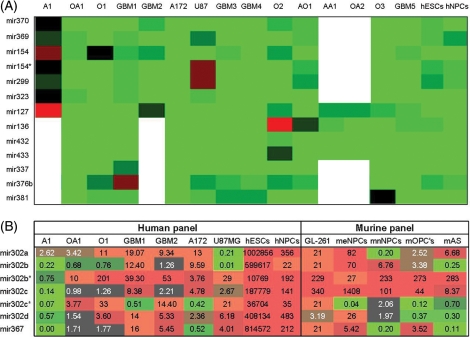
Heat maps displaying the expression levels of miRNAs of 2 clusters in gliomas, hESCs and NPCs, relative to human brains. (A) Thirteen miRNAs within the miRNA cluster in the Dlk1-Dio3 genomic region 14q32.31 in 15 gliomas, hNPCs and hESCs (white cell represents untested). The expression values ranged between +10 log2 to −10 log2. (B) The ES-specific cluster, mir367-302, on chromosome 4q25 (fold change is specified in each cell) in human (left panel) and murine (right panel) tissues comprises gliomas and precursor cells.
Unclustered miRNAs in the Shared Profile
In addition to the previously discussed miRNA clusters, we also identified unclustered miRNAs that shared similar expression in gliomas and SCs (Fig. 1A, Table 1). This group included mir21, which has been previously found to be related to the pathogenesis of gliomas,10 as well as both mir124 and mir137, which both demonstrate a significantly reduced expression in gliomas in our study as well as in a previous study.7
Fifteen miRNAs Showed Disparate Expression in SCs and Gliomas
Fifteen of the 71 miRNAs that showed similar trend of expression in gliomas and SCs compared with normal brain displayed disparate expression (P < .05) between gliomas and SC (Table 1, bold and underlined). Ten of these miRNAs belong to the 2 SC specific clusters mir367-302 and mir371-373. These clusters demonstrated a significantly higher expression in hESCs, when compared with gliomas (range: 223–190,000 fold). The expression of these clusters in hNPCs was only moderately higher than the expression level in gliomas (range: 3–99-fold) (Fig. 3B, Table 1, bold and underlined). The fact that these miRNA gene families have murine homologues with similar genomic organization and expression patterns11 prompted us to further explore the expression of these miRNAs in mice. To that end, we determined the expression pattern of these miRNAs in the mouse glioma GL261 line and in neural cells obtained from the following developmental stages: meNPCs, mnNPCs, mOPCs, and mAs. We found that the miRNA expression pattern was similar in the mouse and human gliomas. Furthermore, we found that the expression level of these miRNAs in murine glioma resembled the expression pattern in either meNPCs or mnNPCs (Fig. 1B). Apart from the 10 miRNAs that belong to the SC-specific clusters, the other miRNAs that showed significantly different expression in gliomas versus SC are mir135b, mir141, mir205, mir200C, and mir301a.
We used a PCA to characterize the trends exhibited by the above raw data and to further explore the correlations between the different tissues. We selected PCA because it is a nonparametric analysis that could demonstrate potential clustering in an unsupervised manner, independent of any bias or hypothesis about data probability distribution. Furthermore, PCA allows an easy visualization of similarity and disparity among samples. For these reasons, PCA is usually recommended as an exploratory tool to uncover unknown trends in data. The PCA analysis was performed separately on 2 miRNAs data sets. The first set included all miRNAs except the 15 miRNAs that displayed disparate expression in SCs and gliomas (165 miRNAs) (Fig. 2A). The second set was composed of the 15 miRNAs with dissimilar expression in SC and gliomas (Fig. 2B). This analysis reinforced the validity of the clustering interpretation by demonstrating that both SCs and gliomas are integrated into one principal component, while the normal brain constitutes a separate category (Fig. 2A). Furthermore, the PCA analysis also demonstrated that when only the 15 miRNAs that showed dissimilar expression between gliomas and SCs were incorporated into the analysis, the gliomas integrated together with the normal brain into one category whereas the SCs stood apart (Fig. 2B). This implies that further investigation of these miRNA might provide important clues related to gliomagenesis.
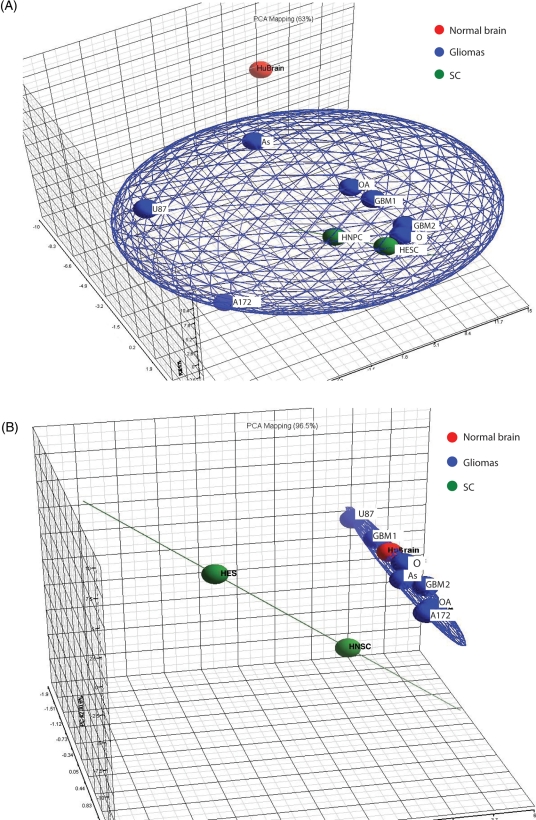
A 3D PCA scatter plot of the miRNA data that characterize the trends exhibited by the expression profiles. Each dot represents a sample as indicated. Each color represents the type of the sample (red, huBrain; blue, gliomas; green, SC). The data are grouped by tissue type. (A) This set of data included all miRNAs except the 15 miRNAs that displayed disparate expression in SCs and gliomas (total of 165 miRNAs). (B) Data composed of the 15 miRNAs showed dissimilar expression between gliomas and stem cells.
Discussion
This study investigated and compared the expression profiles of 180 miRNAs in ESCs, NPCs, gliomas of various grades, and normal adult brains from both humans and mice. Previous studies investigating miRNA profiles in gliomas7,12 or SCs8,11,13 evaluated each tissue separately, using diverse techniques. Thus, the importance and novelty of the current study lies in the direct comparison between miRNA profiles of SCs and gliomas accomplished by one experiment, using a single method.
Both Human and Mouse Gliomas have NPC-like miRNA Expression Signatures
Our results demonstrated that human and mouse glioma cell lines, as well as primary human gliomas, had an NPC-like miRNA expression signature (Figs 1A and and2A).2A). Although our cohort was relatively small, our data is supported by previous miRNA profiling studies performed separately on gliomas7,12 or SCs (including NPCs) in diverse stages of differentiation.8,11,13 Several miRNAs, mir21, mir124, and mir137, shared similar expression in gliomas and SCs (Fig. 1A, Table 1) and have been previously implicated in the pathogenesis of gliomas.7,10 Furthermore, analysis of Sibler's7 raw data demonstrated similar results to those observed in our data (Supplementary Material, Fig. S1). These findings of miRNA expression signature, based on data produced independently in 2 different laboratories, strongly supports the results of our relatively small group of samples. (Supplementary Material, Fig. S1).
We found a similar NPC-like expression signature in cell lines that represent a relatively unified cell population, as well as in unsorted human tumors. This might indicate that most tumor cells, not only cancer SCs, exhibit an SC-like miRNA expression signature. The rationale for this conclusion is based on the previous observation that the fraction of cancer SCs within the tumor bulk is extremely small.14 To conclusively resolve this issue, further evaluation, comparing the miRNA profiles of different cell populations contained within gliomas, is required. We also demonstrated an across-species similarity in the miRNA expression signature of gliomas, independent of whether the tumor arose spontaneously in humans or was induced in mice by the carcinogenic chemical methylcholanthrene. Furthermore, both low-grade and high-grade human gliomas exhibit an NPC-like expression profile. This finding is unexpected considering the phenotypical and genotypical differences between low-grade and high-grade gliomas and it might arise from the small sample size that served for the analysis of each tumor grade, which limits the ability to detect the anticipated differences. It should be noticed that the study was not designed to evaluate differences between tumors of various grades. However, 2 other studies that examined this issue found some differential expressions between GBM and tumors of lower grades.7,15 The first study performed an miRNA array on 8 high-grade gliomas (4 AAs and 4 GBMs) and the other study analysed a single miRNA (mir10b) in 43 gliomas of both low and high-grades. Further studies with larger number of gliomas of each grade should clarify this point in the future.
Half of the miRNAs That Show Similar Expression Patterns in Gliomas and SCs Cluster in Seven Genomic Regions Susceptible to Genetic/Epigenetic Alternations in Various Cancers
This study is the first to demonstrate that half of the miRNAs that are expressed distinctly in gliomas, and exhibit similar expression profiles to hESCs and NPCs, cluster in 7 genomic regions. Most of these miRNA clusters are either involved in cancer development or have been previously connected to SCs. Furthermore, the same 7 genomic regions are frequently susceptible to genetic or epigenetic alternations in different types of malignancies (see Table 2).16,17
The miR17 Family and mir183-182 Cluster
The 3 miRNA clusters related to the miR17 family of miRNAs (Table 2) displayed substantially increased expression in gliomas, hESCs, and hNPCs, as compared with normal brain reference (Fig. 1B, Table 1). The miR17-92 cluster has been reported to have a pro-oncogenic role in a mouse model system of Myc-induced B-cell lymphoma.18,19 Furthermore, lines of evidence link this cluster to cancer pathogenesis, leading to its designation as an “oncomir.” In agreement with our finding that the mir17 family is involved in both cancer and normal development, recent work demonstrated that the mir17-92 and mir106b-25 clusters are also critical for normal development.20–22
The other mir183-182 cluster (Table 2) was also upregulated in human gliomas, hESCs, and hNPCs. This miRNA cluster was previously shown to be upregulated in colorectal cancer,23 but its involvement in gliomagenesis and other cancer pathology deserves further study.
Is the Bipartite Cluster of 7 + 46 miRNAs on Chromosome 14q32.31 the Largest Tumor Suppressor miRNA Cluster?
The large 7 + 46 bipartite miRNA cluster on chromosome 14q32.31 was downregulated in gliomas, ESCs, and NPCs. Further quantification of the expression of 5 additional miRNAs (13 at total) located within this miRNA cluster demonstrated that they were also downregulated in the tested tissues (Fig. 3A). Since all miRNAs in the Dlk1-Dio3 cluster are derived from a single precursor transcript,24 it is likely that the entire cluster is collectively downregulated. There is strong evidence, based on more than 160 publications reported from 1995 to the present (74 reports are summarized in Supplemental Material, Table S2), that this chromosomal region is frequently deleted or genetically altered in both hematopoietic and solid tumors. Such genetic events are more prevalent in high-grade and metastatic tumors than in low-grade tumors. Previous studies demonstrated LOH in this chromosomal region in 57% of GBMs18 and in 68% of Os, where it was either deleted or methylated.25 Our results demonstrated that only 5% of Os and 35% of GBMs showed LOH at this region. As indicated above, the miRNAs within this cluster were uniformly downregulated in all of the gliomas tested. Thus, LOH is certainly not the sole mechanism responsible for the decreased expression. Other genetic or epigenetic mechanisms are likely responsible for silencing this polycistronic cluster in gliomas.
A previous functional complementation study has provided direct evidence for the potential role of the 14q32 region in tumor suppression, as monochromosome transfer of chromosome 14q32 decreased the tumorigenicity of the SLMT-1 S1 esophageal carcinoma cell line.26 Furthermore, 8 of the miRNAs located in the Dlk1-Dio3 region were recently evaluated in epithelial ovarian cancer, where downregulation of these miRNAs was found to be associated with a higher proliferation index and significantly shorter patient survival.27 The abundant evidence accentuating the role of this region in gliomas and other human malignancies raises the possibility that this cluster is the largest polycistronic miRNA tumor suppressor. However, confirmatory data on the specific portion of this miRNA cluster that might be directly involved in human gliomagenesis are still lacking.
The SC-Specific mir367-302 and hsa-mir371/372/373/mmu-mir290 Clusters
The last 2 clusters identified, mir367-302 and hsa-mir-371/372/373/mmu-mir-290 (Table 2), both showed elevated expression in gliomas, hESCs, and hNPCs. The expression pattern of these miRNAs was similar in the mouse glioma GL261 line and in human gliomas. Importantly, in human tissues, these clusters demonstrated significantly higher expression in SCs when compared with gliomas (t-test P ≤ .05) (Fig. 3B, Table 1, bold and underlined). The differences in the expression levels between human gliomas and ESCs were very large, and less pronounced between human gliomas and hNPCs. In the murine samples, the expression level of these glioma miRNAs was within the same range as in meNPCs and mnNPCs (Fig. 3B). Recent work demonstrated that these clusters are strongly expressed in both hESCs8,28 and murine ESCs,2,11,13,29,30 and were thus designated as stem cell-specific miRNAs. Consequently, they are expressed at higher levels in ESCs, and their expression diminishes gradually during neural differentiation (Fig. 3B, murine panel), with a presumably specific pattern of expression at each developmental stage. The similarities in the expression levels of these SC-specific clusters, mir302-367 and mir371-373, in both gliomas and NPCs, as well as the analog expression of these miRNAs in human and mouse gliomas (Fig. 3B) are unlikely to be coincidental. Based on these similarities, it is likely that the origin cells of gliomas are related to NPCs. Of course, there is not enough evidence to rule out the possibility that, following genetic and/or epigenetic alternations, further differentiated progeny are the true glioma-initiating cells that revert to a “stem-like” status. It is also interesting to note that several previous studies have linked the miR372/373 miRNAs to tumorigenesis. It has been shown that miR373 stimulates cancer cell migration and invasion in vitro and in vivo.31 Also, miR372/373 promote tumorigenesis, in combination with oncogenic RAS, at least in part by directly inhibiting the tumor suppressor LATS2.32
In summary, the 7 miRNAs clusters related to the above described miRNA expression profile, have been previously shown to be tightly linked to tumorigenicity in other type of cancers (as summarized in Table 2). Our study shows the first evidence that expression of these miRNAs is also altered in gliomas. Together with the fact that the expression levels of these clusters are similar in gliomas and SCs, our findings may suggest the possibility that these miRNAs have an important role in gliomagenicity.
The miRNAs That Display Disparate Expression Between SC and Gliomas Are Either SC Specific, or They Have Been Previously Associated with Carcinogenesis
Of the 15 miRNAs that displayed disparate expression between SCs and gliomas, 10 were identified as SC-specific miRNAs. The other 5 miRNAs, mir135b, mir141, mir205, mir200C, and mir301a, are of particular interest. It is important to note that all 5 of these miRNAs were previously found to be involved in cancer.33–36 A prior study in colorectal cancer demonstrated that miR135a and b suppress the expression of adenomatous polyposis coli (APC) and induce downstream WNT pathway activity.33 Although APC is a key tumor suppressor gene,37 its role in gliomas, as well as the related function of mir135b, deserves further studies in brain tumors.
The miRNA-200 family members (mir141 and mir200c) were previously explored in ovarian carcinoma. It was demonstrated that high expression of these miRNA-200 family members correlated significantly with poor prognosis.38 Furthermore, miRNA-200 family members, together with mir205, were shown to be important in the epithelial–mesenchymal transition. This transition is characterized by loss of cell adhesion, repression of E-cadherin expression, and increased cell mobility. Similar to our results in gliomas, it was demonstrated that, relative to normal tissues, these miRNAs are usually upregulated in cancer in other tumor types. This transition was not observed in invasive breast cancer cell lines with a mesenchymal phenotype or in regions of metaplastic breast cancer specimens lacking E-cadherin where expression of these miRNAs was lost.35 Although these miRNAs were moderately upregulated in our study in gliomas when compared with normal brain, they were expressed at significantly lower levels in gliomas when compared with SCs (Table 1). More investigations are necessary to elucidate the possible mechanistic role of these miRNAs in gliomas.
In summary, we have shown that the miRNA signature of gliomas is reminiscent of the miRNA signature of SCs, particularly NPCs. About half of the miRNAs expressed in the shared profile cluster in 7 genomic regions. These regions are particularly prone to genetic and/or epigenetic aberrations in different types of cancers (eg, LOH of chromosome 14q32.31, or amplification of chromosome 13q31.3). Together, these findings suggest the possibility that NPCs are the origin cells of gliomas, and that aberrations in critical regions might be necessary to maintain the stem cell nature of gliomas. Still, the possibility that the source of gliomas is related to a more differentiated progeny that has reverted to “stem-like” status, due to aberrations in specific regions, cannot be ignored. We also found that the large bipartite cluster of 7 + 46 miRNAs on chromosome 14q32.31 is downregulated in all gliomas, which might imply that it represents the largest tumor suppressor miRNA cluster. Further study is needed to confirm the role of this cluster in gliomas. Finally, we found that 15 miRNAs displayed differences in expression between gliomas and SCs. When these miRNAs were analyzed by PCA, glioma samples integrated with normal brain, whereas SCs were classified separately. This suggests the importance of these miRNAs as clues to gliomagenesis. Hopefully, future studies will shed additional light on the function of these miRNAs in gliomagenesis and will provide evidence as to whether manipulation of the expression of these miRNAs can intervene in the biology of these devastating tumors.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online.
Acknowledgments
We thank Prof. Haim Ovadia for reviewing the manuscript and providing useful comments.
Conflict of interest statement. None declared.
References
Articles from Neuro-Oncology are provided here courtesy of Society for Neuro-Oncology and Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/neuonc/nop061
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/neuro-oncology/article-pdf/12/5/422/16662726/nop061.pdf
Subscription required at ingentaconnect
http://openurl.ingenta.com/content?genre=article&issn=1522-8517&volume=12&issue=5&spage=422
Citations & impact
Impact metrics
Article citations
Multifaceted role of microRNA-301a in human cancer: from biomarker potential to therapeutic targeting.
Cancer Gene Ther, 24 Sep 2024
Cited by: 0 articles | PMID: 39317714
Review
Glioblastoma Therapy: Past, Present and Future.
Int J Mol Sci, 25(5):2529, 21 Feb 2024
Cited by: 12 articles | PMID: 38473776 | PMCID: PMC10931797
Review Free full text in Europe PMC
Y-box protein-1 modulates circSPECC1 to promote glioma tumorigenesis via miR-615-5p/HIP1/AKT axis.
Acta Biochim Biophys Sin (Shanghai), 55(12):1902-1912, 01 Dec 2023
Cited by: 1 article | PMID: 37994157 | PMCID: PMC10753359
miR-431 secreted by human vestibular schwannomas increases the mammalian inner ear's vulnerability to noise trauma.
Front Neurol, 14:1268359, 09 Oct 2023
Cited by: 1 article | PMID: 37885485 | PMCID: PMC10598552
Two Sides of The Same Coin: Normal and Tumoral Stem Cells, The Relevance of In Vitro Models and Therapeutic Approaches: The Experience with Zika Virus in Nervous System Development and Glioblastoma Treatment.
Int J Mol Sci, 24(17):13550, 31 Aug 2023
Cited by: 3 articles | PMID: 37686355 | PMCID: PMC10487988
Review Free full text in Europe PMC
Go to all (90) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome-wide small noncoding RNA profiling of pediatric high-grade gliomas reveals deregulation of several miRNAs, identifies downregulation of snoRNA cluster HBII-52 and delineates H3F3A and TP53 mutant-specific miRNAs and snoRNAs.
Int J Cancer, 137(10):2343-2353, 03 Jun 2015
Cited by: 24 articles | PMID: 25994230
Microarray analysis of the aberrant microRNA expression pattern in gliomas of different grades.
Oncol Rep, 34(1):318-324, 06 May 2015
Cited by: 5 articles | PMID: 25954994
MiRNA expression profiling in human gliomas: upregulated miR-363 increases cell survival and proliferation.
Tumour Biol, 37(10):14035-14048, 06 Aug 2016
Cited by: 15 articles | PMID: 27495233
New advances of microRNAs in glioma stem cells, with special emphasis on aberrant methylation of microRNAs.
J Cell Physiol, 229(9):1141-1147, 01 Sep 2014
Cited by: 16 articles | PMID: 24374932
Review





