Abstract
Free full text

Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignancy largely caused by aberrant activation of the TAL1/SCL, LMO1/2, and NOTCH1 oncogenes. Approximately 30% of T-ALL patients relapse, and evidence is emerging that relapse may result from a failure to eliminate leukemia-initiating cells (LICs). Thymic expression of the Tal1 and Lmo2 oncogenes in mice results in rapid development of T-ALL; and similar to T-ALL patients, more than half the leukemic mice develop spontaneous mutations in Notch1. Using this mouse model, we demonstrate that mouse T-ALLs are immunophenotypically and functionally heterogeneous with approximately 1 of 10 000 leukemic cells capable of initiating disease on transplantation. Our preleukemic studies reveal expansion of Notch-active double-negative thymic progenitors, and we find the leukemic DN3 population enriched in disease potential. To examine the role of Notch1 in LIC function, we measured LIC activity in leukemic mice treated with vehicle or with a γ-secretase inhibitor. In 4 of 5 leukemias examined, Notch inhibition significantly reduced or eliminated LICs and extended survival. Remarkably, in 2 mice, γ-secretase inhibitor treatment reduced LIC frequency below the limits of detection of this assay, and all transplanted mice failed to develop disease. These data support the continued development of Notch1 therapeutics as antileukemia agents.
Introduction
TAL1/SCL activation occurs in 50% to 60% of human T-cell acute lymphoblastic leukemia (T-ALL) patients, and 80% of these patients coexpress LMO proteins.1 Thus, TAL1 and LMO1 or LMO2 are commonly coactivated in human T-ALL, and 40% of these patients also develop mutations in NOTCH1.2 Although most T-ALL patients respond to cytotoxic therapy, 20% to 30% of patients fail induction or relapse, for reasons not well understood. Relapse has been hypothesized to reflect a failure to eliminate the leukemia-initiating cells (LICs), a rare population of leukemic cells that, like normal stem cells, have the ability to self-renew and differentiate. Although clearly demonstrated for AML,3,4 whether ALL is driven by a LIC population remains controversial.
Studies using primary B-ALL patient cells have yielded conflicting results, providing evidence that leukemic cells with a stem cell-like immunophenotype (CD34+, CD38− or CD34+, CD19−) or a more mature B-cell progenitor phenotype (CD34− CD19+) contain the LICs.5–8 A study of high-risk B-ALL patients reveals that the immature and mature B-ALL populations are both capable of transplanting disease to immunodeficient NOD/SCID mice.9 A recent study of T-ALL patients finds the CD34+ CD4− or CD7− populations enriched in cells capable of initiating disease in immunodeficient NOD/SCID mice,10 indicating that this immature population may be enriched in disease potential compared with unsorted human T-ALL cells.
The transplantation of human leukemic cells into immunodeficient NOD/SCID mice has been the standard assay used to examine the functional properties of AML cells. These studies demonstrated that 1 in 104 to 107 leukemic cells were capable of eliciting leukemia in NOD/SCID mice. This assay has also been used for human solid tumors and provided evidence that cancer stem cells contribute to human breast, colon, and brain tumors.11–13 Concern has been raised, however, that xenotransplant does not accurately measure cancer or leukemia stem cell activity because tumors depend on interactions within the microenvironment for their growth and survival. Thus, the xenotransplant assay may simply measure tumor adaptation to an immunodeficient mouse environment.
To avoid issues associated with xenotransplantation, leukemia stem or initiating cell studies turned to mouse models of the disease where dilutions of mouse leukemia/lymphoma cells can be transplanted into histocompatible recipients. When these studies were performed using a Ras-induced model of T-cell lymphoma or the Eμ-Myc model of pre-B/B-cell lymphoma, all recipient mice developed disease, irrespective of the number of cells injected.14 This report questioned the leukemic stem cell hypothesis by showing that most mouse leukemia/lymphoma cells were capable of sustaining tumor growth on transfer into syngeneic recipients. More recent studies using 2 different Notch1-mediated models of T-ALL yielded conflicting results, with one study reporting no evidence of a disease-enriched population and the other suggesting that expression of intracellular mutant Notch1 is sufficient for LIC transformation.15,16 Additional studies are clearly required to determine whether an LIC contributes to T-ALL pathogenesis and to identify the pathway(s) that mediate the survival and extensive proliferative capabilities associated with LICs. These are critical questions that are highly relevant to the design of future targeted therapies for relapsed T-ALL patients.
To determine whether LICs contribute to T-ALL pathogenesis and to examine the role of Notch1 in LIC activity, we used a Tal1/Lmo2 mouse model of T-ALL in which 75% of the mouse T-ALLs develop spontaneous mutations in Notch1. Limiting dilution analyses reveal that a rare LIC population contributes to disease pathogenesis in this mouse T-ALL model. We find a committed thymic progenitor population enriched in disease potential and demonstrate that γ-secretase inhibitor (GSI) treatment reduces and, in some cases, ablates LIC activity. The present study reveals that LICs contribute to mouse T-ALL pathogenesis and provides evidence that Notch1 inhibition will target the LICs.
Methods
Mice
A cohort of Tal1/Lmo2 transgenic mice was generated and monitored daily for the onset of leukemia.17 To generate the Tal/Lmo2/TNR cohort, Tal/Lmo2 mice were mated with TNR/+ mice (STOCK Tg(Cp-EGFP)25Gaia/J no. 005854; The Jackson Laboratory). Tal1/Lmo2/TNR mice are maintained on a mixed background ((C57BL/6J × SJL/J)F2 × FVB/N). To control for differences in genetic background, all preleukemic studies were performed using TNR/+ control littermates. Tal1/Lmo2 leukemic cells or purified subpopulations of leukemic cells were transplanted into syngeneic recipient FVB/N mice (6-8 weeks old, The Jackson Laboratory). Animal care and all animal procedures have been approved by and are in compliance with the University of Massachusetts Medical School Institutional Animal Care and Use Committee. LIC frequency was determined using Poisson distribution statistics and the L-Calc Version 1.1 software program (StemCell Technologies). For the GSI studies, transplanted mice were treated with vehicle or GSI for 3 eeks as described previously.18 Mice were monitored daily for disease development and weighed to monitor GSI-associated toxicity. Kaplan-Meier survival and statistical analyses were performed using GraphPad Prism Version 4.0 software. The hazard ratio and its 95% confidence interval was also measured, comparing the vehicle- and GSI-treated groups and adjusting for the dilutions of leukemic cells, using the Cox proportional hazards model analysis. A 2-sided P < .05 was considered statistically significant.
FACS analysis
Single-cell suspensions of Tal1/Lmo2 leukemic cells were stained with CD4-phycoerythrin (PE)–Cy5 and CD8-PE or with a lineage cocktail consisting of CD4-PE, CD8-PE, B220-PE, GR1-PE, and MAC1-PE (BD Biosciences PharMingen). Lineage-negative cells were then stained with CD44- allophycocyanin (BD Biosciences PharMingen) and CD25-PE-Cy7 (eBioscience). Dead cells were excluded by propidium iodide staining. Flow cytometric analysis and sorting were performed on the FACSCaliber and FACS LSRII (BD Biosciences), respectively. Data were analyzed using FlowJo Version 8.8.6 software (TreeStar).
RNA analysis
RNA was extracted from murine preleukemic thymocytes or leukemic cells using Trizol. cDNA was synthesized using Superscript First-Strand Synthesis System (Invitrogen). To determine the effects of Notch1 target gene expression on preleukemic thymic subsets, cDNA was quantitated using the SYBR Green kit (QIAGEN). Specific c-Myc, Deltex1, and Pre-Tα primers were designed using Primer Express software (Applied Biosystems): c-Myc forward, 5′-CTGTTTGAAGGCTGGATTTCCT-3′; c-Myc reverse, 5′-CAGCACCGACAGACGCC-3′. Deltex1 forward 5′ TGCCTGGTGGCCATGTACT-3′; Deltex1 reverse 5′-GACACTGCAGGCTGCCATC-3′. The copy number obtained for gene of interest was normalized to the copy number for β-actin.
Notch1 sequencing
To determine the Notch1 mutational status, DNA isolated from preleukemic sorted thymic populations or mouse T-ALL cells was amplified by PCR using Pfu DNA polymerase (Stratagene) with primers specific for exon 34 of the Notch1 gene.19 PCR products were run on a 1.5% agarose gel, purified (QIAquick Gel Extraction Kit; QIAGEN), and cloned into the TOPO TA cloning vector (Invitrogen) for sequencing using the universal M13 primers.
Clonality analysis
To determine clonality, rearrangements of the TCR β-chain were assayed by standard qualitative PCR analysis, using Pfu DNA polymerase (Stratagene) and primers specific for mouse TCR Vβ1-Vβ18 genes and constant region as described.20 Vβ1-Vβ18 primers were each paired with the following Vβ constant primer (Vβ-5′-GGCTCAAACAAGGAGACCTTGGGTGG-3′). The amplification was performed using a Stratagene Robocycler Gradient 96 starting with a 2-minute 94°C denaturation, followed by 30 cycles consisting of 20 seconds at 94°C, 12 seconds at 55°C, and 30 seconds at 68°C and a final elongation step of 10 minutes at 68°C. PCR products were purified on a 2% agarose gel, subcloned, and confirmed by sequencing.
Results
DN3/DN4 thymic progenitor population is expanded in preleukemic Tal1/Lmo2 mice
Greater than 40% of T-ALL patients coexpress both TAL1 and LMO1 or LMO2 oncogenes.1 Coexpression of the Tal1 and Lmo1 or Lmo2 oncogenes in murine thymocytes accelerates T-cell leukemogenesis and interferes with thymocyte maturation to the double-positive (DP) stage.21,22 We reproduced these findings by generating Lck-Lmo2 transgenic lines17 and mating these mice with our previously published Lck-Tal1 mice.23
Similar to published results, we observe a significant reduction in the overall thymic cellularity in 4- to 6-week-old preleukemic Tal1/Lmo2 mice compared with littermate controls (Figure 1B). Although all thymocyte subpopulations are detected, preleukemic Tal1/Lmo2 mice have significant increases in the percentage of double-negative (DN) thymic progenitors (Figure 1A; range, 32.4%-63%) compared with littermate controls (range, 1.92%-2.9%; P < .005), suggesting that thymocyte development may be arrested at the DN thymic progenitor stage. The differentiation block was associated with a 2-fold decrease in the percentage of DP thymocytes, as well as a 3-fold decrease in CD4 single-positive (SP) thymocytes (Figure 1A).
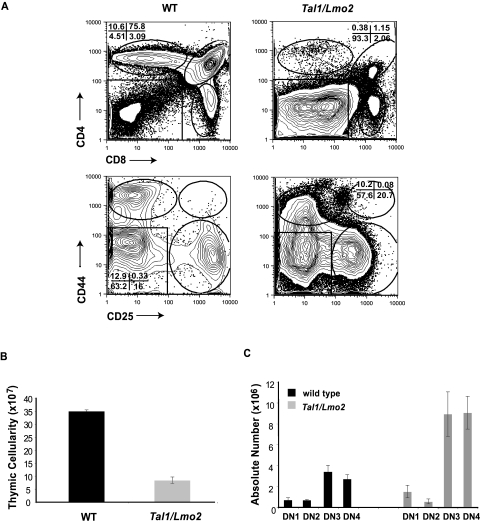
Thymic progenitors are expanded in preleukemic Tal1/Lmo2 mice. (A) Thymocytes from 6-week-old wild-type or preleukemic Tal1/Lmo2 mice were stained with CD4-PE-Cy5 and CD8-PE or with a lineage cocktail consisting of CD4-PE, CD8-PE, B220-PE, GR1-PE, and MAC1-PE. Lineage-negative cells were then stained with CD44-allophycocyanin and CD25-PE-Cy7 and analyzed by flow cytometry. (B) Decreased thymic cellularity in preleukemic Tal1/Lmo2 mice. Thymic cellularity was determined using trypan blue exclusion. (C) The absolute number of DN3 and DN4 progenitors is increased in Tal1/Lmo2 mice. The absolute number in each thymic subset was determined by multiplying the percentage of each thymic subset by the total thymic cellularity.
Further analysis of the DN population in the preleukemic animals revealed an expansion of DN3 and DN4 thymic progenitors. Although the percentage of DN3 and DN4 progenitors between preleukemic and wild-type animals was not significantly different, the absolute number of DN3 and DN4 thymic progenitors increased approximately 2-fold in preleukemic Tal1/Lmo2 animals compared with controls (Figure 1C; P < .05). The differentiation arrest observed in Tal1/Lmo1 and Tal1/Lmo2 animals was greater than that described in animals that expressed Tal1 only,21,24 suggesting that Lmo1 and Lmo2 proteins cooperate with Tal1 to perturb thymocyte development.
Preleukemic Tal1/Lmo2 thymic progenitors exhibit aberrant Notch1 activity
Spontaneous gain-of-function Notch1 mutations are frequently observed in murine T-ALL models.19,25–27 Although it has been suggested that these gain-of-function mutations occur during the preleukemic phase,26,28 the precise nature of the target cell remains unclear. To monitor Notch activity during leukemogenesis, we mated our Tal1/Lmo2 mice with the transgenic notch reporter (TNR) mouse line. The TNR mouse contains a transgene with 4 CSL-binding sites, a minimal SV40 promoter, followed by an enhanced green fluorescent protein (GFP) sequence.29,30 The TNR mouse has been shown to accurately report Notch activity during early thymocyte and neuronal development, angiogenesis, and hair follicle formation.29–32 To confirm that the reporter accurately reflects Notch activity in the thymus, we isolated RNA from GFP+ and GFP− thymocytes and examined Notch1 target gene expression by real-time PCR. We found expression of Deltex1 and c-Myc increased in the GFP+ thymocytes compared with GFP− populations (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), similar to published reports.29
In wild-type mice, we observed the highest percentage of Notch-active cells in DN thymocytes (Figure 2A). We detect increases in Notch activity in DN2 and DN3 progenitors, followed by repression of Notch signaling as cells progressed to DN4 (Figure 2B). We also detect Notch-active cells in the expanded DN population of preleukemic Tal1/Lmo2 animals (Figure 2B). The proportion of GFP+ or Notch-active DN thymocytes in preleukemic Tal1/Lmo2 animals was significantly greater than that observed in control littermates designated TNR/+ (Figure 2A; 46.35% for Tal1/Lmo2/TNR compared with 14.17% for TNR/+; P < .05). No significant increase in Notch-active cells was observed in the DP, CD4SP, or CD8SP populations isolated from preleukemic Tal1/Lmo2 mice (Figure 2A).
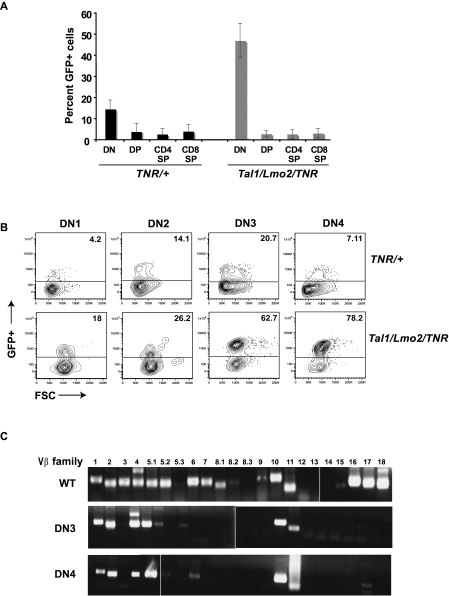
Notch1-active thymic progenitors are expanded in preleukemic Tal1/Lmo2 mice. (A) Six- to 8-week-old TNR/+ or Tal1/Lmo2/TNR mice were killed and thymocytes stained with CD4, CD8 antibodies. DP, SP, and DN populations were analyzed for percentage GFP+ cells by flow cytometry. (B) Lineage-negative cells were stained with CD25 and CD44 antibodies, and GFP+ was analyzed by flow cytometry. The percentage of GFP+ cells in the DN1, DN2, DN3, and DN4 progenitor populations is shown. (C) Using specific primers for Vβ1-18, TCR Vβ mRNA expression was examined in wild-type and sorted GFP+ DN3 and 4 thymic progenitors isolated from preleukemic Tal1/Lmo2 mice. Three mice were examined; one representative experiment is shown. Vertical lines have been inserted to indicate repositioned gel lanes.
Increases in the percentage of GFP+ cells were observed at all DN stages in preleukemic mice (Figure 2A). Preleukemic mice were analyzed at 4 to 6 weeks of age, approximately 50 to 70 days before disease is observed. The increase in GFP+ DN1 and DN2 cells may indicate that a Notch1 mutation occurred in the DN1 progenitor or potentially in the hematopoietic stem cell. However, the preleukemic phase of the disease is consistently associated with DN3/DN4 progenitor expansion and DN1/DN2 precursors are not significantly increased (Figure 1C). Although Notch1 activity is normally down-regulated at the DN4 stage,33 we detected an expansion of GFP+, Notch-active DN3 and DN4 progenitors in the mice predisposed to leukemia (Figure 2B; average increase 52-fold for DN3 and 80-fold for DN4).
The increase in the absolute number of Notch1-active DN3/DN4 progenitors suggested that sustained Notch1 activity might drive progenitor expansion during the preleukemic phase of the disease. To test this possibility, GFP+ DN3 and DN4 cells from preleukemic Tal1/Lmo2 mice were isolated (supplemental Figure 1A) and sequenced for the presence of Notch1 mutations. We detected multiple Notch1 mutations in the DN3 and DN4 progenitors isolated from preleukemic Tal1/Lmo2 mice (Table 1). Similar to what we observed in Tal1-mediated mouse T-ALLs,19 these mutations consisted of point mutations, insertions, or deletions within exon 34 that introduce premature STOP codons or alter the reading frame, resulting in the truncation of PEST regulatory sequences. These mutations are predicted to increase Notch1 protein stability and explain the increased Notch1 reporter activity detected in the preleukemic Tal1/Lmo2 thymic progenitor populations (Figure 2B). Recently, 5′ activating deletions in Notch1 have been found in multiple mouse T-ALL models, including the model used in this study.34,35 These deletions result in expression of a ligand-independent form of activated Notch1; and in some mouse T-ALLs, these deletions may be Rag-mediated.34 Although PEST region mutations were observed (Table 1), 5′ activating deletions were not detected in the preleukemic GFP+ DN3/DN4 progenitors (not shown), raising the possibility that PEST region mutations precede 5′ activating deletions in Notch1 during leukemogenesis.
Table 1
Multiple Notch1 mutations are detected in preleukemic Tal1/Lmo2 thymic progenitors
| Mouse no. | Thymic subset | Nucleotide change | Amino acid change |
|---|---|---|---|
| 9401 | DN3 | 7161 C→G, ins G | 2387 T→R |
| 7873 del TA | 2626 Y→F | ||
| 7542 del C | 2515 I→S | ||
| 7926 del A | 2643 K→N | ||
| 7888 del T | 2631 S→L | ||
| 9401 | DN4 | 7098 del T | 2367 T→H |
| 7272 ins AAGGAAG | 2425 G→R | ||
| 7313 del G | 2438 G→E | ||
| 7264 ins C | 2425 G→A | ||
| 7751 C→T | 2584 A→V | ||
| 7722 del T | 2576 W→G | ||
| 7788 ins G | 2597 Y→I | ||
| 7883 ins T | 2629 L→I | ||
| 9446 | DN3 | 7535 ins G | 2512 P→R |
| 7792 del A | 2599 R→E | ||
| 7844 del T | 2616 L→Y | ||
| 7806 del A | 2603 E→N | ||
| 9446 | DN4 | 7792 del A | 2600 R→E |
| 7752 ins G | 2586 G→W | ||
| 7831 T→C | 2611 F→L | ||
| 9448 | DN3 | 7852 del TC | 2618 S→H |
| 7775 del A | 2594 M→C | ||
| 7869 del T | 2626 Y→I | ||
| 7415 del TCCATGGTCCC | 2472 L→H | ||
| 9448 | DN4 | 7673 T→C | 2558 T→I |
| 7316 ins ACA | 2439 A→D, ins T |
The presence of multiple Notch1 mutations in the preleukemic thymic progenitors led us to hypothesize that mutant Notch1 contributes to the expansion of DN3/DN4 progenitor clones in these mice. This hypothesis predicts that the preleukemic thymus will be oligoclonal in nature and reflect the expansion of multiple progenitor clones. To analyze the clonality of the preleukemic progenitors, wild-type and preleukemic GFP+ DN3 and DN4 progenitors were purified and TCR Vβ gene expression was determined using PCR. In wild-type thymocytes, we detect expression of 15 of the 22 TCR Vβ families examined, confirming the polyclonality of the wild-type thymus (Figure 2C). In each of the 3 Tal1/Lmo2 preleukemic mice examined, the DN3 and DN4 subpopulations expressed 3-6 TCR Vβ families, consistent with an oligoclonal expansion (Figure 2C). Although the TCR Vβ expression patterns differed among the 3 preleukemic mice examined, they were identical between the DN3 and DN4 progenitors within each mouse. The predominance of multiple, identical TCR Vβ expressing clones in the preleukemic DN3 and DN4 populations suggests that the DN3 progenitors maintain their ability to differentiate to the DN4 stage. Thus, in this mouse T-ALL model, the preleukemic phase of the disease is associated with the expansion of multiple Notch1-active thymic progenitors.
Tal1/Lmo2 tumors contain thymic progenitors and the progenitor population is maintained on transplant
To determine whether the thymic progenitors are maintained on leukemic transformation, we stained Tal1/Lmo2 tumors with CD4, CD8 or with a lineage cocktail (CD4, CD8, B220, GR1, and Mac1) and then stained the lineage-negative cells with CD44 and CD25 antibodies. We analyzed 25 Tal1/Lmo2 tumors and found that most (24 of 25) were immunophenotypically heterogeneous (Figure 3A; Table 2). Tumors consisted of undifferentiated DN and differentiated DP or SP cells that maintain expression of the Tal1 and Lmo2 transgenes (not shown). In 23 of the tumors where the DN population was further analyzed, 18 tumors (78%) contained a DN3 or DN3 and DN4 progenitor population (Figure 3A; Table 2). The remaining 5 tumors consisted of DN3 or DN4 progenitors along with DN1 and DN2 progenitors, and one tumor contained a DN2 population only (Table 2). Similarly, a DN population has been noted in other mouse Tal1/Lmo T-ALL models16,21,22 and in 67% of primary human T-ALL samples we have analyzed (J.T. and M.A.K., unpublished data, 2010), suggesting that mouse and human T-ALLs exhibit immunophenotypic heterogeneity.
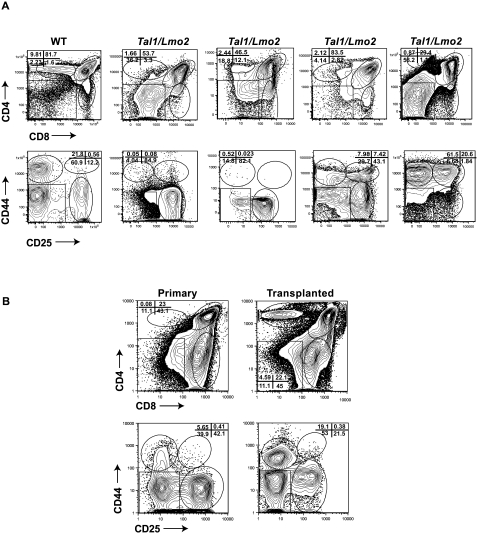
Immunophenotypic heterogeneity among mouse Tal1/Lmo2 tumors. (A) Wild-type thymocytes and 25 Tal1/Lmo2 tumors were stained with CD4 and CD8 antibodies, and the lineage-negative tumor cells were stained with CD25 and CD44 antibodies and analyzed by flow cytometry. Four representative Tal1/Lmo2 tumors are shown. Gating is based on the wild-type control stained with each individual tumor. (B) Tumor-associated thymic progenitors are maintained on transplant. Tal1/Lmo2 tumor cells were then injected via subcutaneous injection into recipient mice. At death, the immunophenotype of the transplanted tumor was reanalyzed by flow cytometry and compared with the primary tumor. Five tumors were analyzed; one representative tumor is shown.
Table 2
Tal1/Lmo2 tumors are immunophenotypically heterogeneous
| Animal no. | Genotype | Phenotype | % DN | DN subtype | Latency |
|---|---|---|---|---|---|
| 1001 | Tal1/Lmo2 5 | DP, CD8SP | 20 | DN3 | 91 |
| 1002 | Tal1/Lmo2 5 | DP, DN | 58 | DN3,4 | 93 |
| 1004 | Tal1/Lmo2 5 | CD8SP | 16 | DN3 | 91 |
| 1007 | Tal1/Lmo2 5 | DP, CD8SP | 18 | DN3 | 124 |
| 1078 | Tal1/Lmo2 5 | CD8SP, DN | 21 | DN3,4 | 100 |
| 1099 | Tal1/Lmo2 5 | DP, DN | 35.9 | DN3,4 | 101 |
| 8115 | Tal1/Lmo2 5 | DP, DN | 27.2 | DN3 | 89 |
| 8129 | Tal1/Lmo2 5 | DP | 2.83 | DN2 (3) | 128 |
| 8159 | Tal1/Lmo2 5 | CD4SP, DN | 20 | ND | 93 |
| 8164 | Tal1/Lmo2 5 | All CD8SP-DN | DN2 | 161 | |
| 8179 | Tal1/Lmo2 5 | DP | 3.15 | DN1,2,3,4 | 119 |
| 8181 | Tal1/Lmo2 5 | DP, DN | 72 | DN3 | 212 |
| 8308 | Tal1/Lmo2 5 | DP, CD8SP | 1.2 | DN4 | 60 |
| 8361 | Tal1/Lmo2 5 | DP | 4 | DN3,4 | 145 |
| 8382 | Tal1/Lmo2 19 | DP, DN | 40 | DN3,4 | 111 |
| 8383 | Tal1/Lmo2 19 | CD8SP | 4.6 | DN3, 4 | 135 |
| 8602 | Tal1/Lmo2 5 | DP, DN, CD8SP | 40 | DN2 (3,4) | 88 |
| 8613 | Tal1/Lmo2 5 | DP | 3 | DN3,4 | 120 |
| 8647 | Tal1/Lmo2 5 | DP | 6 | DN4 | 109 |
| 8653 | Tal1/Lmo2 5 | DP | 4 | DN3 | 109 |
| 8655 | Tal1/Lmo2 5 | DP, CD8SP | 2.73 | DN3,4 | ND |
| 8683 | Tal1/Lmo2 5 | DP | 15.8 | DN1,2 | 109 |
| 8734 | Tal1/Lmo2 19 | DP, CD8SP | 4 | ND | 135 |
| 8757 | Tal1/Lmo2 5 | DP | 19 | DN3,4 | 112 |
| 9053 | Tal1/Lmo2 19 | DP | 6 | DN3,4 | 109 |
ND indicates not determined.
Tumors were designated positive if > 20% of the cells expressed the marker(s).
The thymic progenitors detected within the Tal1/Lmo2 thymic mass could simply reflect the presence of nontransformed thymic progenitors. To determine whether the thymic progenitor population contributes to leukemogenesis, 5 Tal1/Lmo2 tumors were transplanted into syngeneic recipients. The transplanted mice developed disease, and immunophenotyping of the tumors revealed that in all cases the thymic progenitors were maintained (Figure 3B).
To determine whether the immunophenotypic heterogeneity reflects functional differences among mouse T-ALL cells, we performed a series of limiting dilution experiments. Initially, serial dilutions of 3 Tal1/Lmo2 mouse T-ALLs were transplanted into syngeneic mice and recipient mice were monitored for the onset of leukemia. Mouse T-ALL 8129 required injection of between 5000 and 50 000 cells to initiate disease in recipient mice (Figure 4; supplemental Table 1). We also found that injecting fewer leukemic cells from this tumor resulted in an increase in disease latency, as injection of 5 × 106 cells induced disease in approximately 26 days, whereas injection of 50 000 to 5000 cells resulted in an average latency of 41 or 53 days, respectively (Figure 4; supplemental Table 1). The frequency of LICs in this mouse T-ALL is approximately 1:30 735, indicating that the LIC is a rare population. Although an increase in disease latency was also observed when serial dilutions of leukemic blasts from tumor 1002 were transplanted into recipient mice, this tumor appeared to harbor a greater number of LICs. Transplantation of 500 to 5000 leukemic cells resulted in disease in 2 of 8 and 7 of 8 recipient mice, respectively (Figure 4; supplemental Table 1). Thus, the frequency of LICs in mouse T-ALL 1002 is significantly higher than 8129 and estimated to be 1:2209. The third Tal1/Lmo2 leukemia examined 2695, resembled 1002 with the LICs estimated to be 1:4579. We analyzed an additional 4 Tal1/Lmo2 mouse T-ALLs and found the average LIC frequency to be 1:11 549 (supplemental Table 1; range, 1:2209-1:30 735). These data demonstrate that these mouse T-ALLs exhibit functional differences, with between 0.003% and 0.05% of the leukemic cells capable of transplanting disease. An additional feature of this mouse T-ALL model is that the frequency of the LICs is somewhat variable (supplemental Table 1) and may reflect genetic alterations incurred during LIC development.
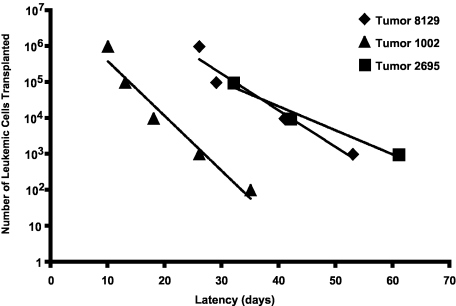
Functional heterogeneity among mouse Tal1/Lmo2 T-ALLs. Leukemic cells from 7 Tal1/Lmo2 mice were transplanted through limiting dilution into syngeneic mice. Three representative Tal1/Lmo2 mouse T-ALLs are shown. Time to leukemia after transplantation of the indicated numbers of cells is shown. LIC frequency for each mouse T-ALL was calculated by applying Poisson statistics using the Limiting Dilution Version 1.1 software (StemCell Technologies).
Tumor-associated progenitors are enriched in disease potential
Whereas the presence of an LIC population has been well documented in AML, whether ALL is driven by a rare LIC remains less clear.5–10 To determine whether tumor-associated thymic progenitors are enriched in disease-initiating potential, the DN3 population from 3 independent Tal1/Lmo2 tumors was sorted, along with DP blasts, and serial dilutions were transplanted into recipient mice. The purity of the DN3 and DP tumor cells ranged between 96% and 99% (supplemental Figure 2). Mice transplanted with unsorted tumor cells developed disease within 24 to 37 days, as expected. Nineteen of 20 mice transplanted with DP cells failed to develop disease, irrespective of the number of DP leukemic blasts injected (Figure 5A). However, 1 of 10 mice transplanted with 1 × 104 DP cells did develop disease. In contrast, 8 of 9 mice transplanted with 104 DN3 cells and 5 of 9 mice injected with 103 DN3 cells developed leukemia within 35 to 60 days (Figure 5A). Collectively, these data suggest that the tumor-associated DN3 thymic progenitors are enriched in leukemic-initiating potential.
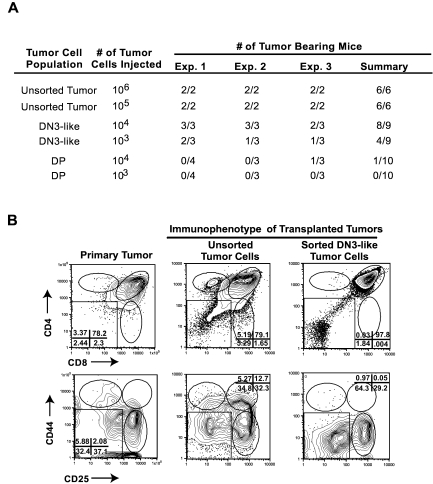
Tumor-associated thymic progenitors are enriched in disease initiating potential. Leukemic cells from Tal1/Lmo2 mice were stained with CD4, CD8 antibodies, and lineage-negative cells were stained with CD25 and CD44 antibodies. (A) Tal1/Lmo2 mouse T-ALL cells were left unsorted or sorted into DN3 or DP populations by flow cytometry. Serial dilutions of unsorted, purified DP, or DN3 cells were transplanted into syngeneic recipients and monitored for disease. Three independent Tal1/Lmo2 tumors were analyzed. (B) Purified DN3 cells recapitulate the leukemic immunophenotype. Leukemic cells were isolated from mice transplanted with unsorted or purified DN3 cells and stained with CD4, CD8 antibodies, and lineage-negative cells were stained with CD25 and CD44 antibodies and analyzed by flow cytometry. Three Tal1/Lmo2 mouse T-ALLs were reexamined; one representative tumor is shown.
It is possible that the tumor-associated DN3 and DN4 cells are altered DP cells that are unable to express CD4 and CD8 coreceptors and do not represent thymic progenitors. The pTα gene encodes the surrogate TCRα protein that associates with the TCRβ chain to form the pre-TCR. pTα mRNA expression increases during DN development, is highest in DN3/DN4 progenitors, and declines in DP thymocytes.36 Therefore, we compared pTα expression levels in the sorted tumor subpopulations from 2 Tal1/Lmo2 tumors. The tumor associated DN3 and DN4 cells expressed higher pTα mRNA levels than the DP leukemic blasts (not shown). The DN3 and/or DN4 progenitors associated with 5 additional mouse T-ALLs were examined for c-Kit and IL7R cell surface expression levels. The transformed progenitors exhibited similar c-Kit and IL7R expression levels as normal thymic DN3 and DN4 cells (not shown), indicating that the transformed progenitors appear to maintain the developmental program and more closely resemble their nontransformed counterparts than differentiated DP thymocytes.
LICs have at least 2 unique features, the ability to self-renew and generate more LICs, while retaining the capacity to differentiate into cells with limited self-renewal potential.37 These features manifest in an immunophenotypically and functionally heterogeneous tumor. To determine whether the tumor associated DN3 cells retain the ability to differentiate into DP and SP tumor cells, we examined the tumors harvested from recipient mice transplanted with purified DN3 tumor cells. In all cases, the purified DN3 tumor cells regenerate the primary tumor and contain undifferentiated DN3- and DN4-like progenitor cells and differentiated DP leukemic cells (Figure 5B). Thus, the tumor-associated DN3 thymic progenitors are enriched in LICs and appear to retain differentiation potential. Although a thymic progenitor population was evident in all the leukemias analyzed, the percentage of DN3 progenitors did not correlate with LIC frequency (supplemental Table 1). These data indicate that the LICs compose a subset of the DN3 cells within a tumor. Thus, additional experiments will be required to purify the LICs from the DN3 population.
Notch1 inhibition targets the LICs in a mouse T-ALL model
The presence of Notch1 mutations and the expansion of Notch1-active thymic progenitors in preleukemic Tal1/Lmo2 mice raise the possibility that a sustained Notch1 signal confers extensive proliferative capabilities on committed thymic progenitors. Although we demonstrated that GSI treatment extends the survival of end-stage leukemic mice,18 effects on LIC frequency or activity were not addressed in the prior study. Because relapse is hypothesized to reflect an inability to eliminate the LIC population, whether LICs are susceptible to Notch inhibitors remains a critical, unanswered question.
To examine the consequences of Notch1 inhibition on mouse LIC activity, we initially treated mouse T-ALL cells in vitro with vehicle or GSI for 48 hours. Serial dilutions of the vehicle- or GSI-treated leukemic cells were then transplanted into syngeneic recipients and treatment was maintained for 3 weeks. GSI was administered by oral gavage using an intermittent dosing regimen we have shown minimizes the gastrointestinal toxicity associated with Notch inhibition.18 We have demonstrated that this GSI dosing regimen reduces Notch1 target gene expression in leukemic cells18 (supplemental Figure 3). Five Tal1/Lmo2 tumors with mutations in Notch1 were examined in this study; and remarkably, complete or partial responses were observed in 4 of 5 mouse T-ALLs treated with GSI (Figure 6). For reasons not yet understood, GSI treatment of mouse T-ALL 1007 had minimal effects on leukemic cell viability (Table 3) and on disease latency and overall survival (Figure 6; log-rank P < .28).
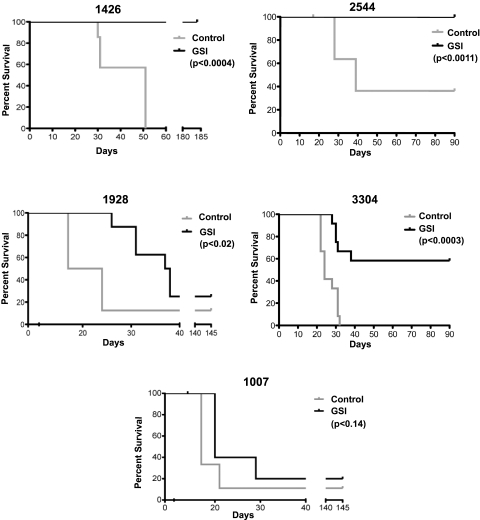
Notch inhibition targets the LIC in Tal1/Lmo2 mouse T-ALLs. Mouse Tal1/Lmo2 T-ALLs with Notch1 mutations were treated with vehicle or GSI for 48 hours in vitro, stained, and analyzed by flow cytometry. Leukemic cells were serially diluted and transplanted into syngeneic mice, and treatment was continued for 3 weeks. GSI was administered using an intermittent dosing regimen,18 and transplanted mice were monitored for disease development. Kaplan-Meier survival curves are shown for 5 mouse T-ALLs. GSI treatment of mouse T-ALLs 1426 and 2544 eliminated LICs, and GSI-treated recipients failed to develop disease (log-rank test: P < .0004 and P < .001, respectively). GSI treatment of 1928 and 3304 reduced LIC activity and significantly extended survival (log-rank test: P < .02 and P < .0003, respectively). GSI treatment had no statistically significant effect on the survival of mice transplanted with leukemic cells from mouse 1007.
Table 3
Summary of GSI responses in vivo
| Mouse T-ALL | 1426 | 2544 | 3304 | 1928 | 1007 |
|---|---|---|---|---|---|
| L-IC frequency | 1:4170 | 1:8731 | ND | 1:8731 | 1:6695 |
| Mean latency (days) | 51 | 39 | 24 | 21 | 17 |
| GSI-induced cell death (%) | 24.1 | 3.8 | 16.4 | 17.5 | 1.1 |
ND indicates not determined.
Transplantation of 105 or 104 vehicle-treated leukemic cells from mouse T-ALLs 1426 or 2544 resulted in disease initiation in all transplanted recipients with an average latency of 31 and 55 days for 1426 and 28 and 39 days for 2544 (Figure 6). Mice transplanted with leukemic cells from mouse 1426 or 2544 treated with GSI failed to develop disease during the 180- or 90-day observation periods (Figure 6; log-rank test, P < .0004 for 1426 and P < .001 for 2544). These mice exhibited no evidence of disease at necropsy and histopathologic examination of tissues failed to detect the presence of leukemic cells. GSI treatment of mice transplanted with leukemic cells from 1928 and 3304 reduced LIC activity approximately 4- to 20-fold, respectively (supplemental Table 2) and significantly extended survival (Figure 6; log-rank test, P < .02 for 1928 and P < .0003 for 3304). Consistent with our published work,18 transplanted mice treated with GSI continued to gain weight throughout the treatment period and exhibited no signs of gastrointestinal toxicity (supplemental Figure 4). This study demonstrates that, in some cases, Notch1 inhibition in vivo reduces LIC activity below the limits of detection of this limiting dilution assay. These findings have clear translational implications and support the continued development of Notch therapeutics for T-ALL patients.
It remains unclear why GSI treatment had no effect on mouse T-ALL 1007. All 5 mouse T-ALLs examined express high levels of intracellular Notch1 and harbor insertions/deletions that result in PEST truncation. These mouse T-ALLs also harbor 5′ activating deletions in Notch1 that result in ligand-independent Notch1 activation34,35 (supplemental Figure 5). In 2 of 5 mouse T-ALLs analyzed, the deletions are probably Rag-mediated (not shown) and further support our finding that the Rag-active DN3 population is important in disease initiation. Mouse T-ALL 1007 fails to respond to GSI treatment potentially because of mutations in Pten or Fbw7.38–41Alternatively, LIC activity in this mouse T-ALL may not depend on Notch1 but rather on the activation of other thymocyte self-renewal pathways potentially mediated by Lmo2.42
Discussion
We hypothesized that deregulated Notch1 activity during the preleukemic phase may confer extensive proliferative properties on committed thymic progenitors. We demonstrate that the DN3 population is enriched in leukemia-initiating activity and provide evidence that Notch inhibition reduces and, in some cases, eliminates LIC activity. These findings have implications for translational research and suggest that Notch therapeutics may prove beneficial for T-ALL patients.
The cancer stem cell model predicts that the heterogeneity observed within most clonal tumors reflects an organizational hierarchy, with some cells having greater self-renewing potential and capability to initiate tumor growth. In the Tal1/Lmo2 mouse T-ALL model, clonal tumors appear hierarchically organized, consisting predominantly of DN3 and/or DN4 thymic progenitors and more differentiated DP and/or SP blasts. This heterogeneity is also observed in infiltrated lymph nodes and tissues and, importantly, is maintained on leukemic transplant. We demonstrate that the tumor-associated DN3 thymic progenitors are enriched in leukemia-initiating potential, whereas the more differentiated DP blasts are not enriched in LIC potential. Our finding that one animal developed disease after transplantation of sorted DP blasts may suggest that LICs reside within each tumor population, albeit at different frequencies. Alternatively, the fact that only 1 of 20 mice transplanted with DP cells developed disease could reflect the presence of contaminating DN3/DN4 progenitors.
Limiting dilution analyses reveal that the LICs are relatively rare and somewhat variable among the mouse T-ALLs examined. The LIC frequencies reported here differ significantly from those recently published for pSIL-TSCL and Lck-Lmo1 transgenic mice.16 The 10- to 100-fold difference in LIC frequency may reflect the transplantation models used to measure LIC activity. Tremblay et al injected limiting dilutions of SCL/LMO1 leukemic cells into adult Rag1-deficient mice, whereas syngeneic recipients were used in the present study. It is conceivable that the Rag1-deficient microenvironment may be more permissive for LIC engraftment because of structural changes associated with immune deficiency. These structural changes could lead to the creation of new progenitor niches or reflect loss of competition for existing progenitor niches within the recipient thymus. Alternatively, the discrepancies in LIC frequency may reflect differences in the strength and/or target cell specificity of the transgene promoters used to drive oncogene expression.
Despite the LIC frequency differences, both studies find the DN3 subpopulation enriched in leukemia initiating potential (Figure 5).16 Tremblay et al also detect reduced LIC activity in the DN4 population of SCL/LMO1 mice and argue that transgenic expression of intracellular mutant NOTCH1, SCL, and LMO1 is sufficient to transform a thymic progenitor into an LIC.16 Although not directly tested here, we predict that the DN4 population associated with Tal1/Lmo2 tumors will also harbor LICs.
The time to disease progression and the estimates of LIC frequency in our model suggest that additional genetic events define the LICs within the DN3 population. The detection of PEST region mutations in endogenous Notch1 in an oligoclonal, thymic progenitor population approximately 60 to 90 days before disease is evident argues that a PEST mutation may not be sufficient for complete LIC transformation in our model. Although 5′ activating deletions in Notch1 were readily observed in these mouse leukemic cells, we were unable to detect an increase in 3′ Notch1 transcripts in preleukemic progenitors (supplemental Figure 5; data not shown). This result suggests that PEST region mutations precede 5′ activating deletions in Notch1.
Mouse LICs remain dependent on sustained Notch1 signals as LIC activity is significantly reduced in 4 of 5 mouse T-ALLs treated with GSIs. The LIC dependence on Notch1 is consistent with the critical role of Notch1 in normal DN3 thymic expansion, survival, and metabolism.43,44 Additional support for NOTCH1 in the maintenance of LIC activity comes from studies by Armstrong et al who expand primary human T-ALL cells on a mouse stromal cell line that expresses the human NOTCH ligand DELTA-like 1.45 GSI treatment of these primary T-ALL cells arrests their growth in vitro and reduces the ability of these primary leukemic cells to initiate disease in NOD/SCID mice.45 Collectively, both studies implicate NOTCH1 in the regulation of LIC maintenance.
Precisely how Notch1 contributes to LIC activity is unknown. The Notch1 target genes c-Myc and Hes1 have been implicated in stem cell self-renewal and thymic progenitor expansion, respectively.46–49 Leukemic or cancer stem cells have been shown to activate gene expression programs similar to those expressed in embryonic stem cells, and c-Myc expression is one of 4 transcription factors capable of reprogramming differentiated adult cells back to a pluripotent state.50 Thus, constitutive Hes1 and/or c-Myc activation may confer “stem cell–like” properties on committed thymic progenitors, and these signals may work in concert with survival pathways activated by Notch1. Although this study reveals a clear LIC dependence on Notch1, our data argue that additional genetic events contribute to complete LIC transformation. The nature of these events will require further purification of the LIC from the DN3 population.
Acknowledgments
The authors thank Dr Chung-Cheng Hsieh (University of Massachusetts Medical) and Dr Donna Neuberg (Dana-Farber Cancer Institute) for advice on experimental design and statistical analyses of the data.
This work (core resources) was supported by the Diabetes Endocrinology Research Center (grant DK32520); (NIH T32 training grant CA130807; J.T.); and the National Institutes of Health (grant CA096899; M.A.K.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.T. and K.C. performed the experiments and made the figures; R.G. provided technical and analytical expertise; T.A. and J.C.A. contributed the data in supplemental Figure 6C; and M.A.K. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michelle A. Kelliher, Department of Cancer Biology, University of Massachusetts Medical School, 364 Plantation St, Worcester, MA 01065; e-mail: [email protected].
References
Articles from Blood are provided here courtesy of The American Society of Hematology
Full text links
Read article at publisher's site: https://doi.org/10.1182/blood-2010-08-300343
Read article for free, from open access legal sources, via Unpaywall:
http://www.bloodjournal.org/content/118/6/1579.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
The role of quiescent thymic progenitors in TAL/LMO2-induced T-ALL chemotolerance.
Leukemia, 38(5):951-962, 29 Mar 2024
Cited by: 1 article | PMID: 38553571 | PMCID: PMC11073972
TAL1 hijacks MYCN enhancer that induces MYCN expression and dependence on mevalonate pathway in T-cell acute lymphoblastic leukemia.
Leukemia, 37(10):1969-1981, 17 Aug 2023
Cited by: 1 article | PMID: 37591943
Overexpression of Lmo2 initiates T-lymphoblastic leukemia via impaired thymocyte competition.
J Exp Med, 220(6):e20212383, 15 Mar 2023
Cited by: 3 articles | PMID: 36920307 | PMCID: PMC10037042
Noncanonical β-catenin interactions promote leukemia-initiating activity in early T-cell acute lymphoblastic leukemia.
Blood, 141(13):1597-1609, 01 Mar 2023
Cited by: 3 articles | PMID: 36315912 | PMCID: PMC10651788
IL-15 Prevents the Development of T-ALL from Aberrant Thymocytes with Impaired DNA Repair Functions and Increased NOTCH1 Activation.
Cancers (Basel), 15(3):671, 21 Jan 2023
Cited by: 2 articles | PMID: 36765626 | PMCID: PMC9913776
Go to all (65) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes.
Genes Dev, 24(11):1093-1105, 01 Jun 2010
Cited by: 82 articles | PMID: 20516195 | PMCID: PMC2878648
SCL, LMO1 and Notch1 reprogram thymocytes into self-renewing cells.
PLoS Genet, 10(12):e1004768, 18 Dec 2014
Cited by: 47 articles | PMID: 25522233 | PMCID: PMC4270438
ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer.
Blood, 117(26):7079-7089, 02 May 2011
Cited by: 67 articles | PMID: 21536859
SCL/TAL1: a multifaceted regulator from blood development to disease.
Blood, 129(15):2051-2060, 08 Feb 2017
Cited by: 74 articles | PMID: 28179281
Review
Funding
Funders who supported this work.
NCI NIH HHS (3)
Grant ID: CA096899
Grant ID: T32 CA130807
Grant ID: R01 CA096899
NIDDK NIH HHS (2)
Grant ID: DK32520
Grant ID: P30 DK032520
 1
1



